Abstract
Oct4 is a transcription factor required for maintaining pluripotency and self-renewal in stem cells. Prior to differentiation, Oct4 must be silenced to allow for the development of the three germ layers in the developing embryo. This fine-tuning is controlled by the nuclear receptors, liver receptor homolog-1 and germ cell nuclear factor. Liver receptor homolog-1 is responsible for driving the expression of Oct4 where germ cell nuclear factor represses its expression upon differentiation. Both receptors bind to a DR0 motif located within the Oct4 promoter. Here, we present the first structure of mouse germ cell nuclear factor DNA binding domain in complex with the Oct4 DR0. The overall structure revealed two molecules bound in a head-to-tail fashion on opposite sides of the DNA. Additionally, we solved the structure of the human liver receptor homolog-1 DNA binding domain bound to the same element. We explore the structural elements that govern Oct4 recognition by these two nuclear receptors.
Keywords: Nuclear receptors, X-ray crystallography, Germ Cell Nuclear Factor, Liver Receptor Homolog-1, Oct4 Regulation
Graphical Abstract

Introduction
The pluripotency of embryonic stem (ES) cells is maintained by a specific group of factors including leukemia inhibitor factor (LIF), and transcription factors Oct4, Sox2, and Nanog [1]. All of these proteins are critical for maintaining precise gene expression profiles in ES cells. Oct4, a member of the POU-domain family is widely recognized as the gatekeeper, preventing ES cell differentiation by maintaining pluripotent gene expression and inhibiting lineage-determining factors [2]. During this time, Oct4 expression must be tightly regulated to ensure proper expression levels. Upon exposure to differentiation cues, such as the presence of retinoic acid (RA), Oct4 and the other pluripotency factors are repressed in a temporal and spatial manner to ensure proper development of the three germ layers. Members of the nuclear receptor superfamily of ligand-regulated transcription factors are responsible for ensuring this specific pattern of Oct4 expression [3, 4].
Nuclear receptors (NR) play key roles in diverse biological processes, including maintaining homeostasis, metabolism, development, and many others [4,5]. These receptors all share the same core domain structure: a N-terminal transactivation domain (NTD), a DNA binding domain (DBD) with two highly conserved zinc fingers, a flexible hinge region, and a ligand binding domain (LBD) that contains an activator function-2 helix that is critical for binding coregulators (Fig. 2a). These receptors bind to palindromic DNA sequences as a monomer, homodimer, or heterodimer to regulate transcription from their target genes [6]. A number of these receptors including, liver receptor homolog-1 (LRH-1, NR5A2), germ cell nuclear factor (GCNF, NR6A1), steroidogenic factor (SF-1, NR5A1), and retinoic acid receptor (RAR, NR1B1) are known to regulate of Oct4 expression by binding to response elements within the promoter regions [3, 7, 8, 9].
Figure 2. GCNF and LRH-1 bind directly to the Oct4 DR0.

(a) Diagram of the GCNF and LRH-1 modular structure. In this study, GCNF DBD-TA (residues 69–180), LRH-1 DBD Ftz-F1 (residues 79–187), and full-length LRH-1 (residues 2–495) were used. (b) GCNF DBD-TA bound to the Oct4 DR0 in a two-site binding mechanism. (c) LRH-1 DBD Ftz-F1 (orange) and full-length LRH-1 (green) bind the Oct4 DR0 in a one-site binding mechanism. Binding data are represented as mean ± s.e.m from three replicates and from three independent fluorescence polarization experiments. (d) Summary of binding data.
LRH-1 regulates Oct4 gene expression by binding DNA as a monomer to a 9 nucleotide recognition element comprised of a YCA followed by a NR half-site (AGGCCR) sequence (Y= pyrimidine, R= purine) [10]. LRH-1 contains a canonical NR DBD composed of two helical zinc fingers, which recognize the NR-halfsite. LRH-1, and all NR5A family members, contain a unique fushi tarazu factor 1 (Ftz-F1) domain located C-terminal to the DBD that has been shown to play a functionally important role as a protein interaction module required for the recruitment of other transcription factors [11–14]. This domain was structurally characterized in the LRH-1 DBD CYP7A1 complex by X-ray crystallography then by NMR for SF-1 [10, 15], and assumes an α-helix fold that packs against body of the zinc-figure domain. Mutations that untether the Ftz-F1 helix from the body of the protein dramatically reduce both DNA binding and transactivation. LRH-1 makes structurally conserved contacts with its DNA half-site through both the canonical DNA-reading helix, located within the zinc finger domain, and the C-terminal extension (CTE), located between the zinc-finger domain and Ftz-F1 domain. The CTE-DNA interactions are influenced by the Ftz-F1 helix which serves to orient the CTE in the DNA minor groove [10].
In early embryonic development, LRH-1 is highly expressed within the inner cell mass and primitive endoderm of the blastocyst, where other NRs are secluded to other cell types [3]. To maintain pluripotency, LRH-1 binds directly to DR0 sequences within the proximal promoter (PP) and proximal enhancer (PE) to drive expression of Oct4 [16]. This regulation via LRH-1 is critical, as loss of LRH-1 results in embryonic lethality at day 6.5. LRH-1−/− embryos also exhibit low Oct4 expression and die before liver development is established. Recently, it has even been shown that LRH-1 can replace Oct4 in the reprogramming of somatic cells into pluripotent stem cells [17]. However, upon signals for differentiation, LRH-1 levels are dramatically reduced while GCNF is recruited to repress Oct4.
