Abstract
Neglected disease drug discovery is generally poorly funded compared with major diseases and hence there is an increasing focus on collaboration and precompetitive efforts such as public–private partnerships (PPPs). The More Medicines for Tuberculosis (MM4TB) project is one such collaboration funded by the EU with the goal of discovering new drugs for tuberculosis. Collaborative Drug Discovery has provided a commercial web-based platform called CDD Vault which is a hosted collaborative solution for securely sharing diverse chemistry and biology data. Using CDD Vault alongside other commercial and free cheminformatics tools has enabled support of this and other large collaborative projects, aiding drug discovery efforts and fostering collaboration. We will describe CDD's efforts in assisting with the MM4TB project.
Keywords: CDD Vault, collaboration, drug discovery, MM4TB, tuberculosis
Introduction
Researchers continually recount the importance of collaboration whether in academia, the pharmaceutical industry or between both of these groups [1–7]. Occasionally, there is also the role of patients and advocates [8,9] which can add to the collaboration. In areas such as neglected diseases where the funding is limited for drug discovery [10] there is a compelling need to collaborate and this might not be limited to drug discovery alone but also other parts of the research and development enterprise [11]. We have highlighted the need for more-competitive collaboration in the industry [12] and alternative business models for drug discovery that balance collaboration, privacy and security [13], and certainly the shift toward public–private partnerships (PPPs) fulfils that gap [14]. These in turn will face the challenge of a growing mountain of data and the need for data mining and collaborative tools [15].
There have also been calls for more cooperation in developing antimicrobials such as the TB Drug Accelerator [16] which uses the Collaborative Drug Discovery (CDD) Vault in this and other collaborations to share chemistry and biology data in a secure web-based environment. This could be perceived as a strength in that collaborative informatics software (such as CDD Vault and other commercial software) underpinning many large-scale collaborations is now virtually ‘invisible’, although it plays a crucial part in ensuring all group members have their data available from anywhere in the world. CDD has been proposed for use by neglected tropical disease researchers [17] as an easy mechanism for eventual public release of data. A further recent example, the European Lead Factory, an Innovative Medicines Initiative (IMI) PPP, has a compound library from seven pharma companies that is made available to academics so they can screen it against their own targets. This program uses the BIOVIA ScienceCloud platform as the core cheminformatics platform [18]. Public database tools have also been created such as BioAssay Research Database (BARD) but they also require some commercial tools to function [19].
We have previously illustrated how drug discovery has increasingly integrated web-based databases and tools and highlighted some of the key tools for linking collaborators [20] as well as the freely available resources that now provide capabilities only previously available in large organizations [21]. Various commercial and publically accessible web-based tools combining elements of chemistry informatics, biology and social networks have been previously reviewed by us in the context of use for intra- and extra-organizational collaborations [22]. In the intervening period we have further developed the CDD Vault software (Box 1) and applied it to many large-scale collaborations for secure sharing of chemistry and biology data. Although we have previously demonstrated the role collaborative software can have in malaria research [22] as part of an integrated drug discovery cycle [23], as well as trypanosomal diseases [24], the focus of this discussion will be on tuberculosis (TB) and it specifically highlights the FP7-funded More Medicines for Tuberculosis (MM4TB) collaboration.
Role in MM4TB
CDD's role in MM4TB primarily involved providing the CDD Vault to over 20 groups (Figure 1). We used a single vault to organize all consortium data in multiple projects, with specific project access to individual participants, which also allowed the data owners to mask structures. For example, the large pharma partners hid the structures but shared the data. This had ramifications when AstraZeneca left the program and also prevented the data from being used for machine learning models, among others. Toward the end of the project the MM4TB CDD Vault contained 130 240 molecules (38 541 without structures), 131 431 batches, 160 protocols, 389 runs and 592 669 readout rows. A second role for CDD was in the area of general cheminformatics support for various target-based projects to help identify compounds for testing within MM4TB. This involved building pharmacophores, docking and machine learning using additional third-party software alongside the CDD Vault. The following examples represent some of the projects undertaken.
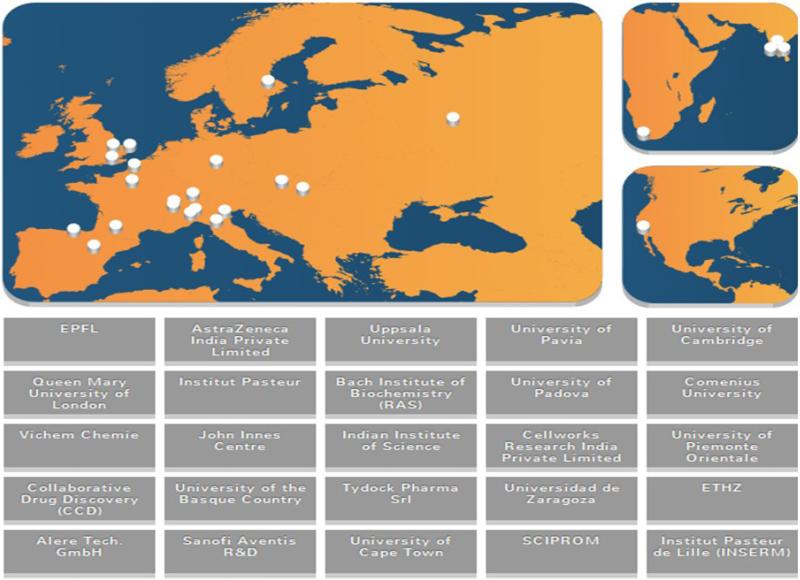
Figure 1.
Map showing the original organizations involved in MM4TB. AstraZeneca India and Sciprom ceased operation during this project. All groups were members of a single CDD Vault.
