Abstract
The process of cellular differentiation requires the distinct spatial organization of the microtubule cytoskeleton, the arrangement of which is specific to cell type. Microtubule patterning does not occur randomly, but is imparted by distinct subcellular sites called microtubule-organizing centers (MTOCs). Since the discovery of MTOCs fifty years ago, their study has largely focused on the centrosome. All animal cells use centrosomes as MTOCs during mitosis. However in many differentiated cells, MTOC function is reassigned to non-centrosomal sites to generate non-radial microtubule organization better suited for new cell functions, such as mechanical support or intracellular transport. Here, we review the current understanding of non-centrosomal MTOCs (ncMTOCs) and the mechanisms by which they form in differentiating animal cells.
Introduction
Microtubules adopt specific spatial arrangements in differentiated cells to perform diverse cellular functions. Early electron microscopy revealed distinct subcellular sites from which microtubules appeared to emanate which were named ‘microtubule-organizing centres’ (MTOCs) [1,2]. Since then, the exact nature of MTOCs has remained somewhat nebulous.
Microtubules have inherent structural polarity, with a dynamic plus end and a comparatively stable and slow-growing minus end [3]. These characteristics of microtubule minus ends are, in part, a function of microtubule structure, but can be influenced in vivo by an association with an MTOC. MTOCs can be broadly defined as sites that localize microtubule minus ends, with functions that include microtubule nucleation, stabilization, and/or anchoring.
The best-studied MTOC is the centrosome, a non-membrane bound organelle composed of two centrioles surrounded by pericentriolar material (PCM). The centrosome is often touted as ‘the major microtubule-organizing center of the cell,’ generating a radial organization of microtubules well suited for the division of genomic material between daughter cells. Microtubules are nucleated and anchored within the PCM in dividing animal cells to generate the classic ‘mitotic halo,’ and similar radial arrays form in migrating animal cells [4] (reviewed in [5,6]). Microtubules can also be anchored at centriolar appendages; an astral interphase array is generated by subdistal appendages decorating the mother centriole and a lattice-like organization is anchored by and potentially nucleated from the basal foot of basal bodies in multiciliated cells for coordinated ciliary beating [7-12]. As we will discuss below, differentiated cells often generate alternative microtubule organization through the reassignment of MTOC function to non-centrosomal sites following cell division.
Non-centrosomal MTOCs (ncMTOCs)
Differentiated animal cells often establish non-centrosomal MTOCs (ncMTOCs) (Figure 1). In many epithelial cells, MTOC function localizes apically, generating microtubules organized along the apical-basal axis; specific examples include C. elegans embryonic intestinal cells, Drosophila tracheal, oocyte follicle, and embryonic epithelial cells, Xenopus neural epithelial cells, mouse cochlear supporting cells, and various mammalian epithelial cell lines [8,13-20]. Cortical MTOCs have also been observed in mouse and C. elegans epidermal cells, and C. elegans germ cells [21-24]. Epithelial microtubule arrays appear to be required for organelle positioning and the initiation of apical-basal polarity [13,19,24,25]. In contrast to in epithelia, MTOCs and microtubules in differentiated muscle cells are organized around nuclei and in the cytoplasm parallel to the long axis of the cell [26,27]. Such microtubule arrays in Drosophila are essential for nuclear positioning and anchoring [28]. In Drosophila oocytes, microtubules grow from the anterior/lateral cortex with plus ends concentrated posteriorly, an arrangement that is critical for directing the localization of mRNAs that establish the embryonic body axis [29] (reviewed in [30]).
Figure 1. Organization of MTOCs and microtubules in a variety of cell types.
Microtubules (red) are organized by MTOCs (blue), the arrangement and localization of which varies with cell type. Drawings are not to scale.
In neurons, microtubules are distributed down the lengths of axons and dendrites and are essential for transport, regeneration, and development (reviewed in [31]). Axonal microtubules are uniformly arranged with their plus ends towards the tip, and dendrite microtubules have mixed polarity in vertebrate neurons or a minus end out orientation in C. elegans and Drosophila [32-34]. A specific ncMTOC in neuronal processes has been elusive. In Drosophila class IV dendritic arborization neurons, Golgi outposts appear to act as ncMTOCs in some dendrites [35]. However, removal of Golgi outposts from dendritic arbors had little effect on microtubule organization [36]. Interestingly, in non-neuronal cells, Golgi and mitochondria have also been reported as MTOCs [37,38]. Microtubules in neurons might also arise from the sides of pre-existing microtubules, a scenario that would provide a polarized template to orient newly forming microtubules [39].
