Abstract
Introduction
Incorporating genetic risk information in electronic health records (EHRs) will facilitate implementation of genomic medicine in clinical practice. However, little is known about patients’ attitudes toward incorporation of genetic risk information as a component of personal health information in EHRs. This study investigated whether disclosure of a genetic risk score (GRS) for coronary heart disease influences attitudes toward incorporation of personal health information including genetic risk in EHRs.
Methods
Participants aged 45–65 years with intermediate 10-year coronary heart disease risk were randomized to receive a conventional risk score (CRS) alone or with a GRS, from a genetic counselor followed by shared decision making with a physician using the same standard presentation and information templates for all study participants. The CRS and GRS were then incorporated into the EHR and made accessible to both patients and physicians. Baseline and post-disclosure surveys were completed to assess whether attitudes differed by GRS disclosure. Data were collected from 2013 to 2015 and analyzed in 2015–2016.
Results
GRS and CRS participants reported similar positive attitudes toward incorporation of genetic risk information in the EHR. Compared with CRS participants, participants with high GRS were more concerned about the confidentiality of genetic risk information (OR=3.67, 95% CI=1.29, 12.32, p=0.01). Post-disclosure, frequency of patient portal access was associated with positive attitudes.
Conclusions
Participants in this study of coronary heart disease risk disclosure overall had positive attitudes toward incorporation of genetic risk information in EHRs, although those who received genetic risk information had concerns about confidentiality.
INTRODUCTION
Little is known about best practices for electronic health record (EHR)-based disclosure of genetic risk for complex diseases to enable precision medicine.1 Including genetic risk information in EHRs is expected to facilitate translation of genomics into clinical practice to improve patient care, but poses new challenges for including personal health information in EHRs.2,3 Patients seek personal health information in their personal health record via a patient portal for self-care and to share with others in their kinship and social networks. To optimize patient participation in precision medicine, it is important to assess the attitudes of patients toward incorporation and use of genetic risk information and other personal health information in their EHR.
A number of prospective cohort and case-control studies have found that a genetic risk score (GRS) can be used to reclassify patients’ disease risk estimates to help individualize preventive measures.4–10 However, to date, the impact of disclosing a GRS in person and placing this information in patients’ EHRs on patients’ attitudes toward incorporation and use of genetic risk information and other personal health information in the EHR has not been studied. Several studies have addressed patients’ attitudes toward EHR use,11–16 but not specifically in the context of GRS disclosure. Assessing patient attitudes may help guide EHR-based preventive measures in precision medicine, in particular for coronary heart disease (CHD).
In this post-hoc analysis of the Myocardial Infarction Genes (MI-GENES) study, the authors hypothesized that, compared with disclosure of conventional risk information alone, EHR-based multi-locus GRS disclosure would influence attitudes toward incorporation and use of genetic risk information and other personal health information in EHRs. It was also hypothesized that this would vary in individuals with high versus low GRS.
METHODS
Study Sample
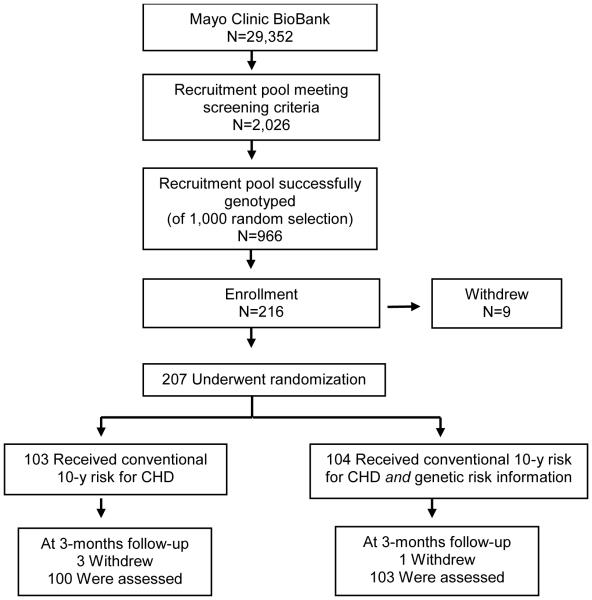
The MI-GENES study design was recently reported17 and is summarized in Figure 1. The Mayo Clinic IRB confirmefd appropriate safeguards and granted ethical approval for this study. Free and informed consent was obtained from each study participant. There were no potential conflicts of interest to disclose to study participants. The study was conducted and data were collected in 2013–2014; data were analyzed in 2015. All study participants (N=203) were residents of Olmsted County, Minnesota, aged 45–65 years, with no history of statin use or CHD, and at intermediate risk for CHD based on a 10-year risk of 5%–20% using the Framingham risk score. Patients were randomized to receive in person at an office visit at Mayo Clinic and then immediately available in their EHR a conventional risk score (CRS)18 for CHD or a CRS and a multi-locus GRS based on 28 CHD variants.19,20 The GRS was stratified as high (H-GRS ≥1.1) or low/average (L-GRS <1.1) risk. Risk was disclosed to all 203 study participants individually in person by the same genetic counselor using a standard presentation and information template, followed by shared decision making individually in person with one of six study physicians using a standard template to determine the need for initiation of statin therapy for high-risk patients. Risk was disclosed to all patients and physicians, along with the genetic counselor and study coordinator, then placed in the EHR for access by all healthcare professionals in their offices at Mayo Clinic, and by patients via their patient portal on their computers at home or on their mobile devices while on the go (Appendix Figure 1). There was no gender bias in the selection of participants (baseline sociodemographic characteristics shown in Table 1).
