Abstract
Purpose
Cell-free DNA (cfDNA) prenatal screening tests have been rapidly adopted into clinical practice, in part due to positive insurance coverage. We evaluated the framework payers used in making coverage decisions to describe a process that should be informative for other sequencing tests.
Methods
We analyzed coverage policies from the 19 largest U.S. private payers with publicly available policies through February 2016, building from the UCSF TRANSPERS Payer Coverage Policy Registry.
Results
All payers studied cover cfDNA screening for detection of trisomies 21, 18 and 13 in high risk, singleton pregnancies, based on robust clinical validity (CV) studies and modeled evidence of clinical utility (CU). Payers typically evaluated the evidence for each chromosomal abnormality separately, although results are offered as part of a panel. Starting in August 2015, 8/19 payers also cover cfDNA screening in average risk pregnancies, citing recent CV studies and updated professional guidelines. Most payers attempted, but were unable to independently assess analytic validity (AV).
Conclusion
Payers utilized the standard evidentiary framework (AV/CV/CU) when evaluating cfDNA screening, but varied in their interpretation of the sufficiency of the evidence. Professional guidelines, large CV studies and decision analytic models regarding health outcomes appeared highly influential in coverage decisions.
Keywords: Non-invasive prenatal testing, next generation sequencing, cell-free DNA, insurance coverage, clinical utility
INTRODUCTION
Screening for fetal chromosomal abnormalities in utero has evolved rapidly over the past two decades. Trisomy 21 (Down syndrome) is the most common genetic cause of human birth defects and provides impetus for the current screening paradigm.1 Traditional screening techniques involve ultrasonography and/or maternal serum markers. However, even in high risk populations, there are considerable rates of false positive results with standard screening, and even some false negative results.2 The idea of capturing placental DNA circulating in maternal blood has always held great promise, and since late 2011, prenatal genetic screening tests based on circulating cell-free DNA (cfDNA) have been offered in the United States. Nevertheless, controversies remain regarding the appropriate use of this technology. While cfDNA screening tests are much better at detecting common fetal aneuploidies than traditional screening methods, cfDNA screening cannot detect structural birth defects nor predict other adverse pregnancy outcomes.3 Conversely, cfDNA screening can potentially detect a broader range of genetic conditions than standard screening such as sex chromosome imbalances or subchromosomal deletions (microdeletions), which have varied newborn, childhood, and adult presentations.
As cfDNA screening tests are not diagnostic, results indicating an increased risk for chromosomal imbalance must be confirmed with invasive testing. Patients must be informed about this limitation, as well as the potential implications of the breadth of genetic information that they may receive. Pre-test counseling by an appropriately knowledgeable provider is recommended; however, many screening delivery models lack dedicated genetic counselors, leaving increasingly complex counselling to providers who may lack necessary time and training, raising concerns about informed patient decision-making.3
Despite these cautionary notes, there has been rapid clinical uptake of cfDNA screening4 due to both features of the test (e.g., sensitivity, specificity) as well as commercial marketing. In the U.S., clinical integration has also been enabled by private insurance coverage5, however, how coverage decisions for cfDNA screening tests are made has not been systematically examined. Previous reviews of payer decision-making criteria for genetic tests broadly revealed that payers use a systematic approach to evaluating underlying evidence regarding safety and effectiveness.6,7 Specifically, a test must demonstrate adequate evidence of analytic validity, clinical validity and clinical utility in order to be considered “medically necessary” and therefore covered.8 Payer coverage decisions often cite publicly available studies and technology assessments to determine whether a new test is accurate and reliable (analytic validity), the result is medically meaningful (clinical validity) and whether results affect clinical decisions and improve health outcomes (clinical utility).
Experts have questioned whether this evidentiary framework is generalizable to next-generation sequencing-based tests because of differences in analytical methods and technology platforms used for testing, as well as the complexity of the results.9 This issue is particularly relevant to cfDNA screening, where the test methodologies are variable, competition has fueled extensive marketing to both patients and providers, and the absence of published evidence on quality control and quality assurance is a significant concern.10 We postulated that careful examination of the evolution of payer coverage decision-making for cfDNA screening would be informative for understanding the current process as well as potential coverage of future related prenatal tests such as genome-wide microdeletions and microduplications, single gene pathogenic variants, and entire fetal genomes. We conducted a systematic assessment of coverage policies for cfDNA screening for the 19 largest private insurers in the US with publically available policies to examine the evidentiary framework for coverage-decision-making, clinical indications for testing, evidence cited, and changes over time among the various payers.
MATERIALS AND METHODS
Search Strategy and Policy Selection
To analyze relevant coverage policies, we utilized the University of California – San Francisco (UCSF) Center for Translational and Policy Research on Personalized Medicine (TRANSPERS) Payer Coverage Registry. This unique registry was developed with a team of collaborators from multiple institutions (UCSF, Tufts Medical Center, American Institutes for Research, and Center for Business Models in Healthcare) with funding from the National Human Genome Research Institute (R01HG007063-S1). The Registry systematically synthesizes payer coverage policies to assess which genetic testing panels are covered or not covered by payers, what factors relevant to coverage decisions are discussed in policies, and how coverage policies vary.
