Abstract
Protein phosphorylation regulates a wide variety of cellular processes. Thus, we hypothesize that single nucleotide polymorphisms (SNPs) that may modulate protein phosphorylation could affect osteoporosis risk. Based on a previous conventional genome-wide association (GWA) study, we conducted a three-stage meta-analysis targeting phosphorylation-related SNPs (phosSNPs) for femoral neck (FN)-, total hip (HIP)-, and Lumbar Spine (LS)-BMD phenotypes. In stage 1, 9,593 phosSNPs were meta-analyzed in 11,140 individuals of various ancestries. Genome-wide significance (GWS) and suggestive significance were defined by α = 5.21×10−6 (0.05/9,593) and 1.00×10−4, respectively. In stage 2, 9 stage 1-discovered phosSNPs (based on α = 1.00×10−4) were in silico meta-analyzed in Dutch, Korean, and Australian cohorts. In stage 3, four phosSNPs that replicated in stage 2 (based on α = 5.56×10−3, 0.05/9) were de novo genotyped in two independent cohorts. IDUA rs3755955 and rs6831280, and WNT16 rs2707466 were associated with BMD phenotypes in each respective stage, and in 3 stages combined, achieving GWS for both FN-BMD (P-value = 8.36×10−10, 5.26×10−10, and 3.01×10−10, respectively) and HIP-BMD (P-value = 3.26×10−6, 1.97×10−6, and 1.63×10−12, respectively). Although in vitro studies demonstrated no differences in expressions of wild-type and mutant forms of IDUA and WNT16B proteins, in silico analysis predicts that WNT16 rs2707466 directly abolishes a phosphorylation site, which could cause a deleterious effect on WNT16 protein, and that IDUA phosSNPs rs3755955 and rs6831280 could exert indirect effects on nearby phosphorylation sites. Further studies will be required to determine the detailed and specific molecular effects of these BMD-associated non-synonymous variants.
Keywords: osteoporosis, human association studies, single nucleotide polymorphism, meta-analysis, Wnt/Beta-catenin/LRPs
INTRODUCTION
Osteoporosis, a complex disease characterized by reduced bone mass, results in microarchitectural deterioration of bone tissue, and increased bone fragility and susceptibility to fracture.(1) It has been estimated that the prevalence of osteoporosis in the United States will increase to >14 million people in 2020,(2) and by 2025 it is projected that there will be >3 million fractures/year in the U.S., costing $25.3 billion annually.(3) A diagnosis of osteoporosis for both males and females is attained when bone mineral density (BMD) is 2.5 standard deviations or more below the young adult mean.(4) BMD, a highly heritable polygenic trait, is the best predictor for skeletal fragility.(5)
Protein phosphorylation represents the most widespread post-translational modification (PTM), which plays a critical role in essential cellular processes, e.g., metabolism, cell signaling, differentiation, and membrane transportation.(6) Large-scale phosphoproteomics studies suggest that more than half of all eukaryotic proteins are phosphorylated.(7) The most common phosphorylation sites in eukaryotes are serine (S), threonine (T), and tyrosine (Y) residues,(8) which are catalyzed by S/T-specific, Y-specific, and dual-specificity protein kinases.(9) Single nucleotide polymorphisms (SNPs) constitute almost 90% of genetic variations in the human genome.(10) Non-synonymous SNPs (nsSNPs), defined as SNPs resulting in amino acid changes that include either missense or nonsense mutations,(11) represent 60% of known disease-causing mutations.(12) Of them, those that create/alter/abolish phosphorylation sites, called phosphorylation-related SNPs (phosSNPs), have been recognized as functional variants for a spectrum of human diseases, e.g., lung cancer (CSF1R rs10079250),(13) prostate cancer (TP53 rs1042522),(13,14) long QT syndrome (KCNH2 rs1805123),(15,16) systemic lupus erythematosus (VEGR2 rs2305948),(17) and tuberculosis (TLR2 rs5743708).(18,19) Each phosphorylation site consists of an acceptor residue surrounded by an evolutionarily conserved motif consisting of 7–12 amino acid residues on either flanking region. Based on the hypothesis that a sequence motif surrounding an acceptor residue represents a main determinant of protein kinase specificity, phosphorylation sites can be predicted in silico, and nsSNPs affecting such sites can be identified. From 91,797 nsSNPs of NCBI dbSNP Build 130, by applying the Group-based Phosphorylation Scoring (GPS) 2.0 program (a kinase-specific phosphorylation site predictor)(20), Ren et al. (2010)(21) identified 64,035 phosSNPs residing in 17,614 human proteins, which were categorized into five distinct types: I, II, III, IV and V based on the different effects they exert on phosphorylation sites.
Among at least 60 loci identified by > 40 previous genome-wide association (GWA) studies and meta-analyses of these studies for osteoporosis, WNT16 locus has been found to be an important genetic determinant of osteoporosis risk.(22) The human WNT16 gene spans ~16 kb from initiation to termination codons, encoding two protein isoforms — WNT16A (40.5 kD) and WNT16B (40.7 kD).(23) As depicted in Supplementary Figure S1, these two WNT16 isoforms have different first exons (i.e., 1a and 1b, respectively), independently controlled by two alternative promoters P1 and P2, respectively.(24) Expression of the WNT16A isoform has been shown to be restricted to the pancreas in humans, whereas WNT16B is expressed in multiple organs.(24) Compared to WNT16A, the role of WNT16B as a key regulator of osteoclastogenesis has been more extensively characterized.(25)
Meta-analyses of GWA studies have significant potentials for detecting subtle genetic effects.(26) However, because conventional GWA studies often include a large number of variants of unknown functional effects, the significance threshold attained by Bonferroni correction becomes overly conservative, producing a high rate of type II error (i.e., β). PhosSNPs are more likely disruptive to protein function than other protein-coding missense mutations.(27) However, such potentially causal missense mutations could be missed by conventional GWA approaches, due to very strict control for type I error (i.e., α). Power to detect disease-causing variants can thus be increased by focusing exclusively on SNPs with higher prior probabilities of functional effects, either as in a whole-exome sequencing(28) approach targeting solely exonic SNPs, or as we apply here, targeting exclusively potentially functional phosSNPs. However, such a functional candidate genomic region approach(29) could be susceptible to a higher rate of false positive results.(30) Therefore, to guard against an inflated α, we employed a three-stage approach, such that those phosSNPs attaining genome-wide significance (GWS) in stage 1 (i.e., GWA discovery) are required to be replicated in independent cohorts of stages 2 and 3, respectively, based on their corresponding Bonferroni-corrected α thresholds.
MATERIALS AND METHODS
A detailed description of study participants, phenotype measurement and modeling, DNA genotyping, quality control (QC), and genotype imputation, association tests, meta-analysis methods, and regional association plots of the three-stage GWA meta-analysis is given in Supplementary Materials and Methods. At stage 1, seven GWA cohorts were included, and a suggestive significance threshold of α = 1.00×10−4 was applied for phosSNP selection. At stage 2 (in silico replication), three GWA cohorts were included, and at stage 3 (de novo genotyping replication), two independent cohorts were included, and at each stage, a Bonferroni-corrected significance threshold was applied.
PhosSNPs in Potential Phosphorylation Sites
The phosSNP-centric GWA meta-analysis focuses exclusively on 9,593 phosSNPs in stage 1 of the conventional GWA meta-analysis.(31) Details about phosSNP selection are given in the Supplementary Materials and Methods.
In Silico Bioinformatics Analyses
(1) Computational Predictions of Phosphorylation Sites Affected by phosSNPs
Phosphorylation sites that could be affected by the three significant phosSNPs — IDUA rs3755955 (R105Q) and rs6831280 (A361T), and WNT16 rs2707466 (WNT16B T263I), were predicted by two commonly used online software programs: NetPhos2.0(32) and NetPhosK1.0.(33) Details about these programs are given in the Supplementary Materials and Methods.