Topo I
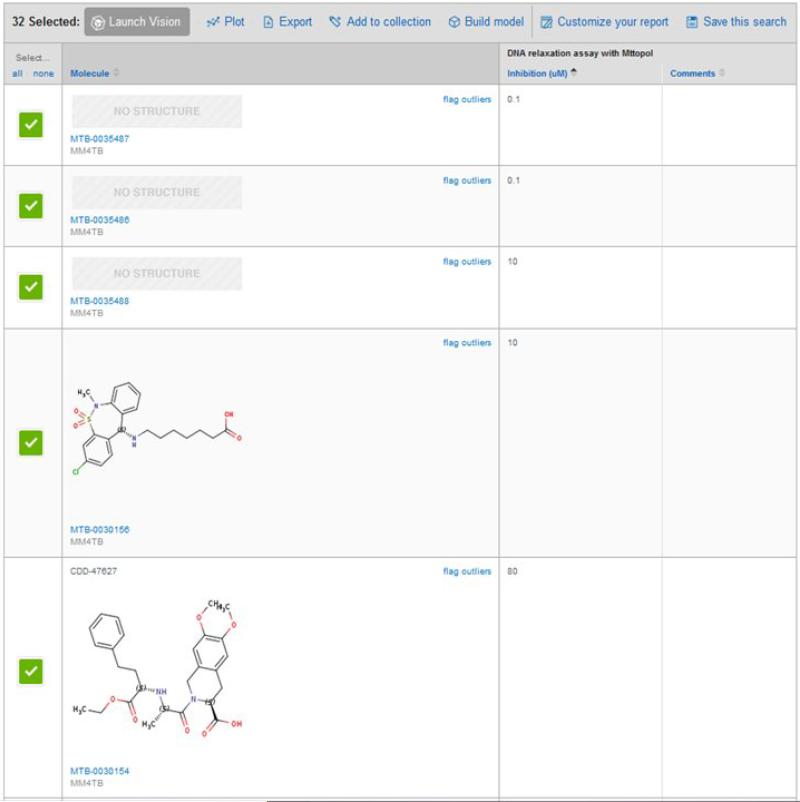
Topoisomerase I (MttopoI) is an essential mycobacterial enzyme and suffers from a shortage of known inhibitors. To identify small-molecule inhibitors of Mttopol a 3D homology model of the enzyme was generated using Escherichia coli topoisomerase I as a template [25]. This model was then used to dock libraries of FDA drugs. Compounds that scored well were then selected for in vitro testing for their inhibitory potential against the DNA relaxation activity of Mttopol. This virtual screening effort resulted in the identification of amsacrine [s1](m-AMSA) – a well-known inhibitor of eukaryotic type II topoisomerases that appears to act by intercalating with DNA against Mttopol [25]. Norclomipramine and imipramine are closely related tricyclic central nervous system active molecules that were also identified in the same way through docking but were found to better stimulate DNA cleavage by MttopoI in vitro (nM inhibitors). These molecules appear to bind to metal-coordinating residues and poison the enzyme [26]. Further, norclomipramine and imipramine inhibit Mycobacterium tuberculosis growth albeit at a higher concentration (60 and 250 μM, respectively). Thus, although these molecules had less desirable whole-cell activity in M. tuberculosis and Mycobacterium smegmatis, they were proposed as an approach to identify further leads. The recently described crystal structure of MttopoI (PDB ID: 5D5H) was also suggested as potentially useful for discovery of poison-type inhibitors that would provide high affinity and selectivity [27]. In our efforts as part of the MM4TB project 639 compounds were tested in vitro by our collaborators. These data were also used to create machine learning models that were further validated and used to select commercially available compounds for testing in vitro. Our collaborators also experimentally demonstrated the inhibition of MttopoI by some of the small-molecule inhibitors identified and showed that the enzyme can be readily targeted for lead molecule development. Figure 2 illustrates how particularly sensitive structures might be in the Vault without structures.
Figure 2.
TopoI data in the MM4TB CDD Vault, demonstrating how some structures can be hidden.
Gyrase B and ThyX
With additional collaborators in MM4TB as well as outside MM4TB we have targeted two essential enzymes in M. tuberculosis that are promising for antibacterial therapy and reported to be inhibited by naphthoquinones. ThyX is an essential thymidylate synthase that has been described as mechanistically and structurally unrelated to the human enzyme [28,29]. DNA gyrase is a DNA topoisomerase present in bacteria and plants but not animals [30]. A combination of cheminformatics (pharmacophore and similarity searching) and in vitro screening was able to identify several new M. tuberculosis ThyX inhibitors (one of which is a drug, idebenone, in Phase III clinical trials for a rare disease) with modest whole-cell activity; and at the same time we were able to show the differences in SAR, with ThyX being much more permissive than GyrB [31].
PyrG
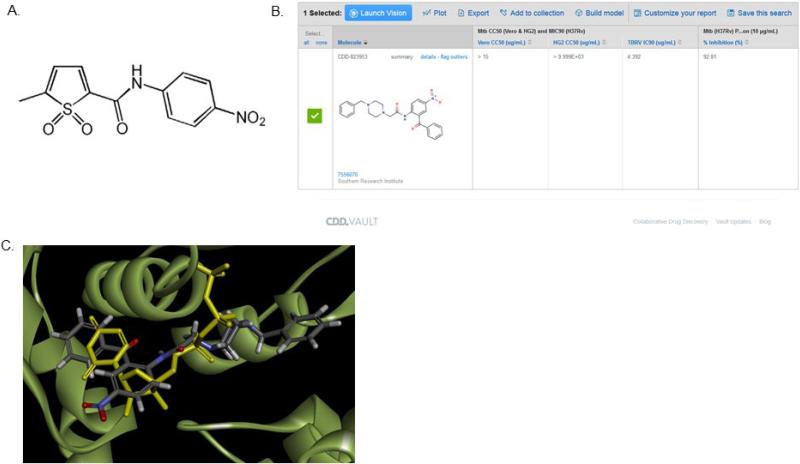
A screen by the MM4TB group of the NIAID library for antitubercular activity identified a new series of molecules, displaying a promising MIC value (0.5 μg/ml; Figure 3a). Isolation of M.-tuberculosis-resistant mutants, genetic validation and biochemical studies identified the main mechanisms of activation and resistance [32]. The molecules were found to be prodrugs activated by the EthA monooxygenase and targeting the PyrG enzyme – a cytidine triphosphate (CTP) synthase catalyzing the ATP-dependent amination of UTP to form the essential pyrimidine nucleotide CTP. This could represent a new and promising TB drug target. We docked the molecule in the crystal structure and also performed substructure and similarity searches to identify additional compounds in the public TB datasets in CDD Vault (Figure 3b). A set of 12 molecules with known M. tuberculosis whole-cell activity were identified, docked in the crystal structure and scored (Figure 3c) before four compounds were tested and one was identified as a weak inhibitor (Ki 88.9 μM, MIC 4.4 μg/ml) of the target. This compound, however, did not require activation [32] by EthA. This represents a prime example of how the combination of the CDD Vault, docking and in vitro testing could be used to narrow down possible compounds to be evaluated experimentally.
Figure 3.
[s14](a) 7947882 (5-methyl-N-(4-nitrophenyl)thiophene-2-carboxamide), (b) substructure search of public M. tuberculosis (Mtb) datasets in CDD Public based on 4-nitroacetanilide retrieved four compounds including CDD-823953. (c) CDD-823953 docked in PyrG crystal structure (LibDock score 106.7) was a weak inhibitor of PyrG (Ki = 88.9 μM).
Library comparisons
Compound library comparisons were performed to evaluate their overlap with the published NIAID/SRI[s2] data [33–36] before testing. We filtered a commercial library of interest for overlap with public TB compounds in the CDD Vault (~340 000 molecules). The library of interest from EPFL[s3] (~53 000 compounds) was uploaded into a Vault with the public TB compounds. In total, 50 445 compounds were imported and found not to be in the public datasets from NIAID/SRI. This compound library was then used for HTS screening in vitro. In addition, this library was also scored with several previously published Bayesian models for the dose–response and cytotoxicity data from the SRI datasets. When ranked by the Bayesian score, some well-known structural motifs were observed (e.g., Mmpl3 inhibitors like those from GSK and Novartis, etc.) [37,38]. This was hence useful to pre-filter compounds that might be less desirable or potentially of less interest based on novelty. A further role for cheminformatics in the MM4TB project was the use of the ADMET machine learning models [39,40] to score compounds for synthesis in the GuaB2 project.