The Registry currently includes coverage policies relevant to whole genome sequencing, whole exome sequencing, and multi-gene panels. The Registry includes data on what panels and testing indications are reviewed, whether panels are covered or not covered, and the evidence and rationales for coverage decisions cited in the policy. The current Registry (Ver.1) includes policies from the five largest US private payers, based on enrollment. These payers represent 128 million enrolled lives.11
For the purposes of this paper, we utilized the Registry and added independent analysis of coverage policies of the top 19 payers (as defined by number of lives covered, covering over 184 million lives) and closely examined policies that pertain to cfDNA screening. We identified such additional policies by searching commercial payers’ respective websites between November 24, 2015 and February 28, 2016 using the terms “pre-natal,” “non-invasive,” “fetal,” “aneuploidy,” “cell-free DNA,” “DNA-based,” “sequencing,” “trisomy,” “Down Syndrome,” “genetic,” and “screening.” Policy titles were screened for applicable terms. The most updated policy as of the February 2016 search date was used in our final analysis. Of note, fourteen of the policies came from Blue Cross Blue Shield-affiliated payers. Each payer’s policy was treated as an independent entity, reflecting their local coverage decision-making procedures.
Data Extraction and Coding
For each included policy, two reviewers independently read and extracted relevant data elements. Relevant information was coded on 28 items, including dates of revision, positive or negative coverage determinations, evidence of analytical and clinical validity and clinical utility, references to professional guidelines and health technology assessments, references to specific test methodologies, and genetic counseling requirements. Data abstraction was limited to the information contained in the coverage policy; original source materials such as published studies were not reviewed. The reviewers convened to resolve any discrepancies.
RESULTS
Policy Identification
Fifteen of 19 payers had a coverage policy exclusively focused on cfDNA screening and four had policies that addressed the broader topic of prenatal screening. No payer had more than one policy that addressed cfDNA screening.
cfDNA screening Coverage by Patient Group and Clinical Condition
In general, coverage policies differentiated between two specific patient subgroups: (1) high risk for having a fetus with a common aneuploidy, or (2) average risk (general obstetric population) (see Table 1 for definitions). They also describe several types of chromosomal abnormalities: Trisomy 21, trisomy 18 (Edwards Syndrome), trisomy 13 (Patau Syndrome), sex chromosome aneuploidies, and microdeletions. All 19 policies covered cfDNA screening as first line assessment for trisomy 21 in high risk women with singleton pregnancies; and if trisomy 21 screening is performed, screening for other common aneuploidies (specifically trisomy 18 and trisomy 13) is also covered (Table 2).
Table 1.
Definitions
|
Table 2.
Variation in Coverage Policies Across Indication, Evidence Base, Use and Technology Platform
| Cover High Riska | Cover Average Riska | Assess Analytical Validity | Assess Clinical Validity | Assess Clinical Utility | Cover Sex Chromosome Aneuploidy or Microdeletions | Include NGS and Array Platforms | Genetic Counseling Recommended | |
|---|---|---|---|---|---|---|---|---|
| Payer 1 | + | + | + | + | − | − | + | + |
| Payer 2 | + | − | + | + | + | − | + | + |
| Payer 3 | + | − | − | + | − | − | + | + |
| Payer 4 | + | + | + | + | + | − | + | + |
| Payer 5 | + | − | − | + | + | − | + | + |
| Payer 6 | + | − | − | − | − | − | + | − |
| Payer 7 | + | − | + | + | − | − | + | + |
| Payer 8 | + | − | − | + | − | − | + | − |
| Payer 9 | + | + | + | + | + | − | + | + |
| Payer 10 | + | + | + | + | + | − | + | + |
| Payer 11 | + | + | + | + | + | − | + | + |
| Payer 12 | + | + | + | + | + | − | + | + |
| Payer 13 | + | + | − | + | − | − | + | − |
| Payer 14 | + | − | + | + | + | − | + | − |
| Payer 15 | + | − | + | + | + | − | + | − |
| Payer 16 | + | − | − | + | + | + | + | + |
| Payer 17 | + | + | − | + | + | − | + | − |
| Payer 18 | + | − | − | + | − | − | + | + |
| Payer 19 | + | − | − | + | − | − | + | − |
| Total | 19 | 8 | 10 | 18 | 11 | 1 | 19 | 12 |
for T21, T18, T13 in singleton pregnancies
However, coverage of cfDNA screening as first line screening for trisomy 21 in average risk women with singleton pregnancies varied by payer. Eight of 19 payers cover cfDNA screening for trisomy 21 in average risk women. No policies cover routine testing for other genetic syndromes, including sex chromosome aneuploidy or microdeletions, with the exception of one policy that covers sex chromosome aneuploidy if testing for trisomy 21 is being performed (Table 2). All but four policies draw direct attention to the fact that cfDNA screening tests are not diagnostic tests, and that no medical decisions should be made with the information without a confirmatory diagnostic (invasive) procedure.