GCNF is an orphan nuclear receptor that was first identified from mouse heart tissue and shows high expression levels in developing germ cells, oocytes, and spermatogenic cells [18]. GCNF comprises its own unique NR superfamily subclass with a DBD that resembles retinoid X receptor (RXR) but a LBD more closely related to COUP transcription factor 2 (COUP-TF). Additionally, GCNF does not have the typical AF-2 helix within its LBD but instead contains a predicted β-sheet that interacts with the transcriptional corepressors nuclear receptor corepressor-1 (NCoR) and nuclear receptor corepressor-2 (NCoR2/SMRT) [7,19]. GCNF represses Oct4 expression by binding directly to a DR0 element within the PP region, the same site as LRH-1 [18–21]. This transrepression of Oct4 is required for cell differentiation as loss of GCNF results in embryonic lethality at day E10.5 from cardiovascular complications and other severe developmental abnormalities [19,22]. DNA binding by GCNF is functionally critical as a deletion of the DBD phenocopies the GCNF−/− mice [23]. GCNF binds the Oct4 DR0 element in the PP as a homodimer and recruits transcriptional corepressors and DNA methyltransferases in order to ultimately silence Oct4 expression [24]. This promoter methylation is maintained well beyond GCNF expression in order to maintain Oct4 silencing [25,26]. This process is depicted in Figure 1.
Figure 1. Schematic representation of differential regulation of Oct4 by LRH-1 and GCNF.
In undifferentiated ES cells, LRH-1 expression is high and drives Oct4 expression by binding to DR0 sequences in the proximal enhancer and promoter. This binding recruits coactivators and the transcriptional machinery to drive gene expression. Upon signals to differentiate, LRH-1 expression is rapidly reduced while GCNF expression is high. GCNF then binds to the DR0 within the proximal promoter to repress Oct4 expression. Binding by GCNF recruits corepressors, such as NCoR, to block Oct4 expression. GCNF also recruits DNA methyltransferases (DMNT) and methyl-binding proteins (MBP) to methylate the Oct4 gene in order to efficiently shut off its expression.
Here, we present the crystal structures of the mGCNF DBD-TA and hLRH-1 DBD-FtzF1 bound to the mOct4 DR0 proximal promoter element. Throughout the rest of this manuscript mGCNF DBD-TA and hLRH-1 DBD-FtzF1 will be referred to as GCNF DBD and LRH-1 DBD, respectively. This work represents the first crystal structure of GCNF and permitted the visualization of the sequence specific contacts that facilitate recognition of this element for both of these orphan NRs.
Results
GCNF and LRH-1 directly bind the mOct4 DR0
To characterize in vitro binding affinity and kinetics of GCNF and LRH-1 we monitored the ability of recombinant GCNF DBD, LRH-1 DBD, and full-length LRH-1 to bind a FAM labeled 16 bp Oct4 DR0 fragment via fluorescence polarization (Fig 2). Intact, GCNF has been shown to bind the Oct4 DR0 as a homodimer via EMSA [18, 19]. Here, GCNF bound the Oct4 DR0 with a two-site binding mechanism with the Kd of the high affinity site at 170 nM and 2.2 μM for the low affinity site (Fig 2c). This apparent two-site binding, rather than cooperative binding, may be due to the lack of the LBD which presumably facilities dimerization [24]. The LRH-1 DBD displayed a one site binding mechanism with an apparent Kd of 60 nM (Fig. 2b). The full-length LRH-1 construct also fit a one site binding curve with the 16 bp Oct4 element with an apparent affinity of 30 nM. The data is summarized in a Figure 2d.
Structural analysis of GCNF and LRH-1 – Oct4 Complexes
To determine how these individual NRs recognize the same Oct4 DR0, we solved crystal structures of each protein-DNA complex. The GCNF-Oct4 16 bp DR0 DNA complex crystallized in the P212121 space group and data were collected to 2.1 Å with 96.2% completeness. The LRH-1-Oct4 DR0 12 bp DNA complex crystallized in the space group P43 and data were collected to 2.2 Å with 99.8% completeness (Table 1).
Table 1.
Data collection and refinement statistics
| GCNF DBD - mOct4 (16bp) | LRH-1 DBD - mOct4 (12bp) | |
|---|---|---|
| Data Collection | ||
| Space Group | P212121 | P43 |
| Cell Dimension | a=53.6, b=69.5, c=84.5 | a=40.9, b=40.9, c=105.1 |
| Resolution (Å) | 2.10 (2.18–2.10) | 50–2.2 (2.28–2.20) |
| Rpim | 6.9 (37.1) | 3.7 (30.0) |
| I/σ | 9.7 (2.0) | 24.3 (2.1) |
| Completeness | 96.2 (80.2) | 99.8 (98.8) |
| Redundancy | 5.2 (3.5) | 7.2 (5.7) |
| Refinement | ||
| Resolution | 2.10 | 2.20 |
| No. Reflections | 18277 | 8789 |
| Rwork/Rfree | 21.6/26.8 | 15.4/19.7 |
| No. Atoms | ||
| Protein | 1244 | 779 |
| DNA | 651 | 485 |
| Water | 43 | 50 |
| B-factors | ||
| Protein | 56.0 | 59.1 |
| DNA | 51.8 | 52.6 |
| Water | 54.6 | 53.4 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.016 |
| Bond angles (°) | 0.57 | 1.65 |
| PDB code | 5KRB | 5L0M |
Data collected from a single crystal; values in parentheses are for the highest-resolution shell
The GCNF structure shows two molecules bound to opposite sides of the DNA in a head-to-tail fashion (Fig. 3a). This orientation on DNA is very similar to the structure of the RXRα-DR1 complex (PDB ID: 1BY4) [27]. The GCNF DBD adopts the canonical NR DBD fold [6], and strong electron density allowed for modeling of the T and A box residues within the C-terminal extension (CTE) for molecule 2. One molecule, colored purple, positions the DNA reading helix into the major groove of the DNA at the first AGGTCA repeat (bases 106–111). This molecule makes three base-specific contacts (Fig. 3b). The Lys96 side chain makes hydrogen bonds to the O6 and N7 positions on guanine 107. Arg101 make additional hydrogen bonds to the on guanine 97 at the N7 position on the other side of the DNA as well as to the backbone phosphate of a thymine 98. The second molecule, colored deep purple, makes similar contacts to the second AGGCTA sequence (bases 112–117) (Fig. 3b). Here, Lys96 again makes hydrogen bonds to the O6 position of guanine 113. Arg101 makes hydrogen bonds to the N7 position on adenine 91 instead of a guanine and also makes additional hydrogen bonds the backbone of thymine 90. These interactions are supported by excellent electron density and additional side chains participating in backbone and water-mediated interactions are highlighted in Figure 3c.
