Abstract
Purpose
To investigate the utility of whole-exome sequencing (WES) to define a molecular diagnosis in patients clinically diagnosed with congenital anomalies of kidney and urinary tract (CAKUT).
Methods
WES was performed in 62 families with CAKUT. WES data were analyzed for Single Nucleotide Variants (SNVs) in 35 known CAKUT genes, putatively deleterious sequence changes in new candidate genes, and potentially disease-associated copy-number variants (CNVs).
Results
In approximately 5% of families, pathogenic SNVs were identified in PAX2, HNF1B, and EYA1. Observed phenotypes in these families expand the current understanding about the role of these genes in CAKUT. Four pathogenic CNVs were also identified using two CNV detection tools. In addition, we found one deleterious de novo SNV in FOXP1 among the 62 families with CAKUT. Database of clinical BMGL laboratory was queried and seven additional unrelated individuals with novel de novo SNVs in FOXP1 were identified. Six of these 8 individuals with FOXP1 SNVs, have syndromic urinary tract defects, implicating this gene in urinary tract development.
Conclusion
We conclude that WES can be used to identify the molecular etiology (SNVs, CNVs) in a subset of individuals with CAKUT. WES can also help identify novel CAKUT genes.
Keywords: CAKUT, WES, FOXP1, PAX2, HNF1B, EYA1
INTRODUCTION
The diagnosis of congenital anomalies of the kidney and urinary tract (CAKUT) is based on the recognition of a broad spectrum of renal and urinary tract malformations which, in aggregate, constitute the most common cause of end-stage renal disease (ESRD) in children. 1,2 CAKUT may result in chronic kidney disease (CKD) which leads to severe impairment of physical and psychosocial development. 3 Socioeconomically, CAKUT poses a substantial economic burden to families and health care systems. CAKUT is a clinically heterogeneous phenotype that encompasses renal agenesis, renal hypo/dysplasia (RHD), multicystic kidney dysplasia (MCDK), cross-fused ectopia, duplex renal collecting system, ureteropelvic junction obstruction (UPJO), mega-ureter, posterior urethral valves (PUV), and vesicoureteral reflux (VUR).
Multiple lines of evidence suggest that genetic factors contribute to CAKUT. This evidence includes familial segregation of CAKUT cases and, the identification of causative genes. 4 Discovery of an underlying genetic etiology facilitates molecular diagnosis and can aid physicians and family members by clarifying associated risks and allowing improved genetic counseling.
Whereas in the past, genetic diagnosis was limited to the analysis of individual candidate genes, whole-exome sequencing (WES) provides an opportunity to arrive at an accurate molecular diagnosis with a single test. 5,6 WES is able to identify Single Nucleotide Variants (SNVs), however recently it has been used to uncover even small CNVs encompassing a single gene or even one exon. 7 Extraction of CNV information from WES data is challenging partly due to the potential artifacts introduced during the exon targeting and amplification steps of WES.8 Moreover, WES can enhance gene discovery for novel potential contributory genes.
Initial reports of clinical WES at a Clinical Laboratory Improvement Amendments (CLIA) certified laboratory indicated a molecular diagnostic rate of 25% for patients referred for genetic evaluation 5,6. However, neurological phenotypes constituted 80% of the patient population in that study. The clinical utility of using WES in common, sporadic birth defects is under active investigation. Current evaluations of CAKUT patients involve diagnostic imaging, however involvement of other organs may go undiagnosed during such evaluations. Here we investigated the utility of WES to define a molecular diagnosis (SNVs and CNVs) in patients clinically diagnosed with CAKUT.
METHODS
Patients and their families were recruited from the pediatric urology and renal diseases clinics at the Texas Children's Hospital, Houston Texas, USA. Inclusion criteria included individuals with non-syndromic forms of CAKUT (as defined above) and individuals with syndromic forms of CAKUT for which a genetic etiology had not been identified. Exclusion criteria included individuals with syndromic forms of CAKUT in which an underlying genetic etiology was known and individuals with non-syndromic and non-familial forms of vesicoureteral reflux (VUR). Therefore, individuals with syndromic features without a known diagnosis were included in the study. The study protocol was approved by the Institutional Review Board for the Protection of Human Subjects at Baylor College of Medicine. Standard procedures were used to recruit subjects for this study. Demographics of the families and phenotypic details of subjects with CAKUT are summarized as Table 1 and Table S1. Blood samples or saliva-based specimens were collected by standard procedures, according to the families' wishes. DNA extraction was performed with a QIAmp kit (QIAGEN) per manufacturer's instructions. DNA was quantified with nanodrop, and 1ug of DNA was used for WES. In familial multiplex cases, WES was performed on the affected available family members most distantly related observed in their respective pedigrees (see Figure 1). Among apparently isolated, singleton cases, we performed WES on the proband and the two apparently unaffected parents (case-parent trios in 20 families), when both parents were available. In all other cases, WES was performed only on the proband.
Table 1.
Demographics and different phenotypes of the 62 families with CAKUT who underwent Whole-Exome Sequencing.
| Age of probands (mean, SD) | 6.5 years, 5.8 years |
| Sex of probands (M/F) | 1.29 |
| Other organ affected (syndromic) | 19 (31%)a |
| Mode of analysis | |
| Familial (more than one affected individual in the family) | 10 (16%) |
| Sporadic: case-parent trios | 20 (32%) |
| Sporadic: single cases | 32 (52%) |
| Diagnosis of probands | |
| Renal dysplasia | 14 (23%) |
| Renal agenesis/hypoplasia | 12 (19%) |
| Posterior urethral valve | 10 (16%) |
| Vesicoureteral reflux (VUR) | 9 (15%) |
| Duplicated collecting system (DCS) | 7 (11%) |
| Ureteropelvic junction obstruction (UPJO) | 5 (8%) |
| Fusion anomaly: horseshoe kidney and crossed fused ectopia | 4 (6%) |
| Anterior urethral valve | 1 (2%) |
| Total number of families | 62 (100%) |
Two more syndromicfamilies were identified after WES results became available; accordingly, total syndromic= 21 (34%)
Figure 1.