(2) Computational Predictions of Functional Impacts of phosSNPs
Functional effects of the three significant phosSNPs — IDUA rs3755955 (R105Q) and rs6831280 (A361T), and WNT16 rs2707466 (WNT16B T263I) were computed using four online software tools: (i) Mutation Assessor,(34) (ii) BLOSUM62,(35) (iii) PMut,(36) and (iv) PANTHER.(37) Details about these tools are given in the Supplementary Materials and Methods.
(3) Computational Prediction of Protein Secondary and Tertiary Structures
Protein secondary and tertiary structures were predicted by Protein Homology/analogy Recognition Engine Version 2.0 (Phyre2).(38,39) The Phyre2 server predicts a protein’s secondary structure based on the amino acid sequence. In brief, this program converts a protein sequence into a hidden Markov model (HMM) based on sequence homologs retrieved from experimentally determined known protein structures using PSI-Blast.(40) The HMM of the query sequence is then scanned against a non-redundant library of HMMs of proteins with experimentally determined structures. The 3D model of the query sequence is then constructed on the basis of alignments between the HMM of query sequence and the HMMs of known structures. Phyre2 program can generate highly accurate models at low sequence identities (e.g., 15–25%).(39)
In Vitro Protein Expression Studies
To assess whether mutant (MUT) alleles of respective phosSNPs, i.e., IDUA rs3755955, rs6831280, and WNT16 rs2707466, could affect protein expression levels in vitro, we designed and constructed plasmid pcDNA3.1-Myc/His vectors harboring either wild-type (WT) or MUT allele of each phosSNP and transfected each of them into Chinese hamster ovary (CHO) cells. Details about cloning and transfection and Western blot analysis are given in the Supplementary Materials and Methods.
RESULTS
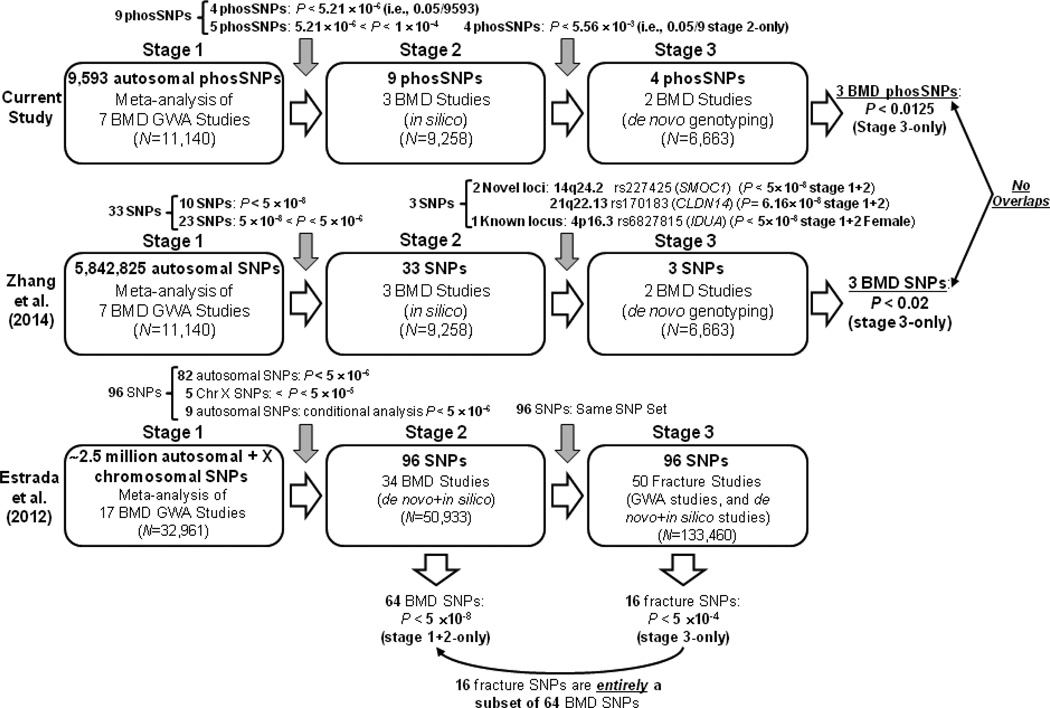
Cohort characteristics at three stages were presented in Supplementary Table S1. A detailed comparison of study designs of current study with those of two previous conventional GWA meta-analysis studies(31,41) is shown in Figure 1. In stage 1, current study restricted association tests to exclusively phosSNPs (~10K), as opposed to the entire set of genotyped and imputed SNPs (~5800K) of previous conventional study.(31) As a result, different SNP sets were selected from stage 1 for stage 2 in silico replication [9 phosSNPs for current study, and none overlapped with 33 SNPs of previous conventional study(31)]. In stage 2, different SNP selection criteria were employed between current study and previous conventional study.(31). Four stage 2-selected phosSNPs (i.e., IDUA rs3755955 and rs6831280, WNT16 rs2707466, and ESPL1 rs56358776) of current study were entirely different from those three stage 2-selected SNPs of previous conventional study(31) (i.e., SMOC1 rs227425, CLDN14 rs170183, and intergenic SNP rs6827815).
FIGURE 1.
Diagrammatic representations of study designs of three-stage GWA meta-analysis of current study (top panel), Zhang et al. (2014)(31) (middle panel), and Estrada et al. (2012)(41) (bottom panel)
Stage 1 (GWA Discovery)
Table 1 presents a comparison of 33 SNPs selected in stage 1 of previous conventional study,(31) with those 9 phosSNPs selected in stage 1 of current study, which include four phosSNPs (located in three gene regions) attaining phosSNP-centric GWS (i.e., α = 0.05/9,593 = 5.21×10−6) — IBSP rs1054627 for FN-BMD in female-specific sample, IDUA rs6831280 and rs3755955 for FN-BMD in gender-combined (i.e., male and female) sample, and WNT16 rs2707466 for HIP-BMD in gender-combined sample, and another five phosSNPs (located in four gene regions) attaining only suggestive significance (i.e., α = 1.00×10−4) — SRMS rs310655 for FN-BMD in gender-combined sample, DNAH8 rs61748601 for HIP-BMD in gender-combined sample, ESPL1 rs56358776 and rs1318648 for LS-BMD in gender-combined and female-specific samples, respectively, and GPATCH1 rs2287679 for FN-BMD in female-specific sample.
Table 1.