Figure 3. Structural Analysis of GCNF - mOct4 Complex.
(a) Overall structure of GCNF DBD (purple) bound to the Oct4 DR0 (gray). GCNF DBD and DNA shown as cartoon in purple and white, respectively and zinc atoms as spheres. Two molecules of GCNF bound to opposite sides of the DNA in a head-to-tail fashion. The DR0 sequence is shown below with arrows denoting the direction of GCNF over the sequence. (b) Each molecule of GCNF makes base-specific contacts with the DR0 (bases in white) mediated by hydrogen bonds (red) made between Arg101 and Lys96. Mesh shows 2Fo-Fc electron density map contoured to 2σ around the DNA bases. (c) Schematic view of protein-DNA interactions. Larger, bold side chains denoted base-contacting side chains. Water molecules are indicated as red spheres.
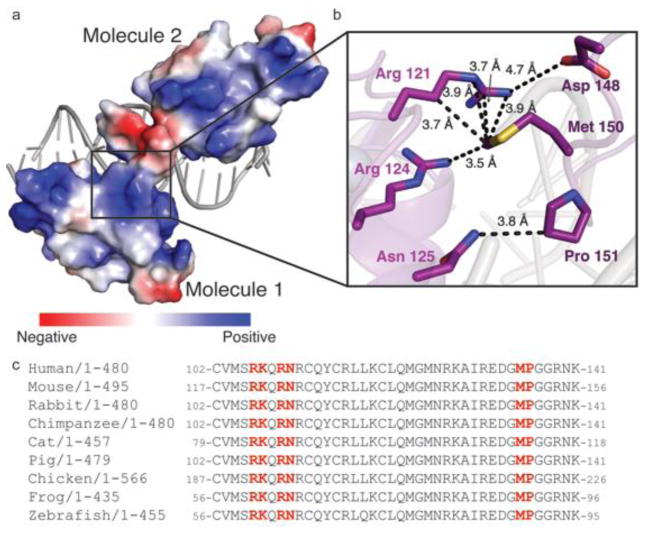
The GCNF CTE, including the T/A box residues, was observed for molecule 2 (residues 141–156). There is strong electron density for these residues, which is shown in Figure 5d. The GCNF CTE dips into the minor groove at the TCAA sequence (bases 109–112) to make additional sequence specific contacts. Arg86 makes hydrogen bonds to the O2 positions on both thymine 109 and cytosine 110. Arg78 makes hydrogen bonds to the polypeptide backbone oxygen at Gly152, which may help lock the CTE into a more stable conformation for interaction with the DNA minor groove. In addition to DNA contacts, the CTE also makes a series of contacts with GNCF molecule 1 via a set of highly conserved residues (Fig 4). The largely negative CTE of molecule 2 rests within the minor groove but also makes contacts with the positive surface of molecule 1(Fig 4a). Namely, Asp148 makes weak electrostatic contacts with Arg121.This intermolecular interface is also supported by a series of hydrophobic contacts driven largely by the conserved Met150, which interacts with Arg121 and Arg124 (Fig 4b,c).
Figure 5. Structural Analysis of LRH-1 - mOct4 Complex.
(a) Overall structure of LRH-1 DBD (orange) bound to the 12 bp Oct4 DR0 (gray) with zinc atoms as spheres. The Oct4 DR0 sequence is shown below with arrows denoting the footprint and orientation of the LRH-1 binding site. (b) LRH-1 makes base-specific contacts with the DR0 (bases in white) mediated by hydrogen bonds (red) made between Glu104, Lys107, Lys 111, Arg112. Mesh shows 2Fo-Fc electron density map contoured to 2σ around the DNA bases. (c) Schematic view of protein-DNA interactions. Larger, bold side chains denoted base-contacting side chains. Water molecules are indicated as red spheres.
Figure 4. Interactions between GCNF molecules.
(a) Electrostatic surface of overall GCNF structure. (b) Zoom in of contacts (black) made between the two molecules. Side chains from molecule 1 are colored purple and dark purple from molecule 2. (c) Sequence alignments from numerous species reveal that the side chains that participate in homodimer formation are highly conserved.
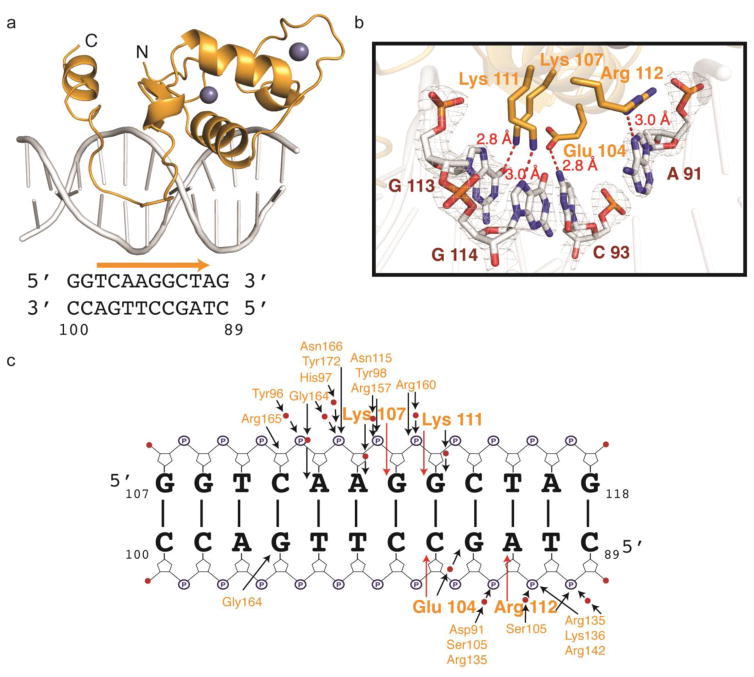
The LRH-1 DBD crystallized as a monomer on a 12 bp Oct4 fragment, with its DNA recognition helix resting in the major groove formed by the DR0 motif, flanked by two conserved zinc fingers (Fig. 5a). This structure is similar to the previously solved LRH-1 – hCYP7A1 complex (PDB ID: 2A66) [10]. The DNA reading helix contains four residues that make distinct base-specific contacts with the Oct4 DR0, including hydrogen bonds between Glu104 and cytosine 93, Lys107 and the O6 atom on guanine 113, Lys111 and the N7 atom on guanine 114, and Arg112 and the N7 atom on adenine 91 (Fig. 5b). In addition to the core DBD there was also density to model the CTE and Ftz-F1 domains. The CTE of hLRH-1 wedges into the minor groove of the DNA duplex (Fig. 6c). Base-specific contacts formed by the CTE include hydrogen bonds between Arg162 and thymine 95 and Arg165 and cytosine 110. Other stabilizing interactions with the phosphate backbone and the water network are shown in Figure 5c.
