Abstract
Acinetobacter baumannii is an important pathogen that causes nosocomial infections generally associated with high mortality and morbidity in Intensive Care Units (ICUs). Currently, little is known about the Quorum Sensing (QS)/Quorum Quenching (QQ) systems of this pathogen. We analyzed these mechanisms in seven clinical isolates of A. baumannii. Microarray analysis of one of these clinical isolates, Ab1 (A. baumannii ST-2_clon_2010), previously cultured in the presence of 3-oxo-C12-HSL (a QS signalling molecule) revealed a putative QQ enzyme (α/ß hydrolase gene, AidA). This QQ enzyme was present in all non-motile clinical isolates (67% of which were isolated from the respiratory tract) cultured in nutrient depleted LB medium. Interestingly, this gene was not located in the genome of the only motile clinical strain growing in this medium (A. baumannii strain Ab421_GEIH-2010 [Ab7], isolated from a blood sample). The AidA protein expressed in E. coli showed QQ activity. Finally, we observed downregulation of the AidA protein (QQ system attenuation) in the presence of H2O2 (ROS stress). In conclusion, most of the A. baumannii clinical strains were not surface motile (84%) and were of respiratory origin (67%). Only the pilT gene was involved in surface motility and related to the QS system. Finally, a new QQ enzyme (α/ß hydrolase gene, AidA protein) was detected in these strains.
Introduction
Quorum Sensing (QS) is a general mechanism used by Gram-negative bacteria to regulate many biological processes, including virulence, competence, conjugation, resistance, motility and biofilm formation [1]. The production and detection of bacterial cell-cell signalling molecules by various species have been linked to the enhanced development of single and multi-species biofilms [2]. A variety of structurally different bacterial cell-cell signalling molecules have been shown to mediate cell-cell communication, including acyl homoserine lactones (AHLs) and autoinducer-2 molecules (AI-2). AHLs have been proposed to mediate intra-species bacterial communication; different species typically only recognize AHLs produced from closely related species [3]. On the other hand, AI-2 has been shown to mediate inter-species signalling [4]. The term AI-2 describes a family of inter-convertible molecules derived from the precursor molecule (4,5-dihydroxy-2,3-pentanedione, DPD) [5]. This precursor molecule is produced or detected by many Gram-positive and Gram-negative bacteria [6].
The QS system in Acinetobacter sp. has been described as homologous to the LuxR receptor (AbaR) and LuxI synthase (AbaI) proteins in Vibrio fisheri [7, 8]. Acyl homoserine lactones (AHLs) are classified on the basis of the length of the acyl chain as short- or long-chain molecules. Many strains of Acinetobacter (63%) produce more than one AHL (≥C10). Moreover, none of the AHL signals can be specifically assigned to any particular species of this genus [9]. In this pathogen, AHL molecules are autoinducers of the QS system involved in motility and biofilm production [10]. Secretion of quorum signals has been associated with multidrug efflux pumps [11, 12]. The AdeFGH efflux pump has recently been related to the synthesis and transport of autoinducers during biofilm formation regulated by the QS system in clinical strains [12]. Overexpression of the AdeABC efflux pump by deletion of the two-component regulatory system, AdeRS, has also been associated with biofilm formation and virulence phenotype in this pathogen [13]. However, little is known about the cascade of genes associated with various mechanisms controlled by the QS system in nosocomial pathogens such as A. baumannii. In an earlier study of the genes involved in QS activation in A. baumannii ATCC 17978, Clemmer et al. observed overexpression of an operon comprising the A1S_0112 to A1S_0118 genes [10].
The Quorum Quenching (QQ) mechanism can effectively interfere with any one of the key processes in QS, and this could potentially be exploited to quench QS and prevent microbial infections (inhibition of motility and biofilm formation) [14]. Naturally occurring QQ mechanisms act by blocking the key steps of QS, such as signal generation, signal accumulation and signal reception. Microorganisms exist in a multi-species, competitive environment and have developed many survival strategies to gain benefits and compete for space, nutrition and ecological niches. One of these, QS interruption, is straightforward because bacteria that produce QQ agents can inhibit the QS-regulated behaviour of competing species and therefore obtain benefits or avoid being killed. An AHL acylase, AmiE, has recently been identified in Acinetobacter sp. strain Ooi24 [15, 16]. However, this QQ mechanism is not well known in clinical isolates of A. baumannii.
In 2016, Vijayakumar et al. analyzed the nature of the clinical isolates of A. baumannii in relation to biofilm formation and motility [17]. These authors concluded that the least motile strains were those obtained from respiratory samples (the main origin of isolates of this pathogen). Interestingly, 67% of the non-motile clinical isolates in this study were of a respiratory nature. The oxygen-rich environment generates reactive oxygen species (ROS) (e.g. superoxide anion [O2-] and hydrogen peroxide [H2O2]), which can dramatically increase damage to cell structures in a process known as oxidative stress. To reduce the potential damage caused by these reactive intermediates, the bacteria possess ROS detoxifying enzymes (SOD and catalase proteins) [18]. Recent studies have suggested that the response to ROS is controlled by the QS system in A. baumannii [19, 20]. In all aerobically-grown microorganisms, the stress induced by ROS such as the superoxide anion radical (O2-) and hydrogen peroxide (H2O2) causes macromolecular damage [21].
In this study, we used microbiological and transcriptomic assays (microarrays and RT-PCR) to analyze the Quorum Sensing/Quenching systems in clinical strains of Acinetobacter baumannii in relation to surface motility. We also studied the involvement of these systems (QS/QQ) in the oxidative stress mechanism (ROS system).
Material and methods
Strains and susceptibility
Seven clinical strains shown by multilocus sequence typing (MLST) to have different allelic profiles or sequence types (STs) and different susceptibility to several antimicrobials were used in this study (Table 1). The mechanisms of resistance to several antimicrobials are shown in Table 2. The genomes of two isolates used in the study have already been sequenced: Acinetobacter baumannii ST-2_clon_2010 (Genbank acc.num. LJHB00000000) [22] and Acinetobacter baumannii strain Ab421_GEIH-2010 (Genbank acc.num CP014266) [23], Ab1 and Ab7 respectively. Both whole genome sequencing studies (WGS) are part of the GEIH-REIPI Spanish Multicenter Acinetobacter baumannii Study II 2000–2010, project PRJNA308422. The abaR and abaI genes were also sequenced in the present study.
Table 1. Clinical isolates of A. baumannii used in this study (molecular typing and antimicrobial resistance).
| Strain | Molecular Typing | Antimicrobial resistance (MIC, mg/L) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IMIPENEM | MEROPENEM | TIGECYCLINE | GENTAMICIN | AMIKACIN | TOBRAMYCIN | CIPROFLOXACIN | COLISTIN | NETILMICIN | ||
| Ab1 | ST-2 | 64 | 64 | 16 | 64 | 4 | 4 | >64 | ≤0.5 | 16 |
| Ab2 | ST-186 | 64 | 64 | ≤0.25 | ≤0.5 | 4 | 0.5 | 16 | ≤0.5 | ≤0.5 |
| Ab3 | ST-52 | 16 | 16 | ≤0.25 | 32 | <2 | >64 | 64 | ≤0.5 | >64 |
| Ab4 | ST-169 | 1 | <0.5 | 0.5 | <0.5 | <2 | 1 | <0.5 | ≤0.5 | ≤0.5 |
| Ab5 | ST-80 | 64 | >64 | 32 | 64 | 128 | 1 | 64 | ≤0.5 | 32 |
| Ab6 | ST-181 | 64 | >64 | 16 | >64 | 128 | <64 | 64 | ≤0.5 | 16 |
| Ab7 | ST-79 | <0.5 | 1 | 8 | >64 | 64 | 64 | 64 | ≤0.5 | 64 |
Table 2. Mechanisms of resistance to several antimicrobials in clinical isolates of A. baumannii.
