Abstract
Few investigations have evaluated the incremental usefulness of tubular injury biomarkers for improved prediction of chronic kidney disease (CKD) progression. As such we measured urinary kidney injury molecule-1, neutrophil gelatinase-associated lipocalin, N-acetyl beta-D-glucosaminidase and liver fatty acid binding protein under highly standardized conditions among 2466 enrollees of the prospective Chronic Renal Insufficiency Cohort Study. Over 9433 person-years of follow-up, there were 581 cases of CKD progression defined as incident end stage renal disease or halving of the estimated glomerular filtration rate. Levels of the urine injury biomarkers, normalized for urine creatinine, were strongly associated with CKD progression in unadjusted Cox proportional-hazard models with hazard ratios in the range of 7 to 15 comparing highest to lowest quintiles. However, after controlling for the serum creatinine-based estimated glomerular filtration rate and urinary albumin/creatinine ratio, none of the normalized biomarkers was independently associated with CKD progression. None of the biomarkers improved upon the high (0.89) C-statistic for the base clinical model. Thus, among patients with CKD, risk prediction with a clinical model that includes the creatinine-based estimated glomerular filtration rate and the urinary albumin/creatinine ratio is not improved with the addition of renal tubular injury biomarkers.
Keywords: urine biomarkers, chronic kidney disease, kidney injury, CKD progression, prediction, proteinuria, microalbuminuria
INTRODUCTION
There is substantial interest in identifying and validating novel biomarkers in chronic kidney disease (CKD) to better identify patients at high risk of rapid loss of renal function.1 One major motivation has been the perceived short-comings of serum creatinine-based estimated glomerular filtration rate (eGFR) or albuminuria which have been criticized as being insensitive and inadequate for identifying high risk patients.1–3
A number of studies have shown that among CKD patients, higher levels of renal tubular injury biomarkers--many discovered in the context of acute ischemia-reperfusion injury and including kidney injury molecule-1 (KIM-1),4 neutrophil gelatinase-associated lipocalin (NGAL),5–8 N-acetyl-beta-D-glucosaminidase (NAG)8, and liver fatty acid binding protein (L-FABP)9–12-- are associated with more rapid loss of renal function.4–6, 8, 10, 12–15 Here we evaluated in a large national prospective study, the Chronic Renal Insufficiency Cohort Study (CRIC), the independent association between these markers and the rate of progression of CKD as well as their incremental usefulness in predicting progression beyond the established markers: eGFR and urinary albumin/creatinine ratio (ACR).
RESULTS
Table 1 shows baseline characteristics of the study population (N=2466). Median [interquartile range] eGFR was 42 [IQR 30, 55] ml/min/1.73m2, median ACR 53.0 [5.6, 503.1] mg/g, median KIM-1/Cr 1399 [758, 2618] ng/g, NGAL/Cr 12.75 [3.89, 46.19] microg/g, NAG/Cr 4.03 [2.41, 7.29] U/g, and L-FABP/Cr 7.31 [1.88, 28.11] microg/g. Patients with higher levels of ACR had higher levels of all biomarkers (and lower eGFR). (Supplementary Table 1 shows baseline characteristics by quintiles of KIM-1/Cr, NGAL/Cr, and NAG/Cr and for categories of L-FABP/Cr. Supplementary Table 2 shows Spearman correlation coefficients among the normalized biomarkers and with ACR and eGFR.)
Table 1.
Baseline patient characteristics (N=2466)
| Age (mean±SD) | 59.5±10.8 years |
| Female N (%) | 1131 (46%) |
| Race/ethnicity N (%) | |
| Non-Hispanic White | 1049 (43%) |
| Non-Hispanic Black | 949 (38%) |
| Hispanic | 378 (15%) |
| Other | 90 (4%) |
| Diabetes Mellitus N (%) | 1226 (50%) |
| Cardiovascular Disease N (%) | 849 (34%) |
| Urinary albumin/Cr ratio (ACR) (median [IQR]) | 53.0 [5.6 – 503.1] mg/g |
| eGFR (mean±SD) | 43.6 ± 17.8 mL/min/1.73 m2 |
| Systolic BP (mean±SD) | 127.1 ± 22.1 mmHg |
| Diastolic BP (mean±SD) | 69.7 ± 12.7 mmHg |
| Body Mass Index (mean±SD) | 32.1 ± 7.7 kg/m2 |
| KIM-1/Cr (median [IQR]) | 1399 [758 – 2618] ng/g |
| NGAL/Cr (median [IQR]) | 12.75 [3.89 – 46.19] mcg/g |
| NAG/Cr (median [IQR]) | 4.03 [2.41–7.29] U/g |
| L-FABP/Cr (median [IQR]) | 7.31 [1.88–28.11] mcg/g |
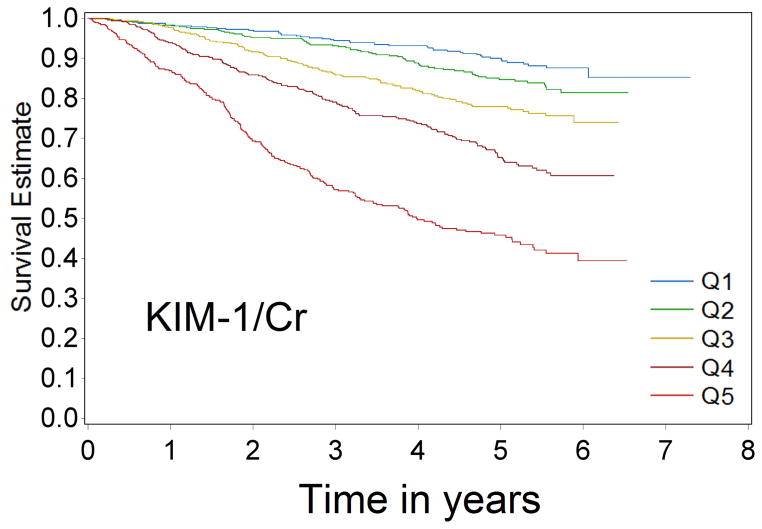
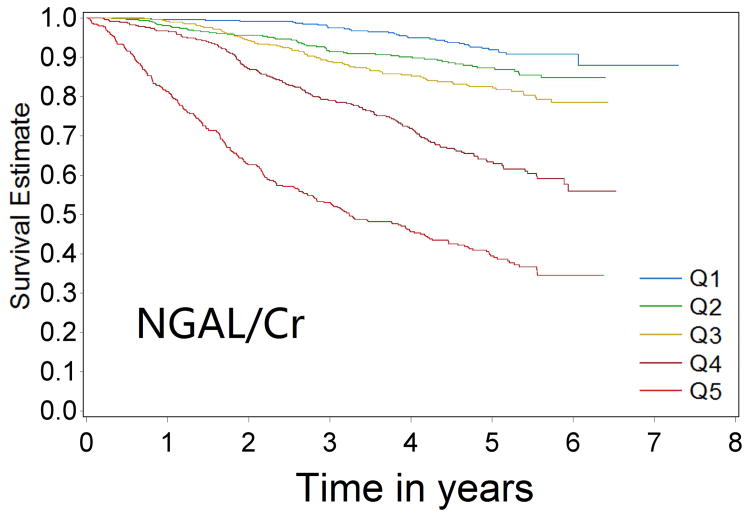
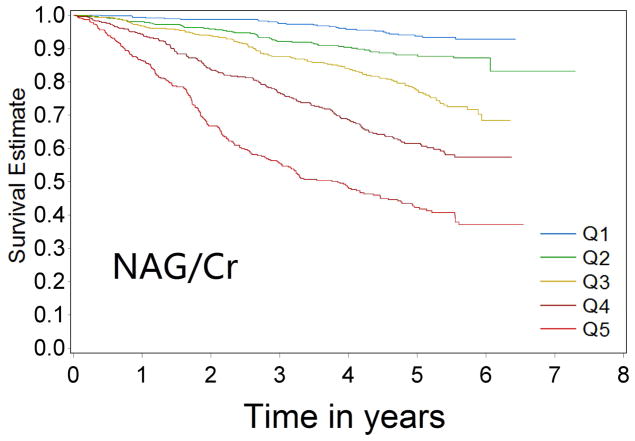
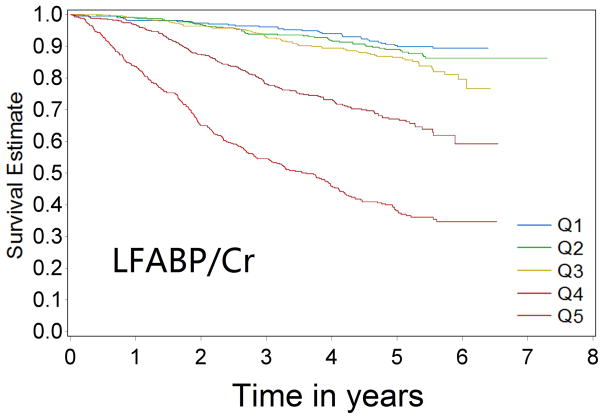
