Abstract
Protein folding in cells occurs in the presence of high concentrations of endogenous binding partners, and exogenous binding partners have been exploited as pharmacological chaperones. A combined mathematical modeling and experimental approach shows that a ligand improves the folding of a destabilized protein by biasing the kinetic partitioning between folding and alternative fates (aggregation or degradation). Computationally predicted inhibition of test protein aggregation and degradation as a function of ligand concentration are validated by experiments in two disparate cellular systems.
Proteins in a physiological milieu can have many binding partners. Surprisingly, little attention has been paid to how such ligands affect protein folding, despite the current interest in in vivo protein folding1-5. In the intracellular environment, proteins must navigate an intersecting folding–aggregation landscape to reach their functional states6. In principle, native ligands as well as pharmacological chaperones—ligands added to cells as potential therapeutics7—can influence this landscape. In the present study, we explore how ligand binding affects folding in the cell by combining mathematical modeling with experiments.
Our model for in vivo protein folding in the presence of ligands is pictured in Fig. 1. In this model, protein is synthesized in an unfolded state (U), which can fold to the native state (N) with forward and reverse rate constants kf and ku. Alternatively, the unfolded state can be degraded (Deg) or aggregate (A). Degradation is irreversible with a rate constant kdeg. We also treat aggregation as being irreversible with a rate constant kagg, although intracellular aggregates can be disaggregated by chaperones or degraded by autophagy8. These and other effects of the cellular protein folding machinery are subsumed into the rate constants in our model. The native state can bind to a ligand (L) to form a complex (N:L) with association and dissociation rate constants ka and kd. Finally, the N or N:L states can be secreted with a rate constant ksec. We assume that there is a large extracellular reservoir of ligand with a constant concentration and that the intracellular ([L]in) and extracellular ([L]out) ligand concentrations are proportional: [L]in = Kio[L]out. The model's rate equations and more detailed discussion of the model are given in Supplementary Results, Supplementary Note.
Figure 1. A model for the partitioning of protein among folding, aggregation, and degradation pathways.
The species in the model are: U, unfolded protein; N, natively folded protein; L, unbound ligand; N:L, ligand bound natively folded protein; Deg, degraded protein; A, aggregated protein; Sec, secreted protein. The total protein synthesized, Ptot, includes all of these states. The rate constants are: σ, protein synthesis rate (μM s−1); kf, folding rate constant (s−1); ku, unfolding rate constant (s−1); ka, protein–ligand association rate constant (μM−1 s−1); kd, protein–ligand dissociation rate constant (s−1); kdeg, degradation rate constant (s−1); kagg, aggregation rate constant (s−1); ksec, secretion rate constant (s−1).
The fraction of protein that remains soluble (i.e., is not degraded and does not aggregate) at a given time, or Fr, is experimentally measureable. Fr will evolve in two stages during a protein expression time course when a ligand is present. It rapidly reaches a first “pseudo-steady state” during which ligand binding is effectively irreversible (ka[N][L] ≫ kd[N:L]). The system then gradually approaches a second, more robust pseudo-steady state as ligand dissociation becomes appreciable. In both of these stages, Fr is given by:
| (1) |
where [A]t, [Deg]t, and [Ptot]t are the concentrations of aggregated, degraded, and total protein synthesized at time t, and B1 = kf/(kagg + kdeg) and B2 = ksec/ku during both pseudo-steady states (see Supplementary Note). However, during the first pseudo-steady state B3 = kaKio/ku, whereas during the second B3 = kaKio/(ku(1+kd/ksec)). When necessary, we will write the former as B3,1 and the latter as B3,2. The Fr vs. [L]out curve shifts to higher ligand concentrations during the second pseudo-steady state because B3,2 < B3,1, indicating that maintaining a given protein level requires more ligand when ligand dissociation is non-negligible. The time needed to transition from the first to the second pseudo-steady state is discussed in the Supplementary Note, but we note that it is greater in systems that do not secrete protein than in systems that do.