| Strain | ß-lactamase | Efflux pump (RND type) | Phenotype of antimicrobial resistance |
|---|---|---|---|
| Ab 1 | OXA-24 (Plasmid harbouring AbKAB TA system) | Overexpression of AdeABC/AdeIJK | Carbapenems, aminoglycosides, quinolones and glycines |
| Ab 2 | OXA-24 (Plasmid harbouring AbKAB TA system) | Not overexpression RND efflux pump | Carbapenems |
| Ab 3 | OXA-58 | Overexpression of AdeABC/AdeFGH | Carbapenems, aminoglycosides and quinolones |
| Ab 4 | - | Not overexpression RND efflux pump | - |
| Ab 5 | OXA-24 (Plasmid harbouring AbKAB TA system) | Overexpression of AdeABC/AdeIJK | Carbapenems, aminoglycosides, quinolones and glycines |
| Ab 6 | OXA-24 (Plasmid harbouring AbKAB TA system) | Overexpression of AdeABC/AdeIJK | Carbapenems, aminoglycosides, quinolones and glycines |
| Ab 7 | - | Absence of AdeABC efflux pump/ Overexpression AdeFGH | Aminoglycosides, quinolones and glycines |
Effect of culture conditions and quorum sensing inhibitors on motility
In view of the different responses shown by A. baumannii and A. nosocomialis to culture medium and Quorum Sensing inhibitors (Mayer et al., 2016, submitted), motility assays were performed in plates containing either Luria–Bertani (Normal LB) medium or modified LB-LN (nutrient depleted) [10]. The Normal LB medium contained (per litre) 10 g tryptone, 5 g yeast extract and 10 g NaCl per litre, while the modified LB contained 2 g tryptone, 1 g yeast extract and 5 g NaCl. Assays were carried out with 0.25% of Difco (BactoTM agar). Motility studies were carried out in the presence of QQ lactonase Aii20J enzyme, furanones and exogenous N-acyl homoserine lactone molecule (3-oxo-C12-HSL 10 uM).
The strains were inoculated in both normal LB and modified LB broth and incubated overnight at 37°C. An aliquot (1μl) of the culture was spotted in the centre of each well containing LB medium with agar and again incubated overnight at 37°C. Migration of the culture was then measured. The isolate was classified as non-motile when the average diameter of the zone of surface motility was <5 mm.
Detection of the quorum quenching phenotype: AHL lactonase assay
All A. baumannii clinical strains were cultured overnight in 5 ml of modified LB broth. The culture was recovered by centrifugation at 3000 g for 10 min, resuspended, washed and finally resuspended in 5 ml of modified LB. Aliquots of the culture (1 ml) were supplemented with N-3-oxo-dodecanoyl-L-homoserine lactone (3-oxo-C12-HSL at a final concentration of 10 μM) and incubated at 37°C in a shaker for 6 h. Aliquots (50 μl) of the resulting supernatant were used to detect AHL degradation in a well diffusion assay in double agar plates, in which Chromobacterium violaceum CV026 was added to soft agar as a biosensor together with C6HSL (5 μM) to detect inhibition of violacein synthesis [24, 25]. An overnight culture of E.coli BL21(DE3)pET28-AidA strain was recovered and incubated in the presence of IPTG (Isopropyl β-D-1-thiogalactopyranoside) for 4 hours to induce expression of the aidA gene, and the medium was then supplemented with 3-oxo-C12-HSL. The diffusion assay was carried out as previously described [24, 25].
Gene expression profiles
Gene expression studies were carried out by RT-PCR and microarray analysis. In both cases, the RNA samples were quantified in a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies). The quality and integrity of the samples were analyzed in an Agilent 2100 Bioanalyzer, with RNA 6000 Nano reagents and RNA Nano Chips (Agilent Technologies). Only samples with an RNA integrity number (RIN) >8 were included.
For RT-PCR analysis of all clinical strains in both types of LB broth, the High Pure RNA Isolation Kit (Roche, Germany) was used to obtain Dnase-treated RNA from late log-phase cultures (OD = 0.4–0.6) in the presence of the QQ lactonase Aii20J at a final concentration of 20 μM [25] (Fig 1). The ROS experiments were carried out with the RNA extracts in the presence of H2O2 for 5 minutes and in the absence of H2O2. The primers and UPL probe (Universal Probe Library-Roche, Germany) used in RT-PCR analysis in all clinical A. baumannii strains are shown in Table 3. These were designed from QS genes of the operon comprising the A1S-0112 to A1S_0118 cluster of A. baumannii ATCC 17978 [10]. We also included the abaR and abaI genes (QS system) [26] as well as genes that encoded efflux pumps [11, 12]. In relation to the QQ system, we analyzed the α/ß hydrolase gene (AidA protein) by using several combinations of primers and probes with variations in the amino acid sequences (S1 Fig). Finally, we studied the ROS response associated with QS/QQ systems. The concentrations of the samples were adjusted to achieve efficiencies of 90%-110% (50 ng of RNA), and all experiments were performed in triplicate (i.e. three RNA extracts). Analysis of controls without reverse transcriptase confirmed the absence of DNA contamination. For each strain, the expression of all genes was normalized relative to the rpoB gene. The normalized expression of each gene of interest was calibrated relative to expression of the reference strains, which were assigned a value of 1.0 (mean relative expression: RE). The following comparisons were made relative to the reference strains: i) QS analysis. A. baumannii clinical isolates of interest due to their surface motility on both types of LB medium (Fig 1) versus strains in the presence of the QQ lactonase Aii20J enzyme (the same strains in the absence of the compounds were used as reference strains); and ii) ROS responses in relation to QS (AbaI protein) and QQ mechanisms (AidA protein) were compared in clinical isolates of A. baumannii in the presence of H2O2 (for 5 minutes) and in the absence of H2O2 (strains with no H2O2 pressure were used as reference strains, CONTROL).
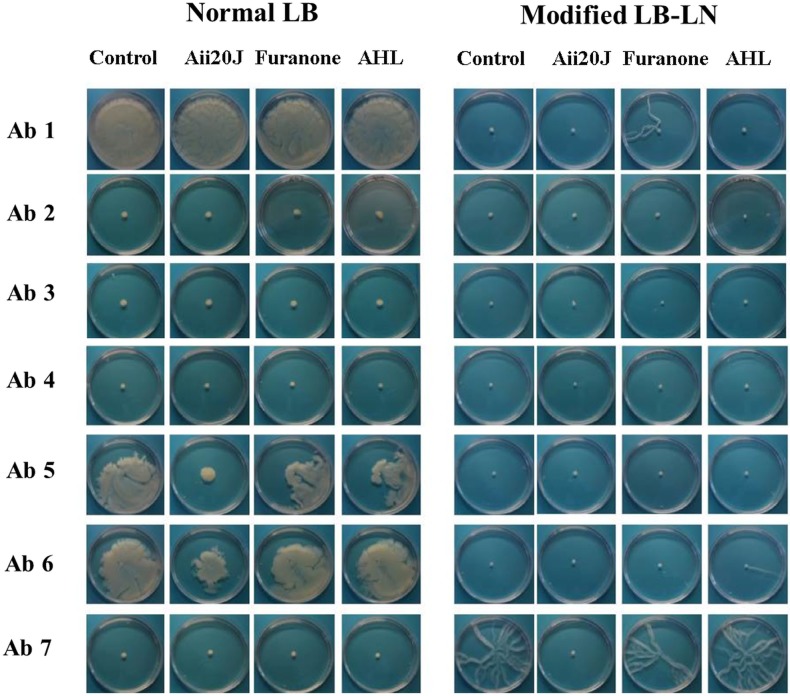
Fig 1. Surface motility of clinical strains of Acinetobacter baumannii on normal LB and modified LB (nutrient depleted).