Figure 6. GCNF and LRH-1 comparison.
(a) Overlay of GCNF and LRH-1 structures. Molecule 2 of GCNF (Dark purple) sites directly on top of the LRH-1 (orange) recognition site. (b) Close up view of the base-specific contacts mediated by these two receptors. LRH-1 makes an additional contact with Lys 107 making hydrogen bonds to guanine 114. (c) LRH-1 CTE has good electron density, mesh shows 2Fo-Fc electron density map contoured to 1σ around the residues. Arg162 and 165 make hydrogen bonds (red) to thymine 95 and cytosine 110. (d) GCNF CTE has good electron density, mesh shows 2Fo-Fc electron density map contoured to 2σ around the residues. Arg86 makes hydrogen bonds to thymine 109 and cytosine 110. Arg78 also folds into the CTE to make hydrogen bonds to Gly152. (e) Sequence alignment of GCNF and LRH-1 CTEs show LRH-1 to contain two additional Arg residues that are used for DNA binding. (f) Mutational analysis of CTE residues on Oct4 binding: GCNF DBD Gly149Arg/Pro151Arg (open purple circles) bound to the Oct4 DR0 with an affinity 20 nM, where WT GCNF DBD (faded closed purple circles) binds with an affinity of 170 nM. LRH-1 DBD Arg160Gly/Arg162Pro (open orange circles) bound with an affinity of 750 nM, where WT LRH-1 DBD (faded closed orange circles) bound with an affinity of 60 nM.
LRH-1 and GCNF differentially recognize the DR0 of the Oct4 PP
To understand how these distinct proteins recognize the same Oct4 DR0 DNA, we compared both the primary amino acid sequence and structure (Fig. 6). Alignment of mGCNF and hLRH-1 DBDs show 44.7% identity with 61.8% similarity. Though human LRH-1 DBD was used, sequence alignments show that human and mouse LRH-1 DBDs are 94.4 % identical, with the critical DNA binding residues being 100% conserved. Mouse GCNF (495 aa) and human GCNF (480 aa) are 95.4% identical, with both the DBD and CTE 100% identical. Furthermore, the core DR0 motif within the Oct4 PP is conserved between mice and humans, suggesting these interactions would be maintained across species.
In order to visualize differences in the structure, we superimposed both to identify how each recognizes the same DNA element (Fig. 6). Unlike, GCNF, LRH-1 binds to DNA as a monomer. While this is uncommon among NRs, other monomeric receptors include SF-1, ERR2, REV-ERB [15,28,29]. Perhaps not surprising, the LRH-1 DBD binds the Oct4 DR0 element with greater affinity than the GCNF DBD, which is lacking the LBDs that are known to participate in receptor dimerization [24]. Superposition of the structures revealed LRH-1 makes four additional base-specific contacts within the major and minor groove. The root-mean-square deviation (r.m.s.d.) between the DBD core domains of LRH-1 (83–154) and GCNF (74–147) is 0.46 A (63 Cα aligned). In the DNA reading helix, LRH-1 displays two additional base-specific contacts with Glu104 and Lys107 making hydrogen bonds to cytosine 93 and guanine 114, respectively (Fig. 5b). The GCNF and LRH-1 CTE share 56.3% identity with 62.5% similarity and a similar backbone position within the DNA minor groove (r.m.s.d 0.51 A; 13 Cα aligned). However, LRH-1 makes two additional H-bonding contacts vs GCNF via Arg160 and 162 (Fig. 5c,e) where GCNF contains glycine and proline residues at those positions (Fig. 5d,e). Mutation of the GCNF CTE glycine and proline to arginine residues (Gly149Arg and Pro151Arg), to mimic the LRH-1 CTE, increased the GCNF DBD affinity from 170 nM to 20 nM (Fig. 5f). Conversely, simultaneous Arg160Gly and Arg162Pro mutations within the LRH-1 CTE reduced binding affinity from 60 nM to 750 nM (Fig. 5f). Taken together, difference within the CTE drive differential affinity for the isolated DBDs for the Oct4 promoter. The difference in affinity between these isolated DBDs should not be used to predict in-cell kinetics; however, it is clear that these proteins compete for Oct4 proximal promoter binding when co-expressed in vivo.
Discussion
The orphan nuclear receptors, LRH-1 and GCNF play a critical role in regulating genes central to embryonic development [3,7]. Notably, LRH-1 and GCNF reciprocally regulate the pluripotency factor, Oct4, by binding to a DR0 response element within the promoter [3,7]. Though many have studied this mechanism, the structural basis for this regulation has yet to be explored. Here, we not only present the first structure of GCNF DBD but also LRH-1 DBD bound to the Oct4 DR0 sequence and examine the sequence-specific contacts that guide Oct4 regulation.