Figure 1. MI-GENES study design.
Notes: The Myocardial Infarction (MI-GENES) study was designed to determine the impact of genetic risk score (GRS) disclosure on clinical and psychosocial outcomes in residents of Olmsted County, Minnesota. Participants aged 45-65 years, with no history of statin use or coronary heart disease (CHD), and at intermediate risk for CHD based on a 10-year risk of 5-20% using the Framingham risk score (or conventional risk score; CRS) received their CRS or their CRS and a GRS based on 28 CHD variants. CRS and GRS were disclosed in conversations with a genetic counselor. This was followed by shared decision-making with a physician, to discuss potential initiation of statin therapy for high-risk patients.
Table 1.
Baseline Descriptive Statistics For Patient Characteristics (N=203)
| Baseline characteristic |
CRS
n=100 |
GRS
n=103 |
p-value |
|---|---|---|---|
| Age, years | 59.4±5.3 | 59.4±4.9 | 0.97 |
| Male sex, n (%) | 49 (49.0%) | 48 (46.6%) | 0.84 |
| Family history of CHD, n (%) | 30 (30.0%) | 25 (24.3%) | 0.45 |
| BMI, kg/m2 | 30.5±7.0 | 30.2±6.1 | 0.73 |
| SBP, mmHg | 130.1±14.2 | 131.9±17.6 | 0.42 |
| College education or higher, n (%) | 67 (67.0%) | 58 (56.3%) | 0.16 |
| GRS | 1.11±0.30 | 1.14±0.29 | 0.54 |
| CRS | 8.48±3.76 | 8.56±4.47 | 0.88 |
CHD, coronary heart disease; CRS, conventional risk score; GRS, genetic risk score; SBP, systolic blood pressure
Measures
Patient EHR attitudes were assessed by survey (Table 2, Appendix Tables 1–2) in person at baseline and 3 months post-disclosure. The majority of attitude statements (Statements 1 and 4–14) were adapted or used from the National Cancer Institute’s Health Information National Trends (HINTS) Surveys (http://hints.cancer.gov). Additional statements were designed to comprehensively address the hypotheses.
Table 2.
Patient Attitudes 3 Three Months Following Risk Disclosure
| Attitudes towards genetic risk information in the EHR |
S
S |
CRS
(n=100) (%) |
GRS
(n=103) (%) |
p- val ue |
|---|---|---|---|---|
| My genetic information should be included in my EHR. | 2 | 88 (88) | 92 (89) | 0.6 6 |
|
| ||||
| Incorporating my genetic information into the EHR will enable tailored medical therapy for my unique genetic make-up. |
3 | 89 (89) | 91 (88) | 0.9 3 |
|
| ||||
| Would agree to automated notifications of genetic risk test results to: Siblings |
4 | 52 (52) | 54 (52) | 0.8 8 |
|
| ||||
| Parents | 4 | 35 (35) | 42 (41) | 0.4 0 |
|
| ||||
| Children | 4 | 65 (65) | 64 (62) | 06 5 |
|
| ||||
| Other relatives | 4 | 26 (26) | 27 (26) | 0.9 4 |
|
| ||||
| Would grant medical insurance companies access to genetic test results. | 5 | 23 (23) | 11 (11) | 0.0 8 |
|
| ||||
| Attitudes towards medical personal health information in the EHR | ||||
|
| ||||
| Details of my family history of medical conditions should be included in my EHR. |
1 | 93 (93) | 89 (86) | 0.2 0 |
|
| ||||
| Doctors and other healthcare providers should be able to share your medical information with each other electronically. |
6 | 96 (96) | 102 (99) |
0.1 2 |
|
| ||||
| Safeguards are in place to protect your medical records from being seen by people who aren’t permitted to see them. |
7 | 93 (93) | 98 (95) | 0.5 8 |
|
| ||||
| Never kept information from healthcare provider due to concerns about privacy or security of personal medical record. |
8 | 98 (98) | 100 (97) |
0.4 4 |
|
| ||||
| Not concerned that an unauthorized person would see medical information sent electronically between healthcare providers. |
9 | 49 (49) | 45 (44) | 0.2 5 |
|
| ||||
| In general, I think that the information I give doctors is safely guarded. | 1 0 |
99 (99) | 102 (99) |
0.8 1 |
|
| ||||
| Confident that I have some say in who is allowed to collect, use and share my private medical information. |
1 1 |
95 (95) | 100 (97) |
0.3 8 |
|
| ||||
| Not concerned that an unauthorized person would see medical information sent by fax between healthcare providers. |
1 2 |
39 (39) | 36 (35) | 0.3 9 |
|
| ||||
| Scientists doing research should be able to review my de-identified medical information. |
1 3 |
94 (94) | 100 (97) |
0.0 7 |
|
| ||||
| Important to be able to get one's own medical information electronically. | 1 4 |
90 (90) | 97 (94) | 0.1 9 |
Notes: Survey attitude statements 1 and 4-14 were taken or adapted from the National Cancer Institute’s Health information National Trends (HINTS) Surveys (http://hints.cancer.gov).