Normal LB contains 10g/L NaCL, 10g/L tryptone and 5g/L yeast extract and the modified LB contains 5g/L NaCL, 2g/L tryptone and 1g/L yeast extract. Inhibition of motility (in motile strains) was analyzed by the QQ enzyme (Aii20J) and molecules with this capacity (furanones and acyl homoserine, AHL).
Table 3. Primers and UPL probes used in this study.
| PRIMERS and PROBES for RT-PCR STUDIES | ||||
|---|---|---|---|---|
| Quorum Sensing | Primer Sequence (5´-3´) | Taqman Probes | Ref | |
| A1S_0115 | Fow | TTGCCGGTTTGAAAAAGACT | 11/CTTCCGC | This study |
| Rev | TAAACGCACTTGGCACCATA | |||
| abaR | Fow | ACCTCTTGTTTGGTCGAGTCA | 96/ACAGGCAG | This study |
| Rev | CGTGCTTCCTCCCAAAAAT | |||
| pilT | Fow | CTTTGGTCTAGTGTGGTCATGC | 102/TGGCTGAG | This study |
| Rev | AAACAAAGTCGCGCAAATG | |||
| bfmS | Fow | TGAAGGAGTCGCTCGACAA | 38/GGAAGCAG | This study |
| Rev | CAGATGCGTCAGAAATCCAAT | |||
| csuD | Fow | CCTGAAAACCACCTAGTCGAA | 60/TGGGGAAG | This study |
| Rev | TTTTACTGGGTGACATTATTACCG | |||
| adeB | Fow | CGAGTGGCACAACTAGCATC | 61/CTGGGCAA | [27] |
| Rev | CCTTGTCTTGGCTGCACTCT | |||
| adeG | Fow | GTCCTGAAATGGTCGTTCGT | 43/CTGCCCCA | [27] |
| Rev | AGCTTCTGCTTGGCTAGATGA | |||
| adeJ | Fow | CCTATTGCACAATATCCAACGA | 119/TTGGTGGT | [27] |
| Rev | AGGATAAGTCGCAGCAATCG | |||
| rpoB | Fow | CGTGTATCTGCGCTTGG | 131/CTGGTGGT | [27] |
| Rev | CGTACTTCGAAGCCTGCAC | |||
| abaI | Fow | CCGCTACAGGGTATTTGTTGAAT | 6FAM-TGGATTCTCTGTCTTGAGCCACGACA-BBQ | This study |
| Rev | GCAGGGAATAGGCATTCCATTG | |||
| Quorum Quenching | Primer Sequence (5´-3´) | Taqman Probes | Ref | |
| aidA | Fow | GGGAACTTCTTTCGGTGGAG | 145/CAGCCACC | This study |
| Rev | AACAGCAGCAAGTCGATTATCA | |||
| Fow | GGGACTTCTTTCGGTGGAG | 145/ CAGCCACC | This study | |
| Rev | GCAGCAAGCCGGTTATCA | |||
| Fow | CCTAACCTTGCATTAGGGCTATTA | 53/TGGCAGAG | This study | |
| Rev | CGGTAAACCACAGGTCGGTA | |||
| PRIMERS for SEQUENCING ANALYSIS | ||||
| Quorum Quenching | Primer Sequence (5´-3´) | Ref | ||
| aidA gene (Putative lactonase) | AidA Fow | ATGGGTAAAAGTCTAAATAA | This study | |
| AidA Rev | CTTGACTGGAACGATG | |||
| AidA FowINT1 | GCCTATGCACGTAGCC | This study | ||
| AidA RevINT1 | GGGGGCAACAGAGTCGG | |||
| AidA FowINT2 | GCGAGATAGCCTGAAT | This study | ||
| AidA FowINT2 | GCCTATGCCCGTAGC | |||
| PRIMERS for CLONING the aidA gene in E.coli strain BL21 (DE3) | ||||
| Primer Sequence (5´-3´) | ||||
| AidA pET Fow (XhoI enzyme restriction site) | GGAATTCCATATGGGTAAAAGTCTAAATAA | |||
| AidA pET Rev (NdeI enzyme restriction site) | GCGGAGCTCTTACTTGACTGGAACGATGCG | |||
Ref, References.
Overexpression of the genes was defined by RE values of ≥ 1.5. The differences in gene expression were analyzed using a Student’s t-test. Differences were considered significant at P<0.05 [27].
In microarray studies (Fig 2), RNA was treated with 3-oxo-C12-HSL (significant signal molecule from QS of P.aeruginosa) at a final concentration of 10 μM [28] for analysis of clinical strain Ab1 (ST-2_clon_2010 isolate). Samples without enzymes and other compounds were used as negative controls. The arrays were designed using eArray (Agilent), on the basis of the Ab1 (ST-2_clon_2010 clinical strain genome) [22], and were carried out by Bioarray Diagnostico Genetico (Alicante, Spain). Labelling was carried out by two-colour microarray-based prokaryote analysis implemented with Fair Play III labeling, version 1.3 (Agilent). In this strain, the QS system was inhibited by 3-oxo-C12-HSL (100 μM) (i.e. inhibition of motility and biofilm formation, Fig 2). Four independent RNA extractions per condition (biological replicates) were used in each experiment. Statistical analysis was carried out using the Bioconductor tool of the RankProd software package for the R computing environment. A gene was considered overexpressed when the ratio of the treated to the untreated preparation was ≥1.5 at P <0.05.
Fig 2.
The figure shows inhibition of motility (A2) and biofilm formation (B) in Ab1 in the presence of 3-oxo-C12-HSL (QQ activity). As a negative control, motility and biofilm formation in Ab1 were studied in the presence of 0.3% dimethyl sulfoxide (DMSO), which is a solvent for 3-oxo-C12-HSL (A1 and B).
Biofilm formation
The biofilm assays were conducted following the procedure described by Álvarez-Fraga L et al. [29]. The strains were grown on LB medium for 18 h at 37°C and used to inoculate 5 mL of LB broth. Cultures were grown at 37°C with shaking. Overnight cultures were pelleted, washed and resuspended in 5 mL of modified LB-LN. A 1:100 dilution of each strain was incubated at 37°C for 48 h under static conditions. Growth of the culture was measured at OD600 to estimate total cell biomass. Biofilm formation was quantified by staining with crystal violet and solubilised with ethanol-acetone. The OD580/OD600 ratio was used to normalize the amount of biofilm formed to the total cell content of each sample tested, to overcome variations due to differences in bacterial growth under several experimental conditions. Eight independent replicates were considered. A student´s test was performed to evaluate the statistical significance of the observed differences between the strains considered.
Sequencing of the α/ß hydrolase gene (AidA) in clinical strains of A. baumannii
The primers listed in Table 3 were used to sequence and amplify the gene that encoded the protein in all clinical strains isolated in this study. To confirm the absence of this gene, we analyzed the genome of Ab7 (A. baumannii strain Ab421_GEIH-2010 genome) [23]. Moreover, the nucleotide sequences of AbaR and AbaI proteins did not reveal any mutations of interest (data not shown).
