Abstract
Objectives
Extracting data from publication reports is a standard process in systematic review (SR) development. However, the data extraction process still relies too much on manual effort which is slow, costly, and subject to human error. In this study, we developed a text summarization system aimed at enhancing productivity and reducing errors in the traditional data extraction process.
Methods
We developed a computer system that used machine learning and natural language processing approaches to automatically generate summaries of full-text scientific publications. The summaries at the sentence and fragment levels were evaluated in finding common clinical SR data elements such as sample size, group size, and PICO values. We compared the computer-generated summaries with human written summaries (title and abstract) in terms of the presence of necessary information for the data extraction as presented in the Cochrane review’s study characteristics tables.
Results
At the sentence level, the computer-generated summaries covered more information than humans do for systematic reviews (recall 91.2% vs. 83.8%, p<0.001). They also had a better density of relevant sentences (precision 59% vs. 39%, p<0.001). At the fragment level, the ensemble approach combining rule-based, concept mapping, and dictionary-based methods performed better than individual methods alone, achieving an 84.7% F-measure.
Conclusion
Computer-generated summaries are potential alternative information sources for data extraction in systematic review development. Machine learning and natural language processing are promising approaches to the development of such an extractive summarization system.
Keywords: Text Summarization, Text Classification, Machine Learning, Data Collection, Review Literature as Topic
Graphical abstract
1. Introduction
Systematic reviews (SR) are important information sources for healthcare providers, researchers, and policy makers. An SR attempts to comprehensively identify, appraise, and synthesize the best available evidence to find reliable answers to research questions (1). The Cochrane Collaboration is an internationally recognized non-profit organization that develops SRs for health-related topics. Cochrane reviews aim to identify and synthesize the highest standard in evidence-based practice (2). Cochrane usage data in 2009 showed that “Every day someone, somewhere searches The Cochrane Library every second, reads an abstract every two seconds, and downloads a full-text article every three seconds.” (3).
The development of systematic reviews has been faulted as resource-intensive and slow (4-6). Data extraction is one of the steps in SR development whose goal is to collect relevant information from published reports to perform quality appraisal and data synthesis, including meta-analysis. Yet, studies have shown that the manual data extraction task has a high prevalence of errors (7, 8). This is partially because of human factors such as limited time and resources, inconsistency, and tedium-induced errors. Computer methods have been proposed as a potential solution to enhance productivity and to reduce errors in SR data extraction.
Boundin et al. (9), Huang et al. (10, 11), and Kim et al. (12) investigated machine learning approaches to classify sentences that contain PICO (Population, Intervention, Control, and Outcome) elements. PICO is a popular framework used to formulate and find answers to clinical questions. Demner-Fushman and Lin (13), Kelly and Yang (14), and Hansen et al. (15) employed rule-based and machine learning approaches to extract PICO and patient related attributes. Those studies extracted information from abstracts, which, while important, are not sufficient for extracting information for SRs. In fact, extraction from full-text reports is the standard requirement in SR development (16). Full-text extraction is more challenging since it has to process much larger chunks of text containing substantial redundancy and noise.
Kiritchenko et al. (17) and de Bruijin et al. (18) developed ExaCT to help extract clinical trial characteristics. ExaCT is considered as one of the most successful full-text extraction systems for clinical elements. Their method first uses a machine learning classifier to select the top five relevant sentences for each element, and then uses hand-crafted weak extraction rules to collect values for each element. ExaCT selects RCT studies from top five core clinical journals that have full-texts available in HTML format. In practice, SRs must select studies outside the top five clinical journals and many study publications are not available in HTML format. Wallace et al. conducted another notable work on extracting relevant sentences from full-text PDF reports to aid SR data extraction (19). The authors used the supervised distant supervision algorithm to rank sentences based on the relevance to PICO elements. Extracting short phrases (or fragments) and measuring sensitivity (or recall) were not their primary focus.
In the present research, we investigated an automatic extractive text summarization system to collect relevant data from full-text publications to support the development of systematic reviews. Text summarization research aims to reduce texts while keeping the most important information. Previous research has generally followed two main approaches: extractive and abstractive (20, 21). Extractive approaches obtain relevant words, phrases, or sentences from the original text sources to construct the summary (22, 23). Abstractive approaches attempt to build a common semantic model and then generate graphs or natural language summaries to describe the model (24, 25). We followed the extractive approach in the present study to automatically collect relevant sentences and phrases from the published PDF manuscripts. Although we focused on common clinical trial data types such as sample size, group size, and PICO values, the technique can be applied to other data types that are used in SR development.