Over 9433 person-years of follow-up, 581 cases of CKD progression were observed (incidence rate 61.6/1000 person-years). In univariable analysis, patients with progressively higher levels of biomarkers were at progressively higher risk of CKD progression for all four biomarkers (Figure 1a–1d Kaplan-Meier curves). Comparing the highest quintile of biomarker concentration with the lowest quintile, the unadjusted Cox model HRs for renal progression were 7.68 (95% confidence interval [CI] 5.61–10.5) for KIM-1/Cr, 12.85 (95% CI 9.02–18.31) for NGAL/Cr, 15.16 (95% CI 10.17–22.59) for NAG/Cr and 10.67(95% CI 7.46–15.25) for L-FABP/Cr (comparing top group with the lowest group, where levels were undetectable)(p-value for trend <0.0001 for all)(Table 2).
Figure 1a–1d.
Kaplan-Meier survival curves for CKD progression by biomarker quintile (all log-rank p-values < 0.0001) (Q1 is lowest quartile and Q5 is highest quartile)
Table 2.
Association between quintiles of normalized biomarker concentrations and the risk of progressive CKD
| Events | Unadjusted Hazard Ratio (95% CI) | Model 1 Hazard Ratio (95% CI): Adjusted for age, sex, race/ethnicity, clinical center |
Model 2 Hazard Ratio (95% CI): Model 1+ACR |
Model 3 Hazard Ratio (95% CI): Model 1+eGFR |
Model 4 Hazard Ratio (95% CI): Model 1+ACR+ eGFR |
Model 5 Hazard Ratio (95% CI): Model 4+DM, CVD, systolic BP, BMI, ACE-I/ARB use, education |
|
|---|---|---|---|---|---|---|---|
| KIM/Cr (ng/g) | |||||||
| ≤ 661 | 48 | Ref | Ref | Ref | Ref | Ref | Ref |
| > 661–1112 | 68 | 1.46(1.01–2.12) | 1.59(1.1–2.31) | 0.89(0.61–1.30) | 1.46(1.00–2.11) | 0.98(0.67–1.43) | 0.93(0.64–1.37) |
| > 1112–1831 | 97 | 2.24(1.59–3.17) | 2.61(1.84–3.70) | 1.11(0.78–1.60) | 2.09(1.47–2.98) | 1.10(0.76–1.58) | 1.10(0.76–1.59) |
| > 1831–2990 | 146 | 3.76(2.72–5.21) | 4.69(3.36–6.55) | 1.31(0.92–1.88) | 2.84(2.02–3.98) | 1.19(0.83–1.70) | 1.22(0.85–1.75) |
| > 2990 | 222 | 7.68(5.61–10.5) | 8.79(6.35–12.17) | 1.38(0.95–2.00) | 5.15(3.71–7.16) | 1.30(0.89–1.89) | 1.36(0.93–2.00) |
| Per SD | 2.26(2.07–2.47) | 2.28(2.08–2.50) | 1.17(1.05–1.31) | 1.88(1.71–2.07) | 1.10(0.98–1.24) | 1.10(0.98–1.24) | |
| NGAL/Cr (mcg/g) | |||||||
| ≤ 2.63 | 35 | Ref | Ref | Ref | Ref | Ref | Ref |
| > 2.63–8.40 | 56 | 1.74(1.14–2.66) | 1.69(1.11–2.59) | 1.28(0.84–1.97) | 1.40(0.92–2.15) | 1.11(0.72–1.70) | 1.18(0.77–1.81) |
| > 8.4–20.2 | 80 | 2.48(1.67–3.69) | 2.43(1.62–3.63) | 1.19(0.79–1.80) | 1.55(1.03–2.32) | 0.85(0.56–1.30) | 0.85(0.56–1.30) |
| >20.2–64.07 | 155 | 5.51(3.82–7.96) | 5.55(3.81–8.09) | 1.66(1.11–2.46) | 2.57(1.75–3.78) | 0.85(0.56–1.28) | 0.84(0.55–1.27) |
| > 64.07 | 255 | 12.85(9.02–18.31) | 13.54(9.37–19.56) | 3.05(2.04–4.56) | 4.29(2.91–6.30) | 0.88(0.56–1.37) | 0.97(0.62–1.52) |
| Per SD | 2.40(2.22–2.59) | 2.39(2.21–2.6) | 1.53(1.38–1.7) | 1.68(1.52–1.85) | 0.95(0.83–1.08) | 0.97(0.84–1.10) | |
| NAG/Cr (U/g) | |||||||
| ≤ 2.08 | 27 | Ref | Ref | Ref | Ref | Ref | Ref |
| > 2.08–3.33 | 53 | 2.07(1.30–3.30) | 2.12(1.33––3.38) | 1.28(0.79–2.07) | 1.69(1.06–2.71) | 1.29(0.79–2.09) | 1.21(0.74–1.98) |
| > 3.33–4.96 | 104 | 4.36(2.85–6.66) | 4.10(2.68–6.27) | 1.40(0.88–2.24) | 2.38(1.55–3.66) | 1.26(0.79–2.00) | 1.23(0.77–1.97) |
| > 4.96–8.36 | 159 | 7.86(5.23–11.83) | 7.49(4.96–11.33) | 1.63(1.02–2.6) | 3.61(2.37–5.5) | 1.42(0.89–2.26) | 1.32(0.82–2.13) |
| > 8.36 | 238 | 15.16(10.17–22.59) | 13.27(8.84–19.91) | 1.45(0.89–2.34) | 6.59(4.37–9.95) | 1.33(0.82–2.15) | 1.24(0.76–2.02) |
| Per SD | 2.05(1.93–2.18) | 2.04(1.90–2.19) | 1.06(0.95–1.19) | 1.88(1.73–2.04) | 1.12(1.00–1.26) | 1.08(0.96–1.22) | |
| L-FABP/Cr (mcg/g) | |||||||
| Undetectable | 34 | Ref | Ref | Ref | Ref | Ref | Ref |
| Below LLD | 48 | 1.27(0.82–1.97) | 1.25(0.80–1.94) | 1.06(0.68–1.64) | 0.99(0.63–1.54) | 0.90(0.58–1.40) | 0.90(0.58–1.41) |
| < 9.68 | 70 | 1.68(1.12–2.54) | 1.55(1.02–2.34) | 1.18(0.78–1.78) | 1.29(0.85–1.95) | 1.16(0.76–1.76) | 1.19(0.78–1.80) |
| ≥ 9.68 – 33.8 | 150 | 4.31(2.96–6.25) | 3.46(2.37–5.05) | 1.36(0.92–2.01) | 1.84(1.25–2.7) | 1.07(0.72–1.57) | 1.07(0.72–1.58) |
| ≥ 33.8 | 279 | 10.67(7.46–15.25) | 8.34(5.79–12.01) | 1.62(1.09–2.40) | 3.08(2.12–4.48) | 0.91(0.61–1.35) | 0.90(0.60–1.34) |
ACE-I: angiotensin converting enzyme inhibitor; ACR: albumin-creatinine-ratio; ARB: angiotensin receptor blocker; BMI: body mass index; BP: blood pressure; DM: diabetes mellitus; LLD: Lower Limit of Detection
Adjusting for demographic factors (and clinical center) did not significantly attenuate the associations (Table 2, Model 1). However, additional adjustment for ACR greatly weakened associations (Table 2, Model 2). Adjusting for eGFR also weakened associations, but to a lesser degree than adjusting for ACR (Table 2, Model 3). After adjusting for both ACR and eGFR, none of the biomarkers were associated with renal disease progression (Table 2, Model 4). There were also no independent associations between the biomarkers and CKD progression in the full model, which also adjusted for clinical parameters such as diabetes status and systolic BP (Table 2, Model 5).