Expression of AidA protein in E.coli strains
DNA isolated from Acinetobacter baumannii strain ST-2_clon_2010 (Ab1) was used to clone the full-length AidA protein. The primers used to clone this gene in E.coli strains are shown in Table 3. These primers included an internal XhoI restriction site (bold), with a stop codon (underlined). The amplified PCR products were digested using NdeI and XhoI (NEB) and purified using a QIAquick PCR Purification Kit (QIAGEN). Finally, they were ligated with T4 DNA ligase (Fermentas) into a modified pET28-a plasmid (Novagen) which includes a human rhinovirus 3C protease cleavage site. Recombinant plasmids were transformed into competent E.coli DH5α cells (Novagen) for DNA production and purification (QIAprep Spin Miniprep Kit, QIAGEN). The integrity of both constructs was verified by sequencing. Finally, the plasmids were transformed into BL21(DE3) pLysS competent cells (Novagen) to yield the E.coli BL21(DE3)pET28-AidA construct.
Results
Surface motility in clinical strains of A. baumannii
The motility data for all clinical strains of A. baumannii are shown in Fig 1. Around 85% of the isolates did not exhibit surface motility on either medium tested. In those strains displaying surface motility, the lactonase Aii20J was the most effective in inducing QQ activity and thus inhibiting the motility (Ab 5 and Ab 6 on normal LB versus Ab 7 on modified LB-LN).
Genes associated with surface motility (activation of the QS system)
We used RT-PCR to study the following QS genes in the clinical strains of A. baumannii of interest in relation to surface motility (shown in Fig 1): A1S_115, abaR, pilT, bfmS, csuD, adeB, adeG and adeJ [10–12].
We observed only one gene, pilT, associated with surface motility on both types of LB medium and involved in the QS system, as indicated by a decrease in expression (RE between 1.5 and 8 times higher) in the presence of the QQ lactonase Aii20J enzyme (Ab 5 on Normal LB and Ab 7 on modified LB-LN). The most strongly expressed gene was A1S_115 in Ab3 (RE > 100 times higher). However, as clinical strain Ab3 did not display surface motility, this gene may be associated with another function.
Finally, in the Ab3 strain, overexpression of the genes that encoded the proteins AdeB (AdeABC) and AdeG (AdeFGH) decreased significantly in presence of the QQ lactonase Aii20J enzyme. In the other clinical strains, the differences were not statistically significant.
Genes involved in inhibition of surface motility (activation of the QQ system)
Stacy et al. used non-native N-acyl homoserine lactones such as 3-oxo-C12-HSL to study attenuation of the QS system (QQ activity) [28]. We carried out microarray analysis of A. baumannii ST-2_clon_2010 (the genome of which has been sequenced) cultured with 3-oxo-C12-HSL, in order to analyze the expression profile of the genes involved in the QQ system in clinical strains of A. baumannii (the solvent used, dimethyl sulfoxide [DMSO] was included as a blank control). We also confirmed activation of the QQ system in this strain with 3-oxo-C12-HSL, by inhibition of surface motility and a significant decrease in biofilm formation (Table 4 and Fig 2).
Table 4. Gene expression in A. baumannii ST-2_clon_2010 (Ab1) revealed by microarray assays in the presence of 3-oxo-C12-HSL.
| GenBank no. Sequencesa | Gene name | Fold change | Function |
|---|---|---|---|
| GI:1056209154 | Alpha/beta hydrolase | 5.01 | Quorum Quenching |
| GI:1056211401 | Acyl-dehydrogenase (Acyl-CoA dehydrogenase) | 4.83 | AHLs synthesis |
| GI:1056209152 | Short-chain dehydrogenase (3-oxoacyl-ACP reductase) | 4.79 | |
| GI:1056211410 | Amp-binding enzyme (Acyl CoA synthase) | 4.31 | |
| GI:1056211399 | Non-Ribosomal Peptide synthase | 3.91 | |
| (Long chain fatty-acid CoA ligase) | |||
| GI:1056209152 | Kr domain protein (3-oxoacyl-ACP reductase) | 3.27 | |
| GI:1056211405 | Acyl-homoserine-lactone synthase | 3.18 | |
| GI:1056211426 | Phosphopantetheine attachment domain | 2.72 | |
| (Beta-ketoacyl-ACP synthetase) | |||
| GI:1056212294 | Glutatione-S-transferase | 2.56 | Detoxification |
| GI:1056211398 | Mmpl family protein (RND transporter) | 1.89 | Efflux pump |
| GI:1056212337 | Outer membrane protein omp38 | 1.61 | OmpA porin |
| GI:1056212369 | Enterocidin EcnA/B family | 1.51 | Stress response |
| GI:1056211397 | Porin | 1.50 | Porin |
Only 13 genes were overexpressed in the presence of 3-oxo-C12-HSL in this A. baumannii strain (GEO database arrays GSE87009). The most strongly expressed gene (5.01) was a gene encoding an α/ß hydrolase enzyme, AidA (putative QQ ENZYME, GI:1056209154). Around 46.15% of the genes that were overexpressed were involved in the synthesis of the acyl-homoserine lactones (AHLs), including AHL synthase (GI:1056211405). Moreover, the results of the microarray analysis revealed overexpression of the genes coding for the following proteins: i) glutathione-s-transferase (DETOXIFICATION, GI:1056212294); ii) RND efflux pump (TRANSPORTER, GI:1056211398); iii) outer membrane protein-OmpA-Like (TRANSPORTER/VIRULENCE, GI:1056212337); iv) entericidin EcnA/B family (STRESS RESPONSE, GI:1056212369); and v) a new PORIN (GI:1056211397) (Table 4).
Detection of the aidA gene in clinical isolates
To confirm the role of AidA as a new QQ enzyme, we studied the presence of this protein in the clinical strains of A. baumannii that did not display surface motility on modified LB-LN (Ab2, Ab3, Ab4, Ab5 and Ab6) and in the only strain displaying surface motility (Ab7). We found the α/ß hydrolase gene encoded an AidA protein (QQ enzyme) in all non-motile isolates (67% from respiratory tract) (Table 5). The AidA enzyme showed variable levels of amino acid in different clinical strains of A. baumannii (S1 Fig). Importantly, the α/ß hydrolase gene was not amplified in strain Ab7 (reference strain displaying surface motility). The absence of this gene was confirmed by sequencing the genome of strain Ab7 (which belongs to the PFGE-HUI-1 clone), which has recently been published as A. baumannii strain Ab421_GEIH-2010 [23]. Other characteristics of this isolate were that it did not have an AdeABC efflux pump and it did not harbour OXA 24 ß-lactamase in a resistance plasmid (Table 5).
Table 5. Detection of the α/ß hydrolase gene (AidA protein) in clinical isolates by PCR.
Features of the isolates used in this study.
| Strain | Quorum Sensing system | Surface Motility | Type of infection |
|---|---|---|---|
| α/ß hydrolase gene, AidA protein: GI:1056209154a | (Modified LB-LN) | ||
| Ab 1 | + | No | Respiratory |
| Ab 2 | + | No | Ulcer |
| Ab 3 | + | No | Respiratory |
| Ab 4 | + | No | Respiratory |
| Ab 5 | + | No | Respiratory |
| Ab 6 | + | No | Exudade |
| Ab 7 | - | Yes | Blood |
a Genome A. baumannii ST-2_clon_2010.
