Abstract
The ability to sequence DNA outside of the laboratory setting has enabled novel research questions to be addressed in the field in diverse areas, ranging from environmental microbiology to viral epidemics. Here, we demonstrate the application of offline DNA sequencing of environmental samples using a hand-held nanopore sequencer in a remote field location: the McMurdo Dry Valleys, Antarctica. Sequencing was performed using a MK1B MinION sequencer from Oxford Nanopore Technologies (ONT; Oxford, United Kingdom) that was equipped with software to operate without internet connectivity. One-direction (1D) genomic libraries were prepared using portable field techniques on DNA isolated from desiccated microbial mats. By adequately insulating the sequencer and laptop, it was possible to run the sequencing protocol for up to 2½ h under arduous conditions.
Keywords: Antarctica, extremophiles, MinION, Nanopore sequencing
INTRODUCTION
In the past several years, both research and commercial enterprises have begun to develop DNA sequencers that can be used in the field for on-site sequencing studies.1–4 Among these is the handheld MinION sequencer from Oxford Nanopore Technologies (ONT) that measures ionic current changes as DNA passes through an array of protein nanopores that are embedded in a special polymer membrane.5 As DNA passes through the nanopore, it causes a characteristic current change for each group of nucleotides, enabling the bases to be sequenced. This process of single molecule, massively parallel real-time sequencing can generate continuous read lengths >100 kb with a total run output of >10 Gb.6 As this instrument allows for direct sequencing of native DNA and avoids PCR bias, it has beneficial applications in many sequencing situations and environments.7 For instance, in extremely cold and arid locations, ambient conditions can preserve fragmented strands of ancient DNA over very long timescales. Strands of ancient DNA recovered from environmental samples are typically only 100–500 bp in length,8 thus the long read lengths (>10 kb) generated by third-generation sequencing techniques can enhance genomic analyses by helping to distinguish DNA that is free floating in the environment from DNA recovered from intact cells.
The objective of this study was to measure performance characteristics of the MinION in a remote location without internet or a laboratory facility, using only local battery power supplied by the laptop through a USB 3.0 cable connection, to determine the feasibility for in situ genomic sequencing in extreme environments, such as the Dry Valleys of Antarctica (Fig. 1).
FIGURE 1.
Map of Antarctica showing the location of the Dry Valleys (A). DNA sequenced in this study was isolated from paleomat samples collected in Victoria Valley, Wright Valley, and Taylor Valley (B). MinION calibrations were performed in the Albert P. Crary Science and Engineering Center at McMurdo Station, as well as on the peak of Hut Point Peninsula on Ross Island (C). Field sequencing runs were performed in the vicinity of Lake Fryxell in Taylor Valley (D).
METHODS
To establish if the ONT MinION and related hardware would function in Antarctic conditions, system and software performance was initially authenticated by running a full-system calibration and flow cell test in the field at −2°C. Once all verifications were satisfactory, field sequencing was conducted by performing library synthesis, flow cell priming, and sequencing in situ at a field site near Lake Fryxell in Taylor Valley (Fig. 1). To gauge the effect of the cold on sequencing performance, we completed a subsequent control run with lambda DNA under laboratory conditions at 4°C, and compared the results with a sequencing run under standard laboratory conditions to establish baseline platform performance.
Further proficiency testing before field deployment ncluded sequencing 2 samples to ensure that the temperature-sensitive reagents shipped to Antarctica retained their efficacy and that extraction methods were capable of producing high-quality DNA. These samples included a modern microbial mat collected from the sediment-water interface of the moat at Lake Vida in Victoria Valley and a desiccated paleomat (ancient microbial biofilm) sample collected from a perched delta above Lake Fryxell in Taylor Valley (Fig. 2). Field sequencing using a MK1B MinION was then performed on DNA isolated from 2 additional paleomats: one from the vicinity of Lake Vida in Victoria Valley and the other from the vicinity of Lake Vanda in Wright Valley.
FIGURE 2.
Modern microbial mat sample from Lake Vanda in Wright Valley, collected at the sediment-water interface beneath 2 cm of ice (A) and a desiccated paleomat from the vicinity of Lake Vida in Victoria Valley, removed from beneath 20 cm of soil (B).