2. Methods
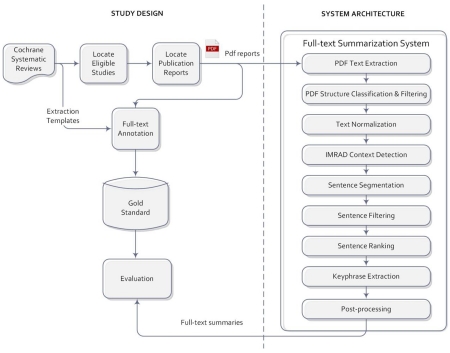
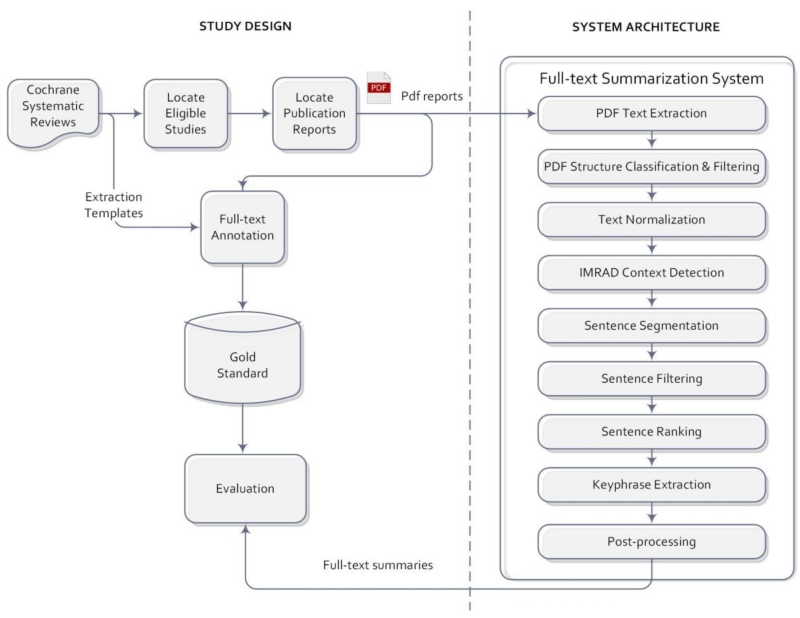
Our study design consisted of three main parts: (1) development of a data extraction gold standard from Cochrane reviews; (2) development of a computer system that can automatically generate summaries at the sentence and fragment levels; and (3) evaluation of system performance in the summarization of clinical trial data elements and a comparison with the study title and abstract. The overall system architecture and study design are summarized in Figure 1.
Figure 1.
System architecture and study design.
2.1. Gold Standard
From the Cochrane Library, we retrieved systematic reviews on the subject “heart and circulation” that were published after October 2014. In each review, we identified the included primary studies and archived publication reports in PDF format. A clinical trial might report partial results in multiple publications during the course of a study. Text summarization of multiple documents is not this study’s primary focus; therefore, we excluded those multiple-report trials from our dataset. We also excluded nonrandomized trials.
To develop the data extraction gold standard, we used the original Cochrane’s data extraction templates as references, reviewing full-text reports to validate and collect synonymous mentions (e.g., synonyms, abbreviations, morphological variations). For each document, we built an extraction template including five data elements: sample size, group size, population, study arm (intervention or control), and outcome. Sample size is defined as the total number of patients enrolled in the study as included in the statistical analysis. Group size is the number of participants in each study group. Sample size can be inferred by summing up all group sizes. Population is defined as the main characteristics of the target population recruited in the study. The population characteristics describe the group of patients sharing the same disease, the same demographics, or that underwent the same medical procedure. Study arm is defined as the name of an interventional or control treatment. We did not distinguish between intervention and control arms since this is a convention not always explicitly mentioned in publications and not every study has a control arm. For instance, groups absent of an intervention treatment or “placebo” treated groups are implicitly classified as control group. Outcome is defined as measurements used to assess a study hypothesis, such as clinical attributes and adverse events. We did not distinguish between the primary and secondary outcomes in individual studies since reviewers might have a different selection of primary outcome for a given systematic review. Table 1 shows examples of the extraction template with data extracted from two publication reports.
Table 1.
Data extraction template with examples extracted from the Cochrane review “Primary prophylaxis for venous thromboembolism in ambulatory cancer patients receiving chemotherapy”.
| Cochrane ID | Klerk 2005 |
|
| |
| Study Title | The effect of low molecular weight heparin on survival in patients with advanced malignancy |
| Sample Size | 302 |
| Group Size | 148 |
| 154 | |
| Population | Advanced Malignancy |
| Study Arm | low molecular weight heparin|Nadroparin |
| Placebo | |
| Outcome | death from any cause|death as a result of any cause|death major bleeding|non-major bleeding|bleeding |
|
| |
| Cochrane ID | Mitchell 2003 |
|
| |
| Study Title | Trend to efficacy and safety using antithrombin concentrate in prevention of thrombosis in children receiving l-asparaginase for acute lymphoblastic leukemia. Results of the PAARKA study. |
| Sample Size | 85 |
| Group Size | 25 |
| 60 | |
| Population | Children acute lymphoblastic leukaemia |
| Study Arm | antithrombin |
| Outcome | symptomatic or asymptomatic thrombotic event|thrombotic event major and minor bleeding|bleeding |
2.2. System overview
We implemented a pipeline of nine stages for data extraction. Overall, the system takes the PDF publication reports as input, and outputs text summaries including a list of relevant sentences for each data element as well as recommended key phrases in each sentence. The nine stages are explained below (see Figure 1):
-
(1)
PDF Text Extraction: We used the open-source tool PDFBox (26) to extract raw text from PDF documents. Text extracted using PDFBox tool has characteristics similar to manually copying-and-pasting text from a PDF reader. Principal structures are lost, and texts are broken into multiple lines of text snippets. However, the essential text order is well-maintained and can be used for natural language processing (NLP) research.