Some organisms, including yeast and higher plants, lack centrosomes altogether, thus microtubule organization by definition is non-centrosomal (Figure 1). Yeast have an analogous structure to the centrosome called the spindle pole body (SPB). Although the SPB is the only MTOC in budding yeast, in fission yeast, ‘interphase MTOCs’ generate non-SPB microtubules in the cytoplasm, on the nucleus, and on other microtubules, and ‘equatorial MTOCs’ organize microtubules around the cell division site at the end of mitosis (reviewed in [40,41]). Finally, cells of higher plants completely lack centrosomes or analogous structures, yet have elaborate cortical microtubule arrays that appear to be largely generated by microtubule-based microtubule nucleation and are required for growth and morphogenesis [42,43]. Despite the wide range of ncMTOCs in many diverse cell types, their composition and mechanisms of assembly are just beginning to be understood.
ncMTOC structure and composition
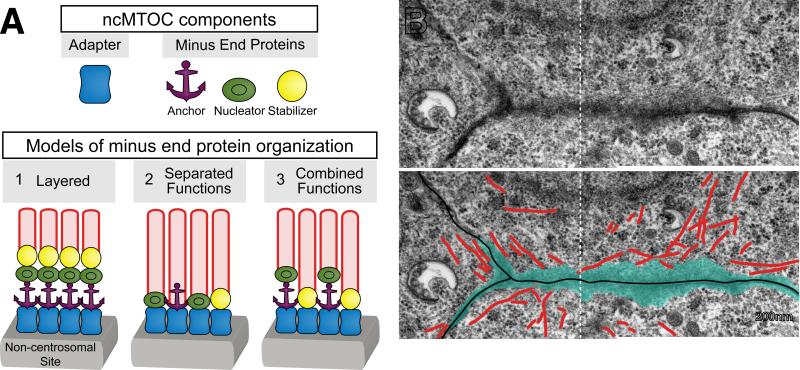
The complexity of the centrosome obscures our understanding of the specific structure or proteins that alone impart MTOC function. Similarly, the structure or composition of an ncMTOC is unclear. ncMTOCs could be layered structures composed of shells of proteins, similar to PCM (reviewed in [44]) or could be composed of discrete populations of microtubules held together by site-specific adapters (Figure 2). In either case, ncMTOCs should in principle contain 1) proteins that interact with microtubule minus ends, and 2) adapter proteins that link these proteins to specific subcellular sites.
Figure 2. ncMTOC structure and composition.
(A) Cartoons depicting ncMTOC components and models for their arrangement at non-centrosomal sites. Cell-type specific adaptors (blue) bound to non-centrosomal sites (grey) interact with microtubule minus end proteins that anchor (purple), nucleate (green) and/or stabilize (yellow) microtubules (red). (1) Minus end proteins might be layered on top of adapters, which function together to sustain microtubules. (2) Alternatively, different minus end proteins could localize to independent minus ends, distributing their function between different populations of microtubules. (3) Finally, minus end proteins might colocalize at the same microtubule ends, functioning together to promote microtubule nucleation, anchoring, and/or stabilization. For example, NOCA-1 and γ-tubulin colocalize on microtubules in the C. elegans larval epidermis, but PTRN-1 does not and functions in a parallel pathway [25]. (B) An electron microscopy image of ncMTOCs (blue) at the apical membrane in C. elegans embryonic intestinal cells. Electron dense material (blue) is visible at the apical surfaces of three cells from which microtubules (red) emanate (partially reproduced from [54]). Note that two separate electron microscopy images have been overlaid (white dotted line).
(1) Microtubule minus end proteins
Unlike the large number of proteins that have been shown to interact with microtubule plus ends, only a handful of minus end-associated proteins has been identified. These proteins act as microtubule nucleators, stabilizers, and anchors, examples of which are discussed below.
Nucleators: γ–tubulin ring complex (γ–TuRC)
Although it is clear that microtubules form spontaneously in the absence of accessory proteins, nucleators help to enhance microtubule assembly and so are key components of MTOCs. γ–tubulin was the first microtubule minus end protein discovered and was shown to play a role in centrosomal microtubule nucleation; a γ–tubulin mutation in Aspergillus nidulans blocks mitotic spindle assembly and γ–tubulin depletion inhibits microtubule growth from the centrosome in vitro [45-48]. γ–tubulin is part of a larger complex, termed the γ–tubulin ring complex (γ–TuRC), which nucleates microtubules and inhibits their minus end growth and depolymerization [49,50]. However, γ–TuRC may not be the only complex involved in microtubule nucleation as microtubules are still nucleated in Drosophila and C. elegans cycling cells after γ–tubulin depletion, albeit at a reduced rate [51-53].
The role of γ–TuRC in nucleating non-centrosomal microtubules has often been inferred from localization observations. For example, γ–tubulin localizes to the cell cortex of C. elegans germ cells, at hemidesmosomes of C. elegans epidermal cells, surrounding nuclei of cultured muscle cells, at the Golgi membrane in RPE1 cells, and along the apical membrane of Drosophila tracheal cells, C. elegans intestinal cells, and Caco-2 and WIF-B epithelial cell lines [13,14,20,23,24,27,37,54]. Microtubules appear to regrow from these sites following induced depolymerization, suggesting that γ–TuRCs might control microtubule nucleation there. Indeed, alterations in γ–tubulin expression suggest that γ–TuRC nucleates non-centrosomal microtubules in the axons and dendrites of neurons, at the nuclear envelope in myotubes, and from Golgi membranes in RPE1 cells [27,35-37,39,55]. Altogether, these data suggest a microtubule nucleation function of γ–TuRC at ncMTOCs, but do not rule out its role in stabilization and/or capping. Regardless of the exact function of γ–TuRC in ncMTOCs, other microtubule minus end proteins must exist because not all minus ends associate with γ–TuRCs [53,55].