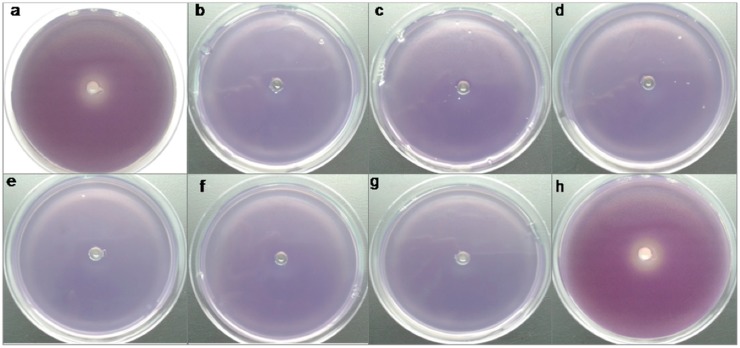
Finally, in order to detect the presence of QQ activity in the A. baumannii strains used in this study, we conducted well diffusion assays with Chromobacterium violaceum CV026 as a biosensor. This bacterium produces violacein in the presence of short-chain AHLs such as C6HSL; however, the presence of long-chain AHLs inhibits violacein production [24]. In all strains except Ab7 (the reference strain exhibiting surface motility) and the control strain, no halo was detected in the plates, and therefore the 3OC12HSL with which the cultures were incubated was not present (Fig 3).
Fig 3. Diffusion assays carried out with the biosensor Chromobacterium violaceum CV026 to detect the presence of QQ activity in the clinical strains of A. baumannii under study.
The presence of a halo indicates inhibition of violacein production by the presence of 3-oxo-C12-HSL and, therefore, the absence of QQ activity (a, h). The absence of a halo indicates QQ activity (b, c, d, e, f, g). a) Control; b) Ab1; c) ab2; d) Ab3; e) Ab4; f) Ab5; g) Ab6; h) Ab7.
Functional characterization of the AidA protein by overexpression in E.coli strains
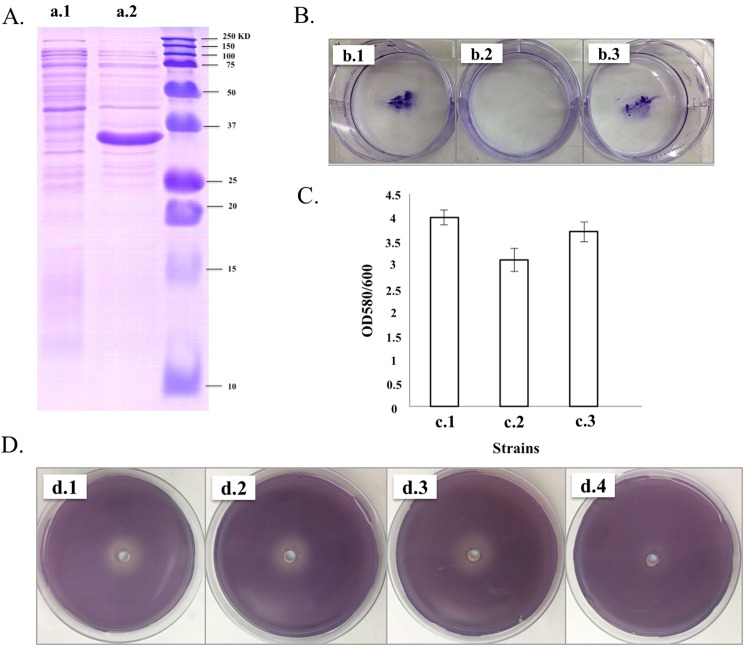
We used E. coli BL21 (DE3) as a model to overexpress the AidA protein (pET28-AidA) (Fig 4A, line a.2). Three QS systems have been described in this model [26]:
Fig 4. Studies with AidA protein overexpressed in E. coli BL21(DE3).
A. SDS PAGE with expression of AidA protein in E. coli BL21(DE3): a.1. E.coli BL21(DE3)pET28 and a.2 E.coli BL21(DE3)pET28-AidA; B. Twitching motility: b.1 E.coli BL21(DE3), b.2 E.coli BL21(DE3)pET28-AidA and b.3 E.coli BL21(DE3)pET28; C. Quantification of biofilm formation by crystal violet staining: c.1 E.coli BL21(DE3), c.2 E.coli BL21(DE3)pET28-AidA and c.3 E.coli BL21(DE3)pET28. The differences were statistically significant (Student's t-test, P <0.05); D. Detection by plate diffusion assay of QQ activity after incubation in presence of 3OC12HSL: d.1. Control (LB); d.2. E. coli BL21(DE3); d.3. E.coli BL21(DE3)pET28; d.4. E.coli BL21(DE3)pET28-AidA.
Unknown (synthase), SdiA (receptor) and 3-oxo-C8-HSL (signal synthesized in others bacteria). This system has been associated with motility and acid resistance. [30].
LuxS (synthase), LsrB (receptor) and AI-2 (signal). Lsr operon expression (AI-2 uptake) [31].
Unknown (synthase), QseC (receptor) and AI-3 (signal synthesized in other bacteria). This system has been implicated in virulence, motility and biofilm formation [32].
We confirmed the quorum quenching activity of this AidA protein by twitching motility studies with these cells in the presence of IPTG (Fig 4B in which E.coli BL21 [DE3] pET28-AidA showed no motility) and quantification of bacterial biofilm (significant decrease in biofilm formation in E.coli BL21 [DE3] pET28-AidA, Fig 4C). The quorum quenching activity of this protein was also detected by diffusion assay in the presence of the biosensor C. violaceum strain CV026: no halo was produced by the supernatant of the strain BL21 [DE3] pET28-AidA incubated with 3-oxo-C12-HSL (Fig 4D).
Relationship between QS/QQ systems and oxidative stress mechanism (ROS response)
We studied the involvement of QS/QQ systems in the ROS response by RT-PCR. In the presence of H2O2, expression of the α/ß hydrolase gene (QQ system) decreased significantly (P<0.05), while the abaI gene (QS system) was significantly overexpressed (P<0.05) (Table 6).
Table 6. Relative expression (RE) under normal conditions relative to the presence of H2O2 (ROS stress) in clinical strains of A. baumannii of genes involved in the QQ system (α/ß hydrolase gene, AidA protein) and the QS system (abaI gene, AHL synthase protein).
| Strain | Quorum Quenching system | Quorum Sensing system | ||
|---|---|---|---|---|
| α/ß hydrolase gene, AidA protein: GI:1056209154a | abaI gene, AHL synthase protein: A1S_0109b | |||
| CONTROL (References) | H2O2 | CONTROL (References) | H2O2 | |
| Ab 1 | 1 | 0.36 | 1 | 1.60 |
| Ab 2 | 1 | 0.88 | 1 | 1.61 |
| Ab 3 | 1 | 0.53 | 1 | 3.58 |
| Ab 4 | 1 | 0.55 | 1 | 1.64 |
| Ab 5 | 1 | 0.69 | 1 | 2.98 |
| Ab 6 | 1 | 0.62 | 1 | 3.73 |
| Ab 7 | - | - | 1 | 1.54 |
a. Genome A. baumannii ST-2_clon_2010 (Ab1).
b. Genome A. baumannii ATCC 17978.
Control: RNA extraction from clinical isolates under normal conditions; H2O2: RNA extractions from clinical isolates after 5 minutes in the presence of H2O2.