-
(2)
Text Classification & Filtering: we used a text classification algorithm (27) to automatically categorize text snippets into five categories (TITLE, ABSTRACT, BODYTEXT, SEMISTRUCTURE, and METADATA). Also from our prior work, we found that filtering semi-structures and publication metadata enhanced efficiency and effectiveness of information extraction (IE) system operating on full-text articles. Therefore, we discarded those non-prose texts at this stage.
-
(3)
Text Normalization: the goal of this step is to translate texts into canonical form. More specifically, we find and replace all numbers in literal expression to numeric format (e.g., “a hundred and three patients” ➔ 103 patients). We developed an acronym normalization module that reads full-text documents, detecting and replacing acronyms to their fully expanded form. The acronym normalization algorithm first checks all parenthetical expressions and the preceding text (e.g., “small cell lung cancer (SCLC)”) for candidate acronym pairs, then uses the pattern of initial letters for validation. This is the most frequently used acronym pattern in biomedical publications. There are elaborated acronym normalization algorithms which can be incorporated to our system (28-30). Future testing and adaptation are needed to select an optimal approach for RCT publications. Acronym normalization increases the clarity of the sentences in manual review and improves performance of the concept mapping approach in a subsequent stage.
-
(4)
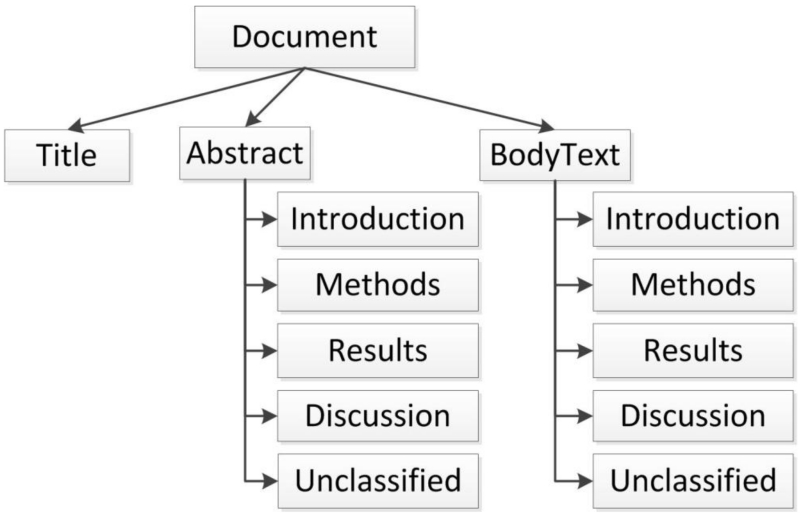
IMRAD Context Detection: This step attempts to assign categories of the common scientific article organizational structure IMRAD (i.e., introduction, methods, results, discussion) to text snippets. We relied on the recognition of common headings in text to assign the IMRAD class to the subsequent snippets until detecting a new heading that triggers a context change. The text snippets are clustered into different context nodes as illustrated in Figure 2.
-
(5)
Sentence Segmentation: We used the Stanford NLP sentence splitter (31) to perform sentence segmentation in different context nodes. Performing sentence segmentation at this later stage has the advantage of knowing the contextual information of the generated sentences. In addition, noisy texts (e.g., tables, figures, author metadata) are filtered in previous stages, which helps to define correct sentence boundaries.
-
(6)
Sentence Filtering: In this step, we attempt to filter all sentences that discuss background knowledge and therefore are not relevant to the extraction goal. We filter sentences having the context INTRODUCTION, sentences containing year and citation expressions, and sentences referring to other studies (e.g., containing phrases such as “these trials”, “et al.”, and “previous studies”).
-
(7)
Sentence Ranking: The goal of this step is to prioritize sentences for each individual data element. We used the Support Vector Machine Regression classifier, Sequential Minimal Optimization, implemented in Weka (32) with a polynomial kernel and default model parameters. To train the regression model, we used 50% of the sentences in the gold standard as the training set. The response variable is set to the number of times the target element appears in a sentence. The predictor variables or features can be divided into three groups:
-
a)
Bag-Of-Terms (BOT) Group: this feature group is based on words, terms, or patterns that appear in the sentence. First, the top 100 most frequent words that are present in relevant sentences (i.e., sentences that contain at least one target element for data extraction) were selected as BOT features. We used the frequency of those words in the sentence to generate a feature vector. Second, we used a binary variable determining whether the sentence contains at least a true-positive mention in the training set. Third, we used regular expression features to capture text patterns that are strong indicators of relevant sentences. For the scope of the present study, we maintained a small set of regular expression features per data element (e.g., SampleSize: “total of \d+ patients”, Outcome: “(end points?|endpoints?| outcomes?) was|were|included”).