Stabilizers: The CAMSAP/Patronin family
In addition to nucleating microtubules, MTOCs stabilize microtubules through an association with minus end stabilizing proteins. The CAMSAP/Nezha/Patronin family, characterized by evolutionarily conserved CKK domains, has been shown to specifically serve this function for non-centrosomal microtubules (reviewed in [56]). Like γ–tubulin, CAMSAP/Patronin has been shown to stabilize and protect microtubule minus ends from depolymerization [57,58]. However, unlike γ–tubulin, CAMSAP/Patronin does not nucleate microtubules or associate with centrosomes, and its depletion specifically reduces non-centrosomal microtubule number and/or organization in cultured cells and in living organisms [19,25,55,58,59]. CAMSAP/Patronin is predicted to form stabilized ‘seeds’ of microtubules for microtubule outgrowth; more specifically, CAMSAP/Patronin stabilizes polymerizing minus ends, slowing the rate of minus end depolymerization and increasing the rate of microtubule plus end outgrowth [15,57,58]. The ability of CAMSAP/Patronin to support microtubule plus end outgrowth may obviate the need for microtubule nucleators at ncMTOCs, and could instead depend on microtubule-severing enzymes to amplify minus ends. This mechanism has been proposed for microtubule outgrowth from Patronin foci at ncMTOCs in Drosophila oocytes [15]. However, the possibility remains that CAMSAP/Patronin stabilizes microtubules nucleated and released by γ–TuRC, as CAMSAP2 localizes to the minus ends of microtubules released from the centrosome in cultured epithelial cells [58].
Anchors: Ninein
Newly nucleated and stabilized microtubules need a mechanism for anchorage at MTOCs. In theory, nucleators and stabilizers could themselves anchor minus ends, but proteins specific for anchoring likely exist. The coiled-coil protein ninein has not been shown to interact directly with microtubule minus ends, but appears to anchor minus ends in many contexts. Ninein was first identified as a centrosomal protein localizing to subdistal appendages of the mother centriole, and thereafter was described to have a microtubule anchoring function: ninein overexpression in mouse fibroblasts inhibited microtubule release from the centrosome and ninein inhibition in U2OS cells resulted in a perturbation in microtubule organization [8,60-62]. Ninein interacts directly with γ–TuRC and potentially recruits it to the centrosome; however, ninein's microtubule anchoring capacity appears to be separable from its ability to localize γ–TuRC to the centrosome [63].
Ninein is also hypothesized to anchor microtubules at non-centrosomal sites. Ninein localizes near microtubule minus ends at apical sites of mouse cochlear cells, at the cell cortex in differentiated mouse epidermal cells, and surrounding nuclei in myotubes [8,21,27]. A putative ninein homologue in C. elegans (NOCA-1) localizes exclusively to ncMTOCs and is important for microtubule organization at these sites [25,64]. Strikingly, noca-1 mutants exhibit severe sterility and gonad morphology defects, and microtubules in adult germ cells are highly disorganized [25]. In epidermal cells, NOCA-1 depletion also perturbs microtubule organization, but to a lesser extent due to an apparent parallel function of PTRN-1 in maintaining these arrays [25].
(2) Site-specific adapters
The attachment of microtubules to a specific subcellular site requires site-specific adapters that interact with minus end proteins (Figure 2A). Several putative adapter proteins have been identified in different cell types, although in most cases a direct link to minus end proteins has not been demonstrated. In Drosophila tracheal cells, the transmembrane protein Piopio is required to localize γ–TuRC to the apical membrane [14]. CAMSAP3/Nezha was originally discovered because of its association with the Zonula Adherens protein PLEKHA7, depletion of which mislocalizes CAMSAP3 from adherens junctions [59]. In Drosophila oocytes, the actin binding protein Short stop recruits Patronin foci to the cortex, which in turn localize microtubules [15]. Finally, in differentiated keratinocytes, ninein localization to desmosomes requires the desmosome component desmoplakin [21].
The interplay between minus end proteins and adapters at ncMTOCs is an open and exciting question in the field. Pairwise localization studies of γ–TuRC, CAMSAP/Patronin, and ninein in vitro and in vivo suggest that their localization is independent of one another and/or non-overlapping, and genetic studies in C. elegans epidermal cells suggest that NOCA-1 and PTRN-1 operate in parallel pathways [15,21,25,59]; however, biochemical studies and higher resolution microscopy will be required to better clarify this issue. It is likely that additional minus end proteins and adapters exist, the discovery of which will provide significant insight into MTOC biology.