Survey statements 2 and 3 were novel, in order to determine participants’ attitudes towards genetic risk information in the EHR. The table reports the numbers and percentages for favorable responses to each statement.
CRS, conventional risk score; EHR, electronic health record; GRS, genetic risk score; genetic risk information, personal genetic risk information; SS, survey statement
Twelve statements taken or adapted from the 2007, 2011, and 2013 HINTS surveys were used on different Likert scales: 1 for strongly agree to 5 for strongly disagree (Statements 1 and 4–5), 1 for strongly agree to 4 for strongly disagree (Statements 10 and 13), 1 for very important to 3 for not at all important and 4 for don’t know (Statements 6 and 14), 1 for very confident to 3 for not at all confident (Statements 7 and 11), 1 for yes and 2 for no (Statement 8), and 1 for very concerned to 3 for not concerned (Statements 9 and 12). Statements 2 and 3 were created to determine participants’ attitudes toward genetic risk information in the EHR and used the following Likert scale: 1 for strongly agree to 5 for strongly disagree. Statements 8–9 and 12 were then reverse coded so that for each statement a higher number indicated more unfavorable EHR attitudes, in keeping with other survey statements. Scores were reported for individual attitude statements.
Frequency of access of the patient portal via the website and mobile app was numerically quantified by counting the number of times each participant logged into their individual patient portal using either the website or the mobile app during a given time period. The total number of logins during the study was designated “TotalLogin.” A pairwise method was used to determine correlation between TotalLogin and the sum of the responses for the four survey statements regarding genetic risk information (Genetic Sum) and the sum of the responses for the nine responses regarding medical personal health information (Medical Sum), with results expressed as r =[calculated correlation] with 95% CI. Direct comparisons of TotalLogin between the CRS and GRS groups without correlation with EHR attitudes will be published in a different MI-GENES post-hoc analysis.
Statistical Analysis
Data were collected from 2013 to 2015 and analyzed in 2015–2016. Survey data were exported from the Research Electronic Data Capture21 software. Analyses were conducted using JMP, version 9.0.2. Statistical significance was determined by a p-value of 0.05. Baseline sociodemographic characteristics for participants were described using basic descriptive statistics. Analyses were performed at individual survey item level, with limited multiple testing. Logistic regression models were then used to compare data at and between visits, with individual EHR attitudes as the outcome variables. All data were adjusted for baseline CRS and GRS, along with sociodemographics (age, sex, family history, and level of education; Table 1), as potential predictors using multivariable logistic regression. The authors assessed whether attitudes differed by GRS disclosure, by H-GRS or L-GRS, or in correlation with EHR access. Data were expressed using ORs or Pearson correlations (r) with 95% CIs.
RESULTS
Attitudes toward incorporation and use of genetic risk information and other personal health information in EHRs were overall positive and similar for participants in both the CRS and GRS groups and remained consistent over time, with the exception of a few statements (Table 2).
Overall, 89% of all study participants felt genetic information should be included in their EHR (Table 2). Similarly, 89% of participants believed that incorporating genetic information into EHRs will help tailor medical therapy. Participants were less enthusiastic about automated notifications of genetic test results to their kinship; for example, 64% were comfortable with notifying their children, and 52% were comfortable notifying their siblings. Only 17% of participants would grant medical insurance companies access to genetic test results.
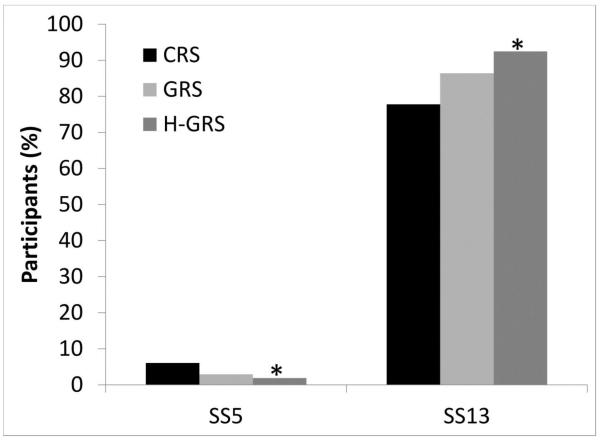
On three of four questions addressing genetic risk information 3 months after risk disclosure, attitudes did not significantly differ by GRS disclosure (Table 2), or by H-GRS versus L-GRS (data not shown). However, H-GRS participants were more likely than CRS participants (OR=3.67, 95% CI=1.29, 12.32, p=0.01) (Figure 2) and trended toward being more likely than L-GRS participants (OR=3.33, 95% CI=0.86, 14.23, p=0.08) to disagree with granting medical insurance companies access to genetic results.