Discussion
The QS system enables bacterial populations to live and proliferate in an environment with effective intercellular communication [33]. In clinical isolates of A. baumannii, little is known about the cascade of genes controlled by this system and associated with various mechanisms, including surface motility (phenotypic expression). In Acinetobacter baumannii ATCC 17978, the A1S-0112 to A1S_0118 operon has been associated with activation of the QS system, and the pilT gene has been related to motility [10]. In this study, only pilT expression (surface motility) was controlled by QS in the LB broth used: normal LB and modified (nutrient depleted) LB-LN.
Amino acid sequences and architecture of the QQ enzymes are diverse [34]. These enzymes have several biological roles: QS-signal clearing in A. tumefaciens [35, 36], recycling of QS signals (Pseudomonas aeruginosa model) in organisms that produce QS molecules [37, 38], detoxification and, finally, disturbance of QS signalling by an organism that does not produce QS signals, but may take advantage of QQ processes, such as the hosts of QS-emitting pathogens (bacterial competition) [39].
Several α/ß hydrolase enzymes have been described in different pathogens [1, 34, 40–42]. In this study, we identified in presence of 3-oxo-AHL, a new QQ enzyme (α/ß hydrolase gene, AidA) which was present in all strains of A. baumannii that did not exhibit surface motility. Moreover, the QQ activity from this protein (inhibition of the motility and biofilm) was confirmed by overexpression in E.coli (which does not produce AHLs). Interestingly, Weiland and collaborators described several QQ enzymes with hydrolytic activity against AHLs and AI-2 signals [43, 44]. Hence, this QQ enzyme (AidA protein) could contribute in bacterial competition, as it is capable of hydrolyzing the signalling molecules mediated between species.
Recent studies suggest that the ROS response is controlled by the QS system in A. baumannii [14, 15]. Several studies have suggested the involvement of the QQ mechanism under ROS response [45, 46]. Veal et al. [47] and other researchers [48] have suggested that members of the glutathione-S-transferase family of proteins are important for protecting cells from oxidative stress. The results of microarray assays showed overexpression of the glutathione-s-transferase and α/ß hydrolase (AidA protein) genes in the Ab1 clinical strain (A. baumannii ST-2_clon_2010) [22] in the presence of 3-oxo-C12-HSL[28]. Moreover, in Deinococcus radiodurans, which is known for its resistance to oxidative stress, the AHL level was “shielded” by QQ enzymes under non-stress conditions (normal conditions), whereas AHLs accumulated when D. radiodurans was exposed to oxidative stress [49]. In the aforementioned study, the synthetic form of the AHL enzyme (DsqI) was immediately induced on exposure to H2O2, while the expression of QQ enzymes began to increase after exposure to H2O2 for about half an hour. The QS system (DqsIR) in this pathogen mediated the adaptive strategy in response to oxidative stress (ROS response) [49]. In the present study, we confirmed the presence of the AidA protein (new QQ enzyme) in all non-motile clinical strains of A. baumannii. The twitching motility and biofilm studies with overexpression of this protein in E.coli BL21(DE3) confirmed its role as a quorum quenching enzyme.
Finally, the AidA protein was downregulated (QQ system attenuation) in the presence of H2O2 (ROS stress), unlike the AbaI protein, which was upregulated in clinical strains of A. baumannii.
In conclusion, we researched the Quorum Sensing/Quenching systems in clinical isolates of A. baumannii. Most of the strains were not surface motile (84%) and were of respiratory origin (67%). Only the pilT gene was involved in surface motility and the QS system in these strains. A new QQ enzyme (α/ß hydrolase gene, AidA protein) was detected by array analysis in the presence of the external signal 3-oxo-C12-HSL. All of the non-motile strains of A. baumannii had the AidA protein (QQ system activation). The function of this protein as a QQ enzyme was confirmed by its expression in E.coli BL21(DE3) strain that produces AI-2 signalling molecules. Finally, the findings confirmed that regulation of ROS stress (presence of H2O2) by the QS/QQ systems in clinical strains of A. baumannii.
Nucleotide sequence accession number
The Ab ST-2_clon GEIH-2010 (Ab1) whole genome shotgun project has been deposited in the DDBJ/ENA/GenBank under accession number LJHB00000000. The version described in this paper is version LJHB01000000, which consists of sequences LJHB01000001 LJHB01000077. The genome sequence of Ab421 GEIH- 2010 strain (Ab7) has been deposited in GenBank under the accession number CP014266. Both WGS studies are part of the II Spanish Multicenter Study. GEIH-REIPI Acinetobacter baumannii 2000–2010 project (PRJNA308422).
Supporting information
(TIF)
Acknowledgments
We are grateful to the following organizations and researchers who participated in the study: Virgen Macarena (Jesús Rodríguez-Baño, Alvaro Pascual, Felipe Fernández Cuenca); Virgen Rocío (Jerónimo Pachón, Jose Miguel Cisneros, José Garnacho, Antonio Gutierrez-Pizarraya, Juan Antonio Márquez-Vácaro); Hospital Marqués de Valdecilla (Luis Martínez-Martínez, María Eliecer Cano, M. Carmen Fariñas); Hospital Clinic (Jordi Vila); Hospital SAS La Línea (Antonio Sánchez-Porto, Gloria Esteban Meruendano, Luis Barbeyto-Vales, Javier Casas-Ciria, Luis Vallejo); Complejo hospitalario de Ourense (Begona Fernández-Pérez, José Carlos Villar-Chao); Hospital Gregorio Maranón (Belén Padilla-Ortega, Emilia Cercenado-Mansilla); Hospital de Navarra (José Javier García-Irure); Hospital Costa del Sol-Marbella (Alfonso del Arco Jiménez); Hospital General de Valencia (Concepción Gimeno-Cardona, Juan Carlos Valía, Núria Tormo-Palop, Vicente Abril, Josefina Rifa, Maria Jesus Martinez-Garcia); Consorci Hospitalari de Vic (Joseph Vilaró-Pujals, Marian Navarro Aguirre, Ana Vilamala); Policlínica Guipúzkoa (José Antonio Jiménez-Alfaro, Carlos Reviejo-Jaca); Hospital Puerta del Mar (Pilar Marín Casanova, Francisca Guerreo, Evelyn Shaw, Virginia Plasencia); Complejo Hospitalario de Soria (Teresa Nebreda-Mayoral, María José Fernández-Calavia, Susana García de Cruz, Carmen Aldea-Mansilla); Hospital Universitario de Alicante (Esperanza Merino de Lucas, Alfredo Zorraquino, Sergio Reus-Bañuls); Hospital Infanta Cristina (Eugenio Garduno-Eseverri, Luis López Sánchez); Hospital Universitario Central de Asturias (Ana Fleites-Gutiérrez, Azucena Rodríguez-Guardado, Alfonso Moreno); Hospital Donostia (José María García-Arenzana Anguera); Complejo Hospitalario Torrecárdenas (Serafín López-Palmero, Manuel Rodríguez-Maresca); Complejo Hospitalario Xeral-Calde Lugo (Fernando García-Garrote, José Varela-Otero, María del Pilar Alonso); Hospital Universitario Reina Sofía de Córdoba (Elisa Vidal-Verdú, Fernando Rodríguez-López); Hospital Universitario