ncMTOC formation
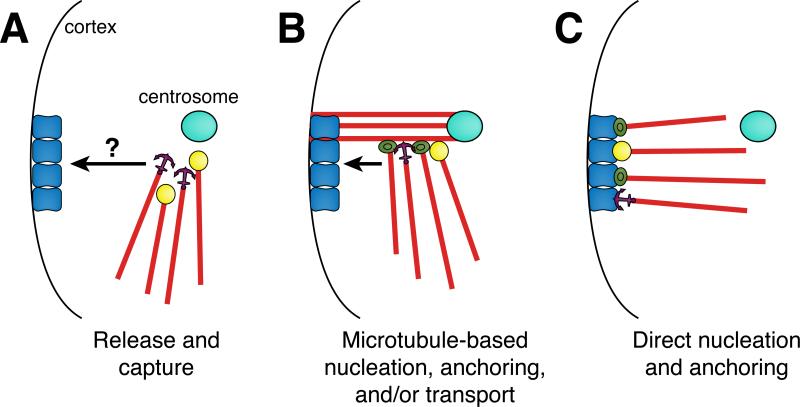
ncMTOC formation requires the attenuation of MTOC function at the centrosome, the designation of a non-centrosomal site, and the proper localization of MTOC components to that site. The mechanisms of ncMTOC site designation are largely unknown and the localization of MTOC components depends on whether the ncMTOC simply anchors or both anchors and nucleates microtubules. Early studies predicted a division of labor between microtubule assembly and localization, suggesting that microtubules in differentiated cells might be nucleated at the centrosome, released, and then captured at non-centrosomal sites (reviewed in [65]) (Figure 3A). This mechanism necessitates the reassignment of anchoring function to a non-centrosomal site coupled with a mechanism for transporting microtubules there. Alternatively, nascent ncMTOCs might form at sites away from the centrosome, such as on residual centrosomal microtubules, which could facilitate transport to non-centrosomal sites (Figure 3B). Finally, ncMTOCs might form independently of centrosomes, with microtubules growing directly from and remaining at non-centrosomal sites (Figure 3C).
Figure 3. Potential mechanisms for ncMTOC formation.
(A) A division of labor model in which microtubules are nucleated at the centrosome, released, and then captured at a non-centrosomal site. Microtubules could be released with anchoring proteins attached or free minus ends could bind to anchoring and/or stabilizing proteins following their release. Microtubules are then transported to a non-centrosomal site via an unknown mechanism and captured by site-specific adapters. (B) Non-centrosomal microtubules could be nucleated, stabilized, and/or anchored from the sides of preexisting centrosomal microtubules and then transported along microtubules to non-centrosomal sites where they would interact with site specific adapters. (C) Microtubule minus end proteins could localize directly to non-centrosomal sites without a centrosome-based intermediate, where they would nucleate, stabilize, and anchor microtubules.
(1) Attenuation of MTOC function at the centrosome
The attenuation of MTOC function at the centrosome can generally be considered as the process of removing microtubule nucleation and/or anchoring potential. The degree to which either of these occurs depends on the cell type. For example, cells exclusively containing ncMTOCs completely inactivate microtubule nucleation and anchoring at the centrosome as is seen in myotubes, rat and Drosophila neurons, and in some C. elegans and Drosophila epithelial cells [27,66,67]. Centrosomes in C. elegans embryonic intestinal cells or Drosophila tracheal cells lose PCM association and the ability to nucleate microtubules [13,14,54,68]. In both cell types, γ–TuRCs move away from the centrosome, and in Drosophila tracheal cells, this release requires the microtubule severing protein spastin. Interestingly, the inactivation of MTOC function at the centrosome in C. elegans embryonic intestinal cells does not appear to be permanent, since centrosomal MTOC function in a differentiated cell can be rapidly reactivated upon fusion with a mitotic cell [54].
Alternatively, attenuation of MTOC function at the centrosome might involve the retention of nucleating potential and the inactivation of anchoring function. Loss of anchoring function at the centrosome has been inferred from localization studies, for example in cochlear and epidermal cells where γ–tubulin is exclusively localized to the centrosome and anchoring proteins such as ninein are localized to non-centrosomal sites [8,21]. Mouse keratinocytes were shown to have distinct microtubule nucleating and anchoring complexes CDK5RAP2-γ–TuRC and Nedd1-γ–TuRC, respectively [69]. During keratinocyte differentiation, centrosomes inactivate anchoring function concomitant with a loss of Nedd1, suggesting that centrosomal MTOC attenuation is partly due to the loss of this factor [69]. Attenuation appears to be coupled to cell cycle exit as serum starvation or CDK inhibitor treatment induced loss of Nedd1–γ–TuRC from the centrosome [69].
(2) Activation of MTOC function at non-centrosomal sites
The attenuation of MTOC function at the centrosome must be paired with the designation and activation of MTOC function at a non-centrosomal site. This process undoubtedly begins with a change in cell state that is permissive for the association of microtubules with new sites. For example, mitotic cytoplasm can rapidly remove the apical ncMTOC in an intestinal epithelial cell, indicating that MTOC location is responsive to cell cycle state [54]. The rapidity of this switch suggests that post-translational modifications might control MTOC location. Activation of MTOC function at non-centrosomal sites can also be coupled to differentiation. For example, in Drosophila tracheal cells, a transcription factor required for tracheal fate specification, Trachealess, and its target, Piopio, are required for apical ncMTOC formation [14]. Whether this requirement for differentiation relies solely on the transcription of specific adaptors or might also involve changes in post-translation modification of MTOC components remains to be tested.
Once the cell has achieved a state permissive for ncMTOC formation, MTOC components need to become properly localized. Cells that use a ‘release and capture’ mechanism must relocate anchoring proteins (Figure 3A). For example, in differentiating mouse cochlear cells, ninein localization is initially restricted to the centrosome early in development, but later localizes to cytoplasmic, and then apical sites [70]. It is tempting to speculate that ninein is released from the centrosome and moved apically, however, live imaging and/or photomarking experiments in differentiating cells would be needed to test this hypothesis. Ninein has been shown to traffic along microtubules in cultured cells lines, suggesting that ncMTOC formation involves ninein transport along centrosomal microtubules to the apical surface [70]. However, this model is complicated by the fact that cells would have to both release anchoring factors from the centrosome and retain microtubules on which to transport them.