Machine learning models for M. tuberculosis
M. tuberculosis in-vitro-based machine learning models
Computational methods have been used across neglected diseases to differing levels and this might reflect the degree of investment that goes toward TB and malaria compared with kinetoplastids and helminths [10]. Computational tools have been increasingly used in the area of TB research. A recent review by Chibale and colleagues [41] described the extensive structure-based and ligand-based approaches used for TB, malaria and trypanosomal disease research, however they did not specifically address machine learning applications. Machine learning techniques have been applied most extensively for genetics and genomics [42] as well as applied to antibacterial drug discovery [43]. A wide variety of machine learning methods have been applied to TB datasets including recursive partitioning (RP Forest [44–47] and RP Single Tree), Support Vector Machines (SVM) [48–50] and Bayesian methods.
Our own work on TB was initially funded by the Bill and Melinda Gates Foundation, which enabled us to curate public datasets on TB. These datasets were subsequently used to analyze the molecular properties of active and inactive compounds, generate machine learning models and test pharmacophore models while also being one of the first groups to describe the distribution of antituberculars in the context of FDA-approved drugs in molecular property space [51] and compounds failing alerts in these datasets [52]. We have taken a similar approach with large-dataset-screened compounds such as the >13 000 malaria hits from GSK calculating descriptors and using SMARTS [s4]filters as alerts [53,54]. We also proposed that many of these malaria datasets could be screened against M. tuberculosis [54].
We have successfully demonstrated the value of machine learning in drug discovery by predicting in advance which subsets of compound libraries collaborators (such as Infectious Disease Research Institute, UMDNJ-NJMS and Southern Research Institute) should screen. In all cases we focused on eight interpretable descriptors and FCFP6 fingerprints. Models that combined bioactivity and cytotoxicity data were used to rank compounds such as the GSK antimalarial dataset [55]. From the top 46 molecules, seven were chosen and five had MIC ≤2 μg/ml, the most active being 0.0625 μg/ml [56]. A second example used two different M. tuberculosis whole-cell models to score three vendor libraries from which 550 compounds were tested and 124 actives identified [57]. A third example filtered a library of >150 000 molecules and tested 48 compounds of which 11 were active [58]. The models achieved screening hit rates of 15–71% for suggested compounds, far higher than the 0.6–1.5% typical for random library HTS screening [57,59,60].
Over the years with these and other collaborators, we have used machine learning approaches with different public and private M. tuberculosis datasets to explore the various algorithms available. Fusion of three dual activity models gave an excellent ROC value with a fourth external dataset from the same laboratory [61]. These models have also been used individually with a testset of 1924 molecules for which cytotoxicity was determined in three cell lines and enrichments of 11.8-fold were observed in the best case [36]. Fusing single point data (bioactivity only) with dual activity data ultimately led to M. tuberculosis models with 345 011 molecules in them but these were no more predictive that the smaller dual activity datasets when tested with external data [62].
More recently we have applied the Bayesian machine learning approach to identify leads and repurpose drugs for Chagas disease [63] and Ebola [64]. All of these efforts have primarily used commercial software as a proof of concept. This in turn led us to the insight that such models are infrequently shared or even accessible to those that do not have access to the underlying software.
We have also reviewed the wide array of TB-related database efforts in the area of pathway tools [65]. By far the biggest area for application of computational approaches is in cheminformatics such as QSAR, pharmacophores, docking and virtual screening. Again we found that there was a disparity in computational model generation, utilization and sharing and little effort in bringing many different approaches together [65] such as combining machine learning with docking [66]. Recently, chemogenomic methods and experimental validation were used to identify two compounds as dihydrofolate reductase (DHFR) inhibitors [67]. Validating such computational approaches experimentally is essential, whether that is similarity searching, pharmacophores [68] or machine learning [69]. These various models also offer an opportunity for drug repurposing when using libraries of FDA-approved drugs [70].
M. tuberculosis in-vivo-based models
With collaborators we have used a similar array of machine learning approaches to model data from the mouse M. tuberculosis efficacy model that have been published over the past 70 years. Models were initially constructed with 773 compounds and used to predict 11 molecules from the literature (eight were correctly predicted) [71]. This work also enabled identification of the gaps in research when few compounds were published in this model [72]. More recently, these models have been updated and used with a testset of 60 molecules. The best M. tuberculosis in vivo models in this case now possess fivefold ROC values >0.7, sensitivity >80% and concordance >60%. These results indicated that Bayesian models using literature in vivo M. tuberculosis data generated by different laboratories in many different mouse models can be predictive and also be used alongside other models to select antitubercular compounds with in vivo efficacy [73].
ADMET models and model sharing
Eventually getting to an in vivo active compound requires good ADMET properties. With the increasing amounts of data generated for properties like microsomal stability, Caco-2 permeability, aqueous solubility, hERG, among others, one can use machine learning approaches to build classification of QSAR models [40]. These models have the potential to be used to score molecules before testing or synthesis. We have long proposed the need to make ADMET data more accessible and facilitate model building [74] and the shift to crowdsourcing for pharmaceutical research [75]. It was suggested that computational models for ADMET properties could ultimately replace the in vitro models [76]. This led to our exploration of using open source computational tools for ADMET properties in collaboration with Pfizer [77]. This in turn provided evidence that they could provide models and model statistics comparable to commercial descriptors and tools. Other groups have been moving in the same direction. For example Tetko et al. used DMSO solubility data for 163 000 molecules from two companies that were analyzed using different descriptors and machine learning methods, they found the most reliable predictions and combining data could increase the accuracy of the models [78]. Sushko et al. then described an online chemical modeling environment (OCHEM) for model development and data storage. This appears to be one of the earliest attempts to share models on the Internet [79].
CDD has created a standardized mechanism (CDD Models) that enables researchers to share models, share predictions from models and create models from distributed, heterogeneous QSAR data, all without needing to divulge the underlying training sets. This was facilitated by embedding standard model building capabilities directly within the CDD Vault and validating the integrated technology. In the process of this work we created a drop-in replacement for the widely used ECFP6 and FCFP6 fingerprints [80] and made the resulting code available to the public as a feature in the Chemical Development Kit (CDK) project under an open source license. These ‘CDD models’ have been applied to several innovative areas including modeling decision making for chemical probes [81] as well as developed ADMET models that leverage publically accessible data from industry and academia [82]. The open source descriptors and Bayesian algorithm have also been used outside of the CDD Vault to create several thousand models with the ChEMBL data, possibly representing the future of using thousands of models to score compounds simultaneously [83]. More recently, a Bayesian binning approach was developed that represents a move to semiquantitative Bayesian models [84]. Overall these combined efforts show how the open source technologies could benefit others and stimulate new technology applications in general.