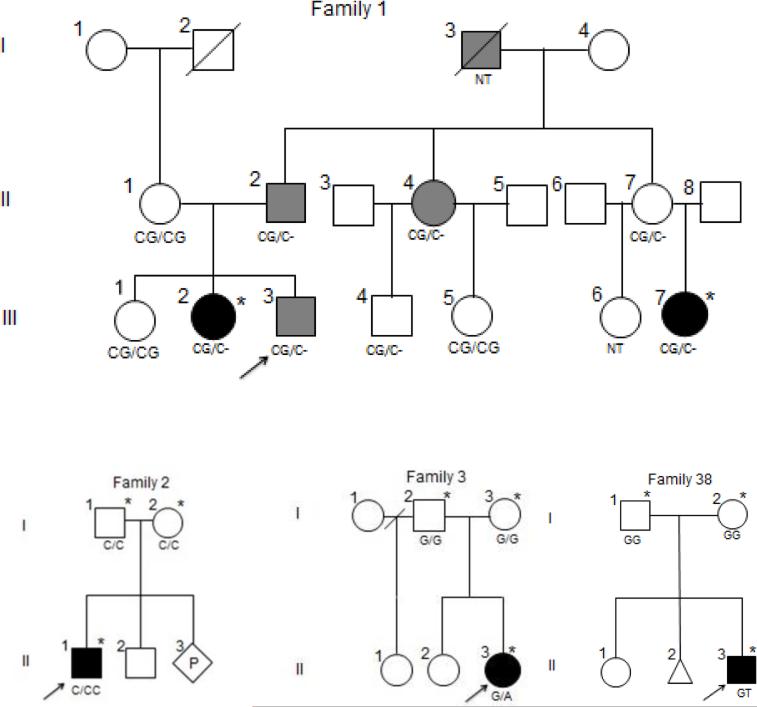
Pedigrees and genotypes of the families with pathogenic SNVs in genes known to cause CAKUT and a novel CAKUT gene (Family 38, FOXP1). * denotes individuals for whom WES were performed. NT means not tested. Family 1. Solid black fill shows renal dysplasia and solid grey fill means proteinuria. CG/CG is normal and CG/C- denotes heterozygous deletion of G (c.70delG) in PAX2. Family 2. Proband has cystic renal dysplasia. C/C is normal and C/CC is heterozygous duplication of C (c.1132dupC) in HNF1B. Family 3. Proband has vesicoureteral reflux and multicystic dysplastic kidney (MCDK). G/G is normal and G/A denotes heterozygous splice site variant (c.867+5G>A) in EYA1. Family 38. Proband has unilateral renal agenesis and hydrocephaly. G/G is normal and G/T denotes heterozygous de novo FOXP1 p.P225T SNV.
Whole-Exome Sequencing analysis
WES analysis started with conversion of raw sequencing data (bcl files) to the fastq format by Casava. Then, the short reads were mapped to a human genome reference sequence (GRCh37) by the Burrows-Wheeler Alignment (BWA). Subsequently, the recalibration was done by GATK,9 and variant calling was performed by the Atlas2 suite. 10 The Mercury pipeline is available in the cloud via DNANexus (http://blog.dnanexus.com/2013-10-22-run-mercury-variant-calling-pipeline/).
SNV prioritization and filtering workflow
After detection of all bi-allelic (homozygous or compound heterozygous) and de novo variants from WES data, we established a SNV prioritization workflow. This included sequential analysis of bi-allelic predicted loss of function variants (stopgain, frameshift indels and splicing); bi-allelic missense variants; de novo truncating variants; and de novo missense variants. Finally, we further examined the shared rare variants among affected family members and parents to detect potential mosaic variants in parents. This SNV prioritization workflow was followed by subsequent filtering of variants based on their frequencies (MAF <= 0.1%) in internal and external databases including Baylor-Hopkins Center for Mendelian Genomics (BHCMG), Exome Aggregation Consortium (ExAC), Exome Variant Server (ESP), 1000 Genome Project, and the Atherosclerosis Risk in Communities Study (ARIC) databases. To retrieve potentially deleterious and conserved missense changes, we utilized various bioinformatics tools including Phylop conservation score and Mutation Taster, SIFT, and PolyPhen-2 prediction scores. Next, these potential rare causative variants were analyzed in terms of: 1) gene function and the associated phenotype in OMIM and Pubmed; 2) gene-associated animal models; 3) tissue expression of the encoded protein; 4) association with already known gene/genes linked to the patient's phenotype in terms of i) gene networks; ii) gene families iii) coexpression; iv) physical protein-protein interaction; v) predicted protein-protein interaction vi) molecular pathways and 5) location of the variant with respect to functional protein domains. The resulting most promising candidate variants were further confirmed and segregated by Sanger sequencing. Finally, the confirmed variants in candidate genes were interrogated in BHCMG and Baylor Miraca Genetics Laboratories (BMGL) databases and/or through GeneMatcher for the identification of additional affected cases with similar phenotypes.