Figure 2. MI-GENES study participants’ EHR attitudes.
Notes: A. At 3 months after initial risk disclosure, high GRS participants (GRS≥1.1) were more likely and GRS participants trended towards being more likely than CRS participants to disagree with granting medical insurance companies access to genetic test results in their EHR (SS5). B. At 3 months after initial risk disclosure, high GRS participants (GRS≥1.1) were less likely and GRS participants trended towards being less likely than CRS participants to disagree with permitting scientists doing research access to participants’ medical information if the information were safely de-identified (SS13). *p-value <0.05.
CRS, conventional risk score; EHR, electronic health record; GRS, genetic risk score; H-GRS, high GRS
The majority of participants felt that family history should be included in EHRs (91%), safeguards were in place to protect EHR confidentiality (94%), doctors should be able to share participants’ medical information with each other electronically (99%), and research scientists should be able to review participants’ de-identified medical information (96%). Most participants felt confident that they have some say in who is allowed to use their private medical information (96%) and believed it was important to have access to one’s medical information electronically (92%). Participants expressed concern that unauthorized individuals might see medical information sent electronically between health professionals (54%), and 63% of participants would be concerned should this information be sent via fax. Only 2% of participants had ever kept information from health professionals owing to privacy concerns.
On nine of ten questions addressing medical personal health information at baseline and 3 months after risk disclosure, attitudes did not significantly differ by GRS disclosure (Table 2) or by H-GRS versus L-GRS (data not shown). However, H-GRS participants were more likely (OR=6.92, 95% CI=1.01, 142.0, p=0.048) (Figure 2), and GRS participants overall trended toward being more likely (OR=4.05, 95% CI=0.91, 28.77, p=0.07) (Table 2), than CRS participants to disagree with research scientists reviewing participants’ safely de-identified information.
The CRS and GRS patients who accessed the patient portal were more likely to have positive attitudes toward incorporation and use of genetic information and other personal health information in EHRs. At 3 months after risk disclosure, increased portal access (higher TotalLogin) significantly correlated with positive attitudes toward the incorporation of medical personal health information (using Medical Sum) in EHRs (r =0.17, 95% CI=0.04, 0.30), but did not significantly correlate with attitudes toward the incorporation of genetic information (using Genetic Sum) in EHRs (r =0.007, 95% CI= –0.13, 0.14). Nevertheless, Medical Sum correlated with Genetic Sum (r =0.37, 95% CI=0.24, 0.48), suggesting a general positive trend toward positive attitudes.
DISCUSSION
Overall, attitudes toward incorporation and use of genetic risk information and other personal health information in the EHR were positive and did not differ significantly by GRS disclosure or by H-GRS or L-GRS, with one exception. After initial risk disclosure, GRS participants expressed greater concern about confidentiality than CRS participants. H-GRS participants were less likely, and GRS participants overall showed a tendency to be less willing, than CRS participants to grant medical insurance companies access to their genetic test results. Nevertheless, H-GRS participants were more likely, and GRS participants overall showed a tendency to be more willing, than CRS participants to permit scientists doing research to review participants’ medical information if the information were safely de-identified.
These results are consistent with other studies utilizing HINTS surveys. In one study, 90% of general respondents valued confidentiality of information in their EHR.24 In another study, most expressed concern about potential data breaches when information from their EHR might be transferred between healthcare professionals by fax (67%) or electronically (65%).22 In other recent studies, only a minority of respondents reported withholding information from a healthcare provider because of security concerns (12% and 13%),22,23 consistent with the present results. In yet another study, 86% of respondents felt it was important for them to have electronic access to their EHR,25 compared with 92% in this study. Of note, in the current study, the frequency of access of the patient portal in CRS and GRS participants post-disclosure was significantly associated with positive attitudes toward medical personal health information in the EHR; there was no significant association with incorporation of genetic risk information.
These results all suggest that patients are interested in engaging with the EHR and are generally amenable to incorporating genetic risk information in the EHR, which will be an essential component of the patient experience in the context of precision medicine. This is in keeping with other studies in which patients had limited concerns about EHR privacy and felt in control of their health self-care, enhancing patient engagement.11–15
Studies suggest that greater patient participation in discussions about EHR use and accessibility could enhance patient trust, irrespective of baseline concerns about privacy and security.12,35 This may particularly be the case for those with a higher level of concern or distrust, such as has been shown among individuals with less computer and health literacy and education, as well as some non-white ethnicities, and others with lower confidence in communicating with doctors.36,37 These populations may benefit from implementation of user-friendly and confidential EHRs for widespread, equitable patient engagement. Indeed, equitable access to EHRs with genomic capability will be important to avoid worsening of healthcare disparities. The inclusion criteria for this study restricted the study participants sample to whites (as a GRS for primarily non-white individuals is not available or validated) and Olmsted County (a county known to have a high college education level and the county host of Mayo Clinic) residents, to maximize opportunity for EHR review and follow-up. Although this is a first step, further studies should more broadly extend genome-wide association studies for CHD susceptibility and subsequent analyses to multiethnic populations.