Comparison of 33 SNPs (previous conventional study(31)) and 9 phosSNPs (current study) selected in stage 1*
| Stage 1 of Zhang et al. (2014)(31) | Stage 1 of Current Study | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Locus | Gene | Phenotype | Analysis | P-value | SNP ID | Locus | Gene | Phenotype | Analysis | P-value |
| rs34920465 | 1p36.12 | ZBTB40 | HIP-BMD | Combined | 3.06×10−9 | --- | --- | --- | --- | --- | --- |
| rs1430740 | 1p31.3 | MIR1262 | LS-BMD | Combined | 8.46×10−9 | --- | --- | --- | --- | --- | --- |
| rs11582394 | 1q32.1 | PLEKHA6 | FN-BMD | Combined | 7.14×10−7 | --- | --- | --- | --- | --- | --- |
| rs11696050 | 2q34 | PTH2R | HIP-BMD | Combined | 3.72×10−8 | --- | --- | --- | --- | --- | --- |
| rs11130082 | 3p21.31 | FYCO1 | LS-BMD | Combined | 9.73×10−7 | --- | --- | --- | --- | --- | --- |
| rs6827815 | 4p16.3 | FGFRL1 | FN-BMD | Combined | 6.01×10−7 | rs6831280 | 4p16.3 | IDUA | FN-BMD | Combined | 1.21×10−6 |
| rs3755955 | 4p16.3 | IDUA | FN-BMD | Combined | 1.80×10−6 | ||||||
| rs17813558 | 4p16.1 | PSAPL1 | FN-BMD | Combined | 9.19×10−7 | --- | --- | --- | --- | --- | --- |
| rs4974930 | 4p14 | WDR19 | HIP-BMD | Combined | 6.20×10−7 | --- | --- | --- | --- | --- | --- |
| rs1463104 | 4q22.1 | HSP90AB3P | LS-BMD | Combined | 7.08×10−7 | rs1054627 | 4q22.1 | IBSP | FN-BMD | Female | 3.54×10−7 |
| rs4703541 | 5q13.2 | ZNF366 | FN-BMD | Combined | 6.52×10−7 | --- | --- | --- | --- | --- | --- |
| rs6894139 | 5q14.3 | MEF2C | FN-BMD | Combined | 2.02×10−9 | --- | --- | --- | --- | --- | --- |
| --- | --- | --- | --- | --- | --- | rs61748601 | 6p21.2 | DNAH8 | HIP-BMD | Combined | 4.83×10−5 |
| rs1871859 | 6q25.1 | C6orf97 | LS-BMD | Female | 5.04×10− | --- | --- | --- | --- | --- | --- |
| rs28529426 | 7p22.1 | FOXK1 | LS-BMD | Combined | 8.12×10−7 | --- | --- | --- | --- | --- | --- |
| rs2529750 | 7p21.1 | PRPS1L1 | LS-BMD | Female | 7.04×10−7 | --- | --- | --- | --- | --- | --- |
| rs10429035 | 7q21.3 | FLJ42280 | HIP-BMD | Combined | 4.24×10−9 | --- | --- | --- | --- | --- | --- |
| rs13242054 | 7q22.3 | PIK3CG | LS-BMD | Combined | 5.83×10−7 | --- | --- | --- | --- | --- | --- |
| rs10242100 | 7q31.31 | WNT16 | HIP-BMD | Combined | 4.63×10−8 | rs2707466 | 7q31.31 | WNT16 | HIP-BMD | Combined | 1.77×10−6 |
| chr8:78734104 | 8q21.12 | PKIA | LS-BMD | Combined | 1.98×10−7 | --- | --- | --- | --- | --- | --- |
| rs4424296 | 8q24.12 | TNFRSF11B | LS-BMD | Combined | 5.94×10−10 | --- | --- | --- | --- | --- | --- |
| rs10868819 | 9q21.12 | KLF9 | FN-BMD | Combined | 6.31×10−7 | --- | --- | --- | --- | --- | --- |
| chr9:87907046 | 9q21.33 | AGTPBP1 | HIP-BMD | Male | 1.88×10−7 | --- | --- | --- | --- | --- | --- |
| rs7025969 | 9q31.3 | ACTL7B | FN-BMD | Female | 7.77×10−8 | --- | --- | --- | --- | --- | --- |
| rs7108738 | 11p15.1 | SOX6 | FN-BMD | Combined | 6.73×10−9 | --- | --- | --- | --- | --- | --- |
| rs4267051 | 11p15.1 | HPS5 | HIP-BMD | Female | 7.97×10−7 | ||||||
| rs525592 | 11q13.2 | LRP5 | LS-BMD | Combined | 9.04×10−7 | --- | --- | --- | --- | --- | --- |
| rs471753 | 11q14.2 | TMEM135 | FN-BMD | Combined | 2.20×10−7 | --- | --- | --- | --- | --- | --- |
| --- | --- | --- | --- | --- | --- | rs1318648 | 12q13.13 | ESPL1 | FN-BMD | Female | 9.87×10−5 |
| rs56358776 | 12q13.13 | ESPL1 | FN-BMD | Combined | 5.36×10−5 | ||||||
| rs9533095 | 13q14.11 | AKAP11 | LS-BMD | Female | 3.96×10−7 | --- | --- | --- | --- | --- | --- |
| rs227425 | 14q24.2 | SMOC1 | LS-BMD | Combined | 2.69×10−7 | --- | --- | --- | --- | --- | --- |
| rs11848357 | 14q32.11 | RPS6KA5 | LS-BMD | Combined | 3.29×10−7 | --- | --- | --- | --- | --- | --- |
| chr16:86714715 | 16q24.1 | FOXL1 | HIP-BMD | Female | 1.86×10−9 | --- | --- | --- | --- | --- | --- |
| rs10756 | 19q12 | C19orf2 | FN-BMD | Combined | 7.60×10−7 | --- | --- | --- | --- | --- | --- |
| --- | --- | --- | --- | --- | --- | rs2287679 | 19q13.11 | GPATCH1 | FN-BMD | Female | 6.59×10−5 |
| rs12481249 | 20q13.33 | OSBPL2 | LS-BMD | Combined | 1.20×10−7 | rs310655 | 20q13.33 | SRMS | FN-BMD | Combined | 3.57×10−5 |
| rs170183 | 21q22.13 | CLDN14 | HIP-BMD | Female | 3.71×10−7 | --- | --- | --- | --- | --- | --- |
Abbreviations: BMD, bone mineral density; FN, femoral neck; HIP, total hip; LS, lumbar spine; phosSNP, phosphorylation-related SNP; SNP, single nucleotide polymorphism. Combined refers to Male and Female. Those SNPs that have attained the conventional genome-wide significance (GWS) (i.e., α = 5.00×10−8) were highlighted in bold font. Shaded regions were matched regions between these two studies, although different SNPs were identified (Note: a region could contain more than one SNP).
Stage 2 (in silico Replication)
In stage 2, the above 9 stage 1-discovered phosSNPs were subject to replication in three in silico independent cohorts. A meta-analysis within stage 2 revealed 6 phosSNPs at Bonferroni corrected α = 5.56×10−3 (i.e., 0.05/9): WNT16 rs2707466 for FN-BMD in gender-combined sample, IBSP rs1054627 for FN-BMD in gender-combined sample, ESPL1 rs1318648 and rs56358776 for LS-BMD in gender-combined sample, and IDUA rs3755955 and rs6831280 for FN-BMD in gender-combined sample. Of these, IBSP encodes a well-known bone matrix protein that is important for bone mineralization(42–44) which, consequently, was not further tested in stage 3. For ESPL1 phosSNPs rs1318648 and rs56358776, neither reached GWS (i.e., 5.21×10−6 ≤ P-values < 1.00×10−4) in stage 1 phosSNP-centric GWA meta-analysis. ESPL1 rs1318648 is a previously known nsSNP suggestively associated with FN- and LS-BMD phenotypes(41), whereas ESPL1 rs56358776 is a novel nsSNP that was not reported in either of two previous conventional studies(41,45), which is in high linkage disequilibrium (LD) with ESPL1 rs1318648 [r2 = 0.798 in 1000 Genomes (1KG) Pilot 1 CEU Population by applying the SNP Annotation and Proxy search (SNAP) tool(46) of Broad Institute]. Therefore, we selected 4 stage 2-replicated phosSNPs — IDUA rs6831280 and rs3755955, WNT16 rs2707466, and potentially novel phosSNP ESPL1 rs56358776, for stage 3 de novo genotyping replication.
Stage 3 (de novo Genotyping Replication)
In stage 3, the above four stage 2-selected phosSNPs identified were subject to further replication by de novo genotyping. Three of these phosSNPs were replicated by stage 3-specific meta-analysis at Bonferroni corrected α = 0.0125 (i.e., 0.05/4). WNT16 rs2707466 was consistently replicated for HIP-, FN-, and LS-BMD phenotypes in gender-combined sample. IDUA rs3755955 and rs6831280 were significantly associated with FN- and HIP-BMD phenotypes in gender-combined sample. ESPL1 rs56358776 was not replicated at this stage (P-value = 0.79, 0.78, and 0.32 in gender-combined sample for FN-, HIP-, and LS-BMD, respectively).