Copy-number variants (CNVs) inference
To identify CNVs, our WES data was analyzed by CoNIFER 11 software and CoNVex algorithms. 12 In CoNVex, as a first step, read depth information from WES data was extracted. Then, the general additive model (GAM) correction method was performed to remove the systemic bias from the read depth information. After this step, Smith-Waterman algorithm was used to infer the CNV state and score the detected CNV regions. Each potential CNV region was assigned a confidence score. We further filtered CoNVex-detected CNV calls by selecting those that have an associated confidence score >=5 and number of probes >= 5. Afterwards, these detected CNV calls by CoNVex were overlapped with CNV calls detected by CoNIFER by using Granges function in R Bioconductor GenomicRanges Package. Overlapping CNVs with previous studies were subjected for validation by aCGH. 13,14 Non-overlapping CNVs were also investigated. Among them, rare CNVs (not present or present with low frequency from Database of Genomic Variants [http://dgv.tcag.ca]) involving genes potentially contributing to kidney abnormalities were selected for array comparative genomic hybridization (aCGH) validation. The flowchart of CNV discovery from WES data is provided (Figure S1a).
SNV and CNV interpretation criteria
SNV interpretation was performed from the most recent guidelines published by America College of Medical Genetics and Genomics (ACMG). 15 Accordingly, only variants that met strict criteria were called pathogenic. CNV interpretation was based on size, gene content, overlap with known disease-associated regions, and phenotype overlap according to ACMG guidelines for postnatal CNV calling. 16
Known CAKUT associated genes 4,17
In this study Codified software (https://www.scienceexchange.com/labs/codified-genomics) was used to search WES data for pathogenic SNVs, variants of uncertain clinical significance (VUS) and benign SNVs in the following 19 dominantly inherited genes reported to be associated with CAKUT: BMP4, BMP7, CDC5L, CHD1L, DSTYK, EYA1, GATA3, HNF1B, KAL1, PAX2, RET, ROBO2, SALL1, SIX1, SIX2, SIX5, SOX17, TNXB, UPK3A. The following recessive CAKUT genes were interrogated for 2 SNVs: AGT, ACE, REN, AGTR1, FRAS1, FREM2, GRIP1, HPSE2, LRP4, and ROR2. In addition, we searched for pathogenic SNVs in the following 6 dominantly inherited genes: GLI3, JAG1, NOTCH2, TFAP2A, TBX18, and WNT4.
Codified Genomics software was used to annotate, filter and prioritize variants. Variants were filtered as previously described. 5 Annotations were generated by Annovar 18 and VEP 19 against the UCSC, RefSeq and Ensembl gene models. Variants and genes were further annotated using dbNSFP, 20 Illumina body map, Uniprot, HPO and OMIM databases amongst others. Variants were prioritized based on patient phenotype similarity to known disease genes, mutation type, and for nonsynonymous variants, predicted deleteriousness.
RESULTS
We performed WES on 112 individuals from 62 families with CAKUT (Table 1 and Table S1). Probands were mostly children and young adults ranging in age from 2 months to 24 years. Thirty-one percent of the probands had more than one organ (other than the kidney and urinary tract) involved, which suggests that these patients potentially harbor a syndromic form of CAKUT. In approximately 16% of the families, WES was performed in a familial mode since more than one individual was affected with CAKUT. The most common phenotypic indications were ‘renal dysplasia’ and ‘agenesis/hypoplasia’.
WES results were interrogated for SNVs in known genes that cause CAKUT as described in methods. Pathogenic SNVs were identified in three known genes (EYA1, HNF1B, and PAX2) in 3 families (approximately 5%) (Table 2). Two of these variants were frameshift variants and one was a splice site variant, each suggesting a loss-of-function mechanism. The frameshift variant in HNF1B is a novel pathogenic allele. Pedigrees of these families are illustrated as Figure 1. Among the pathogenic SNVs identified in these three families, two were de novo from trio analyses, and one was inherited from an affected parent. All selected SNVs identified in the probands and their parents (when available) were confirmed by Sanger sequencing.
Table 2.
Pathogenic SNVs in 35 known genes identified in 62 families with CAKUT by Whole-Exome Sequencing
| Family ID |
Gene | Sex | Ethnicity | Renal phenotype |
Genome build |
Chr: position |
Nucleotide change |
Amino- acid change |
Other organ defects |
Seen in CMG |
ESP EA/AA |
CADD score |
Frequency in ExAc |
Number of LOF in ExAc |
Family segregation |
Comment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PAX2 | M,F | Spanish | RHD | Hg19 | 10: 102509528 | c.70delG | p.G24fs | Optic nerve coloboma (previously undiagnosed) | no | N/R N/R |
35 | 0 | 5 | Yes; multiple individuals, AD | Known pathogenic variant, impacts medical management |
| 2 | HNF1B | M | Mixed Caucasian and African Caribbean | CRD | Hg19 | 17: 36070584 | c.1132dupC | p.Q378fs | Gout, elevated LFTs (recent diagnosis) and increased echogenicity of pancreas (not noted prior to WES) | no | N/R N/R |
35 | 0 | 0 | De novo by WES and Sanger | Novel pathogenic variant, impacts medical management |
| 3 | EYA1 | F | AA | VUR, MCDK | Hg19 | 8: 72183988 | c.867+5G>A | n/a | No | no | N/R N/R |
16.61 | 0 | 4 | De novo by WES and Sanger | Known recurrent pathogenic variant in BOR; MCDK in this patient with VUR likely due to this variant, impacts medical management |
AA African American; AD, autosomal dominant, BOR; Branchio-oto-renal syndrome; CADD, Combined Annotation Dependent Depletion; Chr, chromosome ; CMG, Center for Mendelian Genomics; CRD, Cystic renal dysplasia; ESP, Exome Sequencing Project; ExAc, Exome Aggregation Consortium ; LFT, liver function tests; LOF, loss-of-function; MCDK, Multicystic dysplastic kidney; N/R, none reported, RHD, Renal hypodysplasia; VUR, Vesicoureteral reflux.
Among the three families with pathogenic SNVs, clinical assessment had not identified anomalies of any other organs prior to WES. Importantly, WES elicited further clinical assessment and the delineation of the additional organ system involvement in Families 1 and 2 retrospectively. In Family 3, defects in other organs have not been observed clinically.