A recent HINTS survey publication indicated that those with lower education levels, and African Americans and Asians compared with whites, were less likely to be aware of clinical trials.38 Those with lower education levels, and African Americans compared with whites, felt less positive about the use of their personal medical information for research.38 Correspondingly, those who were aware of clinical trials were more likely to express positive attitudes toward the use of their personal medical information for research.38 In a different study, African Americans and Latinos were found to be less knowledgeable about genetic testing than whites, and were less likely to have financial resources or insurance to facilitate testing.39 Nevertheless, Latinos and African Americans were more likely to express preferences for genetic testing than whites, while holding beliefs that might over-ride those preferences in specific situations.39 Some beliefs were elucidated in another study, which suggested that African Americans were more likely to express concern that genetic research might lead to racial discrimination.40 At the same time, African American respondents pointed out that genetic testing could have benefits for African Americans, by inclusion in research protocols and thereby development of better medical treatments tailored for African Americans.40 All ethnic groups expressed concerns about potential genetic discrimination based on results of genetic research.40 Results from these studies and others imply that those offering genomic testing in research and clinical practice need to do so in the context and understanding of beliefs held by various ethnic groups based on individual and collective experiences, in addition to religious, spiritual, socioeconomic, and sociocultural norms and education.41
In the context of culturally appropriate care, it is important to develop best practices for safe and confidential return and storage of genetic/genomic results in the EHR29–31 in order to implement genomic medicine. For example, to assist providers with limited genomics proficiency and limited access to genetics professionals, clinical decision support can guide patient–physician shared decision making using genetic risk information,32 such as in the MI-GENES study.33
Studies such as this and many others will be useful for implementation of genomics in the clinic. Simultaneously, investigations continue to assess the clinical utility of disclosure of genomic information for complex chronic diseases. For example, a meta-analysis suggested that disclosing single genotype risk estimates for CHD risk factors may not yield changes in lifestyle behavior and outcomes as hoped.27 In the MI-GENES study, H-GRS individuals were more likely to initiate statins in shared decision-making sessions with a physician and consequently reduce cholesterol levels than those who did not receive their GRS and than L-GRS individuals,17 suggesting there may be some benefit to disclosing a GRS in shared decision-making sessions between physician and patient. Study materials and disclosure sessions emphasized the probabilistic nature of the multi-locus GRS, and indicated that studies like this one will help determine the clinical utility of disclosing such a GRS. The shared decision-making sessions in the MI-GENES study were videotaped and checked for quality control with the use of a validated method to avoid variation in the physician response across patients.
Use of EHRs by the patient–physician partnership can increase healthcare quality42,43 and improve patient outcomes.44–46 EHRs equip patients with accurate and personalized medical data readily available for sharing with caregivers and healthcare providers. One in five patients share visit notes with their kinship and social networks, and those who share electronic access report better self-care.44 As such, enabling patient engagement and self-management through patient portals is also predicted to aid implementation.43 Indeed, the present study suggests that patients support engagement with their genetic risk information and other personal health information through the patient portals, if safety and privacy are ensured. Efforts to assure patients of EHR privacy and security measures will be advantageous, with ensured safely controlled access to comprehensive EHRs that protect the patient and maintain confidentiality.26
Limitations
The same genetic counselor disclosed individual CHD risk to all patients in this small study (N=203), using a standard template; this may not be feasible in a “real-world” setting. If patients made decisions on assumed clinically utility of the GRS, this could have influenced participants’ responses. It is also important to note that this probabilistic genetic information differs from more deterministic genetic information with established clinical value, such as BRCA1 mutations. This was explained to patients, and the data may have limited generalizability with respect to the types of genetic data being included in the medical record.
CONCLUSIONS
Health information storage, communication, and exchange systems are being developed worldwide, and will facilitate integration of genomic data into EHRs.47 To help ensure that patients are true partners in implementing genomic medicine for individualized patient care and large-scale precision medicine analyses and trials,48 this study investigated the attitudes of MI-GENES study participants toward the incorporation of genomic information in the EHR. The study has elucidated several patient EHR attitudes in the context of communicating a complex multi-locus GRS for prevention of CHD, and differs from other paradigms operative for monogenic disorders. Participants overall had positive attitudes toward incorporation and use of genetic risk information and other personal health information in safe, private, and confidential EHRs.