Stage 1+2+3 Meta-analysis
Table 2 presents ethnicity-specific and combined meta-analysis results aggregating these 3 stages for stage 1-discovered (α = 1.00×10−4), and stage 2- and 3-replicated (Bonferroni-corrected α = 5.56×10−3 and 0.0125, respectively) phosSNPs — IDUA rs6831280 (A361T), IDUA rs3755955 (R105Q), and WNT16 rs2707466 (WNT16B T263I), respectively. In ethnicity-specific meta-analyses, in Caucasians, all three attained phosSNP-centric GWS (i.e., α = 5.21×10−6) for FN-BMD and only WNT16 rs2707466 attained this threshold for HIP-BMD and in Asians, only WNT16 rs2707466 attained phosSNP-centric GWS for HIP-BMD. The effects of these phosSNPs were consistent between Caucasian and Asian ethnicities. In combined meta-analysis across 3 stages, IDUA rs3755955 was significantly associated with FN- and HIP-BMD phenotypes (P-value = 8.36×10−10 and 3.26×10−6, respectively). Likewise, IDUA rs6831280 was significantly associated with FN- and HIP-BMD phenotypes (P-value = 5.26×10−10 and 1.97×10−6, respectively). Similarly, WNT16 rs2707466 was significantly associated with FN- and HIP-BMD phenotypes (P-value = 3.01×10−10 and 1.63×10−12, respectively). Regional association plots were generated for these three significant phosSNPs — IDUA rs3755955 and rs6831280 (Figure 2), and WNT16 rs2707466 (Figure 3).
Table 2.
GWA Meta-analysis Results of Stage 1+2+3 for BMD-Associated PhosSNPs in Ethnicity-Specific and Combined Cohorts*
| Gene Symbol | SNP ID (WT/MUT Alleles; AA Change) |
PhosSNP Type | Phenotype | ||
|---|---|---|---|---|---|
| FN-BMD | HIP-BMD | LS-BMD | |||
| Caucasian (7 Cohorts) | |||||
| IDUA | rs3755955 (G/A; R105Q) | Type II(+) | 2.38×10−8 | 2.34×10−4 | 0.0188 |
| IDUA | rs6831280 (G/A; A361T) | Type III | 1.98×10−8 | 1.83×10−6 | 0.0366 |
| WNT16 | rs2707466 (C/T; T263I**) | Type I(−) | 1.13×10−7 | 4.54×10−7 | 0.0109 |
| Asian (3 Cohorts) | |||||
| IDUA | rs3755955 (G/A; R105Q) | Type II(+) | 0.03 | 0.0106 | 0.263 |
| IDUA | rs6831280 (G/A; A361T) | Type III | 0.033 | 0.0117 | 0.289 |
| WNT16 | rs2707466 (C/T; T263I**) | Type I(−) | 1.20×10−3 | 1.04×10−6 | 6.67×10−3 |
| Total (12 Cohorts) | |||||
| IDUA | rs3755955 (G/A; R105Q) | Type II(+) | 8.36×10−10 | 3.26×10−6 | 8.50×10−3 |
| IDUA | rs6831280 (G/A; A361T) | Type III | 5.26×10−10 | 1.97×10−6 | 0.0147 |
| WNT16 | rs2707466 (C/T; T263I**) | Type I(−) | 3.01×10−10 | 1.63×10−12 | 1.17×10−4 |
Abbreviations: AA, amino acid; BMD, bone mineral density; FN, femoral neck; HIP, total hip; GWS, genome-wide significance, i.e., P-value < 5.21×10−6; LS, lumbar spine; phosSNP, phosphorylation-related SNP; SNP, single nucleotide polymorphism. PhosSNPs attaining GWS level are highlighted in bold font. Combined refers to Male and Female. PhosSNP types are defined as follows: Type I (−), a non-synonymous (nsSNP) that removes the phosphorylation site; Type II (+), an nsSNP that creates one or multiple adjacent phosphorylation sites; Type III, an nsSNP that induces changes of protein kinase type(s) at adjacent phosphorylation site(s) as defined in Ren et al. (2010),(21) which were predicted by GPS2.0 software.(20)
The amino acid position at WNT16B protein isoform (SWISS-PROT ID: Q9UBV4-1) is indicated.
FIGURE 2.
Regional association plots for chromosome 4p16.3 loci IDUA Exon 3 phosSNP rs3755955 (R105Q), Exon 8 phosSNP rs6831280 (A361T), intergenic SNP rs6827815, and FGFRL1 3’-untranslated region SNP rs4647940 based on RefSeq accession number NG_008103.1 for FN-BMD (most significant phenotype). (A) IDUA rs6831280 with flanking ± 100-kb (B) a zoomed-in view of the center region [indicated by the dashed box of (A)] — IDUA rs6831280 with flanking ± 40-kb. PhosSNPs were highlighted in bond font
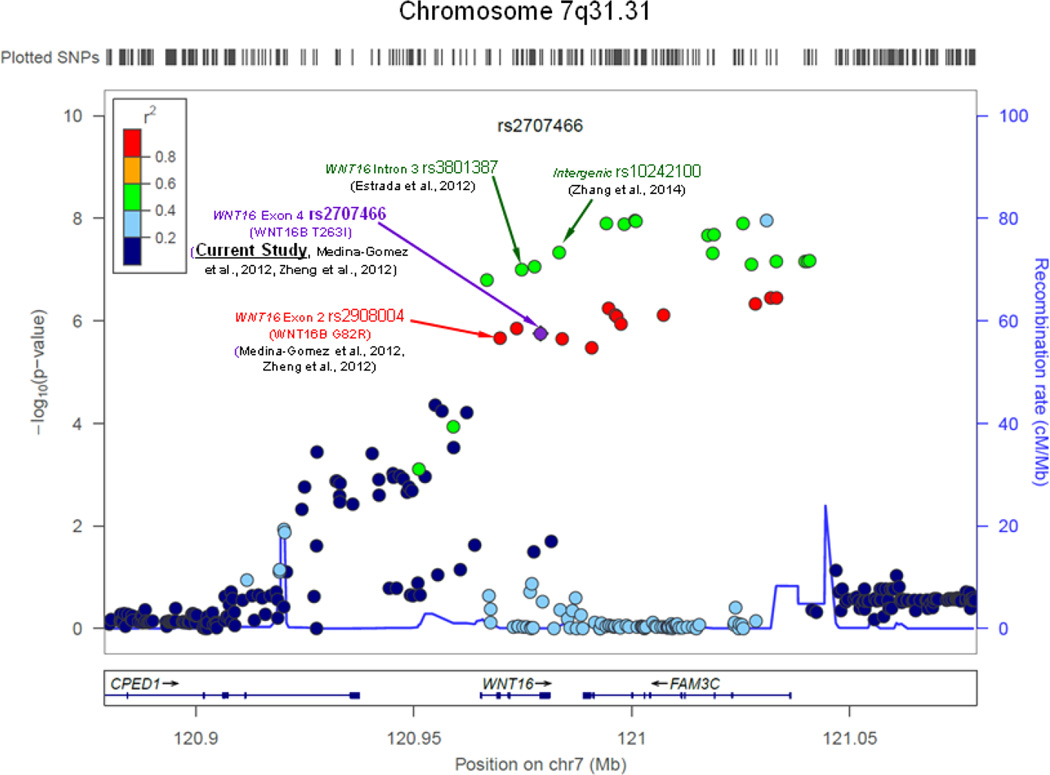
FIGURE 3.
Regional association plot for WNT16 rs2707466 with flanking ± 100-kb for HIP-BMD (most significant phenotype), with chromosome 7q31.31 WNT16 Exon 2 nsSNP rs2908004 (WNT16B, G82R), Intron 3 SNP rs3801387, Exon 4 phosSNP rs2707466 (WNT16B T263I), and intergenic SNP rs10242100 based on RefSeq accession number NG_029242.1 indicated. The phosSNP was highlighted in bond font
Phosphorylation Sites Predicted to be Affected by IDUA and WNT16 PhosSNPs
Based on predictions by NetPhos2.0 and NetPhosK1.0, four phosphorylation sites (have either a NetPhos2.0 score > 0.5 or a NetPhosK1.0 score > 0.5) were predicted by these two in silico bioinformatics tools that could be affected by these three BMD-associated phosSNPs (Table 3). Detailed information on 96 and 54 predicted phosphorylation sites for IDUA and WNT16B were presented in Supplementary Tables S2 and S3, respectively. IDUA phosphorylation sites T98 and S102 were potentially affected by their neighboring phoSNP IDUA rs3755955 (R105Q), whereas IDUA phosphorylation site T366 was potentially affected by a neighboring phoSNP IDUA rs6831280 (A361T). WNT16B phosphorylation site T263 was potentially directly abolished by phoSNP WNT16 rs2707466 (WNT16B T263I). Of them, WNT16B T263 (affected by WNT16 rs2707466) has been experimentally validated to be phosphorylated in vivo,(47) whereas IDUA T98 and S102 (potentially affected by IDUA rs3755955) and T366 (potentially affected by IDUA rs6831280) have yet not been experimentally confirmed.