Types of Clinical Evidence Cited for Coverage Decisions
Analytical Validity
Ten policies explicitly assessed the evidence base for analytic validity (Table 2). The majority of policies reference the conclusions of a 2013 Blue Cross Blue Shield Technology Assessment Report that stated that there are no identified studies which provide direct evidence on analytic validity and that they recognize that clinical sequencing in general is not standardized or regulated by the FDA. A few payers went further and stated conclusions of a 2011 FDA public meeting on massively parallel sequencing that suggested that the trend was for analytic validation procedures to be application specific and therefore not generalizable across tests. The consistent acknowledgement across the ten payer coverage policies was that they lacked access to publicly available data and standards to conduct an independent assessment of analytic validity, but this reflected the current state of sequencing oversight at the time.
Clinical Validity
In contrast, payers cited numerous studies of clinical validity when describing the evidence used to assess cfDNA screening for coverage determinations. An illustrative list of some of the most frequently cited evidence sources is provided in Table 3. All but one coverage policy summarized or referenced published studies of clinical validity for cfDNA screening to detect trisomy 21, trisomy 18 and trisomy 13 in women with high risk singleton pregnancies. All of these studies were observational studies. A minority of policies (four) did not summarize either clinical validity studies or the overall evidence, but they did include clinical validity studies in the policy references.
Table 3.
Evidence Sources Cited in Coverage Policies
| Clinical Validity Studies | Professional Society Guidelines | Technology Assessment Reports | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High Risk | Average Risk | ACOG/SMFM (2015) | ACMG (2013) | ISPD (2015) | NSGC (2012) | California Technology Assessment Foruma | BCBSA TEC Report on T21 or T18/T13b,c | |||||
| Palomaki, 2011, Genet Med | Norton (NICE), 2012, AJOG | Bianchi (MELISSA), 2012, Obstet and Gyn | Nicolaides, 2013, Prenatal Diag | Bianchi, 2014, NEJM | Norton (NEXT), 2015, NEJM | |||||||
| Payer 1 | + | + | + | + | + | + | + | − | + | + | − | − |
| Payer 2 | + | + | + | + | + | − | − | + | − | + | − | + |
| Payer 3 | + | − | + | − | − | − | + | − | − | − | + | + |
| Payer 4 | + | + | + | + | + | + | + | + | − | + | − | + |
| Payer 5 | − | + | + | − | + | + | + | + | − | − | + | + |
| Payer 6 | − | − | − | − | − | − | + | + | − | + | + | + |
| Payer 7 | + | + | + | + | − | + | + | + | − | + | + | − |
| Payer 8 | + | + | − | + | + | − | + | + | − | − | − | + |
| Payer 9 | + | + | + | + | − | + | + | + | + | + | − | + |
| Payer 10 | + | + | + | + | + | + | + | + | − | + | − | + |
| Payer 11 | + | + | + | + | − | + | + | + | + | + | − | + |
| Payer 12 | + | + | + | + | + | + | + | + | + | + | − | + |
| Payer 13 | − | + | − | − | − | − | + | + | − | + | + | + |
| Payer 14 | + | − | − | − | − | − | − | − | − | − | − | − |
| Payer 15 | + | + | + | + | + | + | + | + | − | + | − | + |
| Payer 16 | − | − | − | − | − | − | − | − | − | − | − | + |
| Payer 17 | + | + | + | + | + | + | + | + | − | − | − | + |
| Payer 18 | − | − | − | − | + | + | + | − | − | + | + | + |
| Payer 19 | + | + | − | + | − | − | − | − | − | − | − | + |
| Total | 14 | 14 | 12 | 12 | 10 | 11 | 15 | 13 | 4 | 12 | 6 | 16 |
Walsh J. California Technology Assessment Forum (CTAF). Fetal Aneuploidy Detection by Maternal Plasma DNA Sequencing, Part 2. October 17, 2012.
Blue Cross Blue Shield Technology Evaluation Center (TEC). Noninvasive prenatal cell-free fetal DNA-based screening for aneuploidies other than trisomy 21. Technol Eval Cent Assess Program. 2014 Dec;29(7):1–7.
Blue Cross Blue Shield Technology Evaluation Center (TEC). Sequencing-based tests to determine fetal Down syndrome (trisomy 21) from maternal plasma DNA. Technol Eval Cent Assess Program. 2013 Apr;27(10):1–6.
While 18 policies address clinical validity in high risk singleton pregnancies, only 14 specifically addressed the clinical validity evidence in average risk pregnancies. All but two of those 14 note that the evidence of clinical validity is less robust in this population, primarily because there are fewer available studies. Payers differed in their interpretation of the sufficiency of these clinical validity data in average risk women, as demonstrated by only 8/14 payers determining that the data supporting the use of cfDNA screening in average risk women were sufficient to consider the tests medically necessary.