For example, other types of datasets that could be made more accessible in this way include ADMET datasets relevant to M. tuberculosis drug discovery. For example, mouse liver microsomal (MLM) stability studies are the initial cell-based model system used to assess metabolic stability in academia and industry for many diseases including M. tuberculosis. Perryman et al. have collated published assays on MLM half-life from PubChem; and reformatted and curated them to create a training set with ~900 molecules [85]. These data were then used to generate machine learning models assessed with internal cross-validation and external tests with a published set of antitubercular compounds and another independent validation with an additional large diverse set of compounds. It was found that ‘pruning out’ the moderately unstable and moderately stable compounds from the training set produced models that displayed superior predictive power for identifying compounds that have a half-life ≥1 h in MLM stability studies[s5]. To date, this represents the largest publically accessible MLM dataset and suggests that the pruning strategy could be useful elsewhere [85]. One could speculate that toxicity models for properties like the potassium channel hERG (which is known to be a particular issue for TB drugs like bedaquiline [86] and shows QT prolongation in dogs and human trials at high doses) could perhaps be avoided by using machine learning models to optimize this property relative to the target pharmacophore. Although this might be difficult we are not aware of this being attempted to date.
Target prediction using TB Mobile
Various studies have developed methods for predicting targets for compounds in M. tuberculosis. The TBDrugome [87] was an early example that used binding sites of TB structures to identify molecules. A recent approach developed models of all the pockets in M. tuberculosis targets to create the ‘pocketome’ [88] from which several approved drugs were described alongside their inferred targets. In both cases it appears these approaches were not experimentally validated, which, if they were, would certainly provide some degree of confidence in using these approaches.
In silico target fishing is an emerging technology that predicts the targets of compounds on the basis of chemical structure by using information from biologically annotated chemical databases. The CDD Vault can currently be mined using similarity and substructure screening, and contains public datasets with over 300 000 compounds screened against M. tuberculosis, as well as manually curated datasets for >7000 published molecules. This information has helped in finding compounds already tested with some TB activity, which could be similar to hits of interest to MM4TB (Figure 3).
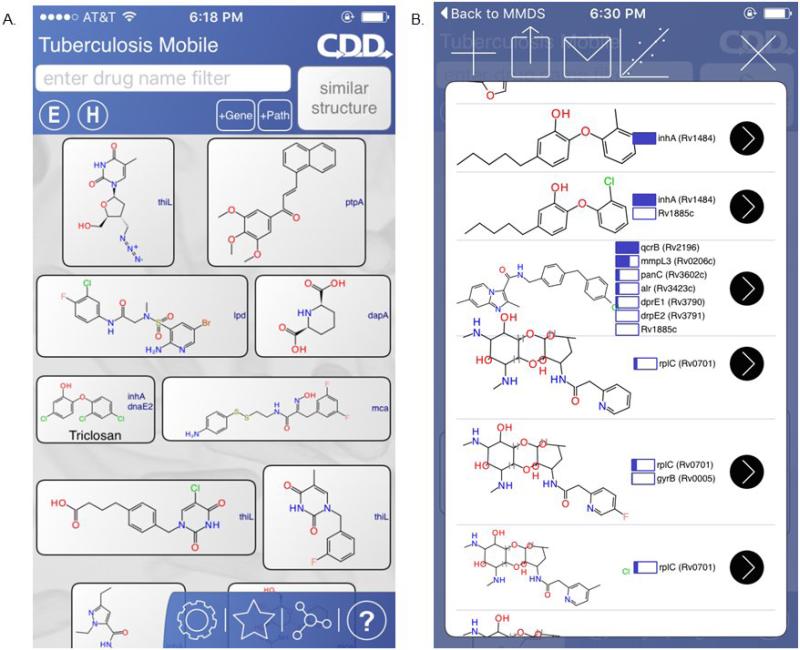
We have made some efforts to bridge bioinformatics and cheminformatics by combining pathway analysis with pharmacophores and machine learning to identify compounds with activity against M. tuberculosis [89]. This also led to the realization that some of the data collected on compounds and targets in M. tuberculosis could be used to create a mobile app called TB Mobile (Figure 4) so that the data could be available in a different accessible format [90]. This in turn has provided a platform for testing various technologies such as the open Bayesian method and ECFP6 fingerprints [80], which are now used in TB Mobile to predict potential TB targets.
Figure 4.
(a) TB Mobile entry page listing structures and targets. (b) Prediction page showing imported compounds and Bayesian scores in app.
Future predictions
Dark matter TB compounds
Several reviews have summarized the difficulties in finding compounds active against M. tuberculosis [91], resulting in few promising candidate compounds and suggesting recent whole-cell screens have identified targets that can form the basis of target- or pathway-based approaches [92]. There have also been calls to expand the chemical diversity and molecular target space [93]. Probably what we also need to do is explore the dark chemical matter [94]; that is, compounds that have never shown biological activity in HTS. Perhaps one way to tease out these compounds is to assess them in combinations looking for synergy. This might be too time-consuming and resource-intensive unless it could be done computationally. For example, Wildenhain[s6] et al. [95] described a large-scale chemical–genetic undertaking, using machine learning to identify synergistic pairs of drug-like molecules. This new study could represent an early example in their use to predict synergism [96]. There is certainly also a growing array of large-scale HTS combination studies [97] and tools for data visualization and exploration [98,99]. New regimens are urgently needed for drug-susceptible and drug-resistant M. tuberculosis and compounds such as PBTZ169 have been suggested as attractive candidates, showing that a combination with bedaquiline and pyrazinamide was more efficacious in mouse than the standard treatment with three drugs [100]. As a caveat to consider, translation of in vitro synergy to in vivo efficacy might not always be clear, as demonstrated with the spectinamide 1599 [101]. Therefore, efforts to combine in silico, in vitro and in vivo synergy prediction might be worthwhile.
Collaborative data sharing
We and others have recently described [15] privacy concerns with data and efforts to find data-sharing methods as well as examples of companies comparing their compound libraries (e.g., Bayer and Schering [102], Bayer and AstraZeneca [103] and Pfizer to the literature [s7][104]). Published efforts have also been reviewed on sharing relevant chemical information about screening data that leave structures blinded, which could open the door for increased collaboration. Swamidass and co-workers recently proposed different approaches to secure sharing of molecules [105], using scaffold networks for compounds demonstrated they do not convey information to reveal chemical structure [105]. A second approach from these researchers uses a method of measuring the overlap between two private datasets using an algorithm that constructs a private dataset's shareable summary (cryptoset [106]), then overlap of private datasets is achieved by comparing these. Other companies have shared anonymized match-pair [107] data for the purpose of improving ADMET optimization of lead compounds [108]. Development of such technologies can be integrated into future versions of collaborative software tools and might help broaden their scope. By default all data or models in CDD Vault always remain private, with options for researchers to share subsets of data or models selectively and securely if and when desired.