Table 3.
In Silico Predicted Phosphorylation Sites of Three PhosSNPs Associated with BMD Phenotypes*
| Gene Symbol |
Predicted Phosphorylation Site [represented by PSP(7,7)] |
PhosSNP ID (WT/MUT Alleles; AA Change) |
PhosSNP Type | NetPhos2.0 Score (prediction) |
NetPhosK1.0 Score (prediction) |
|---|---|---|---|---|---|
| IDUA |
HWLLELVTTRGSTGQ AA Pos: 98 |
rs3755955 (G/A; R105Q) | Type II(+) | 0.417 (Probable) | 0.68 (Yes) |
| IDUA |
ELVTTRGSTGQGLSY AA Pos: 102 |
rs3755955 (G/A; R105Q) | Type II(+) | 0.987 (Yes) | 0.51 (Yes) |
| IDUA |
PFTQRTLTARFQVNN AA Pos: 366 |
rs6831280 (G/A; A361T) | Type III | 0.600 (Yes) | 0.51 (Yes) |
| WNT16 |
SIQISDKIKRKMRRR AA Pos: 263** |
rs2707466 (C/T; T263I**) | Type I(−) | 0.833 (Yes) | 0.86 (Yes) |
Abbreviations: AA, amino acid; BMD, bone mineral density; NetPhos, Neural Network Phosphorylation Predictor; NetPhosK, Neural Network Phosphorylation Kinase-Specific Predictor; phosSNP, phosphorylation-related SNP. Pos, position; PSP, phosphorylation site peptide; SNP, single nucleotide polymorphism; WT, wild-type. PhosSNP types were defined as in footnote of Table 1. The phosphorylation acceptor residue is underlined, and the phosSNP site (mutant allele shown) is highlighted in bold and italic font. Predicted phosphorylation sites for respective phosSNPs have either a NetPhos2.0 score > 0.5 or a NetPhosK1.0 score > 0.5. GPS2.0 scores were directly extracted from PhosSNP1.0 database,(21) where these scores were greater than their respective thresholds ― 3.26, 2.85, and 2.43 for IDUA protein (SWISS-PROT ID: P35475) positions 98, 102 and 366, respectively, and 4.48 for WNT16B protein (SWISS-PROT ID: Q9UBV4-1) position 263. For NetPhos2.0 and NetPhosK1.0 scores, “Yes”, “Probable” and “No” refer to a score > 0.5, 0.1―0.5, and < 0.1, respectively.
The amino acid position at WNT16B protein isoform (SWISS-PROT ID: Q9UBV4-1) is indicated.
Predicted Functional Impacts of IDUA and WNT16 PhosSNPs
As shown in Supplementary Table S4, although IDUA rs6831280 (A361T) and rs3755955 (R105Q) were predicted to have no (Mutation Assessor and BLOSUM62 scores) or low (PMut and PANTHER scores) functional impacts, WNT16 rs2707466 (WNT16B T263I) showed highest Mutation Assessor score (0.705; nearly reaching a “low impact” threshold 0.80), lowest BLOSUM62 score of (−1.00; indicative of “evolutionarily less acceptable”), highest PMut pathogenicity score (0.3099; indicative of a “moderate pathogenicity”), and lowest PANTHER subSPEC score (−1.92476; indicative of a deleterious effect corresponding to a highest deleteriousness probability Pdeleterious = 0.25441). Further, evolutionary analysis by multiple sequence alignment method revealed that a 27-amino-acid peptide (−14 — +12) surrounding the T263 phosphorylation site is conserved across three mammalian species — human, mouse and rat (Supplementary Figure S1), supporting a likely functional significance of this phosSNP. Based on these bioinformatics prediction results, we further assessed the potential impact of WNT16 rs2707466 (WNT16B T263I) on WNT16B secondary and tertiary structures.
Predicted Secondary and Tertiary Structures of WT and MUT alleles for WNT16 PhosSNP
The secondary and tertiary structures of protein isoforms encoded by WT and MUT alleles for WNT16 rs2707466 (WNT16B T263I) predicted by Phyre2 server are presented in Supplementary Figures S2 and S3, respectively. With respect to secondary structures, this phosSNP (i.e., T263 residue) is located in a disordered region (indicated by a tract of “?” symbols) typical for a phosphorylation site,(48) downstream of a predicted β-strand (SIQISDK) for either isoform (Supplementary Figure S2) with potential functional effects (Supplementary Table S4). A comparison of the local 3D structures between WT and MUT isoforms near the T263 residue clearly shows different spatial patterns (Supplementary Figure S3, dashed boxes).
Effects of IDUA and WNT16 PhosSNPs on Protein Stability
In CHO cells, Western blot results showed that, at the protein level, IDUA rs6831280 (A361T) and rs3755955 (R105Q) MUT alleles were expressed at equivalent levels compared with the IDUA WT allele (Panel As for Supplementary Figures S4 and S5, respectively). The WNT rs2707466 (WNT16B T263I) MUT allele was also expressed at equivalent levels compared with the WNT16 WT allele (Panel B for Supplementary Figures S4 and S5, respectively). Overall, the protein expression of the MUT allele is equivalent to that of the WT allele for each of these three phosSNPs, suggesting that their influences of protein phosphorylations could be important, rather than on expression levels per se.
DISCUSSION
In human genome, nsSNPs account for 60% of mutations that cause diseases.(12) However, not all nsSNPs lead to a functional impact. Therefore, it is essential to select only those nsSNPs that are most plausible causal variants. Our study is unique in associating phosSNPs affecting the most common type of PTM with BMD phenotypes by taking a three-stage approach to protect against an inflated false positive rate. Beyond detecting genetic association, we also performed in silico and in vitro functional characterizations of identified significant nsSNPs. At stage 1, four chromosomal loci i.e., 4p16.3 (IBSP), 4q22.1 (IDUA), 7q31.31 (WNT16), and 20q13.33 (GPATCH1), were detected by both the current and conventional studies,(31) but were represented by totally different SNPs, and for 4q22.1 and 20q13.33, represented by different genes. At 7q31.31, the previous study detected association with intergenic SNP rs10242100 (with no apparent functional significance) near WNT16 gene, which is in moderate LD with the WNT16 SNP rs2707466 detected by our current study [r2 = 0.462 in 1KG Pilot 1 CEU Population by applying SNAP tool(46)]. Overall, three phosSNPs (IDUA rs6831280 and rs3755955, and WNT16 rs2707466), were discovered in stage 1 and were independently replicated in stages 2 and 3, respectively. In ethnicity-specific meta-analyses, their effects were consistent in subgroups of Caucasian and Asian ancestries, and statistical significances were greater in Caucasian than in Asian samples in part because of a larger Caucasian sample size (Table 2). In combined stage 1+2+3 meta-analysis, all three phosSNPs reached conventional GWS for FN-BMD, and WNT16 rs2707466 attained conventional GWS for HIP-BMD also. By applying NetPhos2.0 and NetPhosK1.0, 96 and 54 predicted phosphorylation sites in IDUA and WNT16B proteins, respectively (Supplementary Tables S3 and S4). IDUA encodes a glycosyl hydrolase that hydrolyzes the terminal alpha-L-iduronic acid residues of two glycosaminoglycans, dermatan sulfate and heparan sulfate(49). Wang et al. (2010)(50) created Idua-W392X mouse model, and found that 35-week-old homozygous Idua-W392X mice showed a 24% increase in femur BMD, and bone abnormalities such as thickening of the zygomatic arch and aberrations in the length and width of the femur were also observed(50).