Cells with ncMTOCs capable of both microtubule nucleation and anchoring inactivate these functions at the centrosome and transfer them to the non-centrosomal site. Imaging studies in C. elegans suggest that this task is accomplished through the hand-off of a physical ‘plume’ of γ–TuRCs and microtubules from the centrosome to the membrane, potentially coupling centrosome inactivation and ncMTOC establishment [13]. Plume formation requires the centrosome, microtubules, and the conserved polarity protein PAR-3 [13]. Interestingly, γ–TuRCs and microtubules still form following PAR-3 depletion but at apparently random locations, indicating that in the absence of a specific ‘landing pad’, these factors coalesce at inappropriate sites [13,71]. γ–TuRC proteins also move from the centrosome to the apical surface of Drosophila tracheal cells [14]. As in C. elegans, the initiation of this process requires microtubules, however, in both systems microtubules become dispensable for apical γ–TuRC localization later in development [13,14]. The exact role of microtubules in ncMTOC formation is unclear. Residual centrosomal microtubules may serve as a site for nascent ncMTOC formation and its subsequent transport (Figure 3B). Consistently, microtubule-based microtubule nucleation has been observed in the mitotic spindle, in vitro, and in plant epidermal cells, and is postulated to occur in axons and dendrites of cultured mature neurons [39,42,72,73]. In these contexts, the protein complex augmin is thought to mediate nucleation from existing microtubules. Alternatively, microtubule nucleating factors might be directly recruited to non-centrosomal sites, nucleating microtubules, which in turn help transport additional nucleating, stabilizing, and/or anchoring factors (Figure 3C).
Concluding remarks
Although ncMTOCs are likely found in the majority of differentiated cells in vivo, we are just beginning to understand their composition and assembly. Indeed, we still have much to learn about how cells select and activate non-centrosomal sites as MTOCs, and about ncMTOC composition and function. As non-centrosomal microtubules are critically important for cell function, further studies of ncMTOCs will enhance our basic understanding of cell differentiation. Furthermore, MTOC activity is implicated in human disease. Hyperactive MTOC function at the centrosome has been linked to several types of epithelial cancers and is a hallmark of breast tumors [74-76]. Additionally, enhanced microtubule nucleation at the centrosome is linked to invasive cell behavior [77]. Thus, understanding how cells control their microtubule organization might give us new insight into diseases such as cancer.
Acknowledgements
We would like to thank Maria Sallee and James Nelson for helpful discussions and critical feedback on the manuscript. The research in the Feldman lab is supported by a March of Dimes Foundation Basil O'Connor Starter Scholar Research Award and an NIH New Innovator Award [NIH DP2 GM119136-01]. A.S. is supported by a training grant from the NIH [2T32GM007276].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pickett-Heaps JD. The evolution of the mitotic apparatus: an attempt at comparative ultrastructural cytology in dividing plant cells. Cytobios. 1969;1:257–280. [Google Scholar]
- 2.Porter KR. Cytoplasmic Microtubules and Their Functions. CIBA Foundation Symp. In: Wolstenholme GEW, O'Connor MJ, editors. Principles of Biomolecular Organizations. A. Churchill Ltd.; London: 1966. pp. 308–345. [Google Scholar]
- 3.Voter WA, Erickson HP. he kinetics of microtubule assembly. Evidence for a two-stage nucleation mechanism. Journal of Biological Chemistry. 1984;259:10430–10438. [PubMed] [Google Scholar]
- 4.Gould RR, Borisy GG. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. The Journal of Cell Biology. 1977;73:601–615. doi: 10.1083/jcb.73.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodruff JB, Wueseke O, Hyman AA. Pericentriolar material structure and dynamics. Phil. Trans. R. Soc. B. 2014;369:20130459–20130459. doi: 10.1098/rstb.2013.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etienne-Manneville S. Microtubules in cell migration. Ann. Rev. Cell Dev. Biol. 2013;29:471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- 7.Chrétien D, Buendia B, Fuller SD, Karsenti E. Reconstruction of the centrosome cycle from cryoelectron micrographs. Journal of structural biology. 1997;120:117–133. doi: 10.1006/jsbi.1997.3928. [DOI] [PubMed] [Google Scholar]
- 8.Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. Journal of Cell Science. 2000;113:3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- 9.Reed W, Avolio J, Satir P. The cytoskeleton of the apical border of the lateral cells of freshwater mussel gill: structural integration of microtubule and actin filament-based organelles. Journal of Cell Science. 1984;68:1–33. doi: 10.1242/jcs.68.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Sandoz D, Chailley B, Boisvieux Ulrich E, Lemullois M, Laine MC, Bautista Harris G. Organization and functions of cytoskeleton in metazoan ciliated cells. Biology of the Cell. 1988;63:183–193. doi: 10.1016/0248-4900(88)90057-3. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell DR. Eukaryotic Membranes and Cytoskeleton. Springer; New York: 2007. The Evolution of Eukaryotic Cilia and Flagella as Motile and Sensory Organelles. pp. 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clare DK, Magescas J, Piolot T, Dumoux M, Vesque C, Pichard E, Dang T, Duvauchelle B, Poirier F, Delacour D. Basal foot MTOC organizes pillar MTs required for coordination of beating cilia. Nature Communications. 