Sample Collection and DNA Isolation
Samples of modern microbial mats and desiccated paleomats were collected using sterile Whirl-Pak bags (Nasco, Fort Atkinson, WI, USA) and sterile, DNA-free utensils. Samples were placed in coolers with dry ice or a liquid nitrogen-primed cryoshipper, transported to McMurdo Station, and stored at −80°C until processing. Genomic DNA (gDNA) was extracted from microbial mat samples using a beta metagenomics DNA extraction kit (Omega Bio-tek, Norcross, GA, USA) and the PowerSoil Extraction Kit (Mo Bio Laboratories, Qiagen, Carlsbad, CA, USA).
Library Preparation
Sequencing libraries were synthesized from microbial gDNA using the 1D library protocol (ONT)9 at the Albert P. Crary Science and Engineering Center, McMurdo Station, for initial sequencing and validation runs. Although 2D library synthesis kits produce higher-quality reads than 1D, software to acquire 2D sequence data offline is not yet available. Furthermore, the protocol for 1D sequencing library synthesis is faster and more suitable for use in the field than the 2D protocol. To construct a 1D sequencing library, gDNA was added to FRM buffer (ONT), gently mixed by inversion, and incubated sequentially at 30°C and 75°C for 1 min each. RAD buffer (ONT) and 0.2 μl Blunt/TA Ligase Master Mix (New England Biolabs, Ipswich, MA, USA) were added and incubated for 5 min at 20°C to complete the library. Libraries synthesized in the laboratory and the field were sequenced immediately following preparation.
Library preparation in the field required additional considerations. Aliquots of all reagents were prepared in the lab before departing by helicopter to the field site. An insulated Thermos water bath was used for all field incubations, and the water temperature was monitored using a thermometer during library synthesis.
Laboratory and Field Sequencing
The MinION was prepared for sequencing by first assembling the MinION and v9.4 SpotON MinION flow cell and performing a quality control test on the flow cell using MinKNOW 1.4 software (ONT). The flow cell was subsequently primed and loaded with the sequencing library according to the manufacturer’s recommended protocol. Sequencing was initiated using a laptop equipped with a stand-alone version of MinKNOW 1.4 software, capable of operating without internet connectivity. Base-calling on raw FAST5 files, each of which contains the aggregate signal measurements for a sequenced DNA molecule, is usually performed on an online server and not locally on a computer physically linked to the sequencer.6, 10 During the base-calling process, reads are classified into “pass” and “fail” categories based on whether base-calling attempt was successful. Reads in the pass category were composed of high-quality reads6, 10 and were used to evaluate library prep, sequencer, and software performance in the field.
Although MinKNOW software is well established for Microsoft operating systems, this field work also tested and demonstrated the first use of the stand-alone MinKNOW software on a Macintosh operating system, intimating its expansion onto Linux-based platforms.
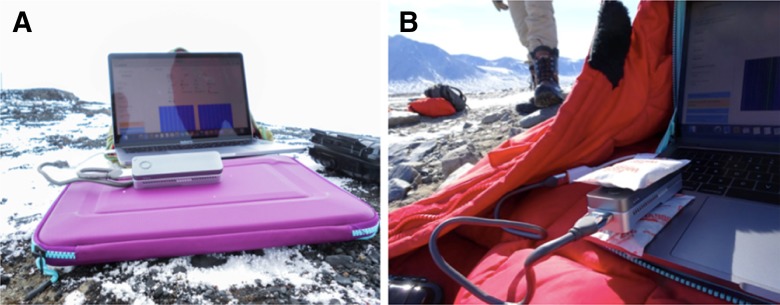
As a result of the cold ambient temperatures in Taylor Valley (2°C), it was anticipated that the sequencer might not be able to maintain proper operating temperature and the laptop battery could lose charge more rapidly because of added drain due to cold conditions. To compensate for this loss and maintain sequencer performance, the temperature of the sequencer and laptop was regulated using hand warmers and insulating materials, all while ensuring the air vents on the MinION remained clear (Fig. 3). During the sequencing runs, the operating temperature of the MinION was monitored continuously.
FIGURE 3.
The ONT MinION MK1B performing a calibration test on Hut Point Peninsula (left), as well as a sequencing run in the field in Taylor Valley (right).