Equation (1) predicts that Fr increases with [L]out until it reaches the following limit:
| (2) |
Thus, at very high ligand concentrations, a protein's fate depends on folding kinetics (kf) but not folding thermodynamics. In addition, the fact that Fr,max is determined by the ratio kf/(kagg+kdeg) illustrates that analyzing Fr vs. [L] curves using our model generally yields information on rate constant ratios. This circumstance arises because Fr measures the partitioning of protein molecules among their possible fates at pseudo-steady states, which is a function of relative, not absolute, rates.
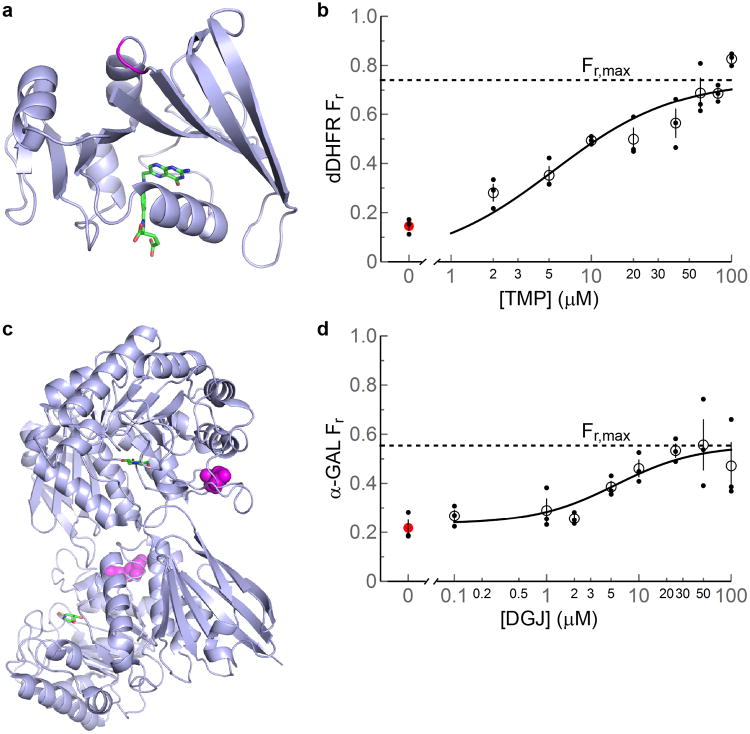
To test the predictions of equation (1), we first studied a mutant of the E. coli protein dihydrofolate reductase (dDHFR) expressed in E. coli in the presence of trimethoprim (TMP), a DHFR ligand9. The mutation in question was a -Gly-Gly- insertion between K106 and A107 in a surface loop (Fig. 2a). This mutation destabilizes DHFR and makes it aggregation-prone but leaves TMP binding largely unperturbed (Kd = 37 ± 6 nM compared to 9-15 nM for wild type DHFR9; Supplementary Fig. 1). In vitro, dDHFR aggregation is efficiently, though not completely, inhibited by an equimolar concentration of TMP (Supplementary Fig. 2).
Figure 2. The proteins studied and plots of fraction soluble protein remaining (Fr) vs. ligand concentration.
(a) Wild type E. coli DHFR with bound folate (colored by atom) (PDB ID: 7DFR24). The site of the -Gly-Gly- insertion in the dDHFR mutant is in magenta. (b) Fr vs. [TMP] (expressed for 1.5 h at 37 °C). The fit of equation (1) to the data is shown (solid curve) with B1(kf/(kagg + kdeg)), = 2.9 ± 0.8, B3,1 = kaKio/ku = 0.048 ± 0.025 μM−1, and R2 = 0.76 (0.84 after correction for measurement error). (c) Dimeric wild type human α-GAL with bound DGJ (colored by atom) (PDB ID: 3S5Y25). The R301Q mutation site is in magenta. (d) Fr vs. [DGJ] (expressed for 24 h at 37 °C). The fit of equation (1) to the data is shown (solid curve) with B1 = 1.2 ± 0.2, B2 (ksec/ku) = 0.33 ± 0.11, B3,2 = kaKio/(ku(1+kd/ksec)) = 0.14 ± 0.10 μM−1, and R2 = 0.59 (0.88 after correction for measurement error). In (b) and (d), data from triplicate measurements are shown as smaller filled circles; the dashed line represents the maximum value of Fr according to equation (2); the value of Fr at [ligand] = 0 is shown as a filled red circle; means are shown as open circles; and error bars represent the standard error of the mean. The consistency of observed standard errors argues for similar underlying variability. Sample sizes were chosen based on errors observed in previous studies10.
