Abstract
Aiming to identify genomic variants associated with osteoporosis, we performed a genome-wide association meta-analysis of bone mineral density (BMD) at Ward's triangle of the hip in 7,175 subjects from 6 samples. We performed in silico replications with femoral neck, trochanter, and inter-trochanter BMDs in 6,912 subjects from the Framingham heart study (FHS), and with forearm, femoral neck and lumbar spine BMDs in 32,965 subjects from the GEFOS summary results. Combining the evidence from all samples, we identified 2 novel loci for areal BMD: 1q43 (rs1414660, discovery p=1.20×10−8, FHS p=0.05 for trochanter BMD; rs9287237, discovery p=3.55×10−7, FHS p=9.20×10−3 for trochanter BMD, GEFOS p=0.02 for forearm BMD, nearest gene FMN2) and 2q32.2 (rs56346965, discovery p=7.48×10−7, FHS p=0.10 for inter-trochanter BMD, GEFOS p=0.02 for spine BMD, nearest gene NAB1). The two lead SNPs rs1414660 and rs56346965 are eQTL sites for the genes GREM2 and NAB1 respectively. Functional annotation of GREM2 and NAB1 illustrated their involvement in BMP signaling pathway and in bone development. We also replicated three previously reported loci: 5q14.3 (rs10037512, discovery p=3.09×10−6, FHS p=8.50×10−3, GEFOS p=1.23×10−24 for femoral neck BMD, nearest gene MEF2C), 6q25.1 (rs3020340, discovery p=1.64×10−6, GEFOS p=1.69×10−3 for SPN-BMD, nearest gene ESR1) and 7q21.3 (rs13310130, discovery p=8.79×10−7, GEFOS p=2.61×10−7 for spine BMD, nearest gene SHFM1). Our findings provide additional insights that further enhance our understanding of bone development, osteoporosis, and fracture pathogenesis.
Keywords: hip Ward's triangle, bone mineral density, genome-wide association study, meta-analysis, GREM2, NAB1
Introduction
Osteoporosis, the most common metabolic skeletal disorder, is characterized by low bone mass and micro-architectural deterioration of bone tissue. People with osteoporosis are predisposed to fragility fracture. Thus osteoporosis which affects over 200 million people worldwide [1], confers substantial morbidity and mortality on the human population [2].
Osteoporosis is diagnosed by low bone mineral density (BMD), which is also the standard predictor of osteoporotic fracture. Though influenced by environmental factors, BMD is a highly heritable trait with heritability ranging from 0.5 to 0.8 [3]. A number of genome-wide association studies (GWASs) and their meta-analyses have been conducted for BMD, and dozens of genomic loci have been identified [4–14]. However, the cumulative effects of these identified loci account for only approximately 6% of total BMD variation [11]. Thus, the majority of genetic factors influencing BMD are still awaiting identification.
Fragility fractures occur relatively frequently at the hip and lumbar spine. The standard approach for measuring BMD at these high risk sites utilizes dual-energy X-ray absorptiometry (DXA). Typical DXA scans of the hip measure BMDs at multiple sub-regions, including the femoral neck, trochanter, inter-trochanter, and Ward's triangle. Among these, the femoral neck is the most studied sub-region because of its dominant clinical relevance. For gene-mapping purposes, however, sub-regions other than the femoral neck may also provide valuable information. These sub-regions are more homogeneous than total hip, and may thus provide new insights into the identification of responsible genes for BMD. Importantly, BMDs at these sub-regions are correlated, but not perfectly, to that at the femoral neck.
Ward's triangle is a radiolucent area between principle compressive, secondary compressive and primary tensile trabeculae in the femoral neck. Within the femoral head, it is usually the area with the lowest BMD. Consequently, BMD measured at Ward's triangle may have important implications for osteoporosis. For example, Yoshihashi et al. [15] reported that BMD at Ward's triangle is a sensitive indicator of osteoporosis, particularly in men. Importantly, however, studies to identify genetic loci associated with osteoporosis have rarely studied BMD at Ward’s triangle.
In the present study, aimed to identify additional BMD loci, we utilized samples from diverse ancestries to perform a large-scale trans-ethnic genome-wide association meta-analysis for BMD at Ward's triangle. We also replicated the discovery findings in the Framingham heart study (FHS) and in the large GEFOS summary results.
Materials and Methods
Discovery samples
The discovery stage was a trans-ethnic GWAS meta-analysis of multiple diverse ancestral samples. We collected 6 GWAS samples for meta-analysis of Ward's triangle (WT-) BMD. Three samples were from the in-house studies and the other 3 were accessed through the database of genotype and phenotype (dbGAP). All samples were approved by the respective institutional ethics review boards, and all participants provided written informed consent. Details of the samples were described previously [13]. Briefly, the first sample comprised 1,000 unrelated subjects of European ancestry from the Omaha osteoporosis study (OOS). The second sample comprised 2,286 unrelated subjects of European ancestry from the Kansas City osteoporosis study (KCOS). The third sample comprised 1,627 unrelated subjects of Chinese Han ancestry from the China osteoporosis study (COS). The fourth sample was the Indiana fragility study (IFS) that was accessed through the dbGAP. The IFS is a cross-sectional cohort comprising 1,493 premenopausal sister pairs of European ancestry. Both the fifth and sixth samples were from the Women’s health initiative (WHI) observational study that was accessed through the dbGAP too. The WHI is a partial factorial randomized and longitudinal cohort with >12,000 genotyped women aged 50–79 years, of African-American or Hispanic ancestry [16]. The fifth sample comprised 845 subjects of African-American ancestry (WHI-AA), and the sixth sample comprised 446 subjects of Hispanic ancestry (WHI-HIS).