RESULTS AND DISCUSSION
The performance calibration conducted on the tip of Hut Point Peninsula, using a configuration cell and the offline MinKNOW software, indicated that both hardware and software functioned properly in an Antarctic field situation. Crucially, the MinION reached and maintained the desired temperature long enough for successful calibration despite snow and cold (−5°C) conditions (Table 1).
TABLE 1.
Metadata and 1D sequencing run performance for the Antarctic tests
| Sample | Description | Sequencing date | Ambient T | Conditions | Sequencing location | Total # reads | Pass/fail ratio | Longest pass read, bp | Mean pass read length, bp | Median pass read length, bp | Mean Q score | Duration of run |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | lambda Phage DNA | Nov. 23, 2016 | RT | S. S. Johnson lab | Georgetown University | 6100 | 0.761 | 79,414 | 7763 | 4910 | 8.1 | 3 h |
| Control | lambda Phage DNA | Feb. 24, 2017 | 4°C | S. S. Johnson lab | Georgetown University | 4525 | 0.021 | 15,922 | 3764 | 2178 | 8.1 | 50 min |
| Calibration | Configuration cell | Dec. 10, 2016 | −5°C | Light snow, overcast | Hut Point | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Environmental | Paleomat, Lake Fryxell | Dec. 15, 2016 | RT | Crary lab | McMurdo Station | 46,936 | 0.121 | 90,183 | 562 | 301 | 5.7 | 10 h |
| Environmental | Modern mat, Lake Vida | Dec. 17, 2016 | RT | Crary lab | McMurdo Station | 18,761 | 0.272 | 171,106 | 3874 | 2067 | 7.8 | 8 h |
| Environmental | Paleomat, Lake Vanda | Dec. 19, 2016 | −1°C | Sunny, slight wind | Taylor Valley | 6026 | 0.035 | 22,128 | 777 | 484 | 5.1 | 50 mina |
| Environmental | Paleomat, Lake Vida | Dec. 20, 2016 | 2°C | Sunny | Taylor Valley | 573 | 0.139 | 21,357 | 3449 | 1238 | 4.6 | 2.5 h |
Run terminated for evacuation in advance of impeding weather.
In all sequencing runs, including the controls, there were more fail reads—those that could not be base called successfully—than those categorized as high-quality pass reads (e.g., pass/fail ratio <1; Table 1). However, the pass/fail ratio—the proportion of pass reads divided by the proportion of fail reads—dramatically increased with increased ambient temperature for control runs using lambda DNA [0.761 compared with 0.021 for room temperature (RT) and 4°C conditions, respectively]. Maximum and median read length also decreased from RT to 4°C conditions, yet the mean Q score of the pass reads remained unchanged (Table 1). It is not clear whether the decrease in proportion of high-quality reads is a result of a decline of software base-calling or sequencer performance under cold temperatures.
During the 4°C control run, the MinION was unable to achieve operating temperature and initiate sequencing without insulation. However, sequencing commenced as soon as the MinION was placed into an insulated cryo-glove that allowed airflow through the vents, ∼20 min into the experiment. The number of reads, maximum read length, and the pass/fail ratio from the cold control run proved similar to those achieved on environmental samples in the field (Table 1).
As previously observed in the control sequencing runs using lambda DNA, longer maximum read lengths were achieved by sequencing at RT (Crary Laboratory) than in the field when sequencing gDNA from Antarctic microbial mats (Table 1). As expected, the Q scores for environmental gDNA were lower than for control lambda DNA, yet they are in line with those reported in other studies using 1D sequencing.6
Even though longer maximum read lengths were achieved under laboratory conditions, the differences observed in read-length distributions did not correlate with ambient temperature, as microbial mats from Lake Vida yielded higher median (>1 kb) and mean (>3 kb) read lengths than DNA sequences from the other 2 samples (<1 kb mean and <500 bp median lengths). These differences are likely a result of variation in the quality of the DNA templates.
Although the duration of our field-sequencing runs was shorter than typical for most laboratory runs, these proof-of-principle concept results demonstrate that preparing and sequencing DNA libraries are feasible on future sampling expeditions, particularly ones requiring prolonged stays in the field. The preservation of battery power in the extreme cold remains a challenge, but longer sequencing runs can be enabled by switching among laptops with fully charged batteries and/or using an electrical generator.