In the absence of TMP, dDHFR is mostly aggregated (Fr = 0.15 ± 0.02) after expression in E. coli BL21 (DE3) cells for 1.5 hours at 37 °C (Fig. 2b; Supplementary Fig. 3a), unlike wild type DHFR, which does not aggregate when overexpressed10. Titration with TMP increases Fr but dDHFR still partially aggregates even at [TMP] = 100 μM (Fig. 2b; Supplementary Fig. 3a). This behavior is consistent with equations (1) and (2). In addition, equation (1) fits well to the data in Fig. 2b, arguing that our model captures the essential features of the system. Arresting dDHFR synthesis by adding chloramphenicol in the presence of excess TMP did not change Fr after 1 h, consistent with our assumption that disaggregation is negligible (Supplementary Fig. 4a).
Interpreting the fit of equation (1) to the data in Fig. 2b requires knowing whether the dDHFR expression system is in the first or second pseudo-steady state, or somewhere in between. Two experiments suggest that the system is in the first pseudo-steady state. First, arresting synthesis with chloramphenicol and shifting to medium lacking TMP led to only a modest change in Fr after 1 h, arguing that dDHFR did not rapidly re-equilibrate from the ligand-bound to the free state (Supplementary Fig. 4b). Second, measurements of Fr vs. ligand concentration at the 0.5 h and 1 h time points were similar to those at the 1.5 h time point (Supplementary Fig. 4c), consistent with ligand-bound dDHFR accumulating as a fixed proportion of the total amount of protein synthesized (which increased roughly linearly with time: [dDHFR] = 1.5 mM, 2.1 mM and 3.3 mM at 0.5, 1 and 1.5 h). Both findings are consistent with irreversible ligand binding being a reasonable assumption at the 1.5 h time point. See the Supplementary Note for further discussion.
Of the parameters from the fit of equation (1), B1 = kf/(kagg + kdeg) = kf/kagg = 2.9 ± 0.8 is of particular interest because it reveals that folding is about three times faster than aggregation (E. coli BL21 lack Lon protease, so kdeg ∼ 0). According to equation (2), this ratio controls the maximum rescue of dDHFR by TMP. No matter how high the concentration of TMP is, some dDHFR will be diverted to aggregation before TMP binding can rescue it; for dDHFR, that portion is about 25% (Fig. 2b). See the Supplementary Note for further discussion of these points.
As a second test of our model, we chose human α-galactosidase A (α-GAL) (Fig. 2c) expressed in HEK-293T cells in the presence of 1-deoxygalactonojirmycin (DGJ; migalastat), a galactose mimic that binds in α-GAL's active site. Destabilized α-GAL mutants can cause Fabry disease because they are often degraded by ER associated degradation (ERAD) before they can be exported from the ER11, leading to a loss-of-function. DGJ increases the activity of α-GAL mutants despite being an inhibitor because it stabilizes α-GAL in the ER but is out-competed by natural α-GAL substrates in the lysosome7,11,12. Note that α-GAL self-associates to form a dimer; in our model the effects of this process are subsumed into ku (see Supplementary Note).
We tested the effect of DGJ on the intracellular levels of FLAG-tagged R301Q α-GAL (which causes a mild form of Fabry disease13) that had been transfected into HEK-293T cells and expressed for 24 h. To determine the total amount of α-GAL synthesized, we used the proteasome inhibitor lactacystin to inhibit ERAD in one sample (note that since total protein levels are of interest in this experiment, any α-GAL aggregation caused by proteasome inhibition would be immaterial). Since no α-GAL was detected in the media, Fr is the ratio of the protein level at a given DGJ concentration to that of the lactacystin sample.