The C-statistic for the base clinical model was 0.890 (95% CI 0.878–0.901)(Table 3). Consistent with the null findings from regression models, none of the biomarkers led to a change in the C-statistic when added to the base clinical model (Table 3). Furthermore, albuminuria (and eGFR) remained a strong independent risk factor for CKD progression in models that simultaneously included all the injury biomarkers and ACR, even when ACR was not modeled with a spline term (Supplementary Table 3).
Table 3. C-statistic for the base clinical model and incremental change with addition of novel biomarkers in the full study sample and intermediate risk population.
The base clinical model included age, sex, race/ethnicity, clinical center, ACR, eGFR, DM, CVD, systolic BP, BMI, ACE-I/ARB use, and education. Log-transformed biomarker results are expressed per log standard deviation
| Model | Full study sample | Intermediate risk¶ |
|---|---|---|
| Base clinical model | 0.890 | 0.764 |
| Base clinical model + log(KIM-1/Cr) | 0.890 | 0.774 |
| Base clinical model + log(KIM-1) | 0.891 | 0.779 |
| Base clinical model + log(NGAL/Cr) | 0.890 | 0.766 |
| Base clinical model + log(NGAL) | 0.891 | 0.762 |
| Base clinical model + log(NAG/Cr) | 0.890 | 0.768 |
| Base clinical model + log(NAG) | 0.890 | 0.775 |
| Base clinical model + L-FABP/Cr* | 0.890 | 0.762 |
| Base clinical model + L-FABP** | 0.890 | 0.766 |
L-FABP/Cr was divided in to those with undetectable levels, those with absolute levels below the lower limit of detection, and then into tertiles of the normalized biomarker
LFABP was divided in to those with undetectable levels, those with absolute levels below the lower limit of detection, and then into tertiles of the raw biomarker
Intermediate risk population had eGFR ≥60 ml/min/1.73m2 and ACR ≥30 mg/g Cr; or eGFR 45–59 ml/min/1.73m2 and ACR 0–300 mg/g Cr; or eGFR 30–44 ml/min/1.73m2 and ACR <30 mg/g Cr
In sensitivity analysis examining biomarker levels not normalized for urine creatinine, higher urine concentrations of KIM-1 and NAG were independently associated with CKD progression the fully adjusted model (Supplementary Table 4). However, the change to the C-statistic was minimal (C-statistics for models with raw KIM-1 and NAG concentrations added were both 0.891)(Table 3).
When we limited our study population to patients deemed at intermediate risk for CKD progression based on Kidney Disease: Improving Global Outcomes (KDIGO) risk stratification, the C-statistic for the base clinical model was lower, 0.764 (95% CI 0.698–0.833). The change in the C-statistic with normalized biomarkers was greatest with KIM-1/Cr with area under the ROC curve of 0.774 (Table 3). Attempts at combining information from biomarkers (e.g. by creating a composite score or via principal component analysis) did not result in further improvement of the C-statistic (data not shown).
DISCUSSION
In this large U.S. study of individuals with CKD, we observed that higher levels of renal tubular injury biomarkers were strongly associated with risk of subsequent CKD progression, with unadjusted hazard ratios on the order of 7 to 15 comparing highest to lowest quintile of biomarker levels. However, after adjusting for conventional risk factors, none of these urine biomarkers normalized to creatinine was independently associated with CKD progression and, accordingly, none improved risk stratification.
Although numerous studies have demonstrated associations between different classes of biomarkers and loss of renal function in CKD patients, few investigations have evaluated the incremental utility of specific biomarkers. Some prior studies did not capture information regarding albuminuria16 or did not have albuminuria measurements concurrent with the biomarkers measurements17, 18 or did not analyze albuminuria beyond a dichotomous17 or semi-quantitatively fashion.18 eGFR was usually not modeled flexibly (e.g. with a spline) to optimize its contribution to prediction.16–19 The conclusions from this study are consistent with the one prior publication from CRIC, which was limited to urine NGAL.5, 20 In that study, NGAL was measured in urine obtained as part of a 24-hour collection that could have been stored at study participant’s home for as long as a week, raising concern that protein degradation may have biased findings towards the null.5 In the current study, we analyzed urine samples that were rapidly processed after voiding under conditions fully controlled by the research team.21 In addition, we expanded our biomarker panel to include KIM-1, NAG and L-FABP.