Concluding remarks and future outlook
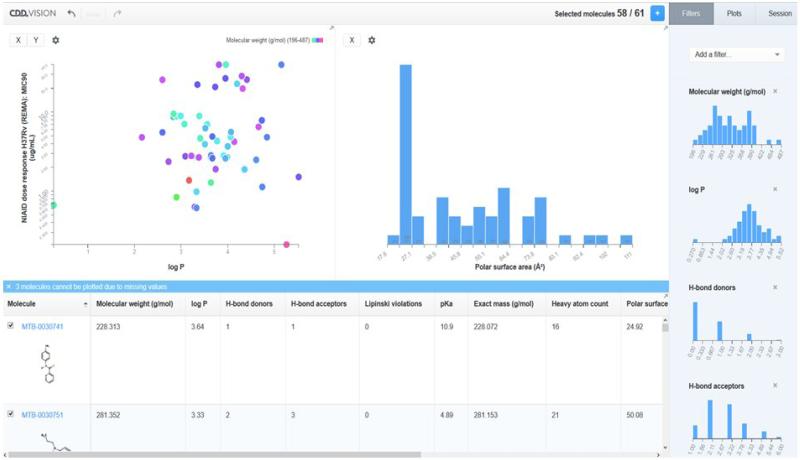
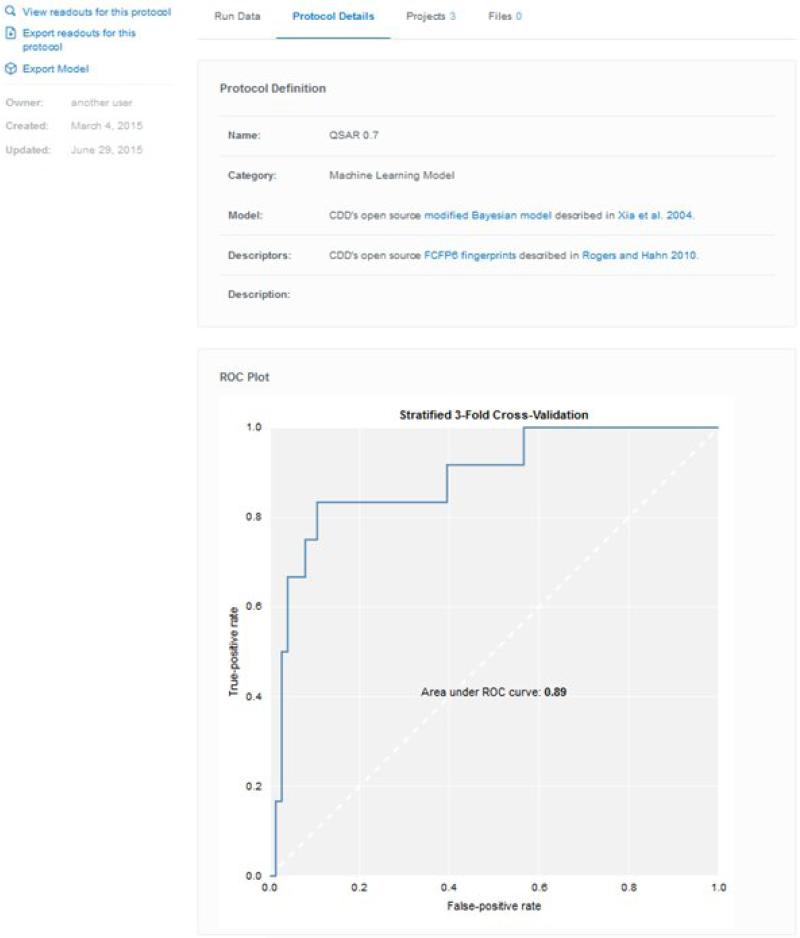
Large-scale collaborations like MM4TB benefitted from the use of collaborative software because it provides a central repository that is accessible to those with the correct permissions. The data are automatically backed up, secure and of course do not require high-level technical expertise to use for uploading, mining or visualizing. As the technology evolves organically, modules will be added (such as an E-laboratory notebook) in the same way we have added inventory, visualization (Figure 5) and machine learning models (Figure 6) to core activity and registration functionality. This represents a strong foundation for future collaborations like MM4TB which will help TB drug discovery and beyond.
Figure 5.
Analysis of MM4TB HTS data alongside calculated properties in CDD Vision.
Figure 6.
An example of a CDD Model created with data for ThyX using >70% inhibition as the cutoff for activity.
Highlights.
There is increasing focus on collaboration and precompetitive efforts such as public–private partnerships (PPPs)
We describe the More Medicines For Tuberculosis project and the role of collaborative software
We describe how different cheminformatics tools were used to identify compounds for testing against multiple targets
We review the literature on how machine learning approaches have been applied to tuberculosis
We propose how collaborative tools will develop in future
Box 1. CDD technical details.
The servers that host CDD Vault sit behind a hardware firewall allowing in only HTTP(S) connections from the Internet. All HTTP requests are redirected to HTTPS, providing transport confidentiality from the user's browser to the server, and session cookies are never transmitted over HTTP or accessible to JavaScript. Production, testing and development environments are all physically distinct. Additional software firewalls on every server provide ‘defense in depth’. All data in the system and application codes are encrypted and backed up nightly onsite and to a redundant site in Europe. CDD retains the full daily backups for 1 month, and retains monthly backups for 2 years. CDD's secure infrastructure has passed multiple big pharma audits and received formal NIH (Federal Information Security Management Act) FISMA compliance and accreditation.
Acknowledgments
The work was partially supported by a grant from the European Community's Seventh Framework Program (grant 260872, MM4TB Consortium) to CDD. TB Mobile was supported by Award Number 2R42AI088893-02 ‘Identification of novel therapeutics for tuberculosis combining cheminformatics, diverse databases and logic based pathway analysis’ from the National Institutes of Allergy and Infectious Diseases. The Bayesian model software within CDD was developed with support from Award Number 9R44TR000942-02 ‘Biocomputation across distributed private datasets to enhance drug discovery’ from the NIH NCATS. The CDD TB has been developed thanks to funding from the Bill and Melinda Gates Foundation (Grant#49852 ‘Collaborative drug discovery for TB through a novel database of SAR data optimized to promote data archiving and sharing’). We sincerely acknowledge our many colleagues, collaborators and advocates who have contributed to the development of CDD over the years and our collaborators on the TB projects described. From the MM4TB: Anthony Maxwell, Valerie Mizrahi, Adwait Anand Godbole, János Pato, Valakunja Nagaraja, Marco Bellinzoni, Maria Rosalia Pasca, Giovanna Riccardi, Rita Szekely and Tanya Parish. From other TB projects: Joel Freundlich, Alexander Perryman, Erin Bradley, Robert Reynolds, Hannu Myllykallio, Carolyn Talcott, Malabika Sarker and Allen Casey. BIOVIA™ is kindly acknowledged for providing Discovery Studio to SE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
AMC consults and other authors are employees of CDD. SE was a consultant for CDD.