Understanding the DNA-binding properties of transcription factors is required to understand their biological function. Initial gel mobility shift assays show GCNF binds direct repeats of the AGGTCA sequence with no spacer or to extended half-sites of TCAAGGTCA sequence as a homodimer, though GCNF bound to the DR0 with higher affinity than the half-sites [18,19]. Deletion studies show that removal of the ligand-binding domain had no effect of DNA binding and in vivo removal of this domain did not affect transrepression [30, 31]. Additionally, N-terminal domain deletions still bound DNA to the same levels of WT GCNF [20]. In contrast, removal of the DBD in vivo phenocopies the GCNF complete knockout mice [23]. Additionally, the DBD and the CTE are strictly required to bind DNA making them necessary to repress Oct4 [20,30]. For this reason, we purified the minimal residues required for DNA recognition (Fig. 2A).
We show that purified GCNF DBD binds to the Oct4 DR0 via a two-site binding mechanism with a high and low affinity binding event (Fig. 2c). In contrast, intact in vitro translated full-length protein binds DNA as a homodimer [15, 20, 30]. The overall structure shows two GCNF molecules bound to opposite sides of the DNA and the sister DBDs only weakly interact (Fig. 3,4). This structure is similar to retinoic X receptor bound to a DR1 sequence [27]. The GCNF DBD sequence has been shown to be most similar to the RXR DBD, which requires an intact LBD for receptor dimerization; therefore, it is likely that strong homodimerization of GCNF also requires the LBD [19,24]. Dimerization motifs have been proposed in the DBD, CTE, and LBD. Structural analysis reveals that the CTE makes a number of hydrophobic contacts with the other GCNF molecule (Fig. 4). Additionally, the side chains that participate in homodimerization are very well conserved (Fig. 4c). Therefore, structural studies of the GCNF LBD or full-length will be required complete the understanding of GCNF:DNA recognition [24, 30].
LRH-1 is required to maintain Oct4 expression [3]. Monomeric LRH-1 interacts with extended half-site sequences in both the proximal promoter and proximal enhancer to activate Oct4 expression prior to differentiation [16]. The only reported structures of NR5A DBDs are the x-ray structure of the LRH-1-hCYP7A1 complex (PDB ID: 2A66) and the NMR solution structure of SF-1 in complex with a 9 bp fragment of inhibin-α (PDB ID: 2FF0; 16 conformers) [10,15]. While SF-1 and LRH-1 bind the same response elements, previous data show that SF-1 is not expressed at detectable levels in ES cells [3]. We show that purified LRH-1 DBD and full-length LRH-1 bind the Oct4 DR0 with high affinity as a monomer positioned over the extended LRH-1 recognition sequence (Fig. 2b,d, 5).
Comparing the GCNF and LRH-1 structures revealed that their respective response elements directly overlap in the DR0 of the PP. (Fig. 6). LRH-1 is highly expressed in ES cells when Oct4 expression is up regulated [3]. However, upon signals to differentiate, both LRH-1 and Oct4 expression is rapidly decreased [3, 28]. During this time GCNF expression is high (Fig. 1) [7]. LRH-1 has an apparent higher affinity in vitro, potentially due to additional side chain – DNA base interactions (Fig. 5,6). Mutational analysis of GCNF CTE to mimic the LRH-1 CTE shows an increased affinity for the Oct4 DR0 and the high LRH-1 affinity can be drastically reduced when the Arg residues of the CTE are mutated to the GCNF glycine and proline residues at the same positions (Fig. 5f). Since binding was only being tested with isolated domains, this result is not surprising. Kinetic studies with intact GCNF may reveal similar affinities for the element. Yet, it is possible that GCNF will not have to compete for binding to the DR0 during embryonic development due to the inverse expression patterns of LRH-1 and GCNF (Fig 6) [3,18].
Mutational analyses of C-terminal extensions from numerous nuclear receptors such as SF-1, RXR, LRH-1, and GCNF have shown this region to be critical for DNA recognition and transcriptional regulation [10, 24, 32, 33]. The GCNF structure shows electron density for the CTE within molecule 2 (Fig 5d). Removal of these residues from GCNF prevents DNA binding, highlighting their importance [19]. Furthermore, DNA sequence analysis revealed that the TCA sequence preceding the second direct repeat, that forms the minor groove CTE interaction, is most important for GCNF to bind DNA [19, 20]. Here, GCNF uses an Arg residue to make two base-specific contacts within the minor groove (Fig 5d). PISA analysis of the GCNF structure reveals that the average gain on complex formation for molecule 2 and DNA is −9.1 kcal/mol (complex score 1.0) but with the CTE removed it is −5.2 kcal/mol (complex score 0.190) and has a much weaker complex score (1.0 being highly favorable complex) [34]. Like GCNF, the LRH-1 CTE also dips into the minor groove to make additional base-specific contacts (Fig 5c).
Structure alignments of GCNF, LRH-1, RXR, and SF-1 bound to DNA elements show high conservation among the overall DBD structures. The r.m.s.d. of LRH-1 and GCNF on the Oct4 DR0 is 0.57 A (76 Cα aligned), while RXRα and GCNF is 0.66 A (66 Cα aligned) and SF-1 and LRH-1 is 1.12 A (88 Cα aligned). Though GCNF is cited as most similar to RXR, the CTE regions are very different. When overlaid, these structures show that the RXRα CTE does not rest in the DNA minor groove but trails off away from the DNA. Furthermore, sequence alignments of GCNF126–141 and RXRα201–216 only have 27% identity and 40% similarity. These differences also set GCNF apart in its own subclass of the nuclear receptor superfamily. LRH-1152–167 and SF-179–94 on the other hand, are 100% identical. Aligned structures show that both of these proteins’ CTEs rest in the minor groove of DNA leading into NR5A conserved Ftz-F1 helices. The NMR solutions of SF-1 show that, although the CTE adopts multiple conformations, all make contacts in the minor groove, as in both LRH-1 structures (PDB ID: 2A66 and 5L0M). Interestingly, replacing the GCNF CTE with that from LRH-1/SF-1, induces a monomeric DNA binding preference likely by removing conserved GCNF intermolecular contacts (i.e. Met150) and increasing half-site affinity [24]. In agreement, the WT GCNF binds to the Oct4 DR0 with an apparent two-site binding curve but the GCNF Gly142Arg/Pro151Arg mutant binds the same element using a one-site binding mechanism (Fig. 5f). As these mutations now mimic the LRH-1 and SF-1 CTE, this result is not surprising. Structural studies using intact LRH-1 and GCNF would shed deep insight Oct4 recognition; however, it is clear that the CTEs of these receptors play a critical role not only in sequence specificity but oligomierization on DNA [3].