The initial diagnosis of Family 1 with p.G24fs SNV in PAX2 was a familial form of renal dysplasia and membranous nephropathy. After the familial variant was identified and in recognition of the current understanding of the phenotype of patients with PAX2 variants (Renal-Coloboma syndrome, [MIM: 120330]), the first degree relatives were referred to an ophthalmologist with expertise in the diagnosis and management of genetic disorders. This clinical evaluation revealed optic nerve colobomata and other congenital optic nerve abnormalities in those first degree relatives who were proven to be variant carriers.
After the initial clinical diagnosis of cystic renal dysplasia (CRD) in Family 2, and in light of the WES results (p.Q378fs in HNF1B), the patient's clinical presentation was further reviewed. The patient had a recent diagnosis of gout and elevated liver function tests (LFTs). The patient also had increased echogenicity of the pancreas (one of known signs of HNF1B variants) noted previously by abdominal ultrasound, whose significance became apparent after the genetic analyses. A known recurrent de novo intronic variant (c.867+5G>A) was identified in EYA1 in the proband of Family 3, who has VUR and multicystic dysplastic kidney (MCDK). Both parents were negative for this variant based on both trio WES and Sanger sequencing.
In this study, we also defined variants of uncertain clinical significance (VUSs) in 19 dominantly inherited known CAKUT genes (Table S2). In this cohort, SNVs were not identified in SIX1, SOX17, GATA3, or UPK3A. Benign SNVs and VUSs were identified in BMP7, CDC5L, CHD1L, SALL1, SIX5, SIX2, ROBO2, BMP4, KAL1, TNXB, RET, PAX2, EYA1, and DSTYK (Table S2). Further allele frequencies and prediction data for all SNVs identified in known CAKUT genes in this cohort are summarized as Table S3. Probability of Loss-of-Function score (pLI) is also provided in this table. The closer pLI score approaches to 1 (unity), the more LoF (loss-of-function) intolerant the gene appears to be (http://exac.broadinstitute.org). We attempted to confirm all VUSs with Sanger sequencing. Details of confirmation are provided in Table S2.
Novel CAKUT gene identification: Forkhead Box P1 (FOXP1)
Trio analyses, consisting of WES in the proband and both biological parents to evaluate for new mutations, were performed in 20 families. We confirmed relationship (paternity and maternity) in the trios by review of the de novo SNVs in each family. There was no proband with more than expected number of de novo SNVs (>2) in the coding exonic region of the genome, well within the expected rate of 1.20 × 10−8 per nucleotide per generation. 21 We identified a de novo SNV (p.P225T) in FOXP1 [MIM: 605515] in a proband with hydrocephaly and unilateral renal agenesis (Family 38) (Figure 1). This patient was enrolled initially into this study at age 4 months. Later, the patient manifested delay in gross motor and speech development. In addition, he was diagnosed with strabismus and left optic atrophy. The pedigree of this family is shown as Figure 1 (Family 38).
We next attempted to identify other subjects/families with variants in FOXP1. The database of Whole Genome Laboratory (WGL) at BMGL was queried for other de novo SNVs in FOXP1. We identified seven more de novo SNVs in this gene among approximately 5000 patients (Table 3). Relationship (paternity and maternity) were confirmed by inheritance of rare SNVs from each parent in cases 3-8. In case 2, paternity was confirmed by inheritance of rare SNVs from the father. Maternity, however could not be genetically confirmed per consent, and was verified by pregnancy history.
Table 3.
FOXP1 single nucleotide variants (SNVs) identified in one individual from this cohort (Family 38) and 7 additional (cases 2-8 from clinical WES database) individuals with pathogenic novel de novo SNVs. Six out of eight cases have syndromic form of kidney and genitourinary tract defects. This strongly suggests an important role for FOXP1 in Kidney and GU tract development.
| Case Number | Genome build | Chr: position | SNV | Protein change | CADD score | Frequency in ExAc | Number of LOF in ExAc | Segregation | Phenotype |
|---|---|---|---|---|---|---|---|---|---|
| Case 1 (Family 38) | Hg19 | 3: 71090675 | c.C673A | p. P225T | 24.7 | 0 | 2 | De Novo | Hydrocephaly, GDD and unilateral renal agenesis |
| Case 2 | Hg19 | 3: 71027085 | c.1240_1241del | p.L414fs | 35 | 0 | 2 | De Novo | Hydrocephaly, brain atrophy, seizure, DD, VSD, neurogenic bladder, hydronephrosis, tethered cord, and behavior problems |
| Case 3 | Hg19 | 3: 71090502 | c.844-845del | p.V283fs | 35 | 0 | 2 | De Novo | Bilateral ventricular enlargement of brain, ID, DD, hypotonia, horseshoe kidney, UDT, hypospadias, and facial dysmorphic features |
| Case 4 | Hg19 | 3: 71021784 | c. 1574G>A | p. R525Q | 34 | 0 | 2 | De Novo | GDD, mega cisterna magna, seizure, hypotonia, concealed penis, retractable testis, and skeletal abnormalities |
| Case 5 | Hg19 | 3: 71021815 | c.1543C>G | p.H515D | 27.7 | 0 | 2 | De Novo | GDD, hip contractures, dysmorphic features, short stature, mild scoliosis, abnormally positioned thumbs, dysmorphic features, and UDT |
| Case 6 | Hg19 | 3: 71019958 | c.1653-2A>T | N/A | 24.8 | 0 | 2 | De Novo | DD, ataxia, mild distal arthrogryposis, ASD, dysmorphic facial features, strabismus, vertical nystagmus and snoring, right duplicated collecting system, and trabeculation of the posterior urinary bladder wall |
| Case 7 | Hg19 | 3: 71027034 | c.1291_1292del | p.T431fs | 35 | 0 | 2 | De Novo | Macrocephaly, GDD, ID, dysmorphic features, hyperextensibility, joint contractures (3rd finger camptodactyly), droopy eyelids, and two cafe au lait spots |
| Case 8 | Hg19 | 3: 71021735 | c.1606_1622dup | p.F541fs | 35 | 0 | 2 | De Novo | Structural brain abnormalities, GDD, aggressive behavior and hyperactivity, hyperopic astigmatism, hypotonia, and ankle tightness |
ASD, atrial septal defect; CADD, Combined Annotation Dependent Depletion; Chr, chromosome , DD, developmental delay; ExAc, Exome Aggregation Consortium; GDD, global developmental delay; ID, intellectual disability; LOF, loss-of-function; UDT, undescended testis; VSD, ventricular septal defect
All eight individuals had neurodevelopmental phenotypes consistent with loss-of-function variants in FOXP1 [MIM: 613670]. However, four out of eight individuals also had upper urinary tract defects, and five of them had defects in lower genitourinary (GU) tract, including undescended testis, hypospadias, neurogenic bladder, inter alia. In addition, these patients have brain and heart involvement, which are consistent with the role of FOXP1 in development of these two organs. 22–24 CNS malformations including hydrocephaly and cardiac defects are among the phenotypes of the patients in this study. The genotypes and phenotypes of these individuals (6/8 with upper or lower urinary tract defects) are summarized as Table 3. pLI score of FOXP1 is 1, which suggests this gene is intolerant to loss-of-function variants.