References
- 1.Bhinder B, Djaballah H. Drug discovery and repurposing at Memorial Sloan Kettering Cancer Center: chemical biology drives translational medicine. ACS Chem. Biol. 2014;9:1394–1397. doi: 10.1021/cb500479z. [DOI] [PubMed] [Google Scholar]
- 2.Robertson GM, Mayr LM. Collaboration versus outsourcing: the need to think outside the box. Future Med. Chem. 2011;3:1995–2020. doi: 10.4155/fmc.11.160. [DOI] [PubMed] [Google Scholar]
- 3.Craddock S. Precarious connections: making therapeutic production happen for malaria and tuberculosis. Soc. Sci. Med. 2015;129:36–43. doi: 10.1016/j.socscimed.2014.07.039. [DOI] [PubMed] [Google Scholar]
- 4.Dorsch H, et al. Grants4Targets: an open innovation initiative to foster drug discovery collaborations. Nat. Rev. Drug Discov. 2015;14:74–76. doi: 10.1038/nrd3078-c2. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, et al. Racing to define pharmaceutical R&D external innovation models. Drug Discov. Today. 2015;20:361–370. doi: 10.1016/j.drudis.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Jordan AM, et al. Rethinking ‘academic’ drug discovery: the Manchester Institute perspective. Drug Discov. Today. 2015;20:525–535. doi: 10.1016/j.drudis.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Farah SI, et al. Opportunities and challenges for natural products as novel antituberculosis agents. Assay Drug Dev. Technol. 2016;14:29–38. doi: 10.1089/adt.2015.673. [DOI] [PubMed] [Google Scholar]
- 8.Rose DM, et al. Pharmaceutical industry, academia and patient advocacy organizations: what is the recipe for synergic (win-win-win) collaborations? Respirology. 2015;20:185–191. doi: 10.1111/resp.12458. [DOI] [PubMed] [Google Scholar]
- 9.Litterman NK, et al. Collaboration for rare disease drug discovery research. F1000Res. 2014;3:261. doi: 10.12688/f1000research.5564.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponder EL, et al. Computational models for neglected diseases: gaps and opportunities. Pharm. Res. 2013;31:271–277. doi: 10.1007/s11095-013-1170-9. [DOI] [PubMed] [Google Scholar]
- 11.Bunin BA, Ekins S. Academic, commercial, and biodefense case studies for collaborative drug discovery: potential for disrupting drug discovery. In: Chatguturu R, editor. Collaborative Innovation in Drug Discovery: Strategies for Public and Private Partnerships. John Wiley & Sons; 2014. pp. 303–317. [Google Scholar]
- 12.Bingham A, Ekins S. Competitive collaboration in the pharmaceutical and biotechnology industry. Drug Discov. Today. 2009;14:1079–1081. doi: 10.1016/j.drudis.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Bunin BA, Ekins S. Alternative business models for drug discovery. Drug Discov. Today. 2011;16:643–645. doi: 10.1016/j.drudis.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Ekins S, et al. Four disruptive strategies for removing drug discovery bottlenecks. Drug Discov. Today. 2013;18:265–271. doi: 10.1016/j.drudis.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Ekins S, et al. Bigger data, collaborative tools and the future of predictive drug discovery. J. Comput. Aided Mol. Des. 2014;28:997–1008. doi: 10.1007/s10822-014-9762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nathan C. Cooperative development of antimicrobials: looking back to look ahead. Nat. Rev. Microbiol. 2015;13:651–657. doi: 10.1038/nrmicro3523. [DOI] [PubMed] [Google Scholar]
- 17.Pollastri MP. Finding new collaboration models for enabling neglected tropical disease drug discovery. PLoS Negl. Trop. Dis. 2014;8:e2866. doi: 10.1371/journal.pntd.0002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paillard G, et al. The ELF Honest Data Broker: informatics enabling public– private collaboration in a precompetitive arena. Drug Discov. Today. 2016;21:97–102. doi: 10.1016/j.drudis.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Howe EA, et al. BioAssay Research Database (BARD): chemical biology and probe-development enabled by structured metadata and result types. Nucleic Acids Res. 2015;43:D1163–1170. doi: 10.1093/nar/gku1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louise-May S, et al. Towards integrated web-based tools in drug discovery. Touch Briefings Drug Discovery. 2009;6:17–21. [Google Scholar]
- 21.Williams AJ, et al. Free online resources enabling crowdsourced drug discovery. Drug Discovery World. 2009;10:33–38. [Google Scholar]
- 22.Hohman M, et al. Novel web-based tools combining chemistry informatics, biology and social networks for drug discovery. Drug Discov. Today. 2009;14:261–270. doi: 10.1016/j.drudis.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Ekins S, Bunin BA. The Collaborative Drug Discovery (CDD) database. Methods Mol. Biol. 2013;993:139–154. doi: 10.1007/978-1-62703-342-8_10. [DOI] [PubMed] [Google Scholar]
- 24.Ekins S, Bunin BA. Computational approaches and collaborative drug discovery for trypanosomal diseases. In: Jager T, et al., editors. Trypanosomatid Diseases: Molecular Routes to Drug Discovery. Wiley-VCH; 2013. pp. 81–102. [Google Scholar]
- 25.Godbole AA, et al. Inhibition of Mycobacterium tuberculosis topoisomerase I by m-AMSA, a eukaryotic type II topoisomerase poison. Biochem. Biophys. Res. Commun. 2014;446:916–920. doi: 10.1016/j.bbrc.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 26.Godbole AA, et al. Targeting Mycobacterium tuberculosis topoisomerase I by small-molecule inhibitors. Antimicrob. Agents Chemother. 2015;59:1549–1557. doi: 10.1128/AAC.04516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan K, et al. Insights from the structure of Mycobacterium tuberculosis topoisomerase I with a novel protein fold. J. Mol. Biol. 2016;428:182–193. doi: 10.1016/j.jmb.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myllykallio H, et al. An alternative flavin-dependent mechanism for thymidylate synthesis. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 29.Koehn EM, et al. An unusual mechanism of thymidylate biosynthesis in organisms containing the thyX gene. Nature. 2009;458:919–923. doi: 10.1038/nature07973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bush NG, et al. DNA Topoisomerases. In: Böck A, et al., editors. In EcoSal--Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; 2015. [Google Scholar]
- 31.Djaout K, et al. Predictive modeling targets thymidylate synthase ThyX in Mycobacterium tuberculosis. Sci. Rep. 2016;6:27792. doi: 10.1038/srep27792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mori G, et al. Thiophenecarboxamide derivatives activated by EthA kill Mycobacterium tuberculosis by inhibiting the CTP synthetase PyrG. Chem. Biol. 2015;22:917–927. doi: 10.1016/j.chembiol.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ananthan S, et al. High-throughput screening for inhibitors of Mycobacterium tuberculosis H37Rv. Tuberculosis. 2009;89:334–353. doi: 10.1016/j.tube.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maddry JA, et al. Antituberculosis activity of the molecular libraries screening center network library. Tuberculosis. 2009;89:354–363. doi: 10.1016/j.tube.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds RC, et al. High throughput screening of a library based on kinase inhibitor scaffolds against Mycobacterium tuberculosis H37Rv. Tuberculosis. 2012;92:72–83. doi: 10.1016/j.tube.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ekins S, et al. Combining computational methods for hit to lead optimization in Mycobacterium tuberculosis drug discovery. Pharm. Res. 2014;31:414–435. doi: 10.1007/s11095-013-1172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remuinan MJ, et al. Tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide and N- benzyl-6′,7′-dihydrospiro[piperidine-4,4′-thieno[3,2-c]pyran] analogues with bactericidal efficacy against Mycobacterium tuberculosis targeting MmpL3. PLoS One. 2013;8:e60933. doi: 10.1371/journal.pone.0060933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W, et al. Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014;58:6413–6423. doi: 10.1128/AAC.03229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark AM, et al. Open source Bayesian models. 1. application to ADME/Tox and drug discovery datasets. J. Chem. Inf. Model. 2015;55:1231–1245. doi: 10.1021/acs.jcim.5b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ekins S. Progress in computational toxicology. J. Pharmacol. Toxicol. Methods. 2014;69:115–140. doi: 10.1016/j.vascn.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Njogu PM, et al. Computer-aided drug discovery approaches against the tropical infectious diseases malaria, tuberculosis, trypanosomiasis, and leishmaniasis. ACS Infect. Disease. 2016;2:8–31. doi: 10.1021/acsinfecdis.5b00093. [DOI] [PubMed] [Google Scholar]
- 42.Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015;16:321–332. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durrant JD, Amaro RE. Machine-learning techniques applied to antibacterial drug discovery. Chem. Biol. Drug Des. 2015;85:14–21. doi: 10.1111/cbdd.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawkins DM, et al. Analysis of large structure activity data set using recursive partitioning. Quant. Struct. Act. Rel. 1997;16:296–302. [Google Scholar]
- 45.Therneau TM, Atkinson EJ. An introduction to recursive partitioning using the RPART routines. 1997 Available at: http://www.mayo.edu[s8]/research/documents/biostat-61pdf/doc-10026699?_ga=1.249397255.1402737145.1476791219.