-
b)
Context Group: this group includes two features based on the context of the sentence. The document-structure feature is a nominal attribute that takes one of three values: TITLE, ABSTRACT, or BODYTEXT. The IMRAD nominal feature accepts one of four values: INTRODUCTION, METHODS, RESULTS, or DISCUSSION. If the sentence contexts were not determined from the previous steps, they are treated as missing values.
-
c)
Semantic Group: This group uses 15 semantic groups from the Unified Medical Language System (UMLS) (33) as features. We used MetaMap (34) to map sentences to UMLS concepts, from which we map the UMLS sematic types to sematic groups. Then we aggregate and compute the sematic group features based on their frequency.
Based on the three feature groups above, we created four machine learning models for comparing and selecting the best model for each data element: BOT, BOT+Context, BOT+Semantic, and BOT+Context+Semantic.
-
a)
-
(8)
Key phrase Extraction: The goal of this step is to recognize key phrases from the sentences to help reviewers quickly identify parts of the sentence that are relevant to the extraction goal. Based on the type of data element we employed a subset of the following techniques:
-
a)
Regular expression (regex) matching: Since numbers are normalized to numeric expressions, regex pattern matching is an extremely useful technique in the recognition of numeric values. We used a list of regular expressions rules (Table 2) to extract numeric values for sample size and group size. Each regex rule contains context expressions and capturing “groups” referring to target elements. Since each sentence might have a unique way to convey the numeric value, only the best match was considered.
-
b)
Noun phrase chunking and regex matching: For literal-expression data elements (e.g., outcome), applying regular expression matching might detect very long phrases, which is less useful for key phrase identification. Therefore, we performed noun phrase chunking to restrict the matching only to noun phrases of the sentence. To perform the noun phrase chunking, we used the Stanford parser to generate a Penn tree and used the Tregex parser (35) to collect all noun phrase expressions.
-
c)
Concept Mapping & Semantic type restriction: The majority of relevant key terms can be found in medical terminologies. Concept-mapping was found to be an effective approach. We used MetaMap (34) to detect medical terms from the sentence that can be mapped to UMLS concepts. To enhance precision, we restricted the mapping to a few semantic types (Table 2) relevant to the target elements. The selection of optimal sets of semantic types was based on experimental testing on the training set. We maintained an optimal set of semantic types for each data element. Some semantic types are shared by multiple elements.
-
d)
Supplement Dictionary: This approach enables the inclusion of individual literal data element terms not otherwise available in the UMLS controlled terminologies. Since key terms are reused in multiple publication reports, maintaining a good-coverage, element-specific dictionary improves accuracy as well as lowers computational overhead. We added all true positive terms in our training set to the dictionary and matched them against the sentence to extract the candidate terms. Since our training set is relatively small and not representative of the literature as a whole, we still need to combine this method with other generalizable methods.
-
a)
-
(9)
Post-processing: This step filters phrases that are lengthy (> 5 words), phrases contained in other phrases, and phrases contained in a stop list. The stop list was constructed using the top 20 most frequent false-positive terms upon evaluating the system on the training set, which were never recognized as true-positives in all training documents.
Figure 2.
The standard structure organization of text in a scientific article.
Table 2.
Regular expressions and semantic types used for extracting individual elements.
BOUNDARY (B) = (?: and| to| in| with| between| \.| _| ,|$)
| Data element | Extraction Methods |
|---|---|
| Sample Size/ Group Size |
Regular expression:
|
| Population | Semantic type:
|
| Study Arm | Semantic type:
|
| Outcome | Regular expression:
|
Semantic type:
|
2.3. Evaluation Approach
The system creates a ranked list of sentences from which it selects the top N sentences to generate the summary. To evaluate the system performance at the Nth sentence, we used the following metrics:
We selected recall as the primary outcome to emphasize the information coverage of the computer suggested summary. The goal is to enable reviewers to find as much of the needed information in the summary as possible and to reduce the need for conducting manual full-text review for missing information. The evaluation conducted by Wallace et al. (19) was focused on precision of the top ranked sentences, a popular measure in information retrieval and question answering systems. However, recall is the most important metric in judging the quality of an automatic text summarization system in the context of systematic review development.
We grouped together the evaluation of sample size and group size. Sample size might not always be reported explicitly in texts but it can be inferred by summation of all group size values. To account for a true positive sample size or group size, we applied a binary rule: true if the sentence contains a sample size value or all of group size values, and false otherwise.
We tested a hypothesis that text summaries generated by the machine learning classifier retains more information for systematic reviews than title and abstract. Title and abstract, in human-written summaries, are common and popular information sources in SR development. However, those sources might not have sufficient information needed for collecting SR data. This hypothesis evaluates the computer-generated summary against title and abstract, to serve as a potential alternative information source for SR citation screening and data extraction processes.
To test the hypothesis, we collected the corresponding abstracts and titles indexed in MEDLINE. Titles and abstracts were processed by the normalization and segmentation steps described earlier. Then, the abstract/title sentences were compared with system recommended sentences. N is set to number of abstract/title sentences. The same evaluation was applied to both algorithm versions with recall selected a priori as the primary outcome. To test the significance of performance difference, we used the Chi-square test to assess the sample/group size data element, and the Wilcoxon signed rank test to assess other elements. The significance level was set at p < 0.05.