For prenatal conditions beyond the three most common chromosomal aneuploidies, such as microdeletions and sex chromosome aneuploidy, nine and fourteen policies, respectively, cite that data are insufficient for coverage. The one policy that does cover sex chromosome aneuploidy does not clarify the evidence used to support that decision.
Clinical Utility
Eleven of 19 policies explicitly reported the evaluation of clinical utility as part of their coverage determination. Clinical utility was defined by all policies as a change in net health outcomes, with three policies also including a change in “patient management” or “clinical decision-making” as part of their definition. In all cases, only indirect (modeled) evidence of clinical utility is cited. Published technology assessments, which include decision analytic models to demonstrate clinical utility, are common data sources, as were several peer-reviewed modeling papers.12–14 These models all defined outcomes as correctly identifying trisomies while avoiding unnecessary invasive procedures, which carry a risk of miscarriage.
cfDNA screening Coverage by Technology Platform
Three different technology platforms and analysis methodologies underlie the major commercially available tests for cfDNA screening. These include massively parallel whole genome shotgun sequencing, chromosome-selective massively parallel sequencing paired with an array, and single nucleotide analysis only compared against a maternal background reference (Table 4). The results across these 3 different platforms are reported as a risk category (e.g. no aneuploidy detected, aneuploidy suspected or aneuploidy detected) typically with a residual risk.
Table 4.
Technology Platforms and Test Offerings of Four Major NIPS Test Companies
| Company | Test Name | Trisomies | Sex Chromosome Aneuploidy | Microdeletions | Technology Platform | Results Reported |
|---|---|---|---|---|---|---|
| Nateraa | Panorama | 21, 18, 13 | 45,X (Turner syndrome), 47,XXY (Klinefelter syndrome), 47,XXX (Triple X syndrome), 47,XYY (XYY syndrome) | 22q11.2 (DiGeorge syndrome), 1p36 deletion, 5p (Cri-du chat syndrome), 15q11.2 (Angelman and Prader-Willi syndrome) | SNP analysis via sequencing | High Risk, Low Risk |
| Illuminab | Verifi | 21, 18, 16, 13, 9 | 45,X (Turner syndrome), 47,XXY (Klinefelter syndrome), 47,XXX (Triple X syndrome), 47,XYY (XYY syndrome) | 22q11.2 (DiGeorge syndrome), 1p36, 15q11.2 (Angelman, Prader-Willi syndromes) 5p (Cri-du-chat syndrome), and 4p (Wolf-Hirschhorn syndrome) | Genome-wide sequencing | No Aneuploidy Detected, Aneuploidy Detected, or Aneuploidy Suspected (Borderline Value) |
| Sequenomc | MaterniT21 Plus | 22, 21, 18, 16, 13 | 45,X (Turner syndrome), 47,XXY (Klinefelter syndrome), 47,XXX (Triple X syndrome), 47,XYY (XYY syndrome) | 22q11.2 (DiGeorge syndrome), 1p36 deletion, 5p (Cri-du-chat syndrome), 15q11.2 (Angelman and Prader-Willi syndrome) 4p (Wolf-Hirschhorn syndrome), 8q (Langier-Gideon syndrome), 11q (Jacobsen syndrome) | Genome-wide sequencing | Positive, Negative, Additional Finding |
| Ariosad | Harmony | 21, 18, 13 | Yes | No | Targeted sequencing/array | High Risk, Low Risk |
No payer policies distinguish coverage based on whether a test uses a specific technology platform or a specific data interpretation algorithm. Cited clinical studies were typically performed using a single manufacturer’s technology, yet coverage policies cite these single manufacturer studies as rationale for covering cfDNA screening generally across technology and analysis platforms.
Pre- and Post-test Counseling Requirements
Most policies (12/19) include a general statement recommending pre- and post-test counseling, however in no policy is this a pre-authorization requirement. Five of those 12 policies specifically outline that women should be counseled about the risk of a false positive result. The justification for this recommendation is most often professional guidelines such as those by the American College of Obstetrics and Gynecology (ACOG), which recommends formal genetic counseling for cfDNA screening by an individual with experience and expertise in genetic medicine and genetic testing methods (Table 2).
Professional Practice Guidelines
Paralleling the clinical availability of cfDNA screening, numerous professional societies have published clinical practice guidelines to encourage appropriate use of these new tests. This includes a recent combined committee opinion from ACOG and the Society of Maternal Fetal Medicine (SMFM)15, a policy statement from the ACMG16, an updated position statement from the International Society for Prenatal Diagnosis (ISPD)17, and a guideline from the National Society of Genetic Counselors (NSGC)18. These guidelines cover a range of issues from the clinical application of cfDNA screening to appropriate counseling surrounding the test and associated ethical issues.
All but three policies mention at least one of these updated guidelines in their summaries of the evidence reviewed in making their coverage determinations. The most commonly cited source was the committee opinion from ACOG and SMFM, but policies typically mentioned several different sources (Table 3).