As before, Fr for α-GAL increases with increasing [DGJ] until a plateau is reached. Also, equation (1) fits the Fr vs. [DGJ] data well (Fig. 2d; Supplementary Figure 3b). Since no R301Q α-GAL aggregation was observed (kagg = 0), B1 = kf/(kagg + kdeg) = kf/kdeg. Thus, the best-fit value of B1 = 1.2 ± 0.2 indicates that folding and degradation are almost evenly poised. Furthermore, according to equation (2), the maximal rescue for α-GAL R301Q is ∼55% of the protein. See the Supplementary Note for further discussion.
To further probe the role of ligand dissociation in the pharmacological chaperoning of α-GAL, we measured Fr as a function of the concentration of another, weaker binding ligand: α-homogalactonojirimycin (HGJ)14. Since the α-GAL expression system is likely in the second pseudo-steady state (see Supplementary Note), equation (1) predicts that decreases in ka or increases in kd—either of which could cause the lower affinity of HGJ for α-GAL—would shift the Fr vs. [HGJ] response curve to higher ligand concentrations. Consistent with this expectation, HGJ did not affect Fr over the experimentally accessible concentration range (Supplementary Fig. 5). We note, however, that this result could be partly due to HGJ being taken up into cells less efficiently than DGJ.
This study has implications for identifying good candidates for pharmacological chaperoning. Ligands will always increase productive folding because dFr/d[L] must be positive. However, equation (2) shows that the upper limit of Fr is dictated by the relative kinetics of folding vs. degradation and/or aggregation. Pharmacological chaperoning is therefore most effective for thermodynamically unstable but fast folding proteins. For example, proteins fused to destabilized constructs of the fast-folding protein FKBP1215 are efficiently rescued by the FKBP ligand Shield-116. Slow folding proteins, however, benefit less from pharmacological chaperoning, as illustrated by the limited success of pharmacological chaperones as monotherapies for cystic fibrosis17. Cystic fibrosis is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR), a large slow-folding membrane protein. Many CFTR mutants partition almost exclusively to ERAD leading to loss of CFTR function18. Treating cells expressing a disease-associated CFTR mutant (ΔF508 CFTR) with lumacaftor modestly increased the trafficking of CFTR to the cell surface (to 14% of wild type levels)19. Since lumacaftor likely operates as a pharmacological chaperone for CFTR19, this “low ceiling” for its efficacy is an unavoidable consequence of CFTR's slow folding, and it suggests that other strategies could yield better results. For example, stimulating an alternative secretion pathway—and thereby increasing ksec—has shown some promise in a mouse model of ΔF508-CFTR20.
Finally, this study suggests that, like chaperones21, ligands could be “evolutionary buffers” in that their ability to stabilize the native state may allow proteins to evolve via otherwise risky mutations22. An example of naturally occurring binding partners preventing aggregation in vivo is the antibody light and heavy chains, each offsetting the aggregation threat of the other23. However, this protective effect should not extend to mutations that slow down folding. Such mutations could be devastating if the protein's folding rate was already marginal because it would invariably diminish Fr, perhaps leading to a loss-of-function phenotype. Based on this reasoning, one would expect residues in a slow-folding protein's folding nucleus to be strongly conserved.
Online Methods
Cloning, expression, and purification of dihydrofolate reductase
The wild type cysteine-free dihydrofolate reductase (DHFR) gene was a gift from the Matthews lab (University of Massachusetts Medical School, Worcester MA)26. It was subcloned into the pET28 expression vector using NdeI and BamHI cut sites. The -Gly-Gly- insertion between residues K106 and A107 was introduced by site-directed mutagenesis using a QuikChange protocol (Stratagene) yielding the “dDHFR” mutant. The vector was transformed into E. coli BL21 (DE3) cells. Cultures were grown at 37 °C to optical density at 600 nm (OD600) of 0.6, induced with 1 mM final IPTG concentration and expressed at 37 °C for 4 h. The cells were harvested by centrifugation and re-suspended in buffer containing 20 mM HEPES, pH 7.4. The cells were lysed using a microfluidizer, and the protein was purified by refolding from inclusion bodies. Inclusion bodies were dissolved in 8 M urea and subsequently dialyzed to refold the protein, which was further purified using anion exchange chromatography on a DEAE column using a 0 M to 1 M sodium chloride gradient. After elution, the salt was removed by dialysis and the protein was flash frozen and stored at −80 °C until further use.