Evaluation at the fragment-level concerns the ability to highlight key phrases from the sentence. To obtain the sentence corpus, we collected all recommended relevant sentences using our best classifiers from the sentence-level evaluation. Standard information extraction metrics (recall, precision, and f-measure) were measured at the sentence unit. To consider a phrase recommendation as a true positive, an exact match was required for numeric elements (e.g., sample size, group size). Literal elements, phrases of up to five words that contain a correct mention or one of its synonyms, were considered as true positives. We evaluated and compared the performance of the following extraction methods: Regex Matching, Concept Mapping, Supplement Terminology, and a combination of these three methods.
3. Results
The gold standard was composed of 48 publication reports included in 8 systematic reviews. Although all these studies are randomized controlled trials, only 16% of them have posted structured results in ClinicalTrials.gov. The annotation task found 48 sample sizes, 116 group sizes, 53 populations, 99 study arms, and 267 outcomes. Terms that co-referred to the same concept are counted only once. At the sentence-level, 3,166 sentences in the training set were used to train the regression model, and 3,404 sentences in the test set were used for evaluation. At the fragment level, the number of test sentences per data element was as follows: Sample/group size: 39; population: 133; study arm: 225; and outcome: 226. Figure 3 shows an example of a computer-generated summary including topmost relevant sentences and recommended key phrases.
Figure 3.
Example of computer generated summary for the clinical outcome element.
Table 3 shows the results of the evaluation of computer-generated summaries by the four machine learning models. The best ML models were different for specific data elements. The BOT+Context model performed best for sample/group size and population elements; the BOT+Context+Semantic model performed best for the study arm element; and the BOT+Semantic model performed best for the outcome element.
Table 3.
Performance comparison of various machine-learning based summaries and title/abstract summaries.
| Bag-Of-Term (BOT) |
BOT+Context | BOT+Semantic | BOT+Context +Semantic |
|||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Recall | Precision | Recall | Precision | Recall | Precision | Recall | Precision | |
| Sample /Group Size |
83.3 | 14.7 | 91.7 | 15.1 | 75.0 | 13.3 | 79.2 | 13.1 |
| Population | 93.8 | 47.5 | 100 | 50.7 | 100.0 | 47.5 | 95.8 | 52.2 |
| Study Arm | 97.9 | 80.8 | 100 | 84.0 | 95.8 | 81.2 | 100 | 84.9 |
| Outcome | 72.4 | 85.0 | 71.9 | 84.2 | 73.1 | 85.3 | 71.9 | 85.5 |
Best performing ML models for each data element are marked in bold.
Table 4 compares computer generated summaries from our best classifiers and the manually written summaries (title and abstract). On average, summaries generated from our systems achieved 91.2% recall and 59% precision, which is significantly better than title and abstract (recall: +7.4%, p<0.001; precision: +20%, p<0.001).
Table 4.
Performance comparison of machine-generated summaries and title/abstract summaries.
| Machine-generated summaries |
Manual summaries (abstract and title) |
p-value | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Recall | Precision | Recall | Precision | Recall | Precision | |
| Sample /Group Size |
91.7 | 15.1* | 83.3 | 9.0 | 0.186 | 0.002 |
| Population | 100 | 50.7* | 100.0 | 28.7 | NA | 0.004 |
| Study Arm | 100 | 84.9* | 100.0 | 63.4 | NA | <0.001 |
| Outcome | 73.1* | 85.3* | 51.9 | 54.9 | <0.001 | <0.001 |
| Mean | 91.2 * | 59 * | 83.8 | 39 | <0.001 | <0.001 |
Indicates statistically significant improvement over abstract and title. NA indicates the statistical test is not valid to compare two exactly equal groups.
For individual elements, precision reached statistically significant improvement on all data elements, while statistically significant improvement of recall was only achieved on the outcome element. Population and intervention are often reported in an article’s title or abstract. Both baseline and our methods reached perfect recall for those elements. A non-significant difference (+8.4%, p=0.32) was found for the sample/group size element. A subsequent analysis showed that the ML classifier essentially retrieved a group of sentences similar to the abstract and title, but also retrieved relevant sentences from the body-text section.
On the fragment-level (Table 5), the regular expression matching approach achieved an F-measure of 90.3% on Sample/Group Size extraction. This confirms regex matching is the most common and effective approach in extracting numeric values. For other literal elements, the ensemble approach outperformed each individual method. The F-measure was 79.8% for Population, 86.8% for Study Arm, and 81.8% for Outcome. On average, our best extraction methods achieved an F-measure of 84.7%. While recall (95.2%) was satisfactory, the precision (76.6%) could be improved further.
Table 5.