CNV discovery from WES data
CNVs were inferred from WES data as described in Methods. Pathogenic CNVs and CNVs of uncertain clinical significance are summarized as Table 4. A de novo 22q11.1q11.21 triplication was identified in Family 34, in which the proband had syndromic VUR. Thistriplication is proximal to the DiGeorge region, consistent with the gain of genetic material seen with type I supernumerary inv dup(22)(q11), associated with cat eye syndrome25 [MIM: 115470] (Family 34). This patient's phenotype has overlap with Goldenhar or Oculo-Auriculo-Vertebral spectrum (OAV, [MIM: 164210]) and VATER Association [MIM: 192350]. The patient was an 11-year-old Latin American male with short stature, imperforate anus, thumb anomaly, severe gastroesophageal reflux (GERD), VUR, neurogenic bladder, right renal hypoplasia with evidence of scarring/renal damage, bilateral ear tags, ocular Duane anomaly, and left microphthalmia. Patient had normal cognition, although with some difficulty in Mathematics.
Table 4.
Copy-number variants (CNVs) identified (from WES data of 62 families) which are relevant to the patient's phenotype
| Family ID | Chromosomal region | CNV Type | Start (Mb) | End (Mb) | Size (Mb) | Number of genes | Syndrome | Phenotype | Parental Studies | Comments |
|---|---|---|---|---|---|---|---|---|---|---|
| Family 34 | 22q11 | Trp | 16.63 | 18.64 | 2.01 | 33 | Cat eye syndrome | VUR, further details in text (multiple anomalies) | De Novo | Pathogenic |
| Family 39 | 16p13.11 | Dup | 15.12 | 16.29 | 1.17 | 19 | 16p13.11 dup | MCDK, facial dysmorphic features | Unknown | Pathogenic |
| Family 10 | 16p11.2 | Dup | 29.68 | 30.20 | 0.52 | 35 | 16p11.2 dup | Solitary kidney, psychiatric disorder, hypothyroidism | Unknown | Pathogenic |
| Family 31 | 16p11.2 | Del | 28.83 | 29.04 | 0.21 | 13 | 16p11.2 del | VUR, seizure, DD, LD, and optic edema | Unknown | Pathogenic |
| Family 33 | 3q29 | Dup | 197.51 | 197.59 | 0.08 | 2 | - | VUR, cataract, and growth delay | Unknown | VUS |
| Family 25 | 2p24.3 | Del | 15.30 | 15.38 | 0.08 | 1 | - | PUV, heterotaxy | Inherited | VUS |
| Family 21 | 4q35.1 | Dup | 185.99 | 189.11 | 3.12 | 30 | - | PUV | Inherited | VUS |
DD, developmental delay; del, deletion; dup, duplication; LD, learning disability; MCDK, multicystic dysplastic kidney; PUV, posterior urethral valve; trp, triplication; VUR, vesicoureteral reflux; VUS, variant of uncertain clinical significance
Three other pathogenic CNVs were found in regions associated with known syndromes, namely 16p11.2 deletion,16p11.2 duplication, and 16p13.11 duplication. CNVs in all individuals in Table 4 were validated by aCGH. Parental studies were also performed by aCGH. In Family 34, in which samples were available from both parents, the CNV was found to be de novo. The flowchart of copy-number data inference (a), CoNIFER (b) and aCGH data (c) for de novo 22q11 triplication appears as Figure S1.
DISCUSSION
During the past four years, WES has become a powerful clinical test to define both recognized and previously undefined genes and potential variant susceptibilities to establish molecular diagnoses for birth defects. Clinical WES identifies pathogenic SNVs in approximately 25% of pediatric cases (mostly syndromic) that represent diagnostic dilemmas refractory to clinical diagnosis despite previous extensive medical evaluations 5,6. Nevertheless, the utility of WES for molecular diagnosis in isolated birth defects including CAKUT remains uncertain. This study shows that WES can be used in the diagnostic setting to define the molecular defects that underlie CAKUT and to reveal additional insights into the clinical presentation of the disorder. In addition, WES can be used for the identification of new candidate genes.