For IDUA protein, a predicted phosphorylation site T366 could be indirectly affected by IDUA rs6831280 (A361T), a Type III phosSNP, and two predicted phosphorylation sites T98 and S102, could be indirectly affected by IDUA rs3755955 (R105Q), a Type II(+) phosSNP (Table 3). For WNT16B protein, phosphorylation site T263 could be directly abolished by WNT16 rs2707466 (WNT16B T263I), a Type I(−) phosSNP. Of them, only WNT16B T263 has been experimentally validated to be a genuine phosphorylation site by mass spectrometry technology in a phosphoproteomic analysis of human embryonic stem cells in vivo.(47) Whether IDUA T98 and S102, and T366 are actual phosphorylation sites influenced by nearby IDUA phosSNPs rs3755955 (R105Q) and rs6831280 (A361T) remain to be experimentally determined.
WNT16 encodes a member of the wingless-type MMTV integration site family, which has been reported to mediate signaling via both canonical and non-canonical Wnt pathways. Wnt proteins are known to play important roles in vertebrate skeletal development.(51–53) Wnt16 is expressed in osteoid tissue of craniofacial bones during embryonic development in mice, and suppresses osteoblast differentiation through the canonical β-catenin pathway in MC3T3-E1 preosteoblasts.(54) Several GWA meta-analysis studies have demonstrated that WNT16 intron 3 SNP rs3801387,(41) exon 2 rs2908004 (WNT16B G82R) and exon 4 rs2707466 (WNT16B T263I)(55,56) as well as intergenic SNP rs10242100(31) are associated with BMD phenotypes (Figure 3). However, functional roles of non-coding SNPs rs10242100 and rs3801387, which are in almost perfect LD [r2 = 0.915 in 1KG Pilot 1 CEU Population by applying SNAP tool(46)], remain unclear. WNT16 exon 2 rs2908004 (WNT16B G82R) and exon 4 rs2707466 (WNT16B T263I) are shown to be in nearly complete LD [r2 = 0.933 in 1KG Pilot 1 CEU Population by applying SNAP tool(46)], which could represent the same phosphorylation association signal (i.e., WNT16 rs2707466). Consistent with our results, exon 2 nsSNP rs2908004 was significantly associated with upper limbs BMD, lower limbs BMD, as well as skull BMD phenotypes, and is the top signal in the chromosome 7q31.31 region in a GWA meta-analysis of the Avon Longitudinal Study of Parents and their Children and Generation R Study.(57) The phosSNP WNT16 rs2707466 results in a substitution of threonine by isoleucine in both WNT16A (amino acid position 253) and WNT16B (amino acid position 263) isoforms. This phosSNP is predicted to exert a modest impact on protein function (by Mutation Assessor), and to be evolutionarily less acceptable (by BLOSUM62) and moderately deleterious (by PMut and PANTHER) (Supplementary Table S4). Because WNT16B T263 has been experimentally confirmed to be a phosphorylation site in vivo,(47) in silico secondary structure prediction shows that T263 is located in a disordered region (Supplementary Figure S2). This is in agreement with findings of Dephoure et al.,(58) which demonstrated that phosphorylation sites mostly occur in disordered regions, and the addition of a phosphate group to acceptor residue upon phosphorylation can lead to a disorder-to-order transition.(59) Predicted local 3D structures also indicate notable differences between WT and MUT isoforms around T263 phosphorylation site (Supplementary Figure S3). Taken together, it is highly probable that T263, a phosphorylatable residue located in a disordered region of WNT16B protein, acts as a switch for regulating protein-protein interactions,(59) and WNT16 rs2707466, a type I(−) phosSNP that abolishes this phosphorylation site, constitutes a causal variant for BMD phenotype. This is supported by observations of wnt16 null mice, which had significantly reduced total body BMD, thinner cortical bones at the femur midshaft, and reduced bone strength of both the femur and tibia.(55,56). Further, local injection of WNT16B (WT form) could increase BMD, providing direct experimental evidence that WNT16 gene is critical for skeletal development.(25)
There are several limitations of our study. First, 9,593 phosSNPs included in stage 1 (GWA discovery) of current study represent 14.98% of the entire 64,035 phosSNP set. Nevertheless, the original 5,842,825 autosomal SNPs either directly-typed or imputed in stage 1 of conventional GWA study(31) only covered 15.92% of entire 36.7 million human autosomal SNP set(60). Therefore, although these included phosSNPs appear limiting, they constitute a similar proportion of total phosSNPs as the original stage 1 SNP set of previous conventional study(31). Second, our in vitro protein expression experiments of WT and MUT alleles of IDUA rs3755955 (R105Q), rs6831280 (A361T), and WNT16 rs2707466 (WNT16B T263I) only demonstrated relatively equivalent protein expression levels between WT and MUT alleles (Panel Bs of Supplementary Figures S4 and S5, respectively). Additional experiments by applying phospho-specific antibodies could be insightful to reveal whether these phosSNPs truly affect protein phosphorylations either directly (for WNT16 phosSNP rs2707466) or indirectly (for IDUA phosSNP rs6831280 and rs3755955). However, such experiments are time-consuming and the extents of such differences may be challenging to detect, because both IDUA and WNT16B proteins can have multiple phosphorylation sites, and these phosSNPs may only affect 1–2 among them. It also remains to be shown whether a fraction of BMD variation is attributed to impacts of IDUA rs6831280 (A361T) and rs3755955 (R105Q) on their neighboring IDUA putative phosphorylation sites T98, S102 and T366, and to abolishment of WNT16B T263 phosphorylation site by WNT16 rs2707466 (WNT16B T263I). Nevertheless, the study of Moverare-Skrtic et al.(25) clearly demonstrated a pivotal role of WNT16B WT isoform in skeletal development, and phosSNP rs2707466 could indeed play a major functional role in regulating bone metabolism.
The collective findings from our multi-stage phosSNP-centric GWA meta-analysis identified and robustly validated three phosSNPs, IDUA rs6831280 and rs3755955 and WNT16 rs2707466, to be significantly associated with FN- and HIP-BMD. These results could offer new mechanistic insights of causal variants for osteoporosis. Because currently there is a lack of bone-specific phosphorylation maps, for those phosphorylation sites that are impacted by these BMD-associated phosSNPs, more studies are necessary to elucidate whether phosphorylations affected by them are present in these various types of bone cells.
Supplementary Material
Acknowledgments
We are thankful for the generous support and assistance of Dr. Jian Li (Department of Biostatistics and Bioinformatics, Tulane University School of Public Health and Tropical Medicine) in the usage of data of four genome-wide association (GWA) cohorts (i.e., FHS, IFS, WHI-AA, and WHI-HIS) from the National Center for Biotechnology Information (NCBI) database of genotypes and phenotypes (dbGaP). The AOGC thanks the research nurses involved in this study [Ms Barbara Mason and Ms Amanda Horne (Auckland), Ms Linda Bradbury and Ms Kate Lowings (Brisbane), Ms Katherine Kolk and Ms Rumbidzai Tichawangana (Geelong); Ms Helen Steane (Hobart); Ms Jemma Christie (Melbourne); and Ms Janelle Rampellini (Perth)]. The AOGC also acknowledges gratefully technical support from Ms Kathryn Addison, Ms Marieke-Brugmans, Ms Catherine Cremen, Ms Johanna Hadler and Ms Karena Pryce. We thank Rongrong Zhang (Department of Cell and Molecular Biology, Tulane University School of Science and Engineering) for her valuable technical assistance.