Fragment-level performance of various extraction methods.
| Regex Matching | Concept Mapping | Supplement Dictionary | Combined Method | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| R | P | F1 | R | P | F1 | R | P | F1 | R | P | F1 | |
| SampleSize /GroupSize |
93.6 | 87.3 | 90.3 | NA | NA | NA | NA | NA | NA | 93.6 | 87.3 | 90.3 |
| Population | NA | NA | NA | 86.1 | 60.7 | 71.2 | 70.7 | 66.4 | 68.5 | 97 | 67.8 | 79.8 |
| Study Arm | NA | NA | NA | 94.9 | 78.5 | 85.9 | 82.5 | 87.6 | 85 | 96.8 | 78.6 | 86.8 |
| Outcome | 14.0 | 19.7 | 16.4 | 56.4 | 41.9 | 48.1 | 82.4 | 79.5 | 80.9 | 93.5 | 72.6 | 81.8 |
4. Discussion
In this study we developed and evaluated an extractive text summarization system that can support data extraction in the development of systematic reviews. Instead of automatically generating the extraction results, the system generates summaries that humans can review and find relevant information. It is our contention that human involvement will always be necessary in SR development based on NLP methods. We attempted to locate sample size and PICO information from full-text PDF reports, which adds knowledge to previous IE research using simpler, more convenient sources (e.g., MEDLINE abstracts).
In sentence ranking, the best ML model varied for different data elements. Different data elements require a different set of features and optimization techniques. This finding suggest that future studies adopt both element-specific features and generic features to achieve optimal performance. The use of context features and semantic features improved the performance of ML models that use Bag-Of-Term features alone. This finding suggests that rhetorical context and semantic analysis are useful in developing text-based classification models in the SR context. For the primary outcome (recall), the system-generated summaries performed equally (Study Arm and Population) or better (Sample/Group Size and Outcome) than the abstract/title summaries. The majority but not all information can be found in the abstract and title of the studies. Therefore, IE systems supporting the development of SRs need to operate at full-text scale to maximize the comprehensiveness of data extraction (16). In addition, precision was better in full-text reports versus title/abstract for all data elements. Better precision corresponds to a higher number of relevant sentences in the top ranked list. In full-text documents, information can be repeated in multiple sections. The ML system was better in collecting repeated relevant sentences, which offers reviewers multiple sources to confirm the extraction results.
In fragment-level extraction, we proposed three extraction methods: regular expression matching, mapping to UMLS concepts, and element-specific dictionary. Regular expressions are most useful for extracting templates or numerical values. Designing and implementing a regular expression approach requires considerable manual work unless regular expression learning techniques are effectively applied (36, 37). Mapping text to UMLS concepts is one of the extraction methods commonly used in clinical and biomedical NLP studies (38-40). MetaMap tends to perform well in the recognition of texts that can be mapped to medical terms. However, there are more than 3 million UMLS concepts (2015AA Release) and classifying them to the data element of interest is challenging. In this study, we employed a simple semantic type restriction approach to categorize concepts to a specific element. There are other approaches to categorize UMLS concepts such as heuristics using UMLS concept relationships (41), semantic distribution (26), or machine learning (42), which are deserving of additional investigation and optimization to perform well on sample size and PICO elements. Our element-specific dictionary approach was motivated by the fact that the UMLS Metathesaurus might not fully cover medical terms required for SR-specific extraction needs. Element-specific terms are needed to complement the UMLS concept mapping approach. In this study, we utilized true-positive terms that appeared in the training set and that achieved a good coverage (60% recall) on the test set. The experiment results showed that an ensemble approach combining the three methods performed better than any of the individual methods. For PICO elements, the system’s recall was better than precision (95.2% vs. 76.6%), which meets our performance goal. Recall is often more important for semi-automated extraction, since humans are effective at judging whether recommended phrases are true positives; however, humans tend to miss information when screening large amounts of textual contents.
In summary, we demonstrated the feasibility of using machine learning and natural language processing approaches to automatically generate text summaries to aid data collection in SRs. The machine-generated summary has the potential to replace the abstract and title sources when searching for specific data elements.
4.2. Limitations
This study focused on sample size and PICO elements, which are commonly reported in randomized controlled trial studies. There are data elements suggested by the Cochrane Collaboration that were not covered. Some of those elements such as funding sources, study design, and study authors can be easily retrieved from Medline metadata. Data elements such as age, sex distribution, and number of participants in each group are usually reported in table structures, which require a specialized table parsing algorithm. Other elements such as detailed inclusion/exclusion criteria, study duration, randomization method, and blinding can be extracted with an extension of our proposed method. There are other machine learning models, such as linear regression, multilayer perceptron, and Gaussian processes that were not evaluated in this study and could be investigated in future research. For comparison of feature groups, we only used support vector machine regression given its popularity and effectiveness in data mining research (43-45).
4.3. Future work
To fully support the vision of computer-assisted data extraction, the summarization system needs to support diverse systematic review data elements and have an interactive user interface well-integrated into the traditional data extraction workflow. Additional innovative approaches in sentence ranking and phrase extraction can be explored to find optimal strategies for each individual data element. Studies are also needed to assess systematic reviewers’ interaction with the computer-generated summaries, including measures of work efficiency and perceived relevance, completeness, readability, comprehensibility, connectedness, and satisfaction with the text summaries.
5. Conclusion
We presented an extractive text summarization system that can help human reviewers in collecting sample size and PICO values from full-text PDF reports. The system is composed of two main components: sentence ranking and key phrase extraction. In sentence ranking, we demonstrated that using a machine learning classifier on full text to prioritize sentences performed equally or better than screening title and abstract. These findings highlight the potential of using a machine learning approach to replace the traditional abstract screening in searching information for systematic review development. For fragment-level extraction, we showed that using an ensemble approach combining three different extraction methods obtained the best extraction performance. Future research is needed to integrate the system with an effective and usable user interface.