Replication FHS sample
We included the Framingham heart study (FHS) as a replication sample, and accessed it through the dbGAP. FHS is a longitudinal and prospective cohort comprising >16,000 individuals spanning three generations of European ancestry [17].
The FHS datasets accessed through the dbGAP did not include WT-BMD, but instead included BMDs at three other hip sub-regions, including femoral neck, trochanter, and inter-trochanter. Because of the strong correlations between BMDs at WT and the other sub-regions, these three sub-regions were acceptable as replication phenotypes. We identified a total of 6,912 genotyped and phenotyped FHS participants for analysis.
Phenotype measurements and modeling
In the discovery samples, WT-BMD was measured by hip scan with DXA bone densitometer (Lunar Corp., Madison, WI, USA or Hologic Inc., Bedford, MA, USA), following the manufacturer's protocols. Covariates (including gender, age, age2, height and the first five principal components derived from genome-wide genotype data) were screened for significance with the step-wise linear regression model implemented in the R function stepAIC. Raw BMD values were adjusted by significant covariates, and the residuals were normalized by inverse quantiles of standard normal distribution.
In the replication FHS sample, the original generation participants underwent bone densitometry scan by DXA during their 22nd or 24th examinations. The offspring participants underwent DXA scan during their 6th or 7th examinations. The third generation participants underwent scan during their 2nd examination. Raw BMD values were modeled in the same way as that in the discovery samples.
Genotyping and quality control
All GWAS samples were genotyped by high-throughput SNP genotyping arrays (Affymetrix Inc., Santa Clara, CA, USA; or Illumina Inc., San Diego, CA, USA within individual samples), following the manufacturer’s protocols. Quality control (QC) within each sample was implemented at both the individual and SNP levels. At the individual level, sex compatibility was checked by imputing sex from X-chromosome genotype data with PLINK [18]. Individuals of ambiguous imputed sex or of imputed sex inconsistent with reported sex were removed. At the SNP level, SNPs violating the Hardy-Weinberg equilibrium (HWE) rule (p-value<1.0×10−5) were removed. Population outliers were monitored by genotype-derived principal components, and outliers were removed if present.
In the FHS replication sample, which was a familial sample, genotypes presenting the Mendel error were set to missing.
Genotype imputation
GWAS samples were imputed by the 1000 genomes project phase 3 sequence variants (as of May, 2013) [19]. Haplotypes representing 240 individuals of European ancestry, 244 of East Asian ancestry, 319 of African ancestry, and 170 of admixed American ancestry were downloaded from the project download site. Haplotypes of bi-allelic variants, including SNPs and bi-allelic insertions/deletions (indels), were extracted to form reference panels for imputation. As a QC procedure, variants with zero or one copy of minor alleles were removed.
Each GWAS sample was imputed by the respective reference panel with the closest ancestry. Prior to imputation, a consistency test of allele frequency between the GWAS and reference samples was examined with the chi-square test. To correct for potential mis-strandedness, GWAS SNPs that failed the consistency test (p<1.0×10−6) were transformed into the reverse strand. SNPs that again failed the consistency test were removed from the GWAS sample. Imputation was performed with FISH [20], a fast and accurate diploid genotype imputation algorithm that we previously developed.
Individual study association testing
Each GWAS sample was tested for association between normalized phenotype residuals and genotyped and imputed genotypes under an additive mode of inheritance. For unrelated samples, association was examined by the linear regression model with MACH2QTL [21], in which allele dosage was taken as the predictor for the phenotype. For familial samples, a mixed linear model was used to account for genetic relatedness within each pedigree [22].
Meta-analysis
Summary association statistics from individual GWAS samples were combined to perform weighted fixed-effects meta-analysis with METAL [23]. As a QC procedure, only common (MAF>=0.05) and well-imputed (r2>=0.3 in at least two samples) SNPs were included for analysis. Weights were proportional to the inverse variance of regression coefficients. Between-study heterogeneity effects were measured by I2 and Cochran’s Q statistic [24]. Genomic control inflation factor [25] was estimated in each individual study and in meta-analysis of multiple studies.
Replication in the FHS sample
In the FHS replication sample, we first checked if effect direction was consistent with that in the discovery samples. Conditioning on consistent direction, we then reported replication association p-values.
Replication in the GEFOS summary results
The GEFOS consortium recently performed a large scale sequencing-based discovery GWAS of BMD in up to 32,965 participants, and made the summary results publicly available [12]. The released results represented the largest and most powerful analysis to date in the field. The GEFOS results did not include WT-BMD, but instead included BMDs at the forearm, femoral neck and lumbar spine. We examined these alternative sites for association signals of selected SNPs.