In Family 1 (Figure 1 and Table 2), the diagnosis of Renal Coloboma Syndrome (RCS, [MIM: 120330]) was possible only after WES result became available. Prior to WES, the proband was on immunosuppressants for proteinuria; however, after molecular diagnosis management is being altered to a tapering dose of immunosuppressive therapy. This allows avoidance of unnecessary immunosuppression since the etiology of kidney disease is not immunologic in this family. The molecular diagnosis of this family obtained by WES thus affected clinical decision making for the patient and the prognosis and management for family members. These data also expand the phenotype related to PAX2 pathogenic variants as the proband has membranous nephropathy and other family members have proteinuria and renal dysplasia. To date, focal segmental glomerulosclerosis (FSGS) has been reported with PAX2 variants; 26 membranous nephropathy is a novel finding. In Family 2, a novel frameshift pathogenic SNV (p.Q378fs) was identified in HNF1B. This molecular diagnosis concluded by WES further substantiated the clinical phenotype as described in the results, minimizing the necessity for additional diagnostic evaluation. The variant in this family is novel, which adds to our current knowledge of diseases related to HNF1B gene.
The phenotype of Family 3 with the de novo EYA1 variant suggests that underlying genetic predisposition can lead to or at least exacerbate renal pathology in patients with VUR. Variants in EYA1 can cause Branchio-Oto-Renal syndrome (BOR, [MIM: 113650]), an autosomal dominant disorder characterized by sensorineural, conductive, or mixed hearing loss, structural defects of the outer, middle, and inner ear, branchial fistulas or cysts, and renal abnormalities ranging from mild hypoplasia to agenesis. The c.867+5G>A SNV does not affect the invariant splice site: nevertheless, RNA analysis of samples from patients with BOR showed that this SNV affects EYA1 splicing, producing an aberrant mRNA transcript that lacks exon 8 and results in premature termination in exon 9. 27 The proband in this family will be evaluated for hearing impairment, since the SNV in this individual causes BOR. This family provides another example that WES can improve the clinical diagnosis of syndromic forms of CAKUT beyond clinical evaluation alone.
Families 1, 2, and 3 exemplify the effect of WES on the clinical management of the patients and families, since identification of the SNVs in PAX2, HNF1B, and EYA1 respectively warranted further investigations in other organ systems. We identified only a fraction of families (3/62 = 4.8%) with pathogenic SNVs, similar to a recent large study that evaluated 749 individuals from 650 families with CAKUT for variants in 17 known CAKUT genes (6.3%). 28
CAKUT is a clinically heterogeneous clinical spectrum and thus many more genes and causal variants are likely to be identified. Next generation sequencing (NGS) and specifically WES have improved discovery of novel causative genes. 7,29–34 We have identified a novel genes (FOXP1) that likely contribute to the CAKUT phenotype. We found 8 novel different de novo pathogenic SNVs, from both clinical and research WES, in FOXP1 in unrelated individuals. As summarized as Table 3, the phenotypes observed in these individuals suggest a clinical pattern that may be potentially recognizable. Structural brain anomalies (including hydrocephaly), intellectual disability, developmental delay, cardiac defects, hypotonia, behavior problems and renal/GU defects are some of the more common features of this syndrome. Six out of eight individuals in this study (Table 3) have known renal/GU phenotype in addition to other organ involvements. Although de novo disruptions in FOXP1, were recently discovered to cause intellectual disability [OMIM# 613670], 23,24 here we defined a new syndrome which is characterized by hydrocephalus/brain malformation, cognitive impairment, cardiac defects, and CAKUT attributable to a single gene with pleiotropic effect. We recommend that patients with pathogenic or likely pathogenic variants in FOXP1 should have renal ultrasound. Upper tract defects may remain undiagnosed if ultrasound is not performed.
Although FOXP1 has been shown to have important roles in the developmental process of key organs including lung, heart and brain, 22,24,35 there is no data regarding the role of this master transcription factor in kidney and urinary tract development. In this study we showed the role of FOXP1 in CAKUT and lower urinary tract defect. All of the FOXP1 SNVs identified in this study were de novo and novel variants. These variants included frameshift, as documented for other birth defects such as the megacystis microcolon intestinal hypoperistalsis syndrome due to de novo SNVs in ACTG2. 36
Based on previous investigations, CNVs account for about 16% of CAKUT cases. 13 We hypothesized that CNVs underlie a substantive fraction of birth defects in our families as well; therefore, we inferred CNV data with two detection tools. There are some known limitations to CNV discovery from WES data. 37 One primary limitation is a high false positive rate, particularly for small CNVs. We used a stringent approach to identify potentially pathogenic CNVs for validation by aCGH. Approximately 6.5% (4/62) of our cohort have pathogenic CNVs related to the patients’ phenotype.
The four pathogenic CNVs identified (Table 4) are in disease-associated regions and have been evaluated based on ACMG guidelines. Although the fraction of families with pathogenic CNVs (6.5%) is lower than studies designed specifically to identify CNV, we included only known pathogenic CNVs and not copy-number variants of uncertain significance.
Among the most intriguing CNVs identified in this study is triplication of 22q11. Although the patient with proximal 22q11 triplication did not have chromosome analysis to determine if a marker chromosome was present, the gain of euchromatic genetic material is the same as what is seen in cat eye syndrome. Urogenital malformations are present in ~70% of reported individuals with this syndrome and include male and female genital malformations, renal agenesis, hydronephrosis, VUR, dysplastic or cystic kidneys, and bladder defects.38 Individuals with partial gains of the cat eye critical region and renal anomalies have been described, providing evidence that the distal portion of the region, including CECR2, SLC25A18, and ATPV1E1, may be responsible for these features.39 Our patient carried a clinical diagnosis of OAV; however, after uncovering the CNV triplication, most of his features are consistent with cat eye syndrome.
Our findings support the concept that WES could be an adjuvant diagnostic tool even in cases of non-syndromic CAKUT, since the involvement of other organs may be subtle or not manifest at the time of primary evaluation. WES may identify novel candidate genes, as exemplified here, and uncover underlying CNVs that contribute to CAKUT spectrum.