Highlights.
We studied an extractive text summarization system to assist data extraction tasks in systematic reviews.
We extracted sample size, group size, and PICO values automatically from full-text PDF reports.
In information seeking, computer generated summaries are a potential alternative source to human written summaries (based on abstracts and title).
Machine learning and natural language processing approaches are useful in automatic text summarization in the systematic review context.
Acknowledgments
This study was supported in part by grants 1R01LM011416-02, 4R00LM011389-02 and R01-LM010981 from the National Library of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest. The authors have no conflicts of interest to declare.
References
- 1.Cook DJ, Mulrow CD, Haynes RB. Systematic reviews: synthesis of best evidence for clinical decisions. Annals of internal medicine. 1997;126(5):376–80. doi: 10.7326/0003-4819-126-5-199703010-00006. [DOI] [PubMed] [Google Scholar]
- 2.Tong S, Koller D, editors. Restricted bayes optimal classifiers. AAAI/IAAI; 2000. [Google Scholar]
- 3.Dahlmeier D, Ng HT. Domain adaptation for semantic role labeling in the biomedical domain. Bioinformatics. 2010;26(8):1098–104. doi: 10.1093/bioinformatics/btq075. [DOI] [PubMed] [Google Scholar]
- 4.Limited CTC Directors’ reports and financial statements. 2013 [Google Scholar]
- 5.Shojania KG, Sampson M, Ansari MT, Ji J, Doucette S, Moher D. How quickly do systematic reviews go out of date? A survival analysis. Annals of internal medicine. 2007;147(4):224–33. doi: 10.7326/0003-4819-147-4-200708210-00179. [DOI] [PubMed] [Google Scholar]
- 6.Bragge P, Clavisi O, Turner T, Tavender E, Collie A, Gruen RL. The Global Evidence Mapping Initiative: scoping research in broad topic areas. BMC medical research methodology. 2011;11(1):92. doi: 10.1186/1471-2288-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones AP, Remmington T, Williamson PR, Ashby D, Smyth RL. High prevalence but low impact of data extraction and reporting errors were found in Cochrane systematic reviews. Journal of clinical epidemiology. 2005;58(7):741–2. doi: 10.1016/j.jclinepi.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 8.Gotzsche PC, Hrobjartsson A, Maric K, Tendal B. Data extraction errors in meta-analyses that use standardized mean differences. JAMA: the journal of the American Medical Association. 2007;298(4):430–7. doi: 10.1001/jama.298.4.430. [DOI] [PubMed] [Google Scholar]
- 9.Boudin F, Nie J-Y, Bartlett JC, Grad R, Pluye P, Dawes M. Combining classifiers for robust PICO element detection. BMC Med Inform Decis Mak. 2010;10:29. doi: 10.1186/1472-6947-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolpert DH. Stacked generalization. Neural networks. 1992;5(2):241–59. [Google Scholar]
- 11.Huang K-C, Chiang IJ, Xiao F, Liao C-C, Liu CC-H, Wong J-M. PICO element detection in medical text without metadata: are first sentences enough? J Biomed Inform. 2013;46(5):940–6. doi: 10.1016/j.jbi.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Eddy SR. Hidden markov models. Current opinion in structural biology. 1996;6(3):361–5. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 13.Demner-Fushman D, Lin J. Answering clinical questions with knowledge-based and statistical techniques. Computational Linguistics. 2007;33(1):63–103. [Google Scholar]
- 14.Ware M, Mabe M. The STM report: An overview of scientific and scholarly journal publishing. 2015 [Google Scholar]
- 15.Hansen MJ, Rasmussen NO, Chung G. A method of extracting the number of trial participants from abstracts describing randomized controlled trials. J Telemed Telecare. 2008;14(7):354–8. doi: 10.1258/jtt.2008.007007. [DOI] [PubMed] [Google Scholar]
- 16.Higgins JP, Green S. Cochrane handbook for systematic reviews of interventions. Wiley Online Library; 2008. [Google Scholar]
- 17.Kiritchenko S, de Bruijn B, Carini S, Martin J, Sim I. ExaCT: automatic extraction of clinical trial characteristics from journal publications. BMC Med Inform Decis Mak. 2010;10:56. doi: 10.1186/1472-6947-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bruijn B, Carini S, Kiritchenko S, Martin J, Sim I. Automated information extraction of key trial design elements from clinical trial publications; AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium; 2008; pp. 141–5. [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace BC, Kuiper J, Sharma A, Zhu MB, Marshall IJ. Extracting PICO Sentences from Clinical Trial Reports using Supervised Distant Supervision. [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra R, Bian J, Fiszman M, Weir CR, Jonnalagadda S, Mostafa J, et al. Text summarization in the bio-medical domain: A systematic review of recent research. J Biomed Inform. 2014 doi: 10.1016/j.jbi.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pivovarov R, Elhadad N. Automated methods for the summarization of electronic health records. Journal of the American Medical Informatics Association. 2015;22(5):938–47. doi: 10.1093/jamia/ocv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aliguliyev RM. A new sentence similarity measure and sentence based extractive technique for automatic text summarization. Expert Systems with Applications. 2009;36(4):7764–72. [Google Scholar]