Functional annotation
We annotated functional relevance of identified SNPs with HaploReg [26]. HaploReg explores annotations of non-coding genomic variants from multiple functional categories. The information that it uses is from a variety of large experiment projects, such as the Roadmap Epigenomic project [27], the ENCODE project [28], and the GTEx eQTL datasets [29]. The categories include conservation sites, DNase hypersensitivity sites (DHS), transcription factor binding sites (TFBS), promoter sites, enhancer sites, and others. We annotated lead SNPs and their neighbor SNPs with strong LD (r2>0.8).
For candidate genes, we annotated them by constructing gene interaction networks with STRING [30]. STRING uses information based on gene co-expression, text-mining, and others, to construct gene interactive networks.
RESULTS
GWAS meta-analyses
A total of 7,175 subjects from 6 GWAS samples were included in the meta-analysis. Basic characteristics of the samples are listed in Table 1. Seventy-eight per cent of the participants are women. Principal component analysis (PCA) was applied to each individual sample [31], and no population outliers were observed.
Table 1.
Basic characteristics of the discovery samples
| Sample | Source | Anc. | N | Female (%) |
Age | Height (m) |
Weight (kg) |
WT-BMD (g/cm2) |
CV (%) |
Bone densitometer |
|---|---|---|---|---|---|---|---|---|---|---|
| OOS | In-house | EUR | 622 | 48.4 | 60.2(14.2) | 1.70(0.10) | 81.33(17.47) | 0.60(0.17) | 0.28 | Hologic |
| KCOS | In-house | EUR | 2206 | 76.7 | 51.5(13.7) | 1.66(0.08) | 75.02(17.32) | 0.64(0.25) | 0.39 | Hologic |
| COS | In-house | EAS | 1579 | 51.8 | 34.5(13.2) | 1.64(0.08) | 60.12(10.49) | 0.72(0.16) | 0.23 | Hologic |
| IFS | dbGAP | EUR | 1477 | 100.0 | 32.7(7.2) | 1.65(0.06) | 71.65(16.89) | - | - | Lunar |
| WHI-AA | dbGAP | AFR | 845 | 100.0 | 61.2(7.3) | 1.63(0.06) | 82.15(17.31) | 0.66(0.17) | 0.26 | Hologic |
| WHI-HIS | dbGAP | AMR | 446 | 100.0 | 60.1(7.5) | 1.58(0.06) | 72.81(14.77) | 0.57(0.14) | 0.25 | Hologic |
Notes: A total of 6 samples were collected. Three of them were from the in-house studies, and the other 3 were accessed through the dbGAP.
OOS, Omaha osteoporosis study; KCOS, Kansas-city osteoporosis study; COS, Chinese osteoporosis study; IFS, Indiana fragility study; WHI-AA, Women's health initiative African-American sample; WHI-HIS, the WHI Hispanic sample; Anc., ancestral population; EUR, European ancestral population; EAS, East Asian ancestral population; AFR, African ancestral population; AMR, Admixed American ancestral population; WT-BMD, Ward's triangle bone mineral density; CV, coefficient of variations.