The MinION sequencing system’s extremely small size (10.5 × 2.3 × 3.3 cm) and low weight (85 g), combined with its ability to draw power from a laptop while transferring data through a USB connection, makes it far easier to transport than conventional sequencing technologies. The ability to deploy highly portable sequencing technology and generate genomic sequence data in situ has use in a wide variety of difficult-to-access field biology settings and allows for real-time reconnaissance and genomic investigation of candidate sampling sites when time and sampling resources are limited.
In the case of Antarctica, this study marked the first DNA sequencing ever performed in situ on the continent. Teams researching Antarctic genomics often deploy for weeks or months at a time to collect samples, but no genomic metrics are available to gauge the quality of those samples until the return to their home institutions. The ability to gather initial genomic data in remote research settings could lead to real-time adjustments in field season planning and guard against the risks associated with sample transport, such as loss during shipping or thawing en route.
The MinION has also been demonstrated recently to sequence microbial communities in the High Arctic.11 A field laboratory was set up in Svalbard, where an external power supply and an intermittent wi-fi connection enabled a technical demonstration of nanopore-based metagenomics. Genomic DNA samples from a known alpine cryoconite community were re-sequenced alongside DNA extracted in the field laboratory from a cryoconite hole on the slope of an unnamed glacier. Although field extraction methods may have limited the number of sequences successfully base-called, the authors concluded that initial analyses of microbial community composition could serve as a useful prelude to more formal analyses back in the laboratory.11 Their findings suggest that the use of a field laboratory, sheltered from the elements, may be highly advantageous for some field sequencing projects, particularly in the high latitudes.
Applications for the use of MinION sequencing technology in remote environments extend well beyond environmental microbiology. The MinION sequencing system was successfully deployed in April 2015 in Guinea, West Africa, to sequence the Ebola virus genome from RNA isolated from whole blood, allowing for real-time genomic surveillance of the ongoing epidemic.12 This study was among the first to demonstrate the unique use of the MinION system to generate genomic data quickly in real time in a remote and resource-limited setting. Electrical power, while available, was subject to outages and surges, and consistent issues arose regarding access to internet connectivity.12 The fully offline version of the software demonstrated in this study may help to advance the prospects of future in situ surveillance of disease outbreaks in resource-limited settings.
In September 2016, NASA used the MinION to sequence DNA in space for the first time on the International Space Station, sequencing test mixtures of gDNA extracted from Escherichia coli, Mus musculus, and lambda bacteriophage.13 No declines in MinION performance were observable in microgravity, opening possibilities for diagnosing infectious disease and monitoring biologic responses to prolonged spaceflight.13 The continued development of offline nanopore sequencing technology may, one day, allow for remotely telemetered biologic data from the some of our planet’s most inaccessible locales—beneath glaciers or in the deep ocean—as well as locations beyond our planet. Work is under way to develop nanopore sequencing as part of spacecraft instrumentation to search for life on other worlds.14–17 For example, life on Mars, if present, may share common ancestry with organisms on Earth. Sequencing could help to distinguish definitively deeply branching but highly divergent forms of nucleic acid-based life from common clean-room contaminants.
CONCLUSIONS
Data collected for this proof-of-principle sequencing study reveal that the ONT MinION is a promising DNA sequencing platform for use in extreme field conditions. We have validated the offline MinKNOW software for generating 1D reads on environmental samples and demonstrated the success of the code in a remote location. Future studies will benefit from replacing 1D reagents with 2D reagents to generate higher-quality libraries, as well as using ONT’s new VolTRAX instrument, an automated library preparation device.18 As expected, challenges were encountered during sequencing in the field, many of which can be addressed in future expeditions, such as maintaining and ensuring proper library synthesis in the field (a critical step to generating high-quality data), fabricating a well-designed containment system to prevent heat loss, and using high capacity batteries. Areas for future work also include high quality in situ DNA isolation, enabling sequencing that can be coupled to local bioinformatics analyses in the field. In conclusion, the small size and low power requirements of the MinION will likely continue to enable its evolution as a field-deployable DNA sequencing device, opening up new avenues for biologic research in areas where the typical laboratory infrastructure for genomic sequencing is unavailable.