Santiago Compostela (Fernanda Pardo-Sánchez, E. Ferrer-Vizoso, B. Regueiro-Garcia); Hospital Sant Pau (Mercé Gurgui, Roser Pericas,Virginia Pomar); Hospital Galdakao-Usansolo (Pedro María Olaechea-Astigarraga, Rafael Ayarza-Igartua); Hospital Son Dureta (María Dolores Maciá-Romero, Enrique Ruiz de Gopegui-Bordes); Hospital Puerta de Hierro (María Isabel Sánchez-Romero); Hospital Juan Grande (Jesús García-Mata, María José Goyanes, Cristina Morales-Mateos); Hospital San Cecilio (José Hernández-Quero, Trinidad Escobar-Lara); Hospital Sant Joan de Reus (Frederic Ballester-Bastardie, Simona Iftimie, Isabel Pujol-Bajador); Hospital de Motril (María Isabel Galán-Navarro, María Luz Cádiz-Gurrea); Hospital San Agustín (Carmen Amores-Antequera, Montserrat Gómez Purificación Cantudo); Hospital de Granollers (Carmina Martí-Salas, Jordi Cuquet-Peragosa, Antonio Moreno-Flores, Luis Anibarro-García); Hospital de Segovia (Susana Hernando-Real, Pablo A. Carrero-González); Complejo Hospitalario de Pontevedra (María Angeles Pallarés-González, Sergio Rodríguez-Fernández); Hospital de Bellvitge (Miquel Pujol-Rojo, Fe Tubau); Hospital Virgen de la Victoria de Málaga (Enrique Nuno-Alvarez, María Ortega-Torres); Hospital Doctor Moliner (Salvador Giner-Almaraz, María Rosa Roca-Castelló, Manuela Castillo, Elena Hortelano); Hospital 12 de Octubre (Fernando Chaves-Sánchez, Ana García-Reyne); Hospital delMar (Juan Pablo Horcajada-Gallego, Concha Segura); Hospital San Agustín de Avilés (Gema Sierra-Dorado, Raquel Yano-Escudero); Complejo Hospitalario Materno Insular de Gran Canaria (María Elena Dorta-Hung, Cristóbal del Rosario Q). Email for lead author of the GEIH-REIPI (GEMARA-SEIMC) Acinetobacter baumannii 2000–2010 project (PRJNA308422): MA.del.Mar.Tomas.Carmona@sergas.es.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by grant PI13/02390 awarded to MT within the State Plan for R+D+I 2013-2016 (National Plan for Scientific Research, Technological Development and Innovation 2008-2011) and co-financed by the ISCIII-Deputy General Directorate of evaluation and Promotion of Research - European Regional Development Fund "A way of Making Europe" and Instituto de Salud Carlos III FEDER, Spanish Network for the Research in Infectious Diseases (REIPI RD12/0015). M. Tomás was financially supported by the Miguel Servet Research Programme (C.H.U.A Coruña and ISCIII). A. Muras was financially supported by a predoctoral fellowship from the Xunta de Galicia (Plan I2C). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev. 2013;77(1):73–111. 10.1128/MMBR.00046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Irie Y, Parsek MR. Quorum sensing and microbial biofilms. Curr Top Microbiol Immunol. 2008;322:67–84. [DOI] [PubMed] [Google Scholar]
- 3.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–99. 10.1146/annurev.micro.55.1.165 [DOI] [PubMed] [Google Scholar]
- 4.Rickard AH, Palmer RJ, Blehert DS, Campagna SR, Semmelhack MF, Egland PG, et al. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol. 2006;60(6):1446–56. 10.1111/j.1365-2958.2006.05202.x [DOI] [PubMed] [Google Scholar]
- 5.Semmelhack MF, Campagna SR, Federle MJ, Bassler BL. An expeditious synthesis of DPD and boron binding studies. Org Lett. 2005;7(4):569–72. 10.1021/ol047695j [DOI] [PubMed] [Google Scholar]
- 6.Sun J, Daniel R, Wagner-Döbler I, Zeng AP. Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol Biol. 2004;4:36 Epub 2004/09/29. 10.1186/1471-2148-4-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhargava N, Sharma P, Capalash N. Quorum sensing in Acinetobacter: an emerging pathogen. Crit Rev Microbiol. 2010;36(4):349–60. Epub 2010/09/18. 10.3109/1040841X.2010.512269 [DOI] [PubMed] [Google Scholar]
- 8.Kang YS, Jung J, Jeon CO, Park W. Acinetobacter oleivorans sp. nov. is capable of adhering to and growing on diesel-oil. J Microbiol. 2011;49(1):29–34. 10.1007/s12275-011-0315-y [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez RH, Nusblat A, Nudel BC. Detection and characterization of quorum sensing signal molecules in Acinetobacter strains. Microbiol Res. 2001;155(4):271–7. Epub 2001/04/12. 10.1016/S0944-5013(01)80004-5 [DOI] [PubMed] [Google Scholar]
- 10.Clemmer KM, Bonomo RA, Rather PN. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology. 2011;157(Pt 9):2534–44. Epub 2011/06/28. 10.1099/mic.0.049791-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kvist M, Hancock V, Klemm P. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl Environ Microbiol. 2008;74(23):7376–82. 10.1128/AEM.01310-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He X, Lu F, Yuan F, Jiang D, Zhao P, Zhu J, et al. Biofilm Formation Caused by Clinical Acinetobacter baumannii Isolates Is Associated with Overexpression of the AdeFGH Efflux Pump. Antimicrob Agents Chemother. 2015;59(8):4817–25. 10.1128/AAC.00877-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richmond GE, Evans LP, Anderson MJ, Wand ME, Bonney LC, Ivens A, et al. The Acinetobacter baumannii Two-Component System AdeRS Regulates Genes Required for Multidrug Efflux, Biofilm Formation, and Virulence in a Strain-Specific Manner. MBio. 2016;7(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong YH, Wang LY, Zhang LH. Quorum-quenching microbial infections: mechanisms and implications. Philos Trans R Soc Lond B Biol Sci. 2007;362(1483):1201–11. 10.1098/rstb.2007.2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochiai S, Yasumoto S, Morohoshi T, Ikeda T. AmiE, a novel N-acylhomoserine lactone acylase belonging to the amidase family, from the activated-sludge isolate Acinetobacter sp. strain Ooi24. Appl Environ Microbiol. 2014;80(22):6919–25. Epub 2014/08/31. 10.1128/AEM.02190-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bzdrenga J, Daudé D, Rémy B, Jacquet P, Plener L, Elias M, et al. Biotechnological applications of quorum quenching enzymes. Chem Biol Interact. 2016. [DOI] [PubMed] [Google Scholar]
- 17.Vijayakumar S, Rajenderan S, Laishram S, Anandan S, Balaji V, Biswas I. Biofilm Formation and Motility Depend on the Nature of the Acinetobacter baumannii Clinical Isolates. Front Public Health. 2016;4:105 10.3389/fpubh.2016.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassett DJ, Ma JF, Elkins JG, McDermott TR, Ochsner UA, West SE, et al. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34(5):1082–93. [DOI] [PubMed] [Google Scholar]
- 19.Bhargava N, Sharma P, Capalash N. Pyocyanin stimulates quorum sensing-mediated tolerance to oxidative stress and increases persister cell populations in Acinetobacter baumannii. Infect Immun. 2014;82(8):3417–25. Epub 2014/06/04. 10.1128/IAI.01600-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heindorf M, Kadari M, Heider C, Skiebe E, Wilharm G. Impact of Acinetobacter baumannii superoxide dismutase on motility, virulence, oxidative stress resistance and susceptibility to antibiotics. PLoS One. 2014;9(7):e101033 10.1371/journal.pone.0101033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sies H. Oxidative stress: oxidants and antioxidants. Exp Physiol. 1997;82(2):291–5. [DOI] [PubMed] [Google Scholar]