Sample size "N" was reported after QC.
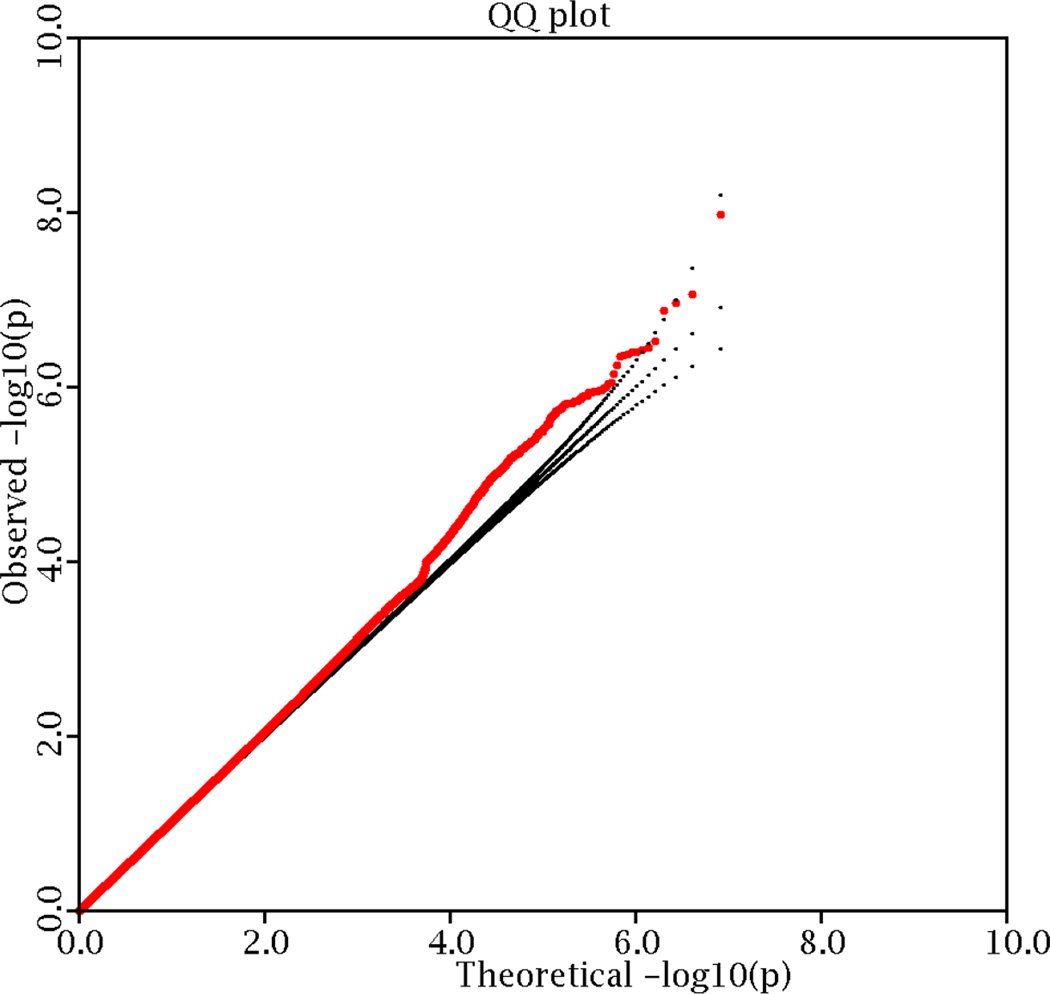
The 1000 genomes sequencing project generated over 10 million qualified SNPs (Supplementary Table 1). After adjusting phenotypes by PCA in each individual study, the genomic control inflation factor of the meta-analysis was 1.03, implying limited effect of potential population stratification. A logarithmic quantile-quantile plot of the meta-analysis test statistics showed a marked deviation in the tail of the distribution, implying the possible existence of true associations (Figure 1).
Figure 1. Logarithmic quantile–quantile (QQ) plot of the discovery GWAS samples.
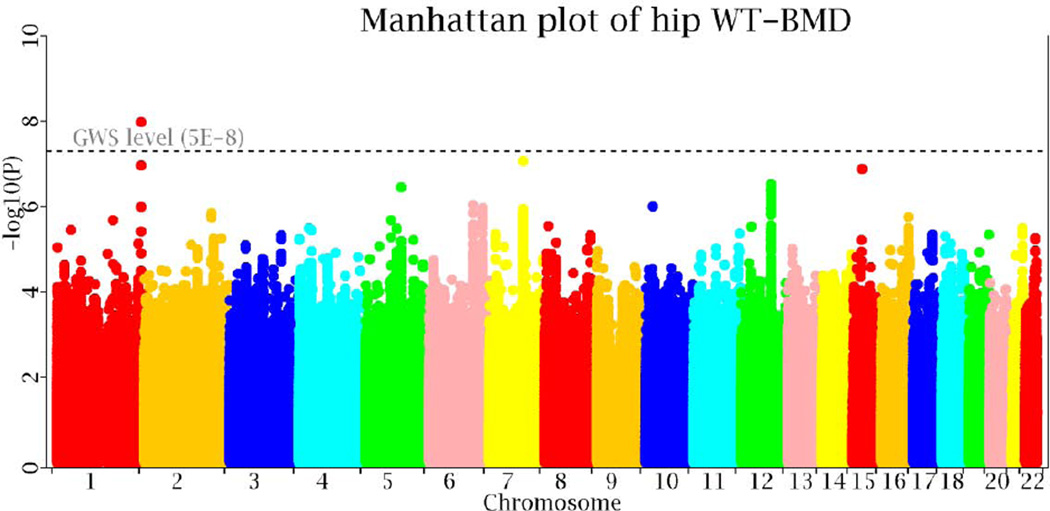
Manhattan plot of the GWAS meta-analyses is displayed in Figure 2. At the genome-wide significance (GWS, 5.0×10−8) level, one locus 1q43 was identified, with the lead SNP being rs1414660 (p=1.20×10−8, I2=38.6%). At a less stringent borderline significance level 5.0×10−6, 16 additional loci were identified (Supplementary Table 2). Four (5q14.3, 6q25.1, 7q21.3 and 16q23) of them have been reported by previous GWAS studies of areal BMD.
Figure 2. Manhattan plot of the discovery GWAS samples.
Replication in the FHS
The lead SNP from each of the above 17 loci was subjected to replication in the FHS sample and in the GEFOS summary results. The FHS sample did not include WT-BMD, but included BMDs at the other three hip sub-regions: femoral neck (FNK), trochanter (TRO) and inter-trochanter (INT). Correlation coefficients between BMDs at WT and the other hip sub-regions are listed in Table 2. There was a strong correlation between BMD’s at WT and the other hip sub-regions, validating the use of these other hip sub-regions in our replication effort.
Table 2.
Correlations between BMDs at the WT and at the other sites
| Correlation coefficients | |||||
|---|---|---|---|---|---|
| Study | WT-FNK | WT-TRO | WT-INT | WT-SPN | WT-FA |
| OOS | 0.88 | 0.68 | 0.68 | 0.53 | 0.54 |
| KCOS | 0.80 | 0.59 | 0.71 | 0.35 | 0.32 |
| COS | 0.87 | 0.79 | 0.73 | 0.65 | 0.53 |
| IFS | 0.88 | 0.79 | - | 0.57 | - |
| WHI-AA | 0.88 | 0.75 | 0.75 | 0.58 | - |
| WHI-HIS | 0.88 | 0.77 | 0.81 | 0.64 | - |
Notes: Within each sample, Pearson's correlation coefficients were calculated for BMDs between pairs of skeletal sites.