- 46.Chen X, et al. Recursive partitioning analysis of a large structure–activity data set using three-dimensional descriptors. J. Chem. Inf. Comput. Sci. 1998;38:1054–1062. [Google Scholar]
- 47.Rusinko A, 3rd, et al. Analysis of a large structure/biological activity data set using recursive partitioning. J. Chem. Inf. Comput. Sci. 1999;39:1017–1026. doi: 10.1021/ci9903049. [DOI] [PubMed] [Google Scholar]
- 48.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. 2001 [s9] [Google Scholar]
- 49.Christianini N, Shawe-Taylor J. Support Vector Machines and Other Kernel-based Learning Methods. Cambridge University Press; 2000. [Google Scholar]
- 50.Heikamp K, Bajorath J. Comparison of confirmed inactive and randomly selected compounds as negative training examples in support vector machine-based virtual screening. J. Chem. Inf. Model. 2013;53:1595–1601. doi: 10.1021/ci4002712. [DOI] [PubMed] [Google Scholar]
- 51.Ekins S, et al. A collaborative database and computational models for tuberculosis drug discovery. Mol. BioSyst. 2010;6:840–851. doi: 10.1039/b917766c. [DOI] [PubMed] [Google Scholar]
- 52.Ekins S, et al. Analysis and hit filtering of a very large library of compounds screened against Mycobacterium tuberculosis. Mol. BioSyst. 2010;6:2316–2324. doi: 10.1039/c0mb00104j. [DOI] [PubMed] [Google Scholar]
- 53.Ekins S, Williams AJ. When pharmaceutical companies publish large datasets: an abundance of riches or fool's gold? Drug Discov. Today. 2010;15:812–815. doi: 10.1016/j.drudis.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 54.Ekins S, Williams AJ. Meta-analysis of molecular property patterns and filtering of public datasets of antimalarial “hits” and drugs. MedChemComm. 2010;1:325–330. [Google Scholar]
- 55.Gamo F-J, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 56.Ekins S, et al. Bayesian models leveraging bioactivity and cytotoxicity information for drug discovery. Chem. Biol. 2013;20:370–378. doi: 10.1016/j.chembiol.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ekins S, et al. Enhancing hit identification in Mycobacterium tuberculosis drug discovery using validated dual-event Bayesian models. PLoS One. 2013;8:e63240. doi: 10.1371/journal.pone.0063240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ekins S, et al. Bayesian models for screening and TB Mobile for target inference with Mycobacterium tuberculosis. Tuberculosis. 2014;94:162–169. doi: 10.1016/j.tube.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ekins S, et al. Bayesian models for screening and TB Mobile for target inference with Mycobacterium tuberculosis. Tuberculosis. 2014;94:162–169. doi: 10.1016/j.tube.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ekins S, et al. Bayesian models leveraging bioactivity and cytotoxicity information for drug discovery. Chem. Biol. 2013;20:370–378. doi: 10.1016/j.chembiol.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ekins S, et al. Fusing dual-event datasets for Mycobacterium tuberculosis machine learning models and their evaluation. J. Chem. Inf. Model. 2013;53:3054–3063. doi: 10.1021/ci400480s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ekins S, et al. Are bigger data sets better for machine learning? Fusing single- point and dual-event dose response data for Mycobacterium tuberculosis. J. Chem. Inf. Model. 2014;54:2157–2165. doi: 10.1021/ci500264r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ekins S, et al. Machine learning models and pathway genome data base for Trypanosoma cruzi drug discovery. PLoS Negl. Trop. Dis. 2015;9:e0003878. doi: 10.1371/journal.pntd.0003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ekins S, et al. Machine learning models identify molecules active against the Ebola virus in vitro. F1000Res. 2016;4:1091. doi: 10.12688/f1000research.7217.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ekins S, et al. Computational databases, pathway and cheminformatics tools for tuberculosis drug discovery. Trends Microbiol. 2011;19:65–74. doi: 10.1016/j.tim.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ekins S, Freundlich JS. Computational models for tuberculosis drug discovery. Methods Mol. Biol. 2013;993:245–262. doi: 10.1007/978-1-62703-342-8_16. [DOI] [PubMed] [Google Scholar]
- 67.Mugumbate G, et al. Mycobacterial dihydrofolate reductase inhibitors identified using chemogenomic methods and in vitro validation. PLoS One. 2015;10:e0121492. doi: 10.1371/journal.pone.0121492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamichhane G, et al. Essential metabolites of M. tuberculosis and their mimics. Mbio. 2011;2:e00301–00310. doi: 10.1128/mBio.00301-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ekins S, Freundlich JS. Validating new tuberculosis computational models with public whole cell screening aerobic activity datasets. Pharm. Res. 2011;28:1859–1869. doi: 10.1007/s11095-011-0413-x. [DOI] [PubMed] [Google Scholar]
- 70.Ekins S, et al. In silico repositioning of approved drugs for rare and neglected diseases. Drug Discov. Today. 2011;16:298–310. doi: 10.1016/j.drudis.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 71.Ekins S, et al. Looking back to the future: predicting in vivo efficacy of small molecules versus Mycobacterium tuberculosis. J. Chem. Inf. Model. 2014;54:1070–1082. doi: 10.1021/ci500077v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ekins S, et al. Minding the gaps in tuberculosis research. Drug Discov. Today. 2014;19:1279–1282. doi: 10.1016/j.drudis.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 73.Ekins S, et al. Machine learning model analysis and data visualization with small molecules tested in a mouse model of Mycobacterium tuberculosis infection (2014–2015). J. Chem. Inf. Model. 2016;56:1332–1343. doi: 10.1021/acs.jcim.6b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ekins S, Williams AJ. Precompetitive preclinical ADME/Tox data: set it free on the web to facilitate computational model building to assist drug development. Lab Chip. 2010;10:13–22. doi: 10.1039/b917760b. [DOI] [PubMed] [Google Scholar]
- 75.Ekins S, Williams AJ. Reaching out to collaborators: crowdsourcing for pharmaceutical research. Pharm. Res. 2010;27:393–395. doi: 10.1007/s11095-010-0059-0. [DOI] [PubMed] [Google Scholar]
- 76.Ekins S, et al. Chemical space: missing pieces in cheminformatics. Pharm. Res. 2010;27:2035–2039. doi: 10.1007/s11095-010-0229-0. [DOI] [PubMed] [Google Scholar]
- 77.Gupta RR, et al. Using open source computational tools for predicting human metabolic stability and additional ADME/TOX properties. Drug Metab. Dispos. 2010;38:2083–2090. doi: 10.1124/dmd.110.034918. [DOI] [PubMed] [Google Scholar]