Supplementary Material
ACKNOWLEDGMENTS
The Myocardial Infarction Genes (MI-GENES) study is registered at ClinicalTrials.gov with number NCT01936675. This study was part of the National Human Genome Research Institute–funded Electronic Records and Genomics Network (U01HG04599 and U01HG006379). The Mayo Clinic Biobank was funded by the Mayo Clinic Center for Individualized Medicine. Use of REDCap was funded by the Center for Clinical and Translational Science grant support (UL1TR000135). No funder or sponsor played any role in the design, conduct, collection, management, analysis, or interpretation of study data; preparation, review, or approval of this manuscript; or any decisions to submit the manuscript for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions are as follows. Study concept and design: Kullo; acquisition, analysis, or interpretation of data: Brown, Jouni, Marroush, Kullo; drafting of the manuscript: Brown; statistical analysis: Brown; revision of the manuscript for important intellectual content and final approval: all authors.
No financial disclosures were reported by the authors of this paper.
REFERENCES
- 1.Collins F, Varmus H. A New Initiative on Precision Medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. http://dx.doi.org/10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kho AN, Rasmussen LV, Connolly JJ, et al. Practical challenges in integrating genomic data into the electronic health record. Genet Med. 2013;15(10):772–778. doi: 10.1038/gim.2013.131. http://dx.doi.org/10.1038/gim.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fein R. Innovate or die!: Genomic data and the electronic health record (EHR) Appl Transl Genom. 2014;3(4):130–131. doi: 10.1016/j.atg.2014.09.007. http://dx.doi.org/10.1016/j.atg.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganna A, Magnusson PK, Pedersen NL, et al. Multilocus genetic risk scores for coronary heart disease prediction. Arterioscler Thromb Vasc Biol. 2013;33(9):2267–2272. doi: 10.1161/ATVBAHA.113.301218. http://dx.doi.org/10.1161/ATVBAHA.113.301218. [DOI] [PubMed] [Google Scholar]
- 5.Thanassoulis G, Peloso GM, Pencina MJ, et al. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012;5(1):113–121. doi: 10.1161/CIRCGENETICS.111.961342. http://dx.doi.org/10.1161/CIRCGENETICS.111.961342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tikkanen E, Havulinna AS, Palotie A, Salomaa V, Ripatti S. Genetic risk prediction and a 2-stage risk screening strategy for coronary heart disease. Arterioscler Thromb Vasc Biol. 2013;33(9):2261–2266. doi: 10.1161/ATVBAHA.112.301120. http://dx.doi.org/10.1161/ATVBAHA.112.301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ripatti S, Tikkanen E, Orho-Melander M, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376(9750):1393–1400. doi: 10.1016/S0140-6736(10)61267-6. http://dx.doi.org/10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brautbar A, Pompeii LA, Dehghan A, et al. A genetic risk score based on direct associations with coronary heart disease improves coronary heart disease risk prediction in the Atherosclerosis Risk in Communities (ARIC), but not in the Rotterdam and Framingham Offspring, Studies. Atherosclerosis. 2012;223(2):421–426. doi: 10.1016/j.atherosclerosis.2012.05.035. http://dx.doi.org/10.1016/j.atherosclerosis.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes MF, Saarela O, Stritzke J, et al. Genetic markers enhance coronary risk prediction in men: the MORGAM prospective cohorts. PLoS One. 2012;7(7):e40922. doi: 10.1371/journal.pone.0040922. http://dx.doi.org/10.1371/journal.pone.0040922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samaan Z, Schulze KM, Middleton C, et al. South Asian Heart Risk Assessment (SAHARA): Randomized Controlled Trial Design and Pilot Study. JMIR Res Protoc. 2013;2(2):e33. doi: 10.2196/resprot.2621. http://dx.doi.org/10.2196/resprot.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schickedanz A, Huang D, Lopez A, et al. Access, interest, and attitudes toward electronic communication for health care among patients in the medical safety net. J Gen Intern Med. 2013;28(7):914–920. doi: 10.1007/s11606-012-2329-5. http://dx.doi.org/10.1007/s11606-012-2329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitropoulous L, Patel V, Scheffler SA, Posnack S. Public attitudes toward health information exchange: perceived benefits and concernss. Am J Manag Care. 2011;17(12):SP111–SP116. Spec No. [PubMed] [Google Scholar]
- 13.Pushpangadan S, Seckman C. Consumer Perspective on Personal Health Records: A Review of the Literature. Online J Nurs Inform. 2015;19(1) [Google Scholar]
- 14.Markle Foundation . Key Findings from Two Surveys of Americans Conducted by Public Opinion Strategies. Alexandria, VA: 2015. Attitudes of Americans Regarding Personal Health Records and Nationwide Electronic Health Information Exchange. [Google Scholar]