Abbreviations, WT, Ward's triangle; FNK, femoral neck; TRO, trochanter; INT, inter-trochanter; SPN, lumbar spine; FA, forearm; "-", not available.
First, we determined whether the effect directions were consistent between the discovery and the FHS samples. For 9 of the 17 SNPs, the effect directions were consistent in all three sub-regions and in the discovery samples. For 5 SNPs, the effect directions were consistent in all three sub-regions, but the directions were opposite to those of the discovery samples. For the remaining 3 SNPs, the effect directions were inconsistent across the three replication sub-regions (Supplementary Table 3).
Next, we checked association signals for the 9 SNPs with consistent directional effects. Three SNPs (rs10037512, rs3020340 and rs13310130) locate in 3 loci (5q14.3, 6q25.1 and 7q21.3) that were reported previously. Among them, only rs10037512 was successfully replicated (p=8.50×10−3 for FNK-BMD). The remaining 6 SNPs (rs1414660, rs56346965, rs3091309, rs79702404, rs8181385 and rs1011728) locate in 6 novel loci for areal BMD. Among them, rs1414660 and rs79702404 were significant for TRO-BMD (p=0.05 and 0.03, respectively). Considering the one-sided nature of the replication test, rs56346965 was weakly associated with INT-BMD (one-sided p=0.05).
Replication in the GEFOS summary results
We further replicated the above 17 SNPs in the larger GEFOS summary results. Two of the lead SNPs, rs1414660 in 1q43 and rs111756027 in 16q23, were not available in the GEFOS summary results. For this analysis, therefore, they were replaced by rs9287237 (discovery p=3.55×10−7) and rs74486092 (discovery p=3.87×10−6).
The GEFOS results did not have WT-BMD, but had BMDs at the forearm (FA), femoral neck (FNK) and spine (SPN). Correlation coefficients between BMDs at the WT and these sites are listed in Table 2.
For 11 of the 17 SNPs, including the 3 SNPs (rs10037512, rs3020340 and rs13310130) locating in 3 loci (5q14.3, 6q25.1 and 7q21.3) that had been reported previously, the effect directions were consistent in all three skeletal sites and in the discovery samples. For 5 of the 17 SNPs, the effect directions were consistent in all three sub-regions, but were opposite to those in the discovery samples. The directional effects for the final SNP were inconsistent across the three replication sub-regions (Supplementary Table 4).
All of the 3 previously reported SNPs were successfully replicated. Specifically, rs10037512 was significant for FA-BMD (p=7.49×10−8) and FNK-BMD (p=1.23×10−24); rs3020340 was significant for FNK-BMD (p=0.02) and SPN-BMD (p=1.69×10−3), and rs13310130 was significant at all three sites (FA-BMD p=0.02, FNK-BMD p=3.03×10−7 and SPN-BMD p=2.61×10−7).
The remaining 8 SNPs with consistent directional effects were not previously reported. Four of them were successfully replicated. Specifically, rs9287237 was significant at all three sites (FA-BMD p=0.02, FNK-BMD p=1.47×10−4, and SPN-BMD p=1.95×10−3); rs56346965 was significant for SPN-BMD (p=0.02); rs3091309 was significant for FNK-BMD (p=0.03) and SPN-BMD (p=8.43×10−3); and rs79702404 was significant for FNK-BMD (p=0.04). One additional SNP, rs8181385, was weakly associated with FA-BMD (two-sided p=0.06, one sided p=0.03).
For FA-BMD (N=8,143), none of the GEFOS samples overlapped with both the discovery samples and the FHS replication sample. Consequently, FA-BMD could serve as a totally independent replication. For FNK- and SPN-BMD, however, the FHS was included in the GEFOS study. To make the GEFOS results as independent as possible, we performed an alternative analysis to derive an association signal in the non-overlapping GEFOS samples based on the GEFOS total sample signal and the overlapping FHS sample signal. Details of the derivation rationale have been previously described [13]. The results of this alternative analysis are listed in Supplementary Table 4. Of the above 4 SNPs, rs79702404 became non-significant in the non-overlapping GEFOS samples.
Combining the results of the discovery samples, the FHS sample and the GEFOS summary results, 6 SNPs (rs1414660, rs9287237, rs56346965, rs10037512, rs3020340 and rs13310130) had strong evidence of association with BMD variation. They were significant at the GWS level, or the suggestive level, in the discovery samples. Additionally, they were nominally replicated for at least one of the sites (FNK-, INT- and TRO-BMDs) in the FHS sample, and for at least one of the sites (FA-, FNK- and SPN-BMDs) in the non-overlapping GEFOS results.