This study reports the use of WES for molecular diagnosis of the genetic contribution to CAKUT. Nearly 5% of individuals with CAKUT have pathogenic SNVs in known key genes that can be uncovered by WES. In addition, 6.5% of these patients have pathogenic CNVs that were extracted from WES data. In some families, organ involvement beyond CAKUT was sought retrospectively, after the review of WES results. We identified previously unrecognized genes and genetic variants (both SNVs and CNVs) in this cohort and expanded the phenotype of several known genes. Pathogenic SNVs in FOXP1 in individuals with GU/Renal phenotype strongly suggests an important role for this gene in urinary tract development.
Supplementary Material
ACKNOWLEDGEMENTS
Supported in part by K12 DK083014 Multidisciplinary K12 Urologic Research Career Development Program scholar and R01 DK078121 from the National Institute of Diabetes and Digestive and Kidney Diseases awarded to DJL. This work was funded in part by grant R01 NS058529 (JRL) from the National Institute of Neurological Disorders and Stroke and U54 HG006542 from the National Human Genome Research Institute/National Heart Lung and Blood Institute to the Baylor Hopkins Center for Mendelian Genomics. Supported in part also by a National Institutes of Human Genome Research (NHGRI) (U54HG003273) awarded to RAG. RAL is a Senior Scientific Investigator of Research to Prevent Blindness whose unrestricted funding supported part of this study.
DNA extraction was performed in laboratory of translational genomics (LTG) by Patricia Hernandez and Gladys Zapata. The analysis of the data was performed by software generously available by Codified Genomics Company. We also express our sincere gratitude to our patients and their families for their willing participation in this study.
Footnotes
Supplementary Data
Supplementary Methods: Whole-Exome Sequencing, Validation of selected SNVs, Validation of selected CNVs by aCGH, References for known CAKUT genes, Accession numbers
Table S1. Description of the cohort (probands of 62 families) in detail with basic demographic, phenotypic information, type of sample (saliva or blood), the summary of the genetic findings, and the pattern of segregation.
Figure S1. Copy-number variants (CNVs) inference from WES data
Table S2. Benign SNVs and variants of uncertain significance (VUS) in 19 CAKUT known genes identified in 62 families with CAKUT using Whole-Exome Sequencing
Table S3. Further allele frequencies and prediction data for the SNVs presented in Tables 2 and S2
REFERENCES
- 1.Sanna-Cherchi S, Ravani P, Corbani V, et al. Renal outcome in patients with congenital anomalies of the kidney and urinary tract. Kidney Int. 2009;76(5):528–533. doi: 10.1038/ki.2009.220. doi:10.1038/ki.2009.220. [DOI] [PubMed] [Google Scholar]
- 2.Mansoor O, Chandar J, Rodriguez MM, et al. Long-term risk of chronic kidney disease in unilateral multicystic dysplastic kidney. Pediatr Nephrol. 2011;26:597–603. doi: 10.1007/s00467-010-1746-0. doi:10.1007/s00467-010-1746-0. [DOI] [PubMed] [Google Scholar]
- 3.Groothoff JW. Long-term outcomes of children with end-stage renal disease. Pediatr Nephrol. 2005;20(7):849–853. doi: 10.1007/s00467-005-1878-9. doi:10.1007/s00467-005-1878-9. [DOI] [PubMed] [Google Scholar]
- 4.Vivante A, Kohl S, Hwang DY, Dworschak GC, Hildebrandt F. Single-gene causes of congenital anomalies of the kidney and urinary tract (CAKUT) in humans. Pediatr Nephrol. 2014;29(4):695–704. doi: 10.1007/s00467-013-2684-4. doi:10.1007/s00467-013-2684-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. http://www.ncbi.nlm.nih.gov/pubmed/24088041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y, Muzny DM, Xia F, et al. Molecular Findings Among Patients Referred for Clinical Whole-Exome Sequencing. JAMA. 2014;312(18):1870–1879. doi: 10.1001/jama.2014.14601. doi:10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karaca E, Harel T, Pehlivan D, et al. Genes that Affect Brain Structure and Function Identified by Rare Variant Analyses of Mendelian Neurologic Disease. Neuron. 2015;88(3):499–513. doi: 10.1016/j.neuron.2015.09.048. doi:10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y, Oldridge DA, Diskin SJ, Zhang NR. CODEX: A normalization and copy number variation detection method for whole exome sequencing. Nucleic Acids Res. 2015;43(6):e39. doi: 10.1093/nar/gku1363. doi:10.1093/nar/gku1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. doi:10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Challis D, Yu J, Evani US, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13(1):8. doi: 10.1186/1471-2105-13-8. doi:10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krumm N, Sudmant PH, Ko A, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22(8):1525–1532. doi: 10.1101/gr.138115.112. doi:10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vijayarangakannan P, Fitzgerald T. Detection of Copy Number Variation from Exomes in the DDD and UK10K Projects. Uk10KOrg. 2012;10(10):7319. http://www.uk10k.org/assets/ashg_vijayarangakannan_etal_2012.pdf. [Google Scholar]