Our findings are consistent with recent reports from a Taiwan cohort of CKD patients,22 and studies of Pima Indians8 and Atherosclerosis Risk in Communities Study enrollees23 where measuring tubular injury biomarkers did not add to the prediction of future renal function decline. Our results differ from those of a study enriched with CKD patients with glomerular disease, which showed that the C-statistic for traditional risk factors for CKD progression (including baseline eGFR and albuminuria) was only 0.758 and the addition of a multipeptide biomarker classifier significantly improved it to 0.831.24 The exact reasons for these discrepant findings are not known but may relate to differences in the patient population and end-point definition. An important observation in the current study is that the C-statistic for the base clinical model was very high at 0.890. The high discriminatory ability of conventional risk factors for CKD progression has been reported25 but not highlighted in prior studies of novel biomarkers for CKD progression. For example, the Chronic Renal Impairment in Birmingham study generated a risk prediction equation for end-stage renal disease (ESRD)–based on sex, serum creatinine, serum phosphate level and ACR–which achieved a C-statistic of 0.87 in the derivation cohort and a C-statistic of 0.91 in an external validation cohort.26 Similarly, a four-variable Kidney Failure Risk Equation (consisting of age, sex, eGFR and ACR) had a C-statistic of 0.91 in the development data set27 and a C-statistics of 0.88–0.90 in validation populations (all with CKD stages 3–5).28 Beyond patients with reduced eGFR, a meta-analysis of general population cohorts reported a C-statistic for predicting ESRD in a model with eGFR, albuminuria, demographics (age, sex, race/ethnicity), and basic clinical parameters (smoking status, history of cardiovascular disease, systolic blood pressure, diabetes, serum total cholesterol concentration, body mass index) of 0.92.29 By comparison, the Framingham equation for coronary heart disease generally performs in the C-statistic range of 0.65–0.80.30, 31 This may be because eGFR and albuminuria are arguably more than just risk factors but actually markers of established kidney disease on the pathway to ESRD.
Our findings underscore the fact that traditional biomarkers such as eGFR and ACR (plus easily available clinical parameters) perform rather well in terms of identifying CKD patients at high risk for future loss of renal function. This has important implications for future studies of CKD biomarkers. It is unlikely that that a C-statistic in the range of 0.9 can be improved upon, or needs improving. Further enhancement in discrimination is unlikely to be achieved regardless of whether future studies are based on particular disease pathways or leverage cutting edge “omic” technologies.
We believe that if novel biomarkers were to improve prediction of CKD progression, they would have to be discovered and validated under circumstances where established and easily assessed clinical risk factors are less predictive. Thus, after completing our pre-planned analyses, we attempted to identify such a population by restricting the range of eGFR and ACR. In our exploratory analysis limited to CKD patients considered intermediate risk by KDIGO, we observed that the C-statistic for our model incorporating traditional risk factors was lower at 0.764, indicative of a greater opportunity for improvement in prediction. Our results in this subgroup suggest that injury biomarkers such as KIM-1 may provide incremental value among these individuals, although the observed magnitude of the change in C-statistic was small. Furthermore, in CRIC, only 44 out of 581 cases of CKD progression were observed among the intermediate risk group (vs. 535 cases among those considered high risk by KGIDO). Thus, any future studies limited to intermediate risk subgroups need to address potential problems of study power, generalizability and public health relevance.
It is interesting to speculate why tubular injury biomarkers did not add incremental usefulness to assessing risk of CKD progression. The association between tubular injury biomarkers and progressive CKD was considerably attenuated after controlling for ACR (more so than after controlling for eGFR; Table 2 Model 2 vs. Model 3). Although albuminuria is often considered solely a marker for glomerular dysfunction,4, 32 it may also be closely linked with tubular injury.33,34, 35 The increased load of filtered albumin (and molecules bound to albumin) reaching the tubular lumen may be directly toxic to tubular cells.36–38 In a model of tubulointerstitial damage in uninephrectomized rats caused by repeated intraperitoneal injections of bovine serum albumin, injury to the proximal tubule from filtered albumin leads to increases in KIM-1 production and release into the urine.39 Alternatively, there are provocative data34, 40–44 that more albumin may be filtered under normal conditions than generally accepted and proximal tubule reclamation rate may be an important determinate of urinary albumin excretion rate. Thus higher ACR may not only be due to altered glomerular permeability but also reflect tubular damage.
We should emphasize that CKD biomarkers have many valuable roles other than for prediction of CKD progression. For example, they can shed important pathophysiological insight into disease pathways,45, 46 which can then lead to novel therapeutic targets or better understanding of drug toxicity. Our results should not be interpreted to suggest that tubular injury is not associated mechanistically with CKD progression.47 Biomarkers may also help distinguish between different subtypes of CKD and provide a non-invasive way to track the state of some diseases that may alternate between periods of active inflammation vs. fibrosis.48, 49 In terms of risk prediction, there are other outcomes besides kidney disease progression that are important for CKD patients, including death, cardiovascular disease events or acute kidney injury.50–52 The base clinical models for these outcomes appear to have lower C-statistics,25 and therefore these biomarkers may be more useful in that context.
The strengths of this study include its large size and national scope, and the simultaneous measurements of multiple biomarkers. In addition, bio-samples were rigorously collected under standardized conditions. Our assay platforms and methodologies were specifically chosen to be state-of-the art and great attention was paid to quality control issues, including creation and adherence to well-annotated standard operating procedures.21 The CRIC study systematically ascertained renal function. In addition to the detailed in-person follow-up and direct determination, ESRD ascertainment in CRIC was supplemented via crosslink with US Renal Data System. Important clinical covariates known to strongly associate with CKD progression were rigorously ascertained and considered in the statistically model. Results were consistent whether biomarkers levels were normalized for urine creatinine concentration or analyzed as absolute urine biomarker concentrations.21
Limitations include the fact that biomarkers were only measured once. There was a relatively long gap between collection of urine and measurement of the biomarkers, but these samples were stored at −80°C for the entire duration.53–57 There is research suggesting that KIM-1 and NGAL are stable over at least 5 years,53 and that there is a minimal amount of KIM-1 and NGAL degradation over several freeze-thaw cycles.53 There are limited published data on the stability of NAG and L-FABP under the same conditions. However, a number of prior biomarker studies have measured these analytes in samples that had been stored for even longer than here.8, 23, 32, 58 Furthermore, the very strong unadjusted associations between urine biomarker levels and clinical outcomes (with hazard ratios in the range of 7 to 15 comparing highest to lowest quintiles) argue against there being substantial degradation which would lead to bias towards the null. Nonetheless, this is a potential limitation of our study, and the results should be interpreted with this caveat. Our results may not be generalizable to all CKD patients such as those with primary glomerular disease,19 polycystic kidney disease or those with multiple myeloma since these were not well represented in or entirely excluded from CRIC. CRIC did not enroll patients receiving active immunosuppression. We do not have renal biopsy data in CRIC study participants to definitely establish cases of CKD due to tubulo-interstitial disease vs. glomerular disease, although we had no a priori hypothesis regarding this distinction. Our findings may also not generalize to populations with higher eGFR.24, 59
In conclusion, urine biomarkers of tubular injury were strongly associated with CKD progression in unadjusted analyses. However there was no independent association after adjusting for eGFR, ACR and other established risk factors for CKD progression. Thus, in a general CKD population, measurement of tubular injury biomarkers does not provide additional information for risk prediction for CKD progression, and do not appear to have a role for purposes such as enrichment of clinical trials or providing additional prognostic information to patients. For many individuals with CKD, existing metrics such as eGFR and ACR are able to correctly identifying those at high risk for progression. If tubular injury biomarkers have a role in risk prediction for CKD progression, it will likely be in subpopulations with more limited ranges of eGFR and albuminuria, such as those deemed intermediate risk according to the KDIGO. These findings should inform the design of future studies of CKD progression.