- 23.Mihalcea R, editor. Graph-based ranking algorithms for sentence extraction, applied to text summarization; Proceedings of the ACL 2004 on Interactive poster and demonstration sessions; Association for Computational Linguistics. 2004. [Google Scholar]
- 24.Fiszman M, Rindflesch TC, Kilicoglu H. Abstraction summarization for managing the biomedical research literature; Proceedings of the HLT-NAACL Workshop on Computational Lexical Semantics; Boston, Massachusetts. 1596442: Association for Computational Linguistics. 2004.pp. 76–83. [Google Scholar]
- 25.Hunter J, Freer Y, Gatt A, Logie R, McIntosh N, Van Der Meulen M, et al., editors. Summarising complex ICU data in natural language; Amia annual symposium proceedings; American Medical Informatics Association. 2008. [Google Scholar]
- 26.Apache PDFBox - A Java PDF Library. 2015 Available from: https://pdfbox.apache.org/
- 27.Bui DDA, Del Fiol G, Jonnalagadda S. PDF text classification to leverage information extraction from publication reports. J Biomed Inform. 2016 doi: 10.1016/j.jbi.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hearst MS. A simple algorithm for identifying abbreviation definitions in biomedical text. 2003 [PubMed] [Google Scholar]
- 29.Osiek BA, Xexéo G, de Carvalho LAV. A language-independent acronym extraction from biomedical texts with hidden Markov models. IEEE Transactions on Biomedical Engineering. 2010;57(11):2677–88. doi: 10.1109/TBME.2010.2051033. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Yoon J. Link-topic model for biomedical abbreviation disambiguation. J Biomed Inform. 2015;53:367–80. doi: 10.1016/j.jbi.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 31.Manning CD, Surdeanu M, Bauer J, Finkel J, Bethard SJ, McClosky D. The Stanford CoreNLP Natural Language Processing Toolkit [Google Scholar]
- 32.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explor Newsl. 2009;11(1):10–8. [Google Scholar]
- 33.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Studies in health technology and informatics. 2001;84(Pt 1):216–20. [PMC free article] [PubMed] [Google Scholar]
- 34.Aronson AR. Metamap: Mapping text to the umls metathesaurus. NLM, NIH, DHHS; Bethesda, MD: 2006. pp. 1–26. [Google Scholar]
- 35.Levy R, Andrew G, editors. Tregex and Tsurgeon: tools for querying and manipulating tree data structures; Proceedings of the fifth international conference on Language Resources and Evaluation; Citeseer. 2006. [Google Scholar]
- 36.Bui DD, Zeng-Treitler Q. Learning regular expressions for clinical text classification. J Am Med Inform Assoc. 2014;21(5):850–7. doi: 10.1136/amiajnl-2013-002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Krishnamurthy R, Raghavan S, Vaithyanathan S, Jagadish HV. Regular expression learning for information extraction; Proceedings of the Conference on Empirical Methods in Natural Language Processing; Honolulu, Hawaii. 1613719: Association for Computational Linguistics. 2008.pp. 21–30. [Google Scholar]
- 38.Soguero-Ruiz C, Hindberg K, Rojo-Alvarez J, Skrovseth SO, Godtliebsen F, Mortensen K, et al. Support Vector Feature Selection for Early Detection of Anastomosis Leakage from Bag-of-Words in Electronic Health Records. 2014 doi: 10.1109/JBHI.2014.2361688. [DOI] [PubMed] [Google Scholar]
- 39.Xu R, Hirano Y, Tachibana R, Kido S. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2011. Springer; 2011. Classification of diffuse lung disease patterns on high-resolution computed tomography by a bag of words approach; pp. 183–90. [DOI] [PubMed] [Google Scholar]
- 40.Bui DD, Jonnalagadda S, Del Fiol G. Automatically finding relevant citations for clinical guideline development. J Biomed Inform. 2015 doi: 10.1016/j.jbi.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuncheva LI. Combining pattern classifiers: methods and algorithms. John Wiley & Sons; 2004. [Google Scholar]
- 42.Beel J, Gipp B, Shaker A, Friedrich N. Research and Advanced Technology for Digital Libraries. Springer; 2010. SciPlore Xtract: Extracting Titles from Scientific PDF Documents by Analyzing Style Information (Font Size) pp. 413–6. [Google Scholar]
- 43.Schäfer U, Kiefer B. Advances in deep parsing of scholarly paper content. Springer; 2011. [Google Scholar]
- 44.Japkowicz N, Stephen S. The class imbalance problem: A systematic study. Intelligent data analysis. 2002;6(5):429–49. [Google Scholar]
- 45.Van Hulse J, Khoshgoftaar TM, Napolitano A, editors. Experimental perspectives on learning from imbalanced data; Proceedings of the 24th international conference on Machine learning; ACM. 2007. [Google Scholar]