- 15.Patel V, Hughes P, Savage L, Barker W. Individuals’ Perceptions of the Privacy and Security of Medical Records and the Sharing of Medical Records between Health Care Providers. ONC Data Brief. 2015;27 [Google Scholar]
- 16.Thornewill J, Dowling AF, Cox BA, Esterhay RJ. Information infrastructure for consumer health: a health information exchange stakeholder study. Am J Prev Med. 2011;40(5 Suppl 2):S123–133. doi: 10.1016/j.amepre.2011.01.010. http://dx.doi.org/10.1016/j.amepre.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Kullo IJ, Jouni H, Austin EE, et al. Incorporating a Genetic Risk Score Into Coronary Heart Disease Risk Estimates: Effect on Low-Density Lipoprotein Cholesterol Levels (the MI-GENES Clinical Trial) Circulation. 2016;133(12):1181–1188. doi: 10.1161/CIRCULATIONAHA.115.020109. http://dx.doi.org/10.1161/CIRCULATIONAHA.115.020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson PW, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97(18):1837–1847. doi: 10.1161/01.cir.97.18.1837. http://dx.doi.org/10.1161/01.CIR.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 19.Kullo I, Jouni H, Austin E, et al. Incorporating a Genetic Risk Score into Coronary Heart Disease Risk Estimates: Effect on LDL Cholesterol Levels (the MIGENES Clinical Trial) Circulation. 2016;133(12):1181–1188. doi: 10.1161/CIRCULATIONAHA.115.020109. http://dx.doi.org/10.1161/CIRCULATIONAHA.115.020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding K, Bailey KR, Kullo IJ. Genotype-informed estimation of risk of coronary heart disease based on genome-wide association data linked to the electronic medical record. BMC Cardiovasc Disord. 2011;11:66. doi: 10.1186/1471-2261-11-66. http://dx.doi.org/10.1186/1471-2261-11-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. http://dx.doi.org/10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agaku IT, Adisa AO, Ayo-Yusuf OA, Connolly GN. Concern about security and privacy, and perceived control over collection and use of health information are related to withholding of health information from healthcare providers. J Am Med Inform Assoc. 2014;21(2):374–378. doi: 10.1136/amiajnl-2013-002079. http://dx.doi.org/10.1136/amiajnl-2013-002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campos-Castillo C, Anthony DL. The double-edged sword of electronic health records: implications for patient disclosure. J Am Med Inform Assoc. 2015;22(e1):e130–140. doi: 10.1136/amiajnl-2014-002804. [DOI] [PubMed] [Google Scholar]
- 24.Beckjord EB, Rechis R, Nutt S, Shulman L, Hesse BW. What Do People Affected by Cancer Think About Electronic Health Information Exchange? Results From the 2010 LIVESTRONG Electronic Health Information Exchange Survey and the 2008 Health Information National Trends Survey. J Oncol Pract. 2011;7(4):237–241. doi: 10.1200/JOP.2011.000324. http://dx.doi.org/10.1200/JOP.2011.000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen KY, Kreps G, Zhu F, Miller S. Consumers' perceptions about and use of the internet for personal health records and health information exchange: analysis of the 2007 Health Information National Trends Survey. J Med Internet Res. 2010;12(4):e73. doi: 10.2196/jmir.1668. http://dx.doi.org/10.2196/jmir.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hood L, Auffray C. Participatory medicine: a driving force for revolutionizing healthcare. Genome Med. 2013;5(12):110. doi: 10.1186/gm514. http://dx.doi.org/10.1186/gm514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollands GJ, French DP, Griffin SJ, et al. The impact of communicating genetic risks of disease on risk-reducing health behaviour: systematic review with meta-analysis. BMJ. 2016;352:i1102. doi: 10.1136/bmj.i1102. http://dx.doi.org/10.1136/bmj.i1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen KD, Roberts JS, Whitehouse PJ, et al. Disclosing Pleiotropic Effects During Genetic Risk Assessment for Alzheimer Disease: A Randomized Trial. Ann Intern Med. 2016;164(3):155–163. doi: 10.7326/M15-0187. http://dx.doi.org/10.7326/M15-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarvik GP, Amendola LM, Berg JS, et al. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet. 2014;94(6):818–826. doi: 10.1016/j.ajhg.2014.04.009. http://dx.doi.org/10.1016/j.ajhg.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hazin R, Brothers KB, Malin BA, et al. Ethical, legal, and social implications of incorporating genomic information into electronic health records. Genet Med. 2013;15(10):810–816. doi: 10.1038/gim.2013.117. http://dx.doi.org/10.1038/gim.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kullo IJ, Jarvik GP, Manolio TA, Williams MS, Roden DM. Leveraging the electronic health record to implement genomic medicine. Genet Med. 2013;15(4):270–271. doi: 10.1038/gim.2012.131. http://dx.doi.org/10.1038/gim.