In vitro trimethoprim binding assay for dDHFR
The in vitro trimethoprim (TMP) binding assay was performed with 50 nM purified dDHFR following the protocol from Watson and co-workers27. Briefly, dDHFR was incubated at 25 °C in phosphate buffer with increasing concentrations of TMP and the degree of quenching of dDHFR tryptophan fluorescence was monitored at 345 nm. Spectra were corrected for any contribution to the fluorescence by TMP by running a parallel titration without protein. Fraction bound (Fbound) was calculated by using the formula below.
The Fbound vs. TMP concentration data were fit to a single site binding model by using non-linear regression as implemented by the “NonlinearModelFit” command in Mathematica 10.4.
Cell growth, protein induction and partitioning for dDHFR expression experiments
For all experiments involving partitioning of dDHFR, E. coli BL21(DE3) cells were grown in LB medium until mid-log phase (OD600 = 0.6). Cells were induced with 1 mM final IPTG concentration for 0.5, 1, or 1.5 h. All growth and induction was performed at 37 °C. The cells were harvested after equalization for OD600 by centrifugation at 4,000 × g for 2 minutes, the media was discarded, and the cells were lysed using BPER-II reagent at room temperature for 10 minutes with 1μg/ml DNase I in the lysis mixture. The sample was then centrifuged at 18,000 × g for 30 minutes to separate insoluble (pellet) and soluble (supernatant) components. The samples were subsequently prepared by boiling in SDS running buffer for 12% SDS-PAGE.
TMP titration in E. coli cultures
TMP was dissolved in 100% methanol and then diluted to the desired concentration in media immediately after the addition of IPTG. Final methanol concentration did not exceed 7% v/v at the end of the experiment. Cells were harvested and partitioned as mentioned above.
Chloramphenicol shut off experiments
Protein was induced for 90 minutes in the presence of 80 μM TMP. A fraction of the cells was harvested before any chloramphenicol was added and a second sample was collected after an hour of incubation with chloramphenicol at a final concentration of 50 μg/ml. The partitioning measurements were performed as described above. For the change of media experiment, a fraction of cells was harvested after the 90 minute induction of DHFR in the presence of 80 μM TMP. The remaining cells were centrifuged for 10 minutes at 4000 × g, washed with equal volume LB centrifuged again and resuspended in LB supplemented with chloramphenicol at 50 μg/ml without any TMP. The second sample was collected after an hour of incubation with chloramphenicol and processed as described above.
In vitro aggregation of dDHFR in the presence and absence of TMP
A solution of dDHFR (20 μM) was prepared in pH 7.0 buffer and divided into two aliquots. To one aliquot was added TMP (20 μM) from a stock solution in methanol (the final methanol concentration was 5% v/v). After incubating the aliquots for 2 days with agitation the turbidities of the samples were measured as optical density at 400 nm.
Mammalian cell growth, and experiments with α-galactosidase
We seeded 10 cm culture dishes with HEK 293T cells and allowed them to reach ∼80% confluency at 37 °C, 5% CO2. We then transfected the cells with the R301Q α-galactosidase pCMV 3xFLAG-14 (R301Q α-GAL) vector using Lipofectamine 2000. At 24 hours post-transfection we trypsinized the cells, and used the resulting suspension to seed 6-well plates for the DGJ titration experiment. The cells were allowed to adhere over a 4-hour incubation period, after which desired amounts of DGJ, HGJ, or lactacystin (20 μM final concentration) were added. After 24 hours the cells were washed thoroughly with PBS, then lysed with ice-cold lysis buffer (1% Triton X-100 in PBS). The lysate was centrifuged at 18,000 × g for 30 minutes and the soluble fraction was used for analysis. The samples were normalized prior to loading using the BCA protein quantification method to ensure that equal amounts of protein were loaded in each case. The western blot was performed using as primary a polyclonal rabbit antibody against human α-galactosidase (GeneTex catalog number GTX 101178).