2014;5:4888. doi: 10.1038/ncomms5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman JL, Priess JR. A Role for the Centrosome and PAR-3 in the Hand-Off of MTOC Function during Epithelial Polarization. Current Biology. 2012;22:575–582. doi: 10.1016/j.cub.2012.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brodu V, Baffet AD, Le Droguen P-M, Casanova J, Guichet A. A developmentally regulated two-step process generates a noncentrosomal microtubule network in Drosophila tracheal cells. Developmental Cell. 2010;18:790–801. doi: 10.1016/j.devcel.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 15**.Nashchekin D, Fernandes AR, Johnston DS. Patronin/Shot Cortical Foci Assemble the Noncentrosomal Microtubule Array that Specifies the Drosophila Anterior-Posterior Axis. Developmental Cell. 2016;38:61–72. doi: 10.1016/j.devcel.2016.06.010. [This paper demonstrates that Patronin and the spectraplakin Short stop (Shot) are required to form cortical ncMTOCs in Drosophila oocytes. Patronin/Shot form cortical foci, from which non-centrosomal microtubules grow. As these foci do not colocalize with γ-tubulin, the authors hypothesize that Patronin/Shot foci capture and protect existing microtubule minus ends.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jankovics F, Brunner D. Transiently Reorganized Microtubules Are Essential for Zippering during Dorsal Closure in Drosophila melanogaster. Developmental Cell. 2006;11:375–385. doi: 10.1016/j.devcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Lee C, Scherr HM, Wallingford JB. Shroom family proteins regulate gamma-tubulin distribution and microtubule architecture during epithelial cell shape change. Development. 2007;134:1431–1441. doi: 10.1242/dev.02828. [DOI] [PubMed] [Google Scholar]
- 18.Tucker JB, Paton CC, Richardson GP, Mogensen MM, Russell IJ. A cell surface-associated centrosomal layer of microtubule-organizing material in the inner pillar cell of the mouse cochlea. Journal of Cell Science. 1992;102:215–226. doi: 10.1242/jcs.102.2.215. [DOI] [PubMed] [Google Scholar]
- 19*.Toya M, Kobayashi S, Kawasaki M, Shioi G, Kaneko M, Ishiuchi T, Misaki K, Meng W, Takeichi M. CAMSAP3 orients the apical-to-basal polarity of microtubule arrays in epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 2016;113:332–337. doi: 10.1073/pnas.1520638113. [This study explores a role for CAMSAP3 in vivo. Intestinal epithelial cells in Camsap3 mutant mice have disorganized microtubule architecture, decreased microtubule density, and misplaced organelles.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meads T, Schroer TA. Polarity and nucleation of microtubules in polarized epithelial cells. Cell Motility and the Cytoskeleton. 1995;32:273–288. doi: 10.1002/cm.970320404. [DOI] [PubMed] [Google Scholar]
- 21.Lechler T, Fuchs E. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. The Journal of Cell Biology. 2007;176:147–154. doi: 10.1083/jcb.200609109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Priess JR, Hirsh DI. Caenorhabditis elegans morphogenesis: The role of the cytoskeleton in elongation of the embryo. Developmental Biology. 1986;117:156–173. doi: 10.1016/0012-1606(86)90358-1. [DOI] [PubMed] [Google Scholar]
- 23*.Quintin S, Wang S, Pontabry J, Bender A, Robin F, Hyenne V, Landmann F, Gally C, Oegema K, Labouesse M. Non-centrosomal epidermal microtubules act in parallel to LET-502/ROCK to promote C. elegans elongation. Development. 2016;143:160–173. doi: 10.1242/dev.126615. [This study investigates the role of microtubules in C. elegans embryonic epidermal cells. In these cells, hemidesmosomes and adherens junctions appear to act as ncMTOCs, recruiting NOCA-1 and γ-tubulin, and growing microtubules following depolymerization. Through genetic analyses, the authors find that microtubules appear to act in parallel to LET-502/ROCK to promote elongation of the embryo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose W. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. The Journal of Cell Biology. 2009;186:229–241. doi: 10.1083/jcb.200902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Wang S, Wu D, Quintin S, Green RA, Cheerambathur DK, Ochoa SD, Desai A, Oegema K. NOCA-1 functions with γ-tubulin and in parallel to Patronin to assemble non-centrosomal microtubule arrays in C. elegans. eLife Sciences. 2015;4:e08649. doi: 10.7554/eLife.08649. [Here, the authors characterize the role of NOCA-1, a putative ninein homologue, in C. elegans epidermal and germ cells. NOCA-1 and γ-tubulin function in a pathway parallel to PTRN-1 to control the assembly of non-centrosomal microtubules in the larval and adult epidermis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tassin AM. Fate of microtubule-organizing centers during myogenesis in vitro. The Journal of Cell Biology. 1985;100:35–46. doi: 10.1083/jcb.100.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bugnard E, Zaal KJM, Ralston E. Reorganization of microtubule nucleation during muscle differentiation. Cell Motility and the Cytoskeleton. 2005;60:1–13. doi: 10.1002/cm.20042. [DOI] [PubMed] [Google Scholar]
- 28.Elhanany-Tamir H, Yu YV, Shnayder M, Jain A, Welte M, Volk T. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. The Journal of Cell Biology. 2012;198:833–846. doi: 10.1083/jcb.201204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theurkauf WE, Smiley S, Wong ML, Alberts BM. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development. 1992;115:923–936. doi: 10.1242/dev.115.4.923. [DOI] [PubMed] [Google Scholar]