METHODS
Study Sample
The CRIC study design and baseline characteristics have been published.60–62 Briefly, adult patients with eGFR 20–70 mL/min/1.73m2 were enrolled throughout the U.S. Important exclusion criteria included polycystic kidney disease, multiple myeloma, or glomerulonephritis on active immunosuppression. Since late 2005, urine samples designed specifically for biomarker studies were collected. Freshly voided urine samples were placed on ice immediately and processed within one hour. Samples were spun at 2000g for 5 minutes in a refrigerated centrifuge; the supernatants were immediately transferred to new screw-top cryovial tubes and frozen at −80°C. All urine aliquots used in this analysis had undergone only one prior freeze-thaw cycle. Samples for this study were from 3,232 participants who were free of ESRD at the time of urine collection. A sample aliquoting error by a commercial vendor resulted in exclusion of 720 participants, leaving 2,512 with available samples--among whom 2466 had valid measurements of all four biomarkers.
Biomarker measurements
The mean duration of storage time between collection and measurement was 7.2±0.5 years. Each urinary biomarker was assayed at a single performance laboratory using a single assay selected based on established track record and expertise,8, 23 sample volume requirement and regulatory considerations. Urinary albumin and creatinine were measured by a kinetic colorimetric assay on a Roche automated analyzer (Indianapolis, IN) and L-FABP was measured by a 2-step sandwich ELISA assay (CMIC, Tokyo, Japan) at the University of Pennsylvania. KIM-1 was measured by a microbead-based sandwich ELISA on a Bioplex-200 platform (Bio-Rad, Hercules, CA) and NAG was measured using an enzymatic assay (Roche, Indianapolis, IN) at Brigham and Women’s Hospital. NGAL was measured by a non-competitive sandwich assay with chemiluminescent signal detection on an ARCHITECT® platform (Abbott Diagnostics, Abbott Park, IL) at University College, Dublin, Ireland. Description of the relevant laboratory Standard Operating Procedures for KIM-1, NAG and L-FABP have been posted online by the CKD Biomarker Consortium.63 For NGAL, all assays were performed on an Abbott ARCHITECT analyzer according to the manufacturer’s instructions.64 Blind replicate samples were included for quality assurance.21 To detect assay drift, for KIM-1, NAG, and L-FABP, control samples were measured during each sample run, and Levey-Jennings plots were used to determine if the results were satisfactory. There was no net movement in the concentrations of any of these biomarkers during the course of the analyses, and it was concluded that there was no drift present. In addition, proficiency samples were run periodically during testing. For NGAL, prior to sample testing, the laboratory performed imprecision testing with satisfactory results, with CV values for all assays/levels well within the recommended tolerances per the manufacturer. Daily QC (duplicates of all controls run every morning before testing) was also conducted. To take into account differences in urine concentration, our primary analysis was based on biomarker levels normalized to urine creatinine concentration (which is also done for all CKD Biomarkers Consortium studies.8, 23) We performed sensitivity analyses analyzing absolute urine biomarker concentrations.
Outcome
The primary outcome was CKD progression, defined as a composite endpoint of incident ESRD (receipt of chronic dialysis or kidney transplant5) or halving of eGFR.65, 66 Follow-up for this analysis was through March 30, 2012. Death and loss to follow-up were considered as censoring events.
Statistical Analyses
We used Cox proportional hazards models to examine time to CKD progression. Biomarkers were analyzed as quintiles and continuously per standard deviation after log transformation. For L-FABP, a significant proportion of the cohort had biomarker levels that were below the lower limit of detection; consequently, we divided the cohort into those with undetectable levels (N=388), those with levels below the lower limit of detection established by the performance laboratory (N=472), and divided the remaining participants into tertiles of normalized biomarker levels. In addition to unadjusted analysis, we conducted a series of multivariable models to better understand the relations among these injury biomarkers, established risk factors and CKD progression:
Model 1: Biomarkers + baseline age, sex, race/ethnicity, clinical center
Model 2: Model 1 + baseline ACR
Model 3: Model 1 + baseline eGFR
Model 4: Model 1 + baseline ACR + baseline eGFR
Model 5: Model 4 + diabetes mellitus (DM), history of cardiovascular disease (CVD), systolic blood pressure (SBP), body mass index (BMI), use of angiotensin converting enzyme inhibitors (ACE-I)/angiotensin receptor blockers (ARB), education. For the two established predictors of progression, eGFR and ACR, due to known non-linear associations with the outcome,5 quadratic splines terms were used.
We plotted the Schoenfeld residuals vs. time and found no evidence that the proportional hazards assumption was violated.
To assess whether the injury biomarkers improved risk reclassification, we compared C-statistics in fully adjusted models with and without each of the four biomarkers.67, 68 The base clinical model included all the variables in Model 5 above except for the biomarker (namely age, sex, race/ethnicity, clinical center, ACR, eGFR, DM, CVD, SBP, BMI, use of ACE-I/ARB, education). Since these analyses were pre-planned, no adjustments were made for multiple testing.
After we obtained our main results, we conducted post hoc additional exploratory analyses in the subgroup of patients deemed at intermediate risk for CKD progression according to the Kidney Disease: Improving Global Outcomes (KDIGO) risk stratification (specifically eGFR ≥60 ml/min/1.73m2 and ACR ≥30 mg/g Cr; or eGFR 45–59 ml/min/1.73m2 and ACR 0–300 mg/g Cr; or eGFR 30–44 ml/min/1.73m2 and ACR <30 mg/g Cr).69
All analyses were carried out using SAS 9.4 (SAS Institute Inc., Cary, NC).