Gel band quantification
SDS-PAGE gels for dDHFR were stained with Coomassie G-250, scanned and analyzed on the LI-COR ODYSSEY CLx and quantified using the associated image studio software (version 4.0)28. Western blots for the α-GAL experiments were similarly analyzed.
Fits of equation (1) to Fr vs. ligand concentration data
Equation (1) was fit to the Fr vs. ligand concentration data in Figures 2b and 2d by using non-linear regression as implemented by the “NonlinearModelFit” command in Mathematica 10.4. The uncorrected R2 values are calculated from fits of equation (1) to all of the individual Fr measurements. The R2 values that are corrected for measurement error are calculated from fits of equation (1) to the Fr measurements after averaging the triplicates. The latter procedure filters out measurement error, but yields exactly the same parameter estimates.
Supplementary Material
Acknowledgments
We thank C. Robert Matthews (University of Massachusetts Medical School, Worcester) for providing us with a plasmid containing the wild type, cysteine-free E. coli DHFR gene. This work was supported by NIH grants GM101644 to L. M. G. and E. T. P. and DK76877 to S. C. G.
Footnotes
Author contributions: K. S. H., L. M. G., E. T. P. and S. C. G. conceived and designed the experiments. K. S. H. performed the experiments with dDHFR. K. S. H., M. C. M., and D. T. D. performed the experiments with R301Q α-GAL. E. T. P. developed the mathematical model. All authors contributed to the experimental design, data analysis, and manuscript preparation.
Competing financial interests: The authors declare no competing financial interests.
Additional information: Any supplementary information, chemical compound information and source data are available in the online version of the paper.
References
- 1.Hingorani KS, Gierasch LM. Comparing protein folding in vitro and in vivo: foldability meets the fitness challenge. Curr Opin Struct Biol. 2014;24:81–90. doi: 10.1016/j.sbi.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monteith WB, Cohen RD, Smith AE, Guzman-Cisneros E, Pielak GJ. Quinary structure modulates protein stability in cells. Proc Natl Acad Sci U S A. 2015;112:1739–42. doi: 10.1073/pnas.1417415112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzman I, Gruebele M. Protein folding dynamics in the cell. J Phys Chem B. 2014;118:8459–70. doi: 10.1021/jp501866v. [DOI] [PubMed] [Google Scholar]
- 4.Powers ET, Powers DL, Gierasch LM. FoldEco: a model for proteostasis in E. coli. Cell Rep. 2012;1:265–76. doi: 10.1016/j.celrep.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bershtein S, Choi JM, Bhattacharyya S, Budnik B, Shakhnovich E. Systems-level response to point mutations in a core metabolic enzyme modulates genotype-phenotype relationship. Cell Rep. 2015;11:645–56. doi: 10.1016/j.celrep.2015.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jahn TR, Radford SE. Folding versus aggregation: polypeptide conformations on competing pathways. Arch Biochem Biophys. 2008;469:100–17. doi: 10.1016/j.abb.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parenti G, Andria G, Valenzano KJ. Pharmacological chaperone therapy: preclinical development, clinical translation, and prospects for the treatment of lysosomal storage disorders. Mol Ther. 2015;23:1138–48. doi: 10.1038/mt.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sontag EM, Vonk WI, Frydman J. Sorting out the trash: the spatial nature of eukaryotic protein quality control. Curr Opin Cell Biol. 2014;26:139–46. doi: 10.1016/j.ceb.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baccanari DP, Daluge S, King RW. Inhibition of dihydrofolate reductase: effect of reduced nicotinamide adenine dinucleotide phosphate on the selectivity and affinity of diaminobenzylpyrimidines. Biochemistry. 1982;21:5068–75. doi: 10.1021/bi00263a034. [DOI] [PubMed] [Google Scholar]