- 30.Johnston DS. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- 31.Kapitein LC, Hoogenraad CC. Building the Neuronal Microtubule Cytoskeleton. Neuron. 2015;87:492–506. doi: 10.1016/j.neuron.2015.05.046. [DOI] [PubMed] [Google Scholar]
- 32.Baas PW, Deitch JS, Black MM, Banker GA. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. PNAS. 1988;85:8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maniar TA, Kaplan M, Wang GJ, Shen K, Wei L, Shaw JE, Koushika SP, Bargmann CI. UNC-33 (CRMP) and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting. Nat. Neurosci. 2012;15:48–56. doi: 10.1038/nn.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stone MC, Roegiers F, Rolls MM. Microtubules have opposite orientation in axons and dendrites of Drosophila neurons. Mol. Biol. Cell. 2008;19:4122–4129. doi: 10.1091/mbc.E07-10-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ori-McKenney KM, Jan LY, Jan Y-N. Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron. 2012;76:921–930. doi: 10.1016/j.neuron.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen MM, McCracken CJ, Milner ES, Goetschius DJ, Weiner AT, Long MK, Michael NL, Munro S, Rolls MM. γ-tubulin controls neuronal microtubule polarity independently of Golgi outposts. Mol. Biol. Cell. 2014;25:2039–2050. doi: 10.1091/mbc.E13-09-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia ARR, McLeod IX, et al. Asymmetric CLASP-Dependent Nucleation of Noncentrosomal Microtubules at the trans-Golgi Network. Developmental Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noguchi T, Koizumi M, Hayashi S. Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr. Biol. 2011;21:805–814. doi: 10.1016/j.cub.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 39*.Sánchez-Huertas C, Freixo F, Viais R, Lacasa C, Soriano E, Lüders J. Noncentrosomal nucleation mediated by augmin organizes microtubules in post-mitotic neurons and controls axonal microtubule polarity. Nature Communications. 2016;7:12187. doi: 10.1038/ncomms12187. [This paper finds that depletion of γ-TuRC and/or augmin in cultured hippocampal neurons results in decreased microtubule density and bundling and disrupts axonal specification, morphogenesis, and transport. As augmin depletion disrupts microtubule polarity in axons, the authors speculate that, similar to its role in the mitotic spindle, augmin directs polarized growth of new microtubules from existing microtubules in mature axons.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang F. Establishment of a cellular axis in fission yeast. Trends in Genetics. 2001;17:273–278. doi: 10.1016/s0168-9525(01)02279-x. [DOI] [PubMed] [Google Scholar]
- 41.Winey M, O'Toole ET. The spindle cycle in budding yeast. Nature Cell Biology. 2001;3:E23–27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]
- 42.Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, Horio T, Hasebe M. Microtubule-dependent microtubule nucleation based on recruitment of γ-tubulin in higher plants. Nature Cell Biology. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura M, Ehrhardt DW, Hashimoto T. Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nature Cell Biology. 2010;12:1064–1070. doi: 10.1038/ncb2110. [DOI] [PubMed] [Google Scholar]
- 44.Mennella V, Agard DA, Huang B, Pelletier L. Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends in Cell Biology. 2014;24:188–197. doi: 10.1016/j.tcb.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oakley CE, Oakley BR. Identification of γ-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- 46.Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of γ-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- 47.Moritz M, Zheng Y, Alberts BM. Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. The Journal of Cell Biology. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joshi HC, Palacios MJ, McNamara L, Cleveland DW. γ-Tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature. 1992;356:80–83. doi: 10.1038/356080a0. [DOI] [PubMed] [Google Scholar]
- 49.Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 50.Wiese C, Zheng Y. A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nature Cell Biology. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- 51.Rogers GC, Rusan NM, Peifer M, Rogers SL. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol. Biol. Cell. 2008;19:3163–3178. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srayko M, Kaya A, Stamford J, Hyman AA. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Developmental Cell. 2005;9:223–236. doi: 10.1016/j.devcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Hannak E, Oegema K, Kirkham M, Gönczy P, Habermann B, Hyman AA. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. The Journal of Cell Biology. 2002;157:591–602. doi: 10.1083/jcb.200202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Yang R, Feldman JL. SPD-2/CEP192 and CDK Are Limiting for Microtubule-Organizing Center Function at the Centrosome. Current Biology. 2015;25:1924–1931. doi: 10.1016/j.cub.2015.06.001. [This study investigates the mechanism by which a cell selects MTOC location. By fusing cells in vivo in C. elegans embryos, the authors report that a mitotic centrosome MTOC state is dominant to the membrane MTOC state of a polarized epithelia cell. Activation of MTOC function at the centrosome appears to be limited by SPD-2 and CDK.] [DOI] [PubMed] [Google Scholar]