Timeline for coverage decisions
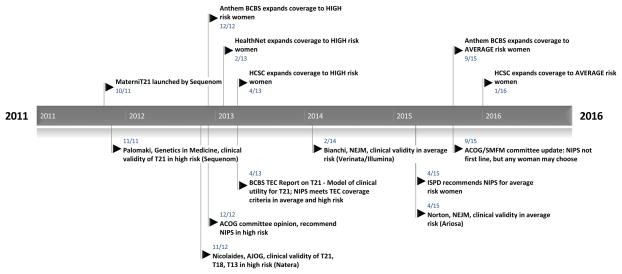
Temporal trends in coverage for high and average risk women are outlined in Figure 1, showing illustrative payer coverage decisions in relationship to either professional guideline changes or publication of key clinical studies. The first positive coverage decisions among these 19 payers was noted in December 2012, temporally following a 2012 recommendation by the ACOG stating that cfDNA screening be offered to women at increased risk of fetal aneuploidy. Policy revision histories are incompletely available, but by mid-2013, at least nine of the 19 payers had specific policies addressing cfDNA prenatal screening with affirmative coverage in high risk singleton pregnancies (data not shown).
Figure 1. Temporal trends in coverage of cell-free DNA prenatal genetic screening tests in women of high and average risk for fetal aneuploidy with professional guideline changes, technology assessments and representative clinical publications.
Five-year timeline of initial cfDNA screening test launch and illustrative payer coverage decisions (all above timeline) with corresponding professional society guideline changes, technology assessment reports and key clinical publications with author, journal and cfDNA screening platform noted (all below timeline); all events with dates of publication (month/year).
As noted in payer policies, over the next two years, large clinical validity studies involving thousands of women were published assessing the performance of cfDNA screening in routine screening of high risk women.13,19–22 Studies were completed using the technology platforms for all four major manufacturers. Subsequently, several large clinical validity studies were undertaken in average risk women.23–25 By mid-2015, following the publication of two large, well-designed, prospective studies in average risk women on two different platforms, the first major prenatal organization, the International Society of Prenatal Diagnosis, released updated guidelines supporting the clinical use of cfDNA screening in screening for trisomy 21 for all risk categories. Later in 2015, two other groups (ACOG and SMFM) released a combined, updated committee opinion reversing their stance that cfDNA screening is inappropriate for average risk women, stating that any patient, regardless of her risk status, may choose cell-free DNA analysis as a screening strategy for common aneuploidies when counseled appropriately on the risks, benefits, and alternatives, though conventional screening methods are still recommended as a first-line given current evidence.26 Coincident with the most recent revision to the ACOG/SMFM opinion, one plan made the decision to expand coverage to average risk pregnancies (Figure 1), citing this updated guideline, followed by 7 more plans within the next 4 months (data not shown).
DISCUSSION
When we systematically evaluated coverage policies for cfDNA screening among the largest 19 private payers with publically available policies, we found that regardless of the specific sequencing platform or test output, the vast majority of payers applied the conventional evaluation framework of analytical validity, clinical validity and clinical utility when assessing evidence for coverage determinations. In particular, the published evidence base supporting the clinical validity of the test in high risk women and professional guideline support for testing were the most common reasons for positive coverage decisions. Technology assessments that included modeled evidence of the clinical utility of testing for trisomies 21, 18 and 13 in high risk women were also important factors in justifying coverage.
Payers had varying interpretations of the adequacy of the clinical validity and clinical utility of cfDNA screening in average risk women (affirmative coverage in eight of 19 policies), despite policies citing similar types of evidence (Table 3). Whenever described, payers had a consistent definition of clinical utility as a “change in net health outcomes,” so the definition was not an obvious source of difference. The only evidence regarding health outcomes associated with cfDNA screening in either high risk or average risk pregnancies was developed from decision analytic models, suggesting that in certain circumstances, a well-conducted model can influence payer decision-making.27 Whether decision analytic models are sufficient evidence of clinical utility in other complex genetic tests is undetermined.27,28
One consistent evidence gap highlighted in coverage policies was the lack of robust analytical validity data, which is not unexpected given that all cfDNA screening tests are laboratory developed tests (LDTs) and public reporting of technical performance data is not required by Clinical Laboratory Improvement Amendments (CLIA), which are federally regulated standards that govern LDTs. Some payers stated that acceptable analytic validity is generally assumed for cfDNA screening based on laboratories meeting quality standards under CLIA. Although many payers acknowledged this evidence gap, it was not ultimately a barrier to coverage. Test performance and reliability is a central issue in the current debate about the FDA’s initiative to regulate LDTs such as cfDNA screening tests.29 While the payers in our analysis do not seem to differentiate among the various test technology platforms or interpretation algorithms currently, future increased regulation and analytical and clinical validity data requirements may lead to differential coverage decisions for specific cfDNA screening methods and platforms based on FDA clearance. This issue seems particularly relevant in the case of screening for trisomies 13 and 18, where the test failure rate may be higher due to low levels of placental cfDNA than in euploid pregnancies.30 Moreover, likely future expansion of cfDNA screening to detect additional genetic conditions will only exacerbate this lack of transparency with respect to analytic validity.