ACKNOWLEDGMENTS
The authors are deeply grateful to Brenda Hall at the University of Maine for her helpful guidance about sampling locations in Antarctica. The authors also thank Michael Micorescu at ONT for software development and technical assistance with the MinION MK1B. The authors also thank the Association of Biomolecular Resource Facilities Metagenomics Research Group and the Extreme Microbiome Project (XMP) team members for their insights and support on this project. This work was supported by the National Science Foundation Office of Polar Programs Award PLR-1620976 to S.S.J. at Georgetown University.
DISCLOSURES
The authors herein declare that this research was conducted in the absence of any financial or commercial interests that could be potentially regarded as a conflict of interest. Some of the flow cells used in this project were given for free to the XMP by ONT.
REFERENCES
- 1.Morey M, Fernández-Marmiesse A, Castiñeiras D, Fraga JM, Couce ML, Cocho JA. A glimpse into past, present, and future DNA sequencing. Mol Genet Metab 2013;110:3–24. [DOI] [PubMed] [Google Scholar]
- 2.Feng Y, Zhang Y, Ying C, Wang D, Du C. Nanopore-based fourth-generation DNA sequencing technology. Genomics Proteomics Bioinformatics 2015;13:4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu DT, Di Carlo D, Doyle PS, et al. Small but perfectly formed? Successes, challenges, and opportunities for microfluidics in the chemical and biological sciences. Chem 2017;2:201–223. [Google Scholar]
- 4.Van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet 2014;30:418–426. [DOI] [PubMed] [Google Scholar]
- 5.Mikheyev AS, Tin MM. A first look at the Oxford Nanopore MinION sequencer. Mol Ecol Resour 2014;14:1097–1102. [DOI] [PubMed] [Google Scholar]
- 6.Camilla LC, Loose M, Tyson JR, et al. MinION Analysis and Reference Consortium: Phase 1 data release and analysis. F1000 Res 2015;4:1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Check Hayden E. Pint-sized DNA sequencer impresses first users. Nature 2015;521:15–16. [DOI] [PubMed] [Google Scholar]
- 8.Willerslev E, Hansen AJ, Rønn R, et al. Long-term persistence of bacterial DNA. Curr Biol 2004;14:R9–R10. [DOI] [PubMed] [Google Scholar]
- 9.Oxford Nanopore Technologies. PCR-based, PCR-free, and rapid barcoding for MinION and PromethION libraries. Available at: https://nanoporetech.com/publications/2016/05/24/pcr-and-pcr-free-barcoding-for-miniontm-and-promethion.
- 10.Lu H, Giordano F, Ning Z. Oxford Nanopore MinION sequencing and genome assembly. Genomics Proteomics Bioinformatics 2016;14:265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards A, Debbonaire AR, Sattler B, et al. Extreme metagenomics using nanopore DNA sequencing: a field report from Svalbard, 78 °N. bioRxiv 2016:073965.
- 12.Quick J, Loman NJ, Duraffour S, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 2016;530:228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castro-Wallace SL, Chiu CY, John KK, et al. Nanopore DNA sequencing and genome assembly on the International Space Station. bioRxiv beta Sept. 27, 2016. Available at: http://biorxiv.org/content/early/2016/09/27/077651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bywaters KF, McKay CP, Davila AF, Quinn RC. In situ life and biosignature detection at Mars analog sites using the Oxford Nanopore Minion sequencer. Biosignature Preservation and Detection in Mars Analog Environments Conference. May 16–18, 2016;No. 1912, id 2014. [Google Scholar]
- 15.Mojarro A, Hachey J, Tani J, et al. SETG: nucleic acid extraction and sequencing for in situ life detection on Mars. 3rd International Workshop on Instrumentation for Planetary Mission. October 24–27, 2016;No. 1980, id 4095. [Google Scholar]
- 16.Carr CE, Mojarro A, Tani J, et al. Advancing the search for extra-terrestrial genomes. 2016 IEEE Aerospace Conference. March 5–12, 2016;No. 16121621. 10.1109/AERO.2016.7500859 [DOI] [Google Scholar]
- 17.Johnson SS, Ellington A, Anslyn E, Graham H, Mahaffy P. Fingerprinting nonterran life. Lunar and Planetary Science Conference XLVIII. March 21–25, 2017;id 2164.
- 18.Oxford Nanopore Technologies. VolTRAX: a rapid, programmable, portable, disposable device for sample and library preparation. Available at: https://nanoporetech.com/publications/voltrax-rapid-programmable-portable-disposable-device-sample-and-library-preparation.