- 55*.Yau KW, van Beuningen SFB, Cunha-Ferreira I, Cloin BMC, van Battum EY, Will L, Schätzle P, Tas RP, van Krugten J, Katrukha EA, et al. Microtubule minus-end binding protein CAMSAP2 controls axon specification and dendrite development. Neuron. 2014;82:1058–1073. doi: 10.1016/j.neuron.2014.04.019. [This paper shows that CAMSAP2 and γ-tubulin are required for non-centrosomal microtubules in mature rat hippocampal neurons. CAMSAP2 and γ-tubulin do not colocalize, suggesting a two-step model in which microtubules are nucleated by γ-tubulin and then stabilized by CAMSAP2.] [DOI] [PubMed] [Google Scholar]
- 56.Akhmanova A, Hoogenraad CC. Microtubule minus-end-targeting proteins. Current Biology. 2015;25:R162–R171. doi: 10.1016/j.cub.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 57.Goodwin SS, Vale RD. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 2010;143:263–274. doi: 10.1016/j.cell.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58*.Jiang K, Hua S, Mohan R, Grigoriev I, Yau KW, Liu Q, Katrukha EA, Altelaar AFM, Heck AJR, Hoogenraad CC, et al. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Developmental Cell. 2014;28:295–309. doi: 10.1016/j.devcel.2014.01.001. [In this study, the authors report that mammalian CAMSAP proteins bind specifically to nascent non-centrosomal microtubule minus ends, stabilize growing minus ends against depolymerization, and can promote microtubule plus end outgrowth. The microtubule severing protein Katanin can bind to CAMSAP proteins and regulate their association with microtubules.] [DOI] [PubMed] [Google Scholar]
- 59.Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135:948–959. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 60.Bouckson-Castaing V, Moudjou M. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. Journal of Cell Science. 1996;109:179–190. doi: 10.1242/jcs.109.1.179. [DOI] [PubMed] [Google Scholar]
- 61.Abal M, Piel M, Bouckson-Castaing V, Mogensen M, Sibarita J-B, Bornens M. Microtubule release from the centrosome in migrating cells. The Journal of Cell Biology. 2002;159:731–737. doi: 10.1083/jcb.200207076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. The Journal of Cell Biology. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. Journal of Cell Science. 2005;118:1565–1575. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- 64.Green RA, Kao H-L, Audhya A, Arur S, Mayers JR, Fridolfsson HN, Schulman M, Schloissnig S, Niessen S, Laband K, et al. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell. 2011;145:470–482. doi: 10.1016/j.cell.2011.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. Journal of Cell Science. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- 66.Stiess M, Maghelli N, Kapitein LC, Gomis-Rüth S, Wilsch-Bräuninger M, Hoogenraad CC, Tolić-Nørrelykke IM, Bradke F. Axon extension occurs independently of centrosomal microtubule nucleation. Science. 2010;327:704–707. doi: 10.1126/science.1182179. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen MM, Stone MC, Rolls MM. Microtubules are organized independently of the centrosome in Drosophila neurons. Neural Dev. 2011;6:38. doi: 10.1186/1749-8104-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu Y, Roy R. Centrosome/Cell cycle uncoupling and elimination in the endoreduplicating intestinal cells of C. elegans. PLoS ONE. 2014;9:e110958. doi: 10.1371/journal.pone.0110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69**.Muroyama A, Seldin L, Lechler T. Divergent regulation of functionally distinct γ-tubulin complexes during differentiation. The Journal of Cell Biology. 2016;213:679–692. doi: 10.1083/jcb.201601099. [This paper reports the existence of distinct γ-TuRC complexes at the centrosome in mouse keratinocytes required for either microtubule nucleation or anchoring. Upon differentiation, centrosomes retain some nucleation capacity through the retention of CDK5RAP2-γ-TuRC, but lose anchoring ability, concomitant with a loss of Nedd1-γ-TuRC complexes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moss DK, Bellett G, Carter JM, Liovic M, Keynton J, Prescott AR, Lane EB, Mogensen MM. Ninein is released from the centrosome and moves bi-directionally along microtubules. Journal of Cell Science. 2007;120:3064–3074. doi: 10.1242/jcs.010322. [DOI] [PubMed] [Google Scholar]
- 71.Achilleos A, Wehman AM, Nance J. PAR-3 mediates the initial clustering and apical localization of junction and polarity proteins during C. elegans intestinal epithelial cell polarization. Development. 2010;137:1833–1842. doi: 10.1242/dev.047647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell. 2013;152:768–777. doi: 10.1016/j.cell.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. The Journal of Cell Biology. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lingle WL, Salisbury JL. Altered Centrosome Structure Is Associated with Abnormal Mitoses in Human Breast Tumors. The American Journal of Pathology. 1999;155:1941–1951. doi: 10.1016/S0002-9440(10)65513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salisbury JL, Lingle WL, White RA. Microtubule nucleating capacity of centrosomes in tissue sections. The Journal of Histochemistry & Cytochemistry. 1999;47:1265–1273. doi: 10.1177/002215549904701006. [DOI] [PubMed] [Google Scholar]
- 76.Pihan GA, Purohit A, Wallace J, Malhotra R, Liotta L, Doxsey SJ. Centrosome defects can account for cellular and genetic changes that characterize prostate cancer progression. Cancer Res. 2001;61:2212–2219. [PubMed] [Google Scholar]
- 77.Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Théry M, Pellman D. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 2014;510:167–171. doi: 10.1038/nature13277. [DOI] [PMC free article] [PubMed] [Google Scholar]