Materials and Methods
Protein expression and purification
The DNA binding domain (DBD) of mouse GCNF (residues 69–180, UniProt Q64249) with a C104S mutation was cloned into a pMCGS7 vector with a 6X-Histidine tag. GCNF was expressed in BL21 (DE3) pLysS E. coli cells. The protein was grown at 37 °C for 2 hrs, then reduced to 20 °C and grown until an OD600 of 0.6, and then induced with 0.78 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Cultures were grown overnight at 20 °C. Cells were lysed in 20 mM Tris-HCl (pH 7.4), 1M NaCl, 25 mM imidazole, and 5% glycerol via sonication. Protein was purified using affinity chromatography (His-Trap FF, GE Healthcare) followed by further purification via gel filtration chromatography. Protein was then concentrated to 2–3 mg/ml in 20 mM Tris-HCL (pH 7.4), 150 mM NaCl, and 5% glycerol, flash frozen in liquid N2, and stored at− −80 °C.
The DBD-FtzF1 domains of human LRH-1 (LRH-1 DBD) were expressed and purified similar to previous [10]. Briefly, cultures were grown in Terrific Broth (TB) at 37 °C to OD600 of 0.6, induced with 0.3 mM IPTG at 18 °C and grown overnight. Fusion protein was purified via affinity chromatography (His-Trap FF, GE Healthcare) and the MBP tag was removed following TEV protease cleavage with an additional pass over His-Trap FF resin. LRH-1 DBD was further purified using gel filtration (Superdex 75; GE Healthcare) equilibrated with 20 mM Tris-HCL (pH 7.4), 150 mM NaCl, and 5% glycerol. Eluted protein was concentrated to 11.5 mg/mL and either flash frozen in liquid N2 and stored at −80 °C or used directly for crystallization experiments.
hLRH-1 has multiple full-length functional isoforms, the canonical sequence (isoform 2; 1–541) was used to number the residues of the crystal structure [10]. Experimentally, isoform 1 (1–495) lacking residues 22–67 in the N-terminal domain, was used to generate a construct (amino acids 2–495) preceded by a TEV protease site, was cloned into the pE-SUMO-Amp vector (LifeSensors) and recombinantly expressed in E. coli BL21 DE3 cells. Cells were grown in LB at 37 °C to OD600 of 0.6, induced with 0.5 mM IPTG at 20 °C and grown overnight. Cells were lysed in 20 mM Tris-HCl (pH 8.5), 0.5 M NaCl, 25 mM imidazole, 2 mM CHAPS, 0.2% Triton-100X and pierce protease inhibitor tablets (Thermo Fisher Scientific) via sonication. Fusion protein was purified by affinity chromatography (His-Trap FF and HiTrap Heparin, GE Healthcare). The His-SUMO fusion tag was removed by incubation with TEV overnight and affinity chromatography used to isolate pure target protein.
Generation of GCNF DBD Gly149Arg/Pro151Arg and LRH-1 DBD Arg160Gly/Arg162Pro
Mutagenesis was performed following the MEGAWHOP protocol. Briefly, megaprimers were generated containing the desired mutations, purified and used for whole plasmid PCR [35]. Mutant proteins were prepared as wild-type.
Sequence Alignments and Analyses
The following sequences were obtained from UniProt: mGCNF (UniProt Q64249-1), hGCNF (Uniprot Q15406-1), mLRH-1 (UniProt P45448-1), hLRH-1 (UniProt O00482-1), hSF-1 (UniProt Q13285-1) and hRXRα (UniProt P19793-1) and aligned using Clustal Omega [36]. Jalview [37] was used for visualization and manipulation of the alignments. The Sequence Manipulation Suite (SMS) was used for percent identity and similarity calculations [38].
Nucleic acid binding assays
Synthesized FAM-labeled nucleic acid duplexes (Integrated DNA Technologies) of mouse Oct4 DR0 (5′ – [FAM] AGAGGTCAAGGCTAGA – 3′) were annealed in a 1 L water bath heated to 90 °C then cooled slowly to room temperature. Fluorescence polarization assays were performed by adding increasing concentrations of GCNF, LRH-1 DBD, or FL LRH-1 (1 nM-50 μM for the DBDs; 1 nM-20 μM for FL hLRH-1) to 10 nM of the FAM-labeled DNA. Reactions were performed in 20 mM Tris-HCL (pH 7.4), 150 mM NaCl, and 5% glycerol. Polarization was monitored on a Biotek Synergy plate-reader at an excitation/emission wavelength of 485/528 nm. The program GraphPad Prism 6 was used to analyze binding data and generate graphs. Binding data were analyzed with an F-test to compare a two-site binding event to a one-site binding event with Hill slope. This test generated an F-statistic and p-value supporting a two-site binding model. These values are represented in Figure 2. Additionally, dissociation values (Kd) and coefficient of determination (rr) are included.