Supplementary Material
*all correlations were significant, with p < 0.0001.
A backwards selection process was used to remove predictors from the following model (which initially included all four injury biomarkers) until all retained predictors had a p-value < 0.05: age, sex, race/ethnicity, clinical center, ACR, eGFR, DM, CVD, systolic BP, BMI, ACE-I/ARB use, education, KIM-1/Cr or KIM-1, NGAL/Cr or NGAL, L-FABP/Cr or L-FABP, NAG/Cr or NAG.
Acknowledgments
Sources of Support: This work was supported by the Chronic Kidney Disease Biomarkers Consortium funded by NIDDK U01DK85649, U01DK085673, U01DK085660, U01DK085688, U01DK085651 and U01DK085689, and by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Funding for the CRIC Study was obtained under a cooperative agreement from National Institute of Diabetes and Digestive and Kidney Diseases (U01DK060990, U01DK060984, U01DK061022, U01DK061021, U01DK061028, U01DK060980, U01DK060963, and U01DK060902). In addition, this work was supported in part by: the Perelman School of Medicine at the University of Pennsylvania Clinical and Translational Science Award NIH/NCATS UL1TR000003, Johns Hopkins University UL1 TR-000424, University of Maryland GCRC M01 RR-16500, Clinical and Translational Science Collaborative of Cleveland, UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research, Michigan Institute for Clinical and Health Research (MICHR) UL1TR000433, University of Illinois at Chicago CTSA UL1RR029879, Tulane University Translational Research in Hypertension and Renal Biology P30GM103337, Kaiser Permanente NIH/NCRR UCSF-CTSI UL1 RR-024131.
We thank Abbott Laboratories for supporting the measurement of urinary NGAL and CMIC for providing control materials for our studies. Abbott Laboratories and CMIC had no role in study design, data collection, data analysis, data interpretation or writing of the report. We also thank E. Cotter at University College Dublin, Dublin, Ireland for performing the urinary NGAL assays.
Footnotes
DISCLOSURE
This study was carried out under the auspices of the CKD Biomarkers Consortium, which was established in 2008 by the National Institute of Diabetes and Digestive and Kidney Diseases to advance the field of CKD biomarker discovery and validation (http://grants.nih.gov/grants/guide/rfa-files/RFA-DK-08-015.html). JVB appears as co-inventor on KIM-1 patents which have been licensed by Partners Healthcare to a number of companies. He has received royalty income from Partners Healthcare. KDL had reagents donated for previous biomarker studies by Abbott and CMIC. CYH had reagents donated for previous biomarker studies by Abbott.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fassett RG, Venuthurupalli SK, Gobe GC, et al. Biomarkers in chronic kidney disease: a review. Kidney Int. 2011;80:806–821. doi: 10.1038/ki.2011.198. [DOI] [PubMed] [Google Scholar]
- 2.Brosius FC, Pennathur S. How to find a prognostic biomarker for progressive diabetic nephropathy. Kidney Int. 2013;83:996–998. doi: 10.1038/ki.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merchant ML. Can the urinary peptidome outperform creatinine and albumin to predict renal function decline? J Am Soc Nephrol. 2015;26:1760–1761. doi: 10.1681/ASN.2014121243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peralta CA, Katz R, Bonventre JV, et al. Associations of urinary levels of kidney injury molecule 1 (KIM-1) and neutrophil gelatinase-associated lipocalin (NGAL) with kidney function decline in the Multi-Ethnic Study of Atherosclerosis (MESA) Am J Kidney Dis. 2012;60:904–911. doi: 10.1053/j.ajkd.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu KD, Yang W, Anderson AH, et al. Urine neutrophil gelatinase-associated lipocalin levels do not improve risk prediction of progressive chronic kidney disease. Kidney Int. 2013;83:909–914. doi: 10.1038/ki.2012.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith ER, Lee D, Cai MM, et al. Urinary neutrophil gelatinase-associated lipocalin may aid prediction of renal decline in patients with non-proteinuric Stages 3 and 4 chronic kidney disease (CKD) Nephrol Dial Transplant. 2013;28:1569–1579. doi: 10.1093/ndt/gfs586. [DOI] [PubMed] [Google Scholar]
- 7.Bolignano D, Lacquaniti A, Coppolino G, et al. Neutrophil gelatinase-associated lipocalin (NGAL) and progression of chronic kidney disease. Clinical journal of the American Society of Nephrology: CJASN. 2009;4:337–344. doi: 10.2215/CJN.03530708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fufaa GD, Weil EJ, Nelson RG, et al. Association of urinary KIM-1, L-FABP, NAG and NGAL with incident end-stage renal disease and mortality in American Indians with type 2 diabetes mellitus. Diabetologia. 2015;58:188–198. doi: 10.1007/s00125-014-3389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Araki S-i, Haneda M, Koya D, et al. Predictive Effects of Urinary Liver-Type Fatty Acid–Binding Protein for Deteriorating Renal Function and Incidence of Cardiovascular Disease in Type 2 Diabetic Patients Without Advanced Nephropathy. Diabetes Care. 2013;36:1248–1253. doi: 10.2337/dc12-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamijo-Ikemori A, Sugaya T, Yasuda T, et al. Clinical significance of urinary liver-type fatty acid–binding protein in diabetic nephropathy of type 2 diabetic patients. Diabetes Care. 2011;34:691–696. doi: 10.2337/dc10-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen SE, Sugaya T, Hovind P, et al. Urinary liver-type fatty acid-binding protein predicts progression to nephropathy in type 1 diabetic patients. Diabetes Care. 2010;33:1320–1324. doi: 10.2337/dc09-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panduru NM, Forsblom C, Saraheimo M, et al. Urinary liver-type fatty acid–binding protein and progression of diabetic nephropathy in type 1 diabetes. Diabetes Care. 2013;36:2077–2083. doi: 10.2337/dc12-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen SE, Andersen S, Zdunek D, et al. Tubular markers do not predict the decline in glomerular filtration rate in type 1 diabetic patients with overt nephropathy. Kidney Int. 2011;79:1113–1118. doi: 10.1038/ki.2010.554. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen SE, Hansen HP, Jensen BR, et al. Urinary neutrophil gelatinase-associated lipocalin and progression of diabetic nephropathy in type 1 diabetic patients in a four-year follow-up study. Nephron Clinical practice. 2011;118:c130–135. doi: 10.1159/000320615. [DOI] [PubMed] [Google Scholar]