- 10.Cho Y, et al. Individual and collective contributions of chaperoning and degradation to protein homeostasis in E. coli. Cell Rep. 2015;11:321–33. doi: 10.1016/j.celrep.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii S, et al. Mutant alpha-galactosidase A enzymes identified in Fabry disease patients with residual enzyme activity: biochemical characterization and restoration of normal intracellular processing by 1-deoxygalactonojirimycin. Biochem J. 2007;406:285–95. doi: 10.1042/BJ20070479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan JQ, Ishii S, Asano N, Suzuki Y. Accelerated transport and maturation of lysosomal alpha-galactosidase A in Fabry lymphoblasts by an enzyme inhibitor. Nat Med. 1999;5:112–5. doi: 10.1038/4801. [DOI] [PubMed] [Google Scholar]
- 13.Sakuraba H, et al. Identification of point mutations in the alpha-galactosidase A gene in classical and atypical hemizygotes with Fabry disease. Am J Hum Genet. 1990;47:784–9. [PMC free article] [PubMed] [Google Scholar]
- 14.Asano N, et al. In vitro inhibition and intracellular enhancement of lysosomal alpha-galactosidase A activity in Fabry lymphoblasts by 1-deoxygalactonojirimycin and its derivatives. Eur J Biochem. 2000;267:4179–86. doi: 10.1046/j.1432-1327.2000.01457.x. [DOI] [PubMed] [Google Scholar]
- 15.Main ER, Fulton KF, Jackson SE. Folding pathway of FKBP12 and characterisation of the transition state. J Mol Biol. 1999;291:429–44. doi: 10.1006/jmbi.1999.2941. [DOI] [PubMed] [Google Scholar]
- 16.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clancy JP, et al. Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax. 2012;67:12–8. doi: 10.1136/thoraxjnl-2011-200393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukacs GL, et al. Conformational maturation of CFTR but not its mutant counterpart (delta F508) occurs in the endoplasmic reticulum and requires ATP. EMBO J. 1994;13:6076–86. doi: 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Goor F, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci U S A. 2011;108:18843–8. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gee HY, Noh SH, Tang BL, Kim KH, Lee MG. Rescue of DeltaF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell. 2011;146:746–60. doi: 10.1016/j.cell.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 21.Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–73. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 22.Gershenson A, Gierasch LM, Pastore A, Radford SE. Energy landscapes of functional proteins are inherently risky. Nat Chem Biol. 2014;10:884–91. doi: 10.1038/nchembio.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corcos D, et al. Immunoglobulin aggregation leading to Russell body formation is prevented by the antibody light chain. Blood. 2010;115:282–8. doi: 10.1182/blood-2009-07-234864. [DOI] [PubMed] [Google Scholar]
- 24.Bystroff C, Oatley SJ, Kraut J. Crystal structures of Escherichia coli dihydrofolate reductase: the NADP+ holoenzyme and the folate.NADP+ ternary complex. Substrate binding and a model for the transition state. Biochemistry. 1990;29:3263–77. doi: 10.1021/bi00465a018. [DOI] [PubMed] [Google Scholar]
- 25.Guce AI, Clark NE, Rogich JJ, Garman SC. The molecular basis of pharmacological chaperoning in human alpha-galactosidase. Chem Biol. 2011;18:1521–6. doi: 10.1016/j.chembiol.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Online Methods
- 26.Ionescu RM, Smith VF, O'Neill JC, Jr, Matthews CR. Multistate equilibrium unfolding of Escherichia coli dihydrofolate reductase: thermodynamic and spectroscopic description of the native, intermediate, and unfolded ensembles. Biochemistry. 2000;39:9540–50. doi: 10.1021/bi000511y. [DOI] [PubMed] [Google Scholar]
- 27.Watson M, Liu JW, Ollis D. Directed evolution of trimethoprim resistance in Escherichia coli. FEBS J. 2007;274:2661–71. doi: 10.1111/j.1742-4658.2007.05801.x. [DOI] [PubMed] [Google Scholar]
- 28.Luo S, Wehr NB, Levine RL. Quantitation of protein on gels and blots by infrared fluorescence of Coomassie blue and Fast Green. Anal Biochem. 2006;350:233–8. doi: 10.1016/j.ab.2005.10.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.