Crystallization, data collection, and structure determination
Crystals of the GCNF-mOct4 (16bp - 5′ – AGAGGTCAAGGCTAGA – 3′) complex were grown by hanging drop vapor diffusion in 0.1M Tris pH 8.5, 20% PEG 3350, 3% glycerol with a 2:1 protein:DNA molar ratio. Crystals were cryo-protected with 0.1M Tris pH 8.5, 30% PEG 3350, and 15% glycerol and flash cooled in liquid N2. The hLRH-1 DBD-mOct4 (12bp duplex - 5′ – GGTCAAGGCTAG – 3′) complex was formed by mixing 1:1.2 molar ratios of protein to DNA and incubating at 25 °C. Crystallization conditions were screened using a Phoenix nanolitre drop dispensing robot (Art Robbins) with a 1:1 protein (5–6 mg/mL) to well solution ratio. Single well-formed crystals appeared overnight at 18 °C in 0.2 M calcium acetate and 20% (w/v) PEG 3350. Larger crystals were grown by hanging drop vapor diffusion in the same solution. Crystals were cryo-protected with an additional 20% (v/v) glycerol and flash cooled in liquid N2.
Data were collected at the 22-ID beamline (Advanced Photon Source, Argonne, IL) and processed using HKL-2000 [39]. The structures were phased using a low-resolution model GCNF-Oct4, previously generated in the lab or the hLRH-1 DBD-CYP7a1 complex (PDB ID: 2A66) [10]. Structure refinement and validation was performed using PHENIX refine software and model building was performed in COOT [40,41]. PDB Redo was used iteratively to optimize refinement parameters and geometry [42]. PyMOL v1.8.2 was used to visualize structures and generate figures (Schrödinger, LLC). Both structures showed good overall geometry with one Ramachandran outlier in the LRH-1 DBD–DNA complex and all other residues (93) in favored or allowed regions and zero Ramachandran outliers in the GCNF DBD–DNA complex and all in favored or allowed regions.
Accession Numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with the accession numbers, 5KRB for GCNF:Oct4 complex and 5L0M for LRH-1:Oct4 complex.
Highlights.
Structural analysis of the pluripotency factor Oct4 Regulation by nuclear receptors GCNF and LRH-1
GCNF and LRH-1 reciprocally regulate Oct4 expression by recognizing the same DNA response element
Solved the first x-ray crystal structure of GCNF revealing it mechanism for DNA recognition
Identified the sequence-specific DNA contacts that allow dual regulation by GCNF and LRH-1
Acknowledgments
X-ray data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members/html. Use of the Advanced Photon source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
Author Contributions
Conceived and designed experiments: ERW MLT EAO. Performed the experiments ERW MLT MNM EAO. Analyzed the data ERW MLT EAO. Contributed to writing the manuscript ERW MLT EAO.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paranjpe SS, Veenstra GJ. Establishing pluripotency in early development. Biochim Biophys Acta. 2015;1849:626–36. doi: 10.1016/j.bbagrm.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu G, Scholer HR. Role of Oct4 in the early embryo development. Cell Regen (Lond) 2014;3:7. doi: 10.1186/2045-9769-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, et al. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Molecular and cellular biology. 2005;25:3492–505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sylvester I, Scholer HR. Regulation of the Oct-4 gene by nuclear receptors. Nucleic Acids Res. 1994;22:901–11. doi: 10.1093/nar/22.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullen EM, Gu P, Cooney AJ. Nuclear Receptors in Regulation of Mouse ES Cell Pluripotency and Differentiation. PPAR Res. 2007;2007:61563. doi: 10.1155/2007/61563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olefsky JM. Nuclear receptor minireview series. The Journal of biological chemistry. 2001;276:36863–4. doi: 10.1074/jbc.R100047200. [DOI] [PubMed] [Google Scholar]
- 7.Fuhrmann G, Chung AC, Jackson KJ, Hummelke G, Baniahmad A, Sutter J, et al. Mouse germline restriction of Oct4 expression by germ cell nuclear factor. Dev Cell. 2001;1:377–87. doi: 10.1016/s1534-5807(01)00038-7. [DOI] [PubMed] [Google Scholar]
- 8.Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–67. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- 9.Barnea E, Bergman Y. Synergy of SF1 and RAR in activation of Oct-3/4 promoter. The Journal of biological chemistry. 2000;275:6608–19. doi: 10.1074/jbc.275.9.6608. [DOI] [PubMed] [Google Scholar]
- 10.Solomon IH, Hager JM, Safi R, McDonnell DP, Redinbo MR, Ortlund EA. Crystal structure of the human LRH-1 DBD-DNA complex reveals Ftz-F1 domain positioning is required for receptor activity. Journal of molecular biology. 2005;354:1091–102. doi: 10.1016/j.jmb.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Yussa M, Löhr U, Su K, Pick L. The nuclear receptor Ftz-F1 and homeodomain protein Ftz interact through evolutionarily conserved protein domains. Mechanisms of Development. 2001;107:39–53. doi: 10.1016/s0925-4773(01)00448-8. [DOI] [PubMed] [Google Scholar]
- 12.Pare JF, Malenfant D, Courtemanche C, Jacob-Wagner M, Roy S, Allard D, Belanger L. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J Biol Chem. 2004;279:21206–16. doi: 10.1074/jbc.M401523200. [DOI] [PubMed] [Google Scholar]
- 13.Takemaru K, Li FQ, Ueda H, Hirose S. Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein. Proc Natl Acad Sci U S A. 1997;94:7251–6. doi: 10.1073/pnas.94.14.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai YN, Zhou Q, Kong YY, Li M, Viollet B, Xie YH, Wang Y. LRH-1/hB1F and HNF1 synergistically up-regulate hepatitis B virus gene transcription and DNA replication. Cell Res. 2003;13:451–8. doi: 10.1038/sj.cr.7290187. [DOI] [PubMed] [Google Scholar]