- 78.Tetko IV, et al. Development of dimethyl sulfoxide solubility models using 163,000 molecules: using a domain applicability metric to select more reliable predictions. J. Chem. Inf. Model. 2013;53:1990–2000. doi: 10.1021/ci400213d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sushko I, et al. Online chemical modeling environment (OCHEM): web platform for data storage, model development and publishing of chemical information. J. Comput. Aided Mol. Des. 2011;25:533–554. doi: 10.1007/s10822-011-9440-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clark AM, et al. New target predictions and visualization tools incorporating open source molecular fingerprints for TB Mobile 2.0. J. Cheminform. 2014;6:38. doi: 10.1186/s13321-014-0038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Litterman N, et al. Computational prediction and validation of an expert's evaluation of chemical probes. J. Chem. Inf. Model. 2014;54:2996–3004. doi: 10.1021/ci500445u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clark AM, et al. Open source bayesian models: 1. Application to ADME/Tox and drug discovery datasets. J. Chem. Inf. Model. 2015;55:1231–1245. doi: 10.1021/acs.jcim.5b00143. [s11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clark AM, Ekins S. Open source bayesian models: 2. Mining A “big dataset” to create and validate models with ChEMBL. J. Chem. Inf. Model. 2015;55:1246–1260. doi: 10.1021/acs.jcim.5b00144. [DOI] [PubMed] [Google Scholar]
- 84.Clark AM, et al. Open source bayesian models: 3. Composite models for prediction of binned responses. J. Chem. Inf. Model. 2016;56:275–285. doi: 10.1021/acs.jcim.5b00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perryman AL, et al. Predicting mouse liver microsomal stability with “pruned” machine learning models and public data. Pharm. Res. 2016;33:433–449. doi: 10.1007/s11095-015-1800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pharmacology/Toxicology NDA review and evaluation Application Number: 204384Orig1s000. Food and Drug Administration; 2012. [s12] [Google Scholar]
- 87.Kinnings SL, et al. The Mycobacterium tuberculosis drugome and its polypharmacological implications. PLoS Comput. Biol. 2010;6:e1000976. doi: 10.1371/journal.pcbi.1000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anand P, Chandra N. Characterizing the pocketome of Mycobacterium tuberculosis and application in rationalizing polypharmacological target selection. Sci. Rep. 2014;4:6356. doi: 10.1038/srep06356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sarker M, et al. Combining cheminformatics methods and pathway analysis to identify molecules with whole-cell activity against Mycobacterium tuberculosis. Pharm. Res. 2012;29:2115–2127. doi: 10.1007/s11095-012-0741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ekins S, et al. TB Mobile: a mobile app for anti-tuberculosis molecules with known targets. J. Cheminform. 2013;5:13. doi: 10.1186/1758-2946-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goldman RC. Why are membrane targets discovered by phenotypic screens and genome sequencing in Mycobacterium tuberculosis? Tuberculosis. 2013;93:569–588. doi: 10.1016/j.tube.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 92.Kana BD, et al. Future target-based drug discovery for tuberculosis? Tuberculosis. 2014;94:551–556. doi: 10.1016/j.tube.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Manjunatha UH, Smith PW. Perspective: challenges and opportunities in TB drug discovery from phenotypic screening. Bioorg. Med. Chem. 2015;23:5087–5097. doi: 10.1016/j.bmc.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 94.Wassermann AM, et al. Dark chemical matter as a promising starting point for drug lead discovery. Nat. Chem. Biol. 2015;11:958–966. doi: 10.1038/nchembio.1936. [DOI] [PubMed] [Google Scholar]
- 95.Wildenhain J, et al. Prediction of synergism from chemical-genetic interactions by machine learning. Cell Systems. 2015;1:383–395. doi: 10.1016/j.cels.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ekins S, Siqueira-Neto JL. Shedding light on synergistic chemical genetic connections with machine learning. Cell Systems. 2015;1:377–379. doi: 10.1016/j.cels.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 97.Mott BT, et al. High-throughput matrix screening identifies synergistic and antagonistic antimalarial drug combinations. Sci. Rep. 2015;5:13891. doi: 10.1038/srep13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lewis R, et al. Synergy Maps: exploring compound combinations using network- based visualization. J. Cheminform. 2015;7:36. doi: 10.1186/s13321-015-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bulusu KC, et al. Modelling of compound combination effects and applications to efficacy and toxicity: state-of-the-art, challenges and perspectives. Drug Discov. Today. 2016;21:225–238. doi: 10.1016/j.drudis.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 100.Makarov V, et al. Towards a new combination therapy for tuberculosis with next generation benzothiazinones. EMBO Mol. Med. 2014;6:372–383. doi: 10.1002/emmm.201303575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bruhn DF, et al. In vitro and in vivo evaluation of synergism between anti- tubercular spectinamides and non-classical tuberculosis antibiotics. Sci. Rep. 2015;5:13985. doi: 10.1038/srep13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schamberger J, et al. Rendezvous in chemical space? Comparing the small molecule compound libraries of Bayer and Schering. Drug Discov. Today. 2011;16:636–641. doi: 10.1016/j.drudis.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 103.Kogej T, et al. Big pharma screening collections: more of the same or unique libraries? The AstraZeneca-Bayer Pharma AG case. Drug Discov. Today. 2013;18:1014–1024. doi: 10.1016/j.drudis.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 104.Tu M, et al. Exploring aromatic chemical space with NEAT: novel and electronically equivalent aromatic template. J. Chem. Inf. Model. 2012;52:1114–1123. doi: 10.1021/ci300031s. [DOI] [PubMed] [Google Scholar]
- 105.Matlock M, Swamidass SJ. Sharing chemical relationships does not reveal structures. J. Chem. Inf. Model. 2014;54:37–48. doi: 10.1021/ci400399a. [DOI] [PubMed] [Google Scholar]
- 106.Swamidass SJ, et al. Securely measuring the overlap between private datasets with cryptosets. PLoS One. 2015;10:e0117898. doi: 10.1371/journal.pone.0117898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Warner DJ, et al. WizePairZ: a novel algorithm to identify, encode, and exploit matched molecular pairs with unspecified cores in medicinal chemistry. J. Chem. Inf. Model. 2010;50:1350–1357. doi: 10.1021/ci100084s. [DOI] [PubMed] [Google Scholar]
- 108.Roche and AstraZeneca launch medicinal chemistry data-sharing consortium to further accelerate drug discovery. 2013 Available at: http://www.astrazeneca.com/Research/news/Article/260613-roche-and-astrazeneca-launch-medicinal-chemistry-datasha[s13]