- 15.Bhavsar NA, Kottgen A, Coresh J, et al. Neutrophil gelatinase-associated lipocalin (NGAL) and kidney injury molecule 1 (KIM-1) as predictors of incident CKD stage 3: the Atherosclerosis Risk in Communities (ARIC) Study. Am J Kidney Dis. 2012;60:233–240. doi: 10.1053/j.ajkd.2012.02.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim WH, Lewis JR, Wong G, et al. Plasma neutrophil gelatinase-associated lipocalin and kidney function decline and kidney disease-related clinical events in older women. Am J Nephrol. 2015;41:156–164. doi: 10.1159/000380831. [DOI] [PubMed] [Google Scholar]
- 17.Looker HC, Colombo M, Hess S, et al. Biomarkers of rapid chronic kidney disease progression in type 2 diabetes. Kidney Int. 2015 doi: 10.1038/ki.2015.199. [DOI] [PubMed] [Google Scholar]
- 18.Hayek SS, Sever S, Ko YA, et al. Soluble urokinase receptor and dhronic kidney disease. N Engl J Med. 2015;373:1916–1925. doi: 10.1056/NEJMoa1506362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ju W, Nair V, Smith S, et al. Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Science translational medicine. 2015;7:316ra193. doi: 10.1126/scitranslmed.aac7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu KD, Yang W, Hsu CY. The value of urinary neutrophil gelatinase-associated lipocalin in risk prediction of renal decline in patients with chronic kidney disease. Kidney Int. 2013;84:217. doi: 10.1038/ki.2013.126. [DOI] [PubMed] [Google Scholar]
- 21.Hsu CY, Ballard S, Batlle D, et al. Cross-disciplinary biomarkers research: lessons learned by the CKD Biomarkers Consortium. Clinical journal of the American Society of Nephrology: CJASN. 2015;10:894–902. doi: 10.2215/CJN.11541114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin HYH, Hwang DY, Lee SC, et al. Urinary neutrophil gelatinase-associated lipocalin and clinical outcomes in chronic kidney disease patients. Clin Chem Lab Med. 2015;53:73–83. doi: 10.1515/cclm-2014-0647. [DOI] [PubMed] [Google Scholar]
- 23.Foster MC, Coresh J, Bonventre JV, et al. Urinary biomarkers and risk of ESRD in the Atherosclerosis Risk in Communities Study. Clinical journal of the American Society of Nephrology: CJASN. 2015;10:1956–1963. doi: 10.2215/CJN.02590315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schanstra JP, Zurbig P, Alkhalaf A, et al. Diagnosis and prediction of CKD progression by assessment of urinary peptides. J Am Soc Nephrol. 2015;26:1999–2010. doi: 10.1681/ASN.2014050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tangri N, Kitsios GD, Inker LA, et al. Risk prediction models for patients with chronic kidney disease: a systematic review. Ann Intern Med. 2013;158:596–603. doi: 10.7326/0003-4819-158-8-201304160-00004. [DOI] [PubMed] [Google Scholar]
- 26.Landray MJ, Emberson JR, Blackwell L, et al. Prediction of ESRD and death among people with CKD: the Chronic Renal Impairment in Birmingham (CRIB) prospective cohort study. Am J Kidney Dis. 2010;56:1082–1094. doi: 10.1053/j.ajkd.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tangri N, Stevens LA, Griffith J, et al. A predictive model for progression of chronic kidney disease to kidney failure. JAMA. 2011;305:1553–1559. doi: 10.1001/jama.2011.451. [DOI] [PubMed] [Google Scholar]
- 28.Tangri N, Grams ME, Levey AS, et al. Multinational assessment of accuracy of equations for predicting risk of kidney failure: A meta-analysis. JAMA. 2016;315:164–174. doi: 10.1001/jama.2015.18202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsushita K, Mahmoodi BK, Woodward M, et al. Comparison of risk prediction using the CKD-EPI equation and the MDRD study equation for estimated glomerular filtration rate. JAMA. 2012;307:1941–1951. doi: 10.1001/jama.2012.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Agostino RB, Sr, Grundy S, Sullivan LM, et al. Validation of the Framingham coronary heart disease prediction scores: results of a multiple ethnic groups investigation. JAMA. 2001;286:180–187. doi: 10.1001/jama.286.2.180. [DOI] [PubMed] [Google Scholar]
- 31.Wang TJ, Gona P, Larson MG, et al. Multiple biomarkers for the prediction of first major cardiovascular events and death. N Engl J Med. 2006;355:2631–2639. doi: 10.1056/NEJMoa055373. [DOI] [PubMed] [Google Scholar]
- 32.Peralta C, Scherzer R, Grunfeld C, et al. Urinary biomarkers of kidney injury are associated with all-cause mortality in the Women’s Interagency HIV Study (WIHS) HIV medicine. 2014;15:291–300. doi: 10.1111/hiv.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tojo A, Endou H. Intrarenal handling of proteins in rats using fractional micropuncture technique. Am J Physiol. 1992;263:F601–606. doi: 10.1152/ajprenal.1992.263.4.F601. [DOI] [PubMed] [Google Scholar]
- 34.Sandoval RM, Wagner MC, Patel M, et al. Multiple factors influence glomerular albumin permeability in rats. J Am Soc Nephrol. 2012;23:447–457. doi: 10.1681/ASN.2011070666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russo LM, Sandoval RM, McKee M, et al. The normal kidney filters nephrotic levels of albumin retrieved by proximal tubule cells: retrieval is disrupted in nephrotic states. Kidney Int. 2007;71:504–513. doi: 10.1038/sj.ki.5002041. [DOI] [PubMed] [Google Scholar]
- 36.Zoja C, Donadelli R, Colleoni S, et al. Protein overload stimulates RANTES production by proximal tubular cells depending on NF-kappa B activation. Kidney Int. 1998;53:1608–1615. doi: 10.1046/j.1523-1755.1998.00905.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Chen J, Chen L, et al. Induction of monocyte chemoattractant protein-1 in proximal tubule cells by urinary protein. J Am Soc Nephrol. 1997;8:1537–1545. doi: 10.1681/ASN.V8101537. [DOI] [PubMed] [Google Scholar]
- 38.Drumm K, Bauer B, Freudinger R, et al. Albumin induces NF-kappaB expression in human proximal tubule-derived cells (IHKE-1) Cell Physiol Biochem. 2002;12:187–196. doi: 10.1159/000066278. [DOI] [PubMed] [Google Scholar]
- 39.van Timmeren MM, Bakker SJ, Vaidya VS, et al. Tubular kidney injury molecule-1 in protein-overload nephropathy. American journal of physiology Renal physiology. 2006;291:F456–464. doi: 10.1152/ajprenal.00403.2005. [DOI] [PubMed] [Google Scholar]
- 40.Wagner MC, Campos-Bilderback SB, Chowdhury M, et al. Proximal tubules have the capacity to regulate uptake of albumin. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2014111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenten V, Menzel S, Kunter U, et al. Albumin is recycled from the primary urine by tubular transcytosis. J Am Soc Nephrol. 2013;24:1966–1980. doi: 10.1681/ASN.2013010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dickson LE, Wagner MC, Sandoval RM, et al. The proximal tubule and albuminuria: really! J Am Soc Nephrol. 2014;25:443–453. doi: 10.1681/ASN.2013090950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarad G, Miner JH. Albuminuria, wherefore art thou? J Am Soc Nephrol. 2009;20:455–457. doi: 10.1681/ASN.2009010075. [DOI] [PubMed] [Google Scholar]