With one exception, payers in our study did not cover cfDNA screening for sex chromosome aneuploidies, and no policies covered cfDNA screening for microdeletions, though almost all test manufacturers offer cfDNA screening for these conditions (Table 4) despite consensus between the payer community and professional societies about the lack of compelling evidence of clinical validity. Thus, they are being promoted without an understanding of the benefits and harms of their use in clinical practice.3,31 While the coverage determinations are clear, cfDNA screening for these additional conditions may still be reimbursed given the lack of granularity in billing (Current Procedural Terminology or CPT) codes used to identify the comprehensiveness of the test performed. As cfDNA screening expands to assess for an increasing number of genetic conditions and competition in test development heightens, this discrepancy will become only more prominent.
What is also apparent is that the largest private payers have responded rapidly to the evolving evidence base for cfDNA screening, issuing new coverage policies within very short time periods of new peer-reviewed publications or major professional guideline updates (Figure 1). The 2015 ACOG/SMFM committee opinion was released a few months prior to the first observed change in payer coverage policies for average risk women, suggesting that payers closely follow professional guideline recommendations. However, half of all policies updated since the release of that committee opinion continued to view the overall evidence for cfDNA screening in average risk pregnancies for trisomy 21 as insufficient (Table 3). As has been observed previously, there is variation in payer coverage decision-making even when the same evidence base is available.32 One explanation may be that the updated clinical guideline did not recommend cfDNA screening as a first line screening tool, but rather removed a prior recommendation against its use in average risk women with singleton pregnancies, a stance that was reiterated in a December 2015 confirmatory guidance statement.33
In addition, although all professional guidelines emphasize the importance of pre-test and post-test counseling, only twelve payers referenced the need for genetic counseling in their coverage policies (with only five highlighting the clear need for patient guidance on potential false positives), and no coverage policies mandated counseling as a requirement for obtaining coverage. It has been shown for other commonly used genetic tests that counseling falls markedly short of what is recommended by professional societies34 due in part to cost and feasibility. This is exacerbated by a shortage of genetic counselors18 and clinical geneticists.35 The potential for misinterpretation of test results is a major concern with the coverage expansion of cfDNA screening to average risk pregnancies. These tests are not diagnostic tests, and false positives have been reported in the literature associated with placental mosaicism, maternal mosaicism, and vanishing twin.36–38 In addition, some patients fail to get an interpretable result due to inadequate circulating fetal DNA (particularly in obese women), failed sequencing, or other problems with the test, and the impact of cfDNA screening on this population is unclear and not addressed in cited models.
Limitations
Our study had several limitations, including that we only analyzed coverage policies from private insurers. As Medicaid covers almost half of births in the US,39 future analyses looking at Medicaid coverage will be informative. In addition, we were limited by the level of detail provided in the coverage policies by each payer and the policies were highly variable in their detail and clarity. Our conclusions may not reflect the actual evidence review processes undertaken by individual payers. Finally, all of the payers that expanded cfDNA screening coverage to average risk pregnancies were Blue Cross Blue Shield plans, though not all of the Blue Cross Blue Shield plans analyzed covered average risk. While each plan makes independent coverage decisions, their actions may be interdependent in ways that are unknown to us as researchers.
Conclusion
Our analysis revealed that cfDNA screening has received broad and rapid private payer coverage in certain indications, unlike other new sequencing-based tests that have faced reimbursement challenges. There are general concerns about the breadth and complexity of massively parallel sequencing-based tests being a barrier to informed coverage decision-making. These concerns were overcome in cfDNA screening by widespread availability of adequate clinical validity and clinical utility evidence in high risk pregnancies. Given expansion of cfDNA screening to other genetic conditions and even the entire fetal genome,40 our analysis of the evidentiary framework used in payer decision-making may serve as an indicator for test developers and researchers regarding likely hurdles to be confronted in payer coverage decisions going forward. One caveat, however, is that the first cfDNA screening indications receiving positive coverage decisions were syndromes like Down syndrome for which there is extensive knowledge about the natural history and management of the condition. As cfDNA screening expands into screening for more clinically heterogeneous genetic conditions, where fewer data are available regarding medical implications and management, the appropriate clinical follow up after a positive test is less clear and the likelihood that modeling studies of clinical utility will suffice for decision-making is undetermined. Moreover, payers continue to evaluate each chromosomal abnormality as a separate test without any evidence of viewing cfDNA screening as a “panel” that has advantages because it can screen for multiple conditions simultaneously. In addition, as use of cfDNA screening continues to grow, appropriate genetic counseling services have been identified as a foreseeable bottleneck, particularly concerning issues of informed decision-making when cfDNA screening is used in low risk populations for a particular condition where even highly specific tests will have high proportions of false positives. Positive coverage of cfDNA screening has enabled expansion of cfDNA screening tests into routine care, but much remains to be learned now about the implementation and outcomes of expanded use of these complex sequencing tests in practice.