Three of these 6 SNPs locate in 3 distinct genomic regions that have been reported previously: 5q14.3 (rs10037512, nearest gene MEF2C), 6q25.1 (rs3020340, ESR1) and 7q21.3 (rs12704871, SHFM1). The remaining 3 SNPs locate into 2 regions that were novel for areal BMD: 1q43 (rs1414660 and rs9287237, FMN2) and 2q32.2 (rs56346965, NAB1). The main results of this study are summarized in Table 3.
Table 3.
Main results of the identified SNPs
| rs# | EA/OA | Locus | Nearest gene |
Discovery samples | Replication FHS sample | Replication GEFOS results | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female (N=5578) | Male (N=1597) | Combined (N=7175) | FNK (N=6908) | TRO (N=6905) | INT (N=6194) | FA (N=8143) | FNK (N=32735) | SPN (N=28498) | ||||||||||||||
| Beta | p | Beta | p | Beta | p | I2(%) | Beta | p | Beta | p | Beta | p | Beta | p | Beta | p | Beta | p | ||||
| Novel | ||||||||||||||||||||||
| rs1414660 | C/T | 1q43 | FMN2 | −0.12 | 4.31×10−7 | −0.10 | 0.02 | −0.12 | 1.20×10−8 | 38.6 | −0.05 | 0.14 | −0.06 | 0.05 | −0.01 | 0.66 | NA | |||||
| rs9287237 | G/T | 1q43 | FMN2 | −0.11 | 5.04×10−6 | −0.08 | 0.05 | −0.11 | 3.55×10−7 | 56.7 | −0.06 | 0.08 | −0.08 | 9.20×10−3 | −0.04 | 0.28 | −0.05 | 0.02 | −0.04 | 1.47×10−4 | −0.04 | 1.95×10−3 |
| rs56346965 | C/A | 2q32.2 | NAB1 | 0.09 | 1.44×10−6 | 0.05 | 0.18 | 0.09 | 7.48×10−7 | 28.4 | 0.00 | 0.83 | −0.01 | 0.44 | −0.03 | 0.10 | 0.01 | 0.37 | 0.01 | 0.07 | 0.02 | 0.02 |
| Previously reported | ||||||||||||||||||||||
| rs10037512 | C/T | 5q14.3 | MEF2C | −0.07 | 2.09×10−4 | −0.10 | 6.99×10−3 | −0.08 | 3.09×10−6 | 4.9 | −0.05 | 8.50×10−3 | 0.00 | 0.95 | −0.03 | 0.14 | −0.09 | 7.49×10−8 | −0.08 | 1.23×10−24 | −0.01 | 0.48 |
| rs3020340 | G/A | 6q25.1 | ESR1 | 0.08 | 2.71×10−4 | 0.12 | 5.15×10−3 | 0.09 | 1.64×10−6 | 14.4 | −0.01 | 0.54 | 0.00 | 0.86 | 0.00 | 0.98 | 0.03 | 0.13 | 0.02 | 0.02 | 0.04 | 1.69×10−3 |
| rs13310130 | G/T | 7q21.3 | SHFM1 | −0.09 | 2.28×10−5 | −0.10 | 4.76×10−3 | −0.09 | 8.79×10−7 | 7.6 | −0.03 | 0.12 | −0.01 | 0.62 | 0.00 | 0.92 | −0.04 | 0.02 | −0.04 | 3.03×10−7 | −0.05 | 2.61×10−7 |
Notes: Alleles were presented as effect allele (EA)/other allele (OA). GEFOS-NO, the GEFOS non-overlapping samples. The GEFOS-NO association signals were derived via the GEFOS and the overlapping FHS association signals.
Functional annotation
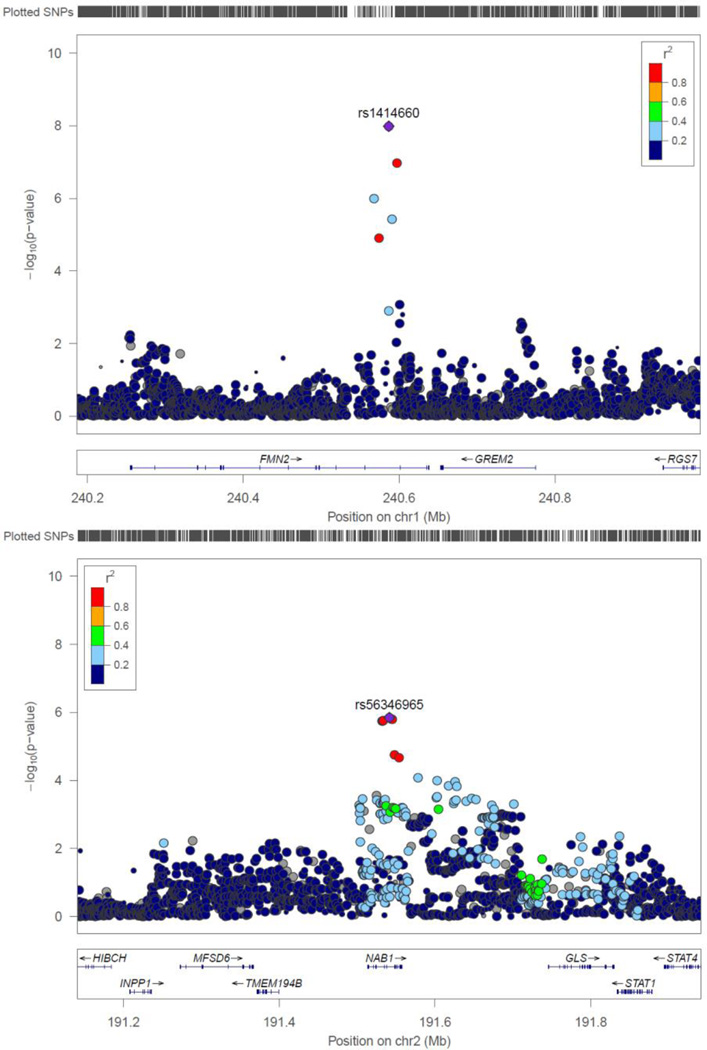