- 44.Haraldsson B, Nystrom J, Deen WM. Properties of the glomerular barrier and mechanisms of proteinuria. Physiol Rev. 2008;88:451–487. doi: 10.1152/physrev.00055.2006. [DOI] [PubMed] [Google Scholar]
- 45.Niewczas MA, Gohda T, Skupien J, et al. Circulating TNF receptors 1 and 2 predict ESRD in type 2 diabetes. J Am Soc Nephrol. 2012;23:507–515. doi: 10.1681/ASN.2011060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gohda T, Niewczas MA, Ficociello LH, et al. Circulating TNF receptors 1 and 2 predict stage 3 CKD in type 1 diabetes. J Am Soc Nephrol. 2012;23:516–524. doi: 10.1681/ASN.2011060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panduru NM, Sandholm N, Forsblom C, et al. Kidney injury molecule-1 and the loss of kidney function in diabetic nephropathy: a likely causal link in patients with type 1 diabetes. Diabetes Care. 2015;38:1130–1137. doi: 10.2337/dc14-2330. [DOI] [PubMed] [Google Scholar]
- 48.Brunner HI, Bennett MR, Mina R, et al. Association of noninvasively measured renal protein biomarkers with histologic features of lupus nephritis. Arthritis Rheum. 2012;64:2687–2697. doi: 10.1002/art.34426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Nagaraja HN, Nadasdy T, et al. A composite urine biomarker reflects interstitial inflammation in lupus nephritis kidney biopsies. Kidney Int. 2012;81:401–406. doi: 10.1038/ki.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu KD, Yang W, Go AS, et al. Urine neutrophil gelatinase-associated lipocalin and risk of cardiovascular disease and death in CKD: results from the Chronic Renal Insufficiency Cohort (CRIC) Study. Am J Kidney Dis. 2015;65:267–274. doi: 10.1053/j.ajkd.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu CY, Ordonez JD, Chertow GM, et al. The risk of acute renal failure in patients with chronic kidney disease. Kidney Int. 2008;74:101–107. doi: 10.1038/ki.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu RK, Hsu CY. Proteinuria and reduced glomerular filtration rate as risk factors for acute kidney injury. Curr Opin Nephrol Hypertens. 2011;20:211–217. doi: 10.1097/MNH.0b013e3283454f8d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuh MP, Nehus E, Ma Q, et al. Long-term stability of urinary biomarkers of acute kidney injury in children. Am J Kidney Dis. 2016;67:56–61. doi: 10.1053/j.ajkd.2015.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van de Vrie M, Deegens JK, van der Vlag J, et al. Effect of long-term storage of urine samples on measurement of Kidney Injury Molecule 1 (KIM-1) and Neutrophil Gelatinase-Associated Lipocalin (NGAL) Am J Kidney Dis. 2014;63:573–576. doi: 10.1053/j.ajkd.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 55.Parikh CR, Butrymowicz I, Yu A, et al. Urine stability studies for novel biomarkers of acute kidney injury. Am J Kidney Dis. 2014;63:567–572. doi: 10.1053/j.ajkd.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nauta FL, Bakker SJL, Heerspink HL, et al. Effect of frozen storage on urinary concentration of kidney damage markers. Am J Kidney Dis. 2012;59:586–589. doi: 10.1053/j.ajkd.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 57.Pennemans V, Rigo JM, Penders J, et al. Collection and storage requirements for urinary kidney injury molecule-1 (KIM-1) measurements in humans. Clin Chem Lab Med. 2012;50:539–543. doi: 10.1515/CCLM.2011.796. [DOI] [PubMed] [Google Scholar]
- 58.Bansal N, Carpenter MA, Weiner DE, et al. Urine injury biomarkers and risk of adverse outcomes in recipients of prevalent kidney transplants: The Folic Acid for Vascular Outcome Reduction in Transplantation Trial. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2015030292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pontillo C, Jacobs L, Staessen JA, et al. A urinary proteome-based classifier for the early detection of decline in glomerular filtration. Nephrology Dialysis Transplantation. 2016;31:38–39. doi: 10.1093/ndt/gfw239. [DOI] [PubMed] [Google Scholar]
- 60.Feldman HI, Appel LJ, Chertow GM, et al. The Chronic Renal Insufficiency Cohort (CRIC) study: design and methods. J Am Soc Nephrol. 2003;14:S148–153. doi: 10.1097/01.asn.0000070149.78399.ce. [DOI] [PubMed] [Google Scholar]
- 61.Lash JP, Go AS, Appel LJ, et al. Chronic Renal Insufficiency Cohort (CRIC) Study: baseline characteristics and associations with kidney function. Clinical journal of the American Society of Nephrology: CJASN. 2009;4:1302–1311. doi: 10.2215/CJN.00070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischer MJ, Go AS, Lora CM, et al. CKD in Hispanics: Baseline characteristics from the CRIC (Chronic Renal Insufficiency Cohort) and Hispanic-CRIC Studies. Am J Kidney Dis. 2011;58:214–227. doi: 10.1053/j.ajkd.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chronic Kidney Disease Biomarker Consortium. [last accessed July 29, 2016];CKD Biomarker Consortium Laboratory SOPs. http://www.ckdbiomarkersconsortium.org/
- 64.Grenier FC, Ali S, Syed H, et al. Evaluation of the ARCHITECT urine NGAL assay: Assay performance, specimen handling requirements and biological variability. Clin Biochem. 2010;43:615–620. doi: 10.1016/j.clinbiochem.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 65.Yang W, Xie D, Anderson AH, et al. Association of kidney disease outcomes with risk factors for CKD: findings from the Chronic Renal Insufficiency Cohort (CRIC) study. Am J Kidney Dis. 2014;63:236–243. doi: 10.1053/j.ajkd.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anderson AH, Yang W, Hsu CY, et al. Estimating GFR among participants in the Chronic Renal Insufficiency Cohort (CRIC) Study. Am J Kidney Dis. 2012;60:250–261. doi: 10.1053/j.ajkd.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harrell FE, Jr, Califf RM, Pryor DB, et al. Evaluating the yield of medical tests. JAMA. 1982;247:2543–2546. [PubMed] [Google Scholar]
- 68.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 69.Kidney Disease: Improving Global Outcomes (KDIGO) CKD Work Group. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int. 2013;(Suppl 3):1–150. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
*all correlations were significant, with p < 0.0001.
A backwards selection process was used to remove predictors from the following model (which initially included all four injury biomarkers) until all retained predictors had a p-value < 0.05: age, sex, race/ethnicity, clinical center, ACR, eGFR, DM, CVD, systolic BP, BMI, ACE-I/ARB use, education, KIM-1/Cr or KIM-1, NGAL/Cr or NGAL, L-FABP/Cr or L-FABP, NAG/Cr or NAG.