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch BM, Kawamoto K. The need for clinical decision support integrated with the electronic health record for the clinical application of whole genome sequencing information. J Pers Med. 2013;3(4):306–325. doi: 10.3390/jpm3040306. http://dx.doi.org/10.3390/jpm3040306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson CL, Jouni H, Kruisselbrink TM, et al. Disclosing genetic risk for coronary heart disease: effects on perceived personal control and genetic counseling satisfaction. Clin Genet. 2016;89(2):251–257. doi: 10.1111/cge.12577. http://dx.doi.org/10.1111/cge.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomines A, Readhead H, Teutsch S. Applications of electronic health information in puublic health: uses, opportunities and barriers. EGEMS (Wash DC) 2013;1(2):1019. doi: 10.13063/2327-9214.1019. http://dx.doi.org/10.13063/2327-9214.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platt J, Kardia S. Public trust in health information sharing: implications for biobanking and electronic health record systems. J Pers Med. 2015;5(1):3–21. doi: 10.3390/jpm5010003. http://dx.doi.org/10.3390/jpm5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel VN, Dhopeshwarkar RV, Edwards A, Barron Y, Sparenborg J, Kaushal R. Consumer support for health information exchange and personal health records: a regional health information organization survey. J Med Syst. 2012;36(3):1043–1052. doi: 10.1007/s10916-010-9566-0. http://dx.doi.org/10.1007/s10916-010-9566-0. [DOI] [PubMed] [Google Scholar]
- 37.Vodicka E, Mejilla R, Leveille SG, et al. Online access to doctors' notes: patient concerns about privacy. J Med Internet Res. 2013;15(9):e208. doi: 10.2196/jmir.2670. http://dx.doi.org/10.2196/jmir.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown M, Moyer A. Predictors of awareness of clinical trials and feelings about the use of medical information for research in a nationally representative U.S. sample. Ethn Health. 2010;15(3):223–236. doi: 10.1080/13557851003624281. http://dx.doi.org/10.1080/13557851003624281. [DOI] [PubMed] [Google Scholar]
- 39.Singer E, Antonucci T, Van Hoewyk J. Racial and ethnic variations in knowledge and attitudes about genetic testing. Genet Test. 2004;8(1):31–43. doi: 10.1089/109065704323016012. http://dx.doi.org/10.1089/109065704323016012. [DOI] [PubMed] [Google Scholar]
- 40.Bates BR, Lynch JA, Bevan JL, Condit CM. Warranted concerns, warranted outlooks: a focus group study of public understandings of genetic research. Soc Sci Med. 2005;60(2):331–344. doi: 10.1016/j.socscimed.2004.05.012. http://dx.doi.org/10.1016/j.socscimed.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 41.Paniagua C, Taylor R. The Cultural Lens of Genomics. Online J Issues Nurs. 2008;13(1) [Google Scholar]
- 42.Ancker JS, Kern LM, Edwards A, et al. Associations between healthcare quality and use of electronic health record functions in ambulatory care. J Am Med Inform Assoc. 2015;22(4):864–871. doi: 10.1093/jamia/ocv030. http://dx.doi.org/10.1093/jamia/ocv030. [DOI] [PubMed] [Google Scholar]
- 43.Kruse CS, Bolton K, Freriks G. The effect of patient portals on quality outcomes and its implications to meaningful use: a systematic review. J Med Internet Res. 2015;17(2):e44. doi: 10.2196/jmir.3171. http://dx.doi.org/10.2196/jmir.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson SL, Mejilla R, Darer JD, et al. Patients who share transparent visit notes with others: characteristics, risks, and benefits. J Med Internet Res. 2014;16(11):e247. doi: 10.2196/jmir.3363. http://dx.doi.org/10.2196/jmir.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ralston JD, Hirsch IB, Hoath J, Mullen M, Cheadle A, Goldberg HI. Web-based collaborative care for type 2 diabetes: a pilot randomized trial. Diabetes Care. 2009;32(2):234–239. doi: 10.2337/dc08-1220. http://dx.doi.org/10.2337/dc08-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross SE, Moore LA, Earnest MA, Wittevrongel L, Lin CT. Providing a web-based online medical record with electronic communication capabilities to patients with congestive heart failure: randomized trial. J Med Internet Res. 2004;6(2):e12. doi: 10.2196/jmir.6.2.e12. http://dx.doi.org/10.2196/jmir.6.2.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milani L, Leitsalu L, Metspalu A. An epidemiological perspective of personalized medicine: the Estonian experience. J Intern Med. 2015;277(2):188–200. doi: 10.1111/joim.12320. http://dx.doi.org/10.1111/joim.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collins FS, Hudson KL, Briggs JP, Lauer MS. PCORnet: turning a dream into reality. J Am Med Inform Assoc. 2014;21(4):576–577. doi: 10.1136/amiajnl-2014-002864. http://dx.doi.org/10.1136/amiajnl-2014-002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lautenbach DM, Christensen KD, Sparks JA, Green RC. Communicating genetic risk information for common disorders in the era of genomic medicine. Annu Rev Genomics Hum Genet. 2013;14:491–513. doi: 10.1146/annurev-genom-092010-110722. http://dx.doi.org/10.1146/annurev-genom-092010-110722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.