We annotated the 3 novel SNPs (rs1414660, rs9287237 and rs56346965) and their neighboring SNPs (LD r2>0.8) in loci 1q43 and 2q32.2. rs1414660 and rs9287237 are in strong LD (r2=0.83) with each other, and with 4 additional neighboring SNPs (r2>0.8, rs953247, rs12044944, rs9659023, and rs9661787). Of these 6 neighboring SNPs, rs1414660 is the only one that has enhancer activity in osteoblast primary cells [27]. It is also the only SNP that is conservative as predicted by both GERP [32] and Siphy [33]. All the 6 SNPs are eQTL sites to the target gene GREM2 in tibial artery and cultured primary fibroblast cell line from fresh skin, while the signals for the lead SNP rs1414660 are the most significant in both tissues (p=5.50×10−9 and 2.04×10−14) [29].
rs56346965 has 7 neighboring SNPs (rs2293765, rs1978273, rs2159819, rs11900804, rs2192008, rs4853727 and rs4853516) with strong LD patterns (r2>0.8). They all locate in a 33 kb genomic region, and all have enhancer activities as predicted by enhancer histone marks H3K4me1 and H3K27ac [27]. rs2293765 and rs2192008 also have promoter activities as predicted by promoter histone mark H3K4me3. These activities are observed in multiple tissues, including primary osteoblast cells. All SNPs are eQTL sites to multiple genes including NAB1, HIBCH, and TMEM194B [29, 34], which locate into the same locus (2q32.2) and are separated by a maximum of 488 kb. A search of GRASP [35] through HaploReg demonstrated that rs4853516 is associated with levels of NAB1 exon expression in peripheral blood mononuclear cells (p=3.18×10−11), precursors of osteoclast cells, which are responsible for bone resorption [36].
New loci/genes associated with areal WT-BMD
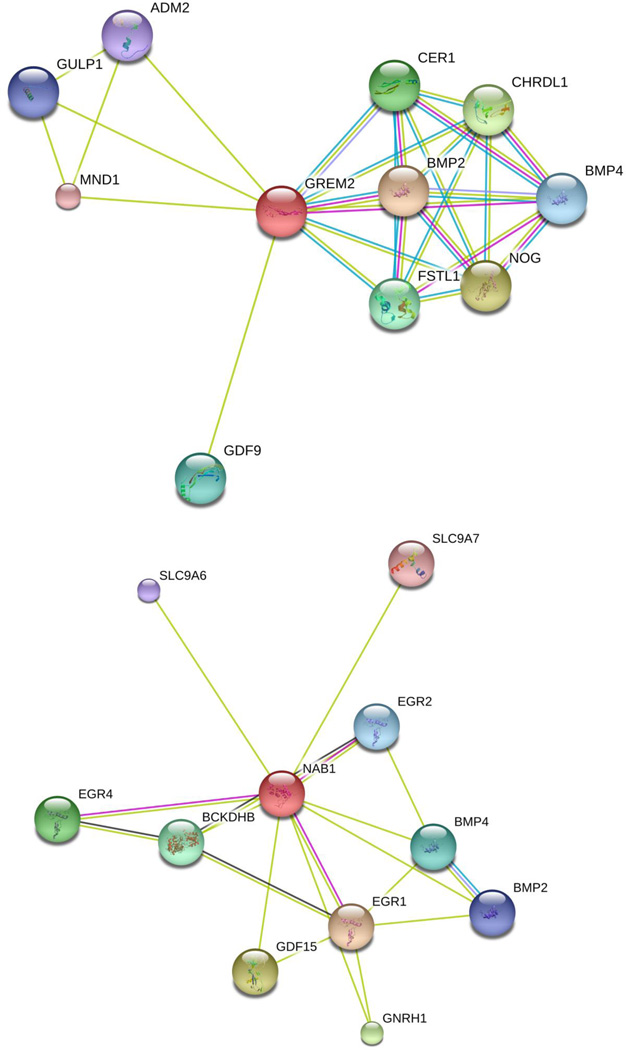
1q43 (GREM2). rs1414660 in 1q43 is a common (MAF=0.29) and imputed SNP with high imputation certainty (r2=0.50–0.99). Allele C at this SNP tended to decrease WT-BMD by 0.12 s.d. per copy. A regional plot of rs1414660 is displayed in Figure 3. It locates into an intron of the FMN2 gene, but is an eQTL site to the GREM2 gene. rs1414660 is 66.2 kb upstream from GREM2. Gene-gene interaction network analysis of GREM2 connected it to two bone morphogenetic protein (BMP) family members, BMP2 and BMP4 (Figure 4). GREM2 encodes a member of the BMP antagonist family. The antagonistic effect of the secreted glycosylated protein encoded by this gene is likely due to its direct binding to BMP proteins [37]. As an antagonist of BMP, this gene may play a role in regulating organogenesis, body patterning, and tissue differentiation.