The study was partially supported by Shanghai Leading Academic Discipline Project (S30501) and a start-up fund from Shanghai University of Science and Technology. The investigators of this work were also supported by grants from NIH (P50AR055081, R01AG026564, R01AR050496, RC2DE020756, R01AR057049, and R03TW008221) and the Franklin D. Dickson/Missouri Endowment and the Edward G. Schlieder Endowment. We thank Dr. Karol Estrada and Dr. Fernando Rivadeneira (both from Department of Internal Medicine and Department of Epidemiology, Erasmus Medical Center, Rotterdam, The Netherlands) for their help with analyzing and delivering the Rotterdam Study data. The Framingham Heart Study (FHS) is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract Number N01-HC-25195). This manuscript was not prepared in collaboration with investigators of FHS and does not necessarily reflect the opinions or views of FHS, Boston University, or NHLBI. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. Funding support for the Framingham Bone Mineral Density datasets was provided by NIH grants R01 AR/AG 41398, R01 AR050066 and R03 AG20321.
Funding support for the Genetic Determinants of Bone Fragility was provided through the National Institute of Aging (NIA) Division of Geriatrics and Clinical Gerontology. Genetic Determinants of Bone Fragility is a genome-wide association studies funded as part of the NIA Division of Geriatrics and Clinical Gerontology. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the NIA Division of Geriatrics and Clinical Gerontology and the NIA Division of Aging Biology. Support for the collection of datasets and samples were provided by the parent grant, Genetic Determinants of Bone Fragility (P01-AG018397). Funding support for the genotyping which was performed at the Johns Hopkins University Center for Inherited Diseases Research was provided by the NIH NIA.
The Women’s Health Initiative (WHI) program is funded by the NHLBI, National Institutes of Health, U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. This manuscript was not prepared in collaboration with investigators of the WHI, has not been reviewed and/or approved by WHI, and does not necessarily reflect the opinions of the WHI investigators or the NHLBI. WHI Population Architecture Using Genomics and Epidemiology (PAGE) is funded through the NHGRI PAGE network (U01 HG004790). Assistance with phenotype harmonization, SNP selection, data cleaning, meta-analyses, data management and dissemination, and general study coordination, was provided by the PAGE Coordinating Center (U01HG004801-01). The FHS datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db:gap through dbGaP accession phs000007.v14.p6. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, of Genetic Determinants of Bone Fragility study was provided by the NIA Division of Geriatrics and Clinical Gerontology and the NIA Division of Aging Biology. The lndiana Fragility Study (IFS) datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db:gap through dbGaP accession phs000138.v2.p1. The WHI datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db:gap through dbGaP accession phs000200.v6.p2.
The Rotterdam study was funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII) and the Municipality of Rotterdam, the Netherlands Organization of Scientific Research NWO Investments (175.010.2005.011 and 911-03-012 to KE, FR and AGU); the Research Institute for Diseases in the Elderly (RIDE) (014-93-015 and RIDE2 to KE, FR and AGU); and the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (050-060-810 to KE, FR and AGU). The Korean Genome Epidemiology Study (KoGES) was funded by the National Genome Research Institute, Korean Center for Disease Control and Prevention (2001-2003-348-6111-221, 2004-347-6111-213 and 2005-347-2400-2440-215 to HJC, CSS, and NHC). The genotype data of KoGES were obtained from the Korean Genome Analysis Project (4845-301), which was funded by a grant from the Korea National Institute of Health (Korea Center for Disease Control, Ministry for Health, Welfare and Family Affairs, Cheongwon, Korea). The National Health and Medical Research Council Project Grant (511132 to AOGC) and the National Health and Medical Research Council (Australia) Career Development Award (569807 to ELD). Funding of AOGC was also received from the Australian Cancer Research Foundation and Rebecca Cooper Foundation (Australia). TN is supported in part by a start-up fund of Tulane University. LZ is partially supported by National Natural Science Foundation of China project (31100902). MAB is funded by a National Health and Medical Research Council (Australia) Senior Principal Research Fellowship. H-W D was partially supported by the Franklin D. Dickson/Missouri Endowment and is supported by the Edward G. Schlieder Endowment.
Footnotes
Author’s roles: Study design: TN, NL, XY, MZ, SX, HWD. Study conduct: TN, NL, XY, MZ, LZ, YFP, SX, HWD. Data collection: HJC, PJL, MAB, HS, HH, SL, XDC, LJT, TLY, YG, NHC, JS, YFG, GCN, RLP, JAE, GJ, PNS, QT, XZZ, ELD, AGU, CSS, SX, HWD. Data Analysis: TN, NL, XY, MZ, LZ, YFP, HH, XF. Materials contribution: SX, HWD. Drafting manuscript: TN, NL, MZ, SX, HWD. Approving final version of manuscript: TN, NL, MZ, MAB, AGU, CSS, SX, HWD. MAB, AGU, CSS, and HWD take responsibility for the integrity of data analysis of their respective conventional genome-wide association studies, TN, LZ, and HWD take responsibility for the integrity of data analysis of phosSNP-centric three-stage meta-analysis, and NL and SL take responsibility for the integrity of data analysis of in vitro experimental studies.
REFERENCES
- 1.Kanis JA. Diagnosis of osteoporosis and assessment of fracture risk. Lancet. 2002;359(9321):1929–1936. doi: 10.1016/S0140-6736(02)08761-5. [DOI] [PubMed] [Google Scholar]
- 2.Foundation NO. America’s Bone Health: The State of Osteoporosis and Low Bone Mass in Our Nation. Washington, DC: National Osteoporosis Foundation; 2002. [Google Scholar]
- 3.Burge R, Dawson-Hughes B, Solomon DH, Wong JB, King A, Tosteson A. Incidence and economic burden of osteoporosis-related fractures in the United States, 2005–2025. J Bone Miner Res. 2007;22(3):465–475. doi: 10.1359/jbmr.061113. [DOI] [PubMed] [Google Scholar]
- 4.Organization WH. Assessment of fracture risk and its application to screening for postmenopausal osteoporosis. Report of a WHO Study Group. 1994 [PubMed]
- 5.Benes H, Weinstein RS, Zheng W, et al. Chromosomal mapping of osteopenia-associated quantitative trait loci using closely related mouse strains. J Bone Miner Res. 2000;15(4):626–633. doi: 10.1359/jbmr.2000.15.4.626. [DOI] [PubMed] [Google Scholar]
- 6.Huang HD, Lee TY, Tzeng SW, et al. Incorporating hidden Markov models for identifying protein kinase-specific phosphorylation sites. J Comput Chem. 2005;26(10):1032–1041. doi: 10.1002/jcc.20235. [DOI] [PubMed] [Google Scholar]
- 7.Olsen JV, Vermeulen M, Santamaria A, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3(104):ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 8.Yan JX, Packer NH, Gooley AA, Williams KL. Protein phosphorylation: technologies for the identification of phosphoamino acids. J Chromatogr A. 1998;808(1–2):23–41. doi: 10.1016/s0021-9673(98)00115-0. [DOI] [PubMed] [Google Scholar]
- 9.Via A, Diella F, Gibson TJ, Helmer-Citterich M. From sequence to structural analysis in protein phosphorylation motifs. Front Biosci (Landmark Ed) 2011;16:1261–1275. doi: 10.2741/3787. [DOI] [PubMed] [Google Scholar]
- 10.Savas S, Ozcelik H. Phosphorylation states of cell cycle and DNA repair proteins can be altered by the nsSNPs. BMC Cancer. 2005;5:107. doi: 10.1186/1471-2407-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haraksingh RR, Snyder MP. Impacts of variation in the human genome on gene regulation. J Mol Biol. 2013;425(21):3970–3977. doi: 10.1016/j.jmb.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 12.Cooper DN, Chen JM, Ball EV, et al. Genes, mutations, and human inherited disease at the dawn of the age of personalized genomics. Hum Mutat. 2010;31(6):631–655. doi: 10.1002/humu.21260. [DOI] [PubMed] [Google Scholar]
- 13.Kang HG, Lee SY, Jeon HS, et al. A Functional Polymorphism in CSF1R Gene Is a Novel Susceptibility Marker for Lung Cancer among Never-Smoking Females. J Thorac Oncol. 2014 doi: 10.1097/JTO.0000000000000310. [DOI] [PubMed] [Google Scholar]