- 15.Little TH, Zhang Y, Matulis CK, Weck J, Zhang Z, Ramachandran A, et al. Sequence-specific deoxyribonucleic acid (DNA) recognition by steroidogenic factor 1: a helix at the carboxy terminus of the DNA binding domain is necessary for complex stability. Mol Endocrinol. 2006;20:831–43. doi: 10.1210/me.2005-0384. [DOI] [PubMed] [Google Scholar]
- 16.Sung B, Do HJ, Park SW, Huh SH, Oh JH, Chung HJ, et al. Regulation of OCT4 gene expression by liver receptor homolog-1 in human embryonic carcinoma cells. Biochem Biophys Res Commun. 2012;427:315–20. doi: 10.1016/j.bbrc.2012.09.049. [DOI] [PubMed] [Google Scholar]
- 17.Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–74. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Chen F, Cooney AJ, Wang Y, Law SW, O’Malley BW. Cloning of a novel orphan receptor (GCNF) expressed during germ cell development. Mol Endocrinol. 1994;8:1434–44. doi: 10.1210/mend.8.10.7854358. [DOI] [PubMed] [Google Scholar]
- 19.Yan ZH, Medvedev A, Hirose T, Gotoh H, Jetten AM. Characterization of the response element and DNA binding properties of the nuclear orphan receptor germ cell nuclear factor/retinoid receptor-related testis-associated receptor. The Journal of biological chemistry. 1997;272:10565–72. doi: 10.1074/jbc.272.16.10565. [DOI] [PubMed] [Google Scholar]
- 20.Cooney AJ, Hummelke GC, Herman T, Chen F, Jackson KJ. Germ cell nuclear factor is a response element-specific repressor of transcription. Biochem Biophys Res Commun. 1998;245:94–100. doi: 10.1006/bbrc.1998.8391. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Wang X, Xu X, Kyba M, Cooney AJ. Germ Cell Nuclear Factor (GCNF) Represses Oct4 Expression and Globally Modulates Gene Expression in Human Embryonic Stem (hES) Cells. The Journal of biological chemistry. 2016;291:8644–52. doi: 10.1074/jbc.M115.694208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung AC, Katz D, Pereira FA, Jackson KJ, DeMayo FJ, Cooney AJ, et al. Loss of orphan receptor germ cell nuclear factor function results in ectopic development of the tail bud and a novel posterior truncation. Molecular and cellular biology. 2001;21:663–77. doi: 10.1128/MCB.21.2.663-677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan ZJ, Chung AC, Xu X, DeMayo FJ, Cooney AJ. The embryonic function of germ cell nuclear factor is dependent on the DNA binding domain. The Journal of biological chemistry. 2002;277:50660–7. doi: 10.1074/jbc.M209586200. [DOI] [PubMed] [Google Scholar]
- 24.Greschik H, Wurtz JM, Hublitz P, Kohler F, Moras D, Schule R. Characterization of the DNA-binding and dimerization properties of the nuclear orphan receptor germ cell nuclear factor. Molecular and cellular biology. 1999;19:690–703. doi: 10.1128/mcb.19.1.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu P, Xu X, Le Menuet D, Chung AC, Cooney AJ. Differential recruitment of methyl CpG-binding domain factors and DNA methyltransferases by the orphan receptor germ cell nuclear factor initiates the repression and silencing of Oct4. Stem Cells. 2011;29:1041–51. doi: 10.1002/stem.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato N, Kondo M, Arai K. The orphan nuclear receptor GCNF recruits DNA methyltransferase for Oct-3/4 silencing. Biochem Biophys Res Commun. 2006;344:845–51. doi: 10.1016/j.bbrc.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Q, Chasse SA, Devarakonda S, Sierk ML, Ahvazi B, Rastinejad F. Structural basis of RXR-DNA interactions. Journal of molecular biology. 2000;296:509–20. doi: 10.1006/jmbi.1999.3457. [DOI] [PubMed] [Google Scholar]
- 28.Gearhart MD, Holmbeck SM, Evans RM, Dyson HJ, Wright PE. Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. Journal of molecular biology. 2003;327:819–32. doi: 10.1016/s0022-2836(03)00183-9. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Q, Khorasanizadeh S, Miyoshi Y, Lazar MA, Rastinejad F. Structural elements of an orphan nuclear receptor-DNA complex. Molecular cell. 1998;1:849–61. doi: 10.1016/s1097-2765(00)80084-2. [DOI] [PubMed] [Google Scholar]
- 30.Borgmeyer U. Dimeric binding of the mouse germ cell nuclear factor. Eur J Biochem. 1997;244:120–7. doi: 10.1111/j.1432-1033.1997.00120.x. [DOI] [PubMed] [Google Scholar]
- 31.Okumura LM, Lesch BJ, Page DC. The ligand binding domain of GCNF is not required for repression of pluripotency genes in mouse fetal ovarian germ cells. PLoS One. 2013;8:e66062. doi: 10.1371/journal.pone.0066062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson TE, Fahrner TJ, Milbrandt J. The orphan receptors NGFI-B and steroidogenic factor 1 establish monomer binding as a third paradigm of nuclear receptor-DNA interaction. Mol Cell Biol. 1993;13:5794–804. doi: 10.1128/mcb.13.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melvin VS, Roemer SC, Churchill ME, Edwards DP. The C-terminal extension (CTE) of the nuclear hormone receptor DNA binding domain determines interactions and functional response to the HMGB-1/-2 co-regulatory proteins. The Journal of biological chemistry. 2002;277:25115–24. doi: 10.1074/jbc.M110400200. [DOI] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Miyazaki K. MEGAWHOP cloning: a method of creating random mutagenesis libraries via megaprimer PCR of whole plasmids. Methods Enzymol. 2011;498:399–406. doi: 10.1016/B978-0-12-385120-8.00017-6. [DOI] [PubMed] [Google Scholar]
- 36.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102, 4. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 37.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–362. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joosten RP, Salzemann J, Bloch V, Stockinger H, Berglund AC, Blanchet C, et al. PDB_REDO: automated re-refinement of X-ray structure models in the PDB. J Appl Crystallogr. 2009;42:376–84. doi: 10.1107/S0021889809008784. [DOI] [PMC free article] [PubMed] [Google Scholar]