- 22.López M, Rueda A, Florido JP, Blasco L, Gato E, Fernández-García L, et al. Genomic Evolution of Two Acinetobacter baumannii Clinical Strains from ST-2 Clones Isolated in 2000 and 2010 (ST-2_clon_2000 and ST-2_clon_2010). Genome Announc. 2016;4(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López M, Álvarez-Fraga L, Gato E, Blasco L, Poza M, Fernández-García L, et al. Genome Sequence of a Clinical Strain of Acinetobacter baumannii Belonging to the ST79/PFGE-HUI-1 Clone Lacking the AdeABC (Resistance-Nodulation-Cell Division-Type) Efflux Pump. Genome Announc. 2016;4(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, et al. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143 Pt 12):3703–11. [DOI] [PubMed] [Google Scholar]
- 25.Mayer C, Romero M, Muras A, Otero A. Aii20J, a wide-spectrum thermostable N-acylhomoserine lactonase from the marine bacterium Tenacibaculum sp. 20J, can quench AHL-mediated acid resistance in Escherichia coli. Appl Microbiol Biotechnol. 2015;99(22):9523–39. 10.1007/s00253-015-6741-8 [DOI] [PubMed] [Google Scholar]
- 26.Castillo-Juárez I, Maeda T, Mandujano-Tinoco EA, Tomás M, Pérez-Eretza B, García-Contreras SJ, et al. Role of quorum sensing in bacterial infections. World J Clin Cases. 2015;3(7):575–98. 10.12998/wjcc.v3.i7.575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rumbo C, Gato E, López M, Ruiz de Alegría C, Fernández-Cuenca F, Martínez-Martínez L, et al. Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2013;57(11):5247–57. 10.1128/AAC.00730-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stacy DM, Welsh MA, Rather PN, Blackwell HE. Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N-Acyl homoserine lactones. ACS Chem Biol. 2012;7(10):1719–28. Epub 2012/08/03. 10.1021/cb300351x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Álvarez-Fraga L, Pérez A, Rumbo-Feal S, Merino M, Vallejo JA, Ohneck EJ, et al. Analysis of the role of the LH92_11085 gene of a biofilm hyper-producing Acinetobacter baumannii strain on biofilm formation and attachment to eukaryotic cells. Virulence. 2016;7(4):443–55. 10.1080/21505594.2016.1145335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soares JA, Ahmer BM. Detection of acyl-homoserine lactones by Escherichia and Salmonella. Curr Opin Microbiol. 2011;14(2):188–93. Epub 2011/03/01. 10.1016/j.mib.2011.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kendall MM, Sperandio V. Quorum sensing by enteric pathogens. Curr Opin Gastroenterol. 2007;23(1):10–5. Epub 2006/11/30. 10.1097/MOG.0b013e3280118289 [DOI] [PubMed] [Google Scholar]
- 32.Moreira CG, Weinshenker D, Sperandio V. QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. Infect Immun. 2010;78(3):914–26. Epub 2009/12/24. 10.1128/IAI.01038-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 25 Netherlands2001. p. 365–404. [DOI] [PubMed] [Google Scholar]
- 34.Grandclément C, Tannières M, Moréra S, Dessaux Y, Faure D. Quorum quenching: role in nature and applied developments. FEMS Microbiol Rev. 2016;40(1):86–116. 10.1093/femsre/fuv038 [DOI] [PubMed] [Google Scholar]
- 35.Carlier A, Chevrot R, Dessaux Y, Faure D. The assimilation of gamma-butyrolactone in Agrobacterium tumefaciens C58 interferes with the accumulation of the N-acyl-homoserine lactone signal. Mol Plant Microbe Interact. 2004;17(9):951–7. 10.1094/MPMI.2004.17.9.951 [DOI] [PubMed] [Google Scholar]
- 36.Chai Y, Tsai CS, Cho H, Winans SC. Reconstitution of the biochemical activities of the AttJ repressor and the AttK, AttL, and AttM catabolic enzymes of Agrobacterium tumefaciens. J Bacteriol. 2007;189(9):3674–9. 10.1128/JB.01274-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang JJ, Han JI, Zhang LH, Leadbetter JR. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2003;69(10):5941–9. 10.1128/AEM.69.10.5941-5949.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang JJ, Petersen A, Whiteley M, Leadbetter JR. Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine lactone acylase of Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2006;72(2):1190–7. 10.1128/AEM.72.2.1190-1197.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang K, Zhang XH. Quorum quenching agents: resources for antivirulence therapy. Mar Drugs. 2014;12(6):3245–82. 10.3390/md12063245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mei GY, Yan XX, Turak A, Luo ZQ, Zhang LQ. AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lactonase. Appl Environ Microbiol. 2010;76(15):4933–42. Epub 2010/06/08. 10.1128/AEM.00477-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schipper C, Hornung C, Bijtenhoorn P, Quitschau M, Grond S, Streit WR. Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl Environ Microbiol. 2009;75(1):224–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krysciak D, Schmeisser C, Preuss S, Riethausen J, Quitschau M, Grond S, et al. Involvement of multiple loci in quorum quenching of autoinducer I molecules in the nitrogen-fixing symbiont Rhizobium (Sinorhizobium) sp. strain NGR234. Appl Environ Microbiol. 2011;77(15):5089–99. 10.1128/AEM.00112-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiland-Bräuer N, Pinnow N, Schmitz RA. Novel reporter for identification of interference with acyl homoserine lactone and autoinducer-2 quorum sensing. Appl Environ Microbiol. 2015;81(4):1477–89. 10.1128/AEM.03290-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiland-Bräuer N, Kisch MJ, Pinnow N, Liese A, Schmitz RA. Highly Effective Inhibition of Biofilm Formation by the First Metagenome-Derived AI-2 Quenching Enzyme. Front Microbiol. 2016;7:1098 Epub 2016/07/13. 10.3389/fmicb.2016.01098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HW, Oh HS, Kim SR, Lee KB, Yeon KM, Lee CH, et al. Microbial population dynamics and proteomics in membrane bioreactors with enzymatic quorum quenching. Appl Microbiol Biotechnol. 2012. Epub 2012/08/01. [DOI] [PubMed] [Google Scholar]
- 46.Gao M, Chen H, Eberhard A, Gronquist MR, Robinson JB, Connolly M, et al. Effects of AiiA-mediated quorum quenching in Sinorhizobium meliloti on quorum-sensing signals, proteome patterns, and symbiotic interactions. Mol Plant Microbe Interact. 2007;20(7):843–56. 10.1094/MPMI-20-7-0843 [DOI] [PubMed] [Google Scholar]
- 47.Veal EA, Toone WM, Jones N, Morgan BA. Distinct roles for glutathione S-transferases in the oxidative stress response in Schizosaccharomyces pombe. J Biol Chem. 2002;277(38):35523–31. 10.1074/jbc.M111548200 [DOI] [PubMed] [Google Scholar]
- 48.Hayes JD, McLellan LI. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radic Res. 1999;31(4):273–300. [DOI] [PubMed] [Google Scholar]
- 49.Lin L, Dai S, Tian B, Li T, Yu J, Liu C, et al. DqsIR quorum sensing-mediated gene regulation of the extremophilic bacterium Deinococcus radiodurans in response to oxidative stress. Mol Microbiol. 2016;100(3):527–41. 10.1111/mmi.13331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.