- 14.Ozeki C, Sawai Y, Shibata T, et al. Cancer susceptibility polymorphism of p53 at codon 72 affects phosphorylation and degradation of p53 protein. J Biol Chem. 2011;286(20):18251–18260. doi: 10.1074/jbc.M110.208587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentile S, Martin N, Scappini E, Williams J, Erxleben C, Armstrong DL. The human ERG1 channel polymorphism, K897T, creates a phosphorylation site that inhibits channel activity. Proc Natl Acad Sci U S A. 2008;105(38):14704–14708. doi: 10.1073/pnas.0802250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Chen S, Zhang L, et al. Protective effect of KCNH2 single nucleotide polymorphism K897T in LQTS families and identification of novel KCNQ1 and KCNH2 mutations. BMC Med Genet. 2008;9:87. doi: 10.1186/1471-2350-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vazgiourakis VM, Zervou MI, Eliopoulos E, et al. Implication of VEGFR2 in systemic lupus erythematosus: a combined genetic and structural biological approach. Clin Exp Rheumatol. 2013;31(1):97–102. [PubMed] [Google Scholar]
- 18.Ogus AC, Yoldas B, Ozdemir T, et al. The Arg753GLn polymorphism of the human toll-like receptor 2 gene in tuberculosis disease. Eur Respir J. 2004;23(2):219–223. doi: 10.1183/09031936.03.00061703. [DOI] [PubMed] [Google Scholar]
- 19.Xiong Y, Song C, Snyder GA, Sundberg EJ, Medvedev AE. R753Q polymorphism inhibits Toll-like receptor (TLR) 2 tyrosine phosphorylation, dimerization with TLR6, and recruitment of myeloid differentiation primary response protein 88. J Biol Chem. 2012;287(45):38327–38337. doi: 10.1074/jbc.M112.375493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7(9):1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren J, Jiang C, Gao X, et al. PhosSNP for systematic analysis of genetic polymorphisms that influence protein phosphorylation. Mol Cell Proteomics. 2010;9(4):623–634. doi: 10.1074/mcp.M900273-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gori F, Lerner U, Ohlsson C, Baron R. A new WNT on the bone: WNT16, cortical bone thickness, porosity and fractures. BoneKEy reports. 2015;4:669. doi: 10.1038/bonekey.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Qiu J, Magrane G, et al. Duplication of C7orf58, WNT16 and FAM3C in an obese female with a t(7;22)(q32.1;q11.2) chromosomal translocation and clinical features resembling Coffin-Siris Syndrome. PLoS One. 2012;7(12):e52353. doi: 10.1371/journal.pone.0052353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fear MW, Kelsell DP, Spurr NK, Barnes MR. Wnt-16a, a novel Wnt-16 isoform, which shows differential expression in adult human tissues. Biochem Biophys Res Commun. 2000;278(3):814–820. doi: 10.1006/bbrc.2000.3852. [DOI] [PubMed] [Google Scholar]
- 25.Moverare-Skrtic S, Henning P, Liu X, et al. Osteoblast-derived WNT16 represses osteoclastogenesis and prevents cortical bone fragility fractures. Nat Med. 2014 doi: 10.1038/nm.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panagiotou OA, Willer CJ, Hirschhorn JN, Ioannidis JP. The power of meta-analysis in genome-wide association studies. Annu Rev Genomics Hum Genet. 2013;14:441–465. doi: 10.1146/annurev-genom-091212-153520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reimand J, Wagih O, Bader GD. The mutational landscape of phosphorylation signaling in cancer. Sci Rep. 2013;3:2651. doi: 10.1038/srep02651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Liu X, Yang BZ, Gelernter J. The role and challenges of exome sequencing in studies of human diseases. Front Genet. 2013;4:160. doi: 10.3389/fgene.2013.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu T, Liu N, Zhao M, et al. Identification of a Novel FGFRL1 MicroRNA Target Site Polymorphism for Bone Mineral Density in Meta-Analyses of Genome-Wide Association Studies. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv144. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3(5):391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L, Choi HJ, Estrada K, et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet. 2014;23(7):1923–1933. doi: 10.1093/hmg/ddt575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294(5):1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 33.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4(6):1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 34.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrer-Costa C, Gelpi JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21(14):3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 37.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70(3):611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 39.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Estrada K, Styrkarsdottir U, Evangelou E, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bogdanova MA, Kostareva AA, Malashicheva AB. P342Lamin A/C mutations associated with different laminopathies affect differentiation of mesenchimal stem cells. Cardiovasc Res. 2014;103(Suppl 1):S62. [Google Scholar]
- 43.Alam I, Padgett LR, Ichikawa S, et al. SIBLING family genes and bone mineral density: Association and allele-specific expression in humans. Bone. 2014;64:166–172. doi: 10.1016/j.bone.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isaac J, Erthal J, Gordon J, et al. DLX3 regulates bone mass by targeting genes supporting osteoblast differentiation and mineral homeostasis in vivo. Cell Death Differ. 2014 doi: 10.1038/cdd.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Li J, Pei YF, Liu Y, Deng HW. Tests of association for quantitative traits in nuclear families using principal components to correct for population stratification. Ann Hum Genet. 2009;73(Pt 6):601–613. doi: 10.1111/j.1469-1809.2009.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brill LM, Xiong W, Lee KB, et al. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5(2):204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 49.Neufeld EMJ. The mucopolysaccharidoses. In: Scriver CRBA, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors; Scriver CRBA, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic and Molecular Basis of Inherited Disease. 8th. New York. NY: McGraw-Hill; 2001. pp. 3421–3452. [Google Scholar]
- 50.Wang D, Shukla C, Liu X, et al. Characterization of an MPS I-H knock-in mouse that carries a nonsense mutation analogous to the human IDUA-W402X mutation. Mol Genet Metab. 2010;99(1):62–71. doi: 10.1016/j.ymgme.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishizeki K, Saito H, Shinagawa T, Fujiwara N, Nawa T. Histochemical and immunohistochemical analysis of the mechanism of calcification of Meckel's cartilage during mandible development in rodents. J Anat. 1999;194(Pt 2):265–277. doi: 10.1046/j.1469-7580.1999.19420265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hartmann C. Skeletal development--Wnts are in control. Mol Cells. 2007;24(2):177–184. [PubMed] [Google Scholar]
- 53.Yang Y. Skeletal morphogenesis during embryonic development. Crit Rev Eukaryot Gene Expr. 2009;19(3):197–218. doi: 10.1615/critreveukargeneexpr.v19.i3.30. [DOI] [PubMed] [Google Scholar]
- 54.Jiang Z, Von den Hoff JW, Torensma R, Meng L, Bian Z. Wnt16 is involved in intramembranous ossification and suppresses osteoblast differentiation through the Wnt/beta-catenin pathway. J Cell Physiol. 2014;229(3):384–392. doi: 10.1002/jcp.24460. [DOI] [PubMed] [Google Scholar]
- 55.Medina-Gomez C, Kemp JP, Estrada K, et al. Meta-analysis of genome-wide scans for total body BMD in children and adults reveals allelic heterogeneity and age-specific effects at the WNT16 locus. PLoS Genet. 2012;8(7):e1002718. doi: 10.1371/journal.pgen.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng HF, Tobias JH, Duncan E, et al. WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. PLoS Genet. 2012;8(7):e1002745. doi: 10.1371/journal.pgen.1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kemp JP, Medina-Gomez C, Estrada K, et al. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS Genet. 2014;10(6):e1004423. doi: 10.1371/journal.pgen.1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dephoure N, Gould KL, Gygi SP, Kellogg DR. Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol Biol Cell. 2013;24(5):535–542. doi: 10.1091/mbc.E12-09-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tyanova S, Cox J, Olsen J, Mann M, Frishman D. Phosphorylation variation during the cell cycle scales with structural propensities of proteins. PLoS Comput Biol. 2013;9(1):e1002842. doi: 10.1371/journal.pcbi.1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Genomes Project C. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.