Acknowledgments
Funding: This work was supported by with funding from the National Human Genome Research Institute (R01HG007063-S1)
Footnotes
Conflicts of Interest: No conflicts of interest are declared for any of the authors.
References
- 1.ACOG Practice Bulletin No. 77: screening for fetal chromosomal abnormalities. Obstet Gynecol. 2007;109(1):217–227. doi: 10.1097/00006250-200701000-00054. [DOI] [PubMed] [Google Scholar]
- 2.Russo ML, Blakemore KJ. A historical and practical review of first trimester aneuploidy screening. Semin Fetal Neonatal Med. 2014;19(3):183–187. doi: 10.1016/j.siny.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregg AR, Van den Veyver IB, Gross SJ, Madankumar R, Rink BD, Norton ME. Noninvasive prenatal screening by next-generation sequencing. Annu Rev Genomics Hum Genet. 2014;15:327–347. doi: 10.1146/annurev-genom-090413-025341. [DOI] [PubMed] [Google Scholar]
- 4.Allyse M, Minear MA, Berson E, et al. Non-invasive prenatal testing: a review of international implementation and challenges. International Journal of Women’s Health. 2015;7:113–126. doi: 10.2147/IJWH.S67124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haymon L, Simi E, Moyer K, Aufox S, Ouyang DW. Clinical implementation of noninvasive prenatal testing among maternal fetal medicine specialists. Prenatal diagnosis. 2014;34(5):416–423. doi: 10.1002/pd.4301. [DOI] [PubMed] [Google Scholar]
- 6.Graf MD, Needham DF, Teed N, Brown T. Genetic testing insurance coverage trends: a review of publicly available policies from the largest US payers. Personalized Medicine. 2013;10(3):235–243. doi: 10.2217/pme.13.9. [DOI] [PubMed] [Google Scholar]
- 7.Hresko A, Haga SB. Insurance coverage policies for personalized medicine. J Pers Med. 2012;2(4):201–216. doi: 10.3390/jpm2040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deverka PA. Pharmacogenomics, evidence, and the role of payers. Public Health Genomics. 2009;12(3):149–157. doi: 10.1159/000189627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deverka PA, Dreyfus JC. Clinical integration of next generation sequencing: coverage and reimbursement challenges. J Law Med Ethics. 2014;42(Suppl 1):22–41. doi: 10.1111/jlme.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norton ME, Rose NC, Benn P. Noninvasive prenatal testing for fetal aneuploidy: clinical assessment and a plea for restraint. Obstet Gynecol. 2013;121(4):847–850. doi: 10.1097/AOG.0b013e31828642c6. [DOI] [PubMed] [Google Scholar]
- 11.AIS. [Accessed February 29, 2016]; http://aishealth.com/health-plan-business/data/enrollment - 1.
- 12.Garfield SS, Armstrong SO. Clinical and Cost Consequences of Incorporating a Novel Non-Invasive Prenatal Test into the Diagnostic Pathway for Fetal Trisomies. Journal of Managed Care Medicine. 2012;15(2) [Google Scholar]
- 13.Palomaki GE, Deciu C, Kloza EM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet Med. 2012;14(3):296–305. doi: 10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohno M, Caughey A. The role of noninvasive prenatal testing as a diagnostic versus a screening tool–a cost-effectiveness analysis. Prenatal diagnosis. 2013;33(7):630–635. doi: 10.1002/pd.4156. [DOI] [PubMed] [Google Scholar]
- 15.Committee opinion no. 640: cell-free DNA screening for fetal aneuploidy. Obstetrics & Gynecology. 2015;126(3):e31–e37. doi: 10.1097/AOG.0000000000001051. [DOI] [PubMed] [Google Scholar]
- 16.Gregg AR, Gross SJ, Best RG, et al. ACMG statement on noninvasive prenatal screening for fetal aneuploidy. Genet Med. 2013;15(5):395–398. doi: 10.1038/gim.2013.29. [DOI] [PubMed] [Google Scholar]
- 17.Benn P, Borrell A, Chiu RW, et al. Position statement from the Chromosome Abnormality Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenatal diagnosis. 2015;35(8):725–734. doi: 10.1002/pd.4608. [DOI] [PubMed] [Google Scholar]
- 18.Devers P, Cronister A, Ormond K, Facio F, Brasinton C. Noninvasive prenatal testing/noninvasive prenatal diagnosis: the position of the National Society of Genetic Counselors (by NSGC Public Policy Committee) NSGC Position Statements. 2012 [Google Scholar]
- 19.Palomaki GE, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med. 2011;13(11):913–920. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 20.Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119(5):890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 21.Norton ME, Brar H, Weiss J, et al. Non-Invasive Chromosomal Evaluation (NICE) Study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;207(2):137, e131–138. doi: 10.1016/j.ajog.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 22.Nicolaides KH, Syngelaki A, Gil M, Atanasova V, Markova D. Validation of targeted sequencing of single-nucleotide polymorphisms for non-invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X, and Y. Prenatal diagnosis. 2013;33(6):575–579. doi: 10.1002/pd.4103. [DOI] [PubMed] [Google Scholar]