- 13.Sanna-Cherchi S, Kiryluk K, Burgess KE, et al. Copy-Number Disorders Are a Common Cause of Congenital Kidney Malformations. Am J Hum Genet. 2012;91:987–997. doi: 10.1016/j.ajhg.2012.10.007. doi:10.1016/j.ajhg.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verbitsky M, Sanna-Cherchi S, Fasel DA, et al. Genomic imbalances in pediatric patients with chronic kidney disease. J Clin Invest. 2015 doi: 10.1172/JCI80877. doi:10.1172/JCI80877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. 2015 doi: 10.1038/gim.2015.30. doi:10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. doi: 10.1097/GIM.0b013e3182217a3a. doi:10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- 17.Nicolaou N, Renkema KY, Bongers EMHF, Giles RH, Knoers NV a. M. Genetic, environmental, and epigenetic factors involved in CAKUT. Nat Rev Nephrol. 2015;(12):1–12. doi: 10.1038/nrneph.2015.140. doi:10.1038/nrneph.2015.140. [DOI] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. doi:10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. doi:10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Jian X, Boerwinkle E. dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32(8):894–899. doi: 10.1002/humu.21517. doi:10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong A, Frigge ML, Masson G, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. doi:10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang S-W, Mislankar M, Misra C, et al. Genetic abnormalities in FOXP1 are associated with congenital heart defects. Hum Mutat. 2013;34:1226–1230. doi: 10.1002/humu.22366. doi:10.1002/humu.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamdan FF, Daoud H, Rochefort D, et al. De novo mutations in FOXP1 in cases with intellectual disability, autism, and language impairment. Am J Hum Genet. 2010;87:671–678. doi: 10.1016/j.ajhg.2010.09.017. doi:10.1016/j.ajhg.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sollis E, Graham SA, Vino A, et al. Identification and functional characterization of de novo FOXP1 variants provides novel insights into the etiology of neurodevelopmental disorder HMG Advance Access. Hum Mol Genet. 2015:1–12. doi: 10.1093/hmg/ddv495. doi:10.1093/hmg/ddv495. [DOI] [PubMed] [Google Scholar]
- 25.McTaggart KE, Budarf ML, Driscoll DA, Emanuel BS, Ferreira P, McDermid HE. Cat eye syndrome chromosome breakpoint clustering: identification of two intervals also associated with 22q11 deletion syndrome breakpoints. Cytogenet Cell Genet. 1998;81(3-4):222–228. doi: 10.1159/000015035. doi:10.1159/000015035. [DOI] [PubMed] [Google Scholar]
- 26.Barua M, Stellacci E, Stella L, et al. Mutations in PAX2 Associate with Adult-Onset FSGS. J Am Soc Nephrol. 2014;25:1942–1953. doi: 10.1681/ASN.2013070686. doi:10.1681/ASN.2013070686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stockley TL, Mendoza-Londono R, Propst EJ, Sodhi S, Dupuis L, Papsin BC. A recurrent EYA1 mutation causing alternative RNA splicing in branchio-oto-renal syndrome: implications for molecular diagnostics and disease mechanism. Am J Med Genet A. 2009;149A(3):322–327. doi: 10.1002/ajmg.a.32679. doi:10.1002/ajmg.a.32679. [DOI] [PubMed] [Google Scholar]
- 28.Hwang D-Y, Dworschak GC, Kohl S, et al. Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int. 2014;85(6):1–5. doi: 10.1038/ki.2013.508. doi:10.1038/ki.2013.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanna-Cherchi S, Sampogna R V, Papeta N, et al. Mutations in DSTYK and dominant urinary tract malformations. N Engl J Med. 2013;369(7):621–629. doi: 10.1056/NEJMoa1214479. doi:10.1056/NEJMoa1214479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saisawat P, Kohl S, Hilger AC, et al. Whole-exome resequencing reveals recessive mutations in TRAP1 in individuals with CAKUT and VACTERL association. Kidney Int. 2014;85(6):1310–1317. doi: 10.1038/ki.2013.417. doi:10.1038/ki.2013.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzaga-Jauregui C, Lupski JR, Gibbs R a. Human Genome Sequencing in Health and Disease. Annu Rev Med. 2012;63(1):35–61. doi: 10.1146/annurev-med-051010-162644. doi:10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vivante A, Kleppa M-J, Schulz J, et al. ARTICLE Mutations in TBX18 Cause Dominant Urinary Tract Malformations via Transcriptional Dysregulation of Ureter Development. Am J Hum Genet. 2015;97(6):291–301. doi: 10.1016/j.ajhg.2015.07.001. doi:10.1016/j.ajhg.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzaga-Jauregui C, Harel T, Gambin T, et al. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. Cell Rep. 2015;12(7):1169–1183. doi: 10.1016/j.celrep.2015.07.023. doi:10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan B, Pehlivan D, Karaca E, et al. Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. 2015;125(2) doi: 10.1172/JCI77435. doi:10.1172/JCI77435.spectrum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D, Morrisey EE. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development. 2007;134:1991–2000. doi: 10.1242/dev.02846. doi:10.1242/dev.02846. [DOI] [PubMed] [Google Scholar]
- 36.Wangler MF, Gonzaga-Jauregui C, Gambin T, et al. Heterozygous de novo and inherited mutations in the smooth muscle actin (ACTG2) gene underlie megacystis-microcolon-intestinal hypoperistalsis syndrome. PLoS Genet. 2014;10(3):e1004258. doi: 10.1371/journal.pgen.1004258. doi:10.1371/journal.pgen.1004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Retterer K, Scuffins J, Schmidt D, et al. Assessing copy number from exome sequencing and exome array CGH based on CNV spectrum in a large clinical cohort. 2014 doi: 10.1038/gim.2014.160. doi:10.1038/gim.2014.160. [DOI] [PubMed] [Google Scholar]
- 38.Rosias PPR, Sijstermans JMJ, Theunissen PMVM, et al. Phenotypic variability of the cat eye syndrome. Case report and review of the literature. Genet Couns. 2001;12:273–282. [PubMed] [Google Scholar]
- 39.Knijnenburg J, van Bever Y, Hulsman LOM, et al. A 600 kb triplication in the cat eye syndrome critical region causes anorectal, renal and preauricular anomalies in a three-generation family. Eur J Hum Genet. 2012;20(9):986–989. doi: 10.1038/ejhg.2012.43. doi:10.1038/ejhg.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.