- 23.Bianchi DW, Parker RL, Wentworth J, et al. DNA sequencing versus standard prenatal aneuploidy screening. The New England journal of medicine. 2014;370(9):799–808. doi: 10.1056/NEJMoa1311037. [DOI] [PubMed] [Google Scholar]
- 24.Norton ME, Jacobsson B, Swamy GK, et al. Cell-free DNA analysis for noninvasive examination of trisomy. The New England journal of medicine. 2015;372(17):1589–1597. doi: 10.1056/NEJMoa1407349. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Gao Y, Jiang F, et al. Non-invasive prenatal testing for trisomies 21, 18 and 13: clinical experience from 146,958 pregnancies. Ultrasound Obstet Gynecol. 2015;45(5):530–538. doi: 10.1002/uog.14792. [DOI] [PubMed] [Google Scholar]
- 26.Committee Opinion No. 640: Cell-Free DNA Screening For Fetal Aneuploidy. Obstet Gynecol. 2015;126(3):e31–37. doi: 10.1097/AOG.0000000000001051. [DOI] [PubMed] [Google Scholar]
- 27.Deverka P, Messner DA, McCormack R, et al. Generating and evaluating evidence of the clinical utility of molecular diagnostic tests in oncology. Genet Med. 2015 doi: 10.1038/gim.2015.162. [DOI] [PubMed] [Google Scholar]
- 28.Khoury MJ, Coates RJ, Evans JP. Evidence-based classification of recommendations on use of genomic tests in clinical practice: dealing with insufficient evidence. Genet Med. 2010;12(11):680–683. doi: 10.1097/GIM.0b013e3181f9ad55. [DOI] [PubMed] [Google Scholar]
- 29.FDA. The Public Health Evidence for FDA Oversight of Laboratory Developed Tests: 20 Case Studies. 2015. [Google Scholar]
- 30.Gil MM, Quezada MS, Revello R, Akolekar R, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol. 2015;45(3):249–266. doi: 10.1002/uog.14791. [DOI] [PubMed] [Google Scholar]
- 31.Allyse M, Chandrasekharan S. Too much, too soon?: Commercial provision of noninvasive prenatal screening for subchromosomal abnormalities and beyond. Genet Med. 2015;17(12):958–961. doi: 10.1038/gim.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trosman JR, Van Bebber SL, Phillips KA. Coverage policy development for personalized medicine: private payer perspectives on developing policy for the 21-gene assay. Journal of oncology practice/American Society of Clinical Oncology. 2010;6(5):238–242. doi: 10.1200/JOP.000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.SMFM Statement: clarification of recommendations regarding cell-free DNA aneuploidy screening. Am J Obstet Gynecol. 2015;213(6):753–754. doi: 10.1016/j.ajog.2015.09.077. [DOI] [PubMed] [Google Scholar]
- 34.Armstrong J, Toscano M, Kotchko N, et al. Utilization and Outcomes of BRCA Genetic Testing and Counseling in a National Commercially Insured Population: The ABOUT Study. JAMA oncology. 2015;1(9):1251–1260. doi: 10.1001/jamaoncol.2015.3048. [DOI] [PubMed] [Google Scholar]
- 35.Markens S. It just becomes much more complicated”: Genetic counselors’ views on genetics and prenatal testing. New Genetics and Society. 2013;32(3) [Google Scholar]
- 36.Wang JC, Sahoo T, Schonberg S, et al. Discordant noninvasive prenatal testing and cytogenetic results: a study of 109 consecutive cases. Genet Med. 2015;17(3):234–236. doi: 10.1038/gim.2014.92. [DOI] [PubMed] [Google Scholar]
- 37.Grati FR, Malvestiti F, Ferreira JC, et al. Fetoplacental mosaicism: potential implications for false-positive and false-negative noninvasive prenatal screening results. Genet Med. 2014;16(8):620–624. doi: 10.1038/gim.2014.3. [DOI] [PubMed] [Google Scholar]
- 38.Snyder MW, Simmons LE, Kitzman JO, et al. Copy-number variation and false positive prenatal aneuploidy screening results. The New England journal of medicine. 2015;372(17):1639–1645. doi: 10.1056/NEJMoa1408408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markus AR, Andres E, West KD, Garro N, Pellegrini C. Medicaid covered births, 2008 through 2010, in the context of the implementation of health reform. Women’s Health Issues. 2013;23(5):e273–e280. doi: 10.1016/j.whi.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Kitzman JO, Snyder MW, Ventura M, et al. Noninvasive whole-genome sequencing of a human fetus. Science translational medicine. 2012;4(137):137ra176. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]