Figure 3. Regional plots of the two lead SNPs.
Regional plots of the discovery samples around rs1414660 (upper) and rs56346965 (lower) are presented.
Figure 4. Interaction network for GREM2 and NAB1.
The figure was plotted by STRING.
2q32.2 (NAB1). The common SNP rs56346965 (MAF=0.38) in 2q32.2 was imputed with very high imputation certainty (r2=0.99–1.00). Allele C at this SNP tended to increase BMD value by 0.09 s.d. per copy. It locates into an intron of the NAB1 gene, and is an eQTL site to the NAB1 gene. NAB1 acts as a transcriptional repressor for zinc finger transcription factors EGR1 and EGR2 [38]. EGR2 impacts osteoclast survival by negatively modulating osteoclast differentiation [39]. Gene-gene interaction networks connect NAB1 to EGR1, EGR2 and EGR4 (Figure 4), and to BMP2 and BMP4, implying that it may play an important role in bone development.
Previously reported genes associated with WT-BMD
We have replicated 3 loci that were reported previously for areal BMD: 5q14.3, 6q25.1 and 7q21.3. The lead SNP rs10037512 in 5q14.3 is 154.8 kb upstream from the MEF2C gene, which was reported by multiple previous GWAS studies [6, 11, 40]. MEF2C is a member of the evolutionarily conserved MADS family of transcription factors, and plays a role in myogenesis. Its activity is modulated by multiple signaling pathways, such as the MAP kinase signaling and calcium signaling pathways [41]. Mice with mutant MEF2C genes in osteocytes had increased bone mass compared to normal controls, implying that the MEF2C may function to control bone mass by regulating osteoclastic bone resorption [42].
The lead SNP, rs3020340 in 6q25.1, locates into an intron of the ESR1 (estrogen receptor 1) gene, which is a well-recognized candidate gene for BMD. As a receptor of estrogen, ESR1 is important for bone metabolism via a variety of mechanisms in osteoblasts, osteocytes and osteoclasts [43].
The lead SNP rs13310130 in 7q21.3 is 6.9 kb upstream from the SHFM1 (split hand/split foot malformation, type 1) gene. Though reported by multiple studies [6, 11, 13], its role in bone metabolism is largely unknown.
DISCUSSION
In this study, we performed a genome-wide association meta-analysis of Wards' triangle BMD in 7,175 participants from 6 GWAS samples. We performed replication studies in the Framingham heart study, and in the largest GEFOS summary results. Combining the evidence of all these samples, we identified two novel loci 1q43 and 2q32.2 for areal BMD, and replicated three previously reported loci, 5q14.3, 6q25.1 and 7q21.3.
Ward's triangle is an anatomic region in the neck of the femur that is formed by the intersection of three trabecular bundles. In densitometry, Ward's triangle is a calculated region of low density in the femoral neck rather than a specific anatomic region. Ward's triangle BMD is a sensitive indicator of osteoporosis. For example, osteoporosis based on World Health Organization criteria (T-score less than −2.5) could be defined by Ward's triangle BMD in 53% of the study subjects [15]. In contrast, it was defined by femoral neck and by lumbar spine BMD in only 22% and 2% of study subjects, respectively.
Though BMD variation is predominantly determined by inheritance, the identified heritability has been relatively small. To enhance gene mapping efficacy, previous studies mainly focused on increasing sample size and expanding the type of tested genomic variants. In the current study, we explored a third option i.e., we expanded the list of tested skeletal sites. Although BMD at these sites are correlated to each other, there are differences. Consequently BMD at each of these sites can be informative for studying the genetic basis of BMD. In previous studies, BMD at the femoral neck and lumbar spine were studied most frequently, while BMD at other skeletal sites were rarely studied.
We replicated the associations of Ward's triangle BMD by analyzing BMD at several relevant related skeletal sites, including the femoral neck, trochanter, inter-trochanter, forearm and spine. Strictly speaking, such replication is indeed cross-validation; its validity is determined by the strength of phenotypic correlation between the primary trait and the replication trait. This replication strategy has been adopted in the identification of common variants for related traits, such as different subtypes of brain tumors [44]. It is important to recognize that, though replication by a related trait may be helpful, weaknesses also exist. First, genetic heterogeneity may occur for the two traits being studied. Second, non-replicability may occur when the genetic effect is specific to the primary trait but not to the replication trait.
The discovery stage of the present study was a trans-ethnic GWAS of samples from diverse ancestral populations. The purpose of including trans-ethnic samples is to maximize sample size and statistical power of association testing, under the hypothesis that phenotypic traits in different ethnic populations may have a common genetic basis. In contrast, the entirety of both replication samples represented populations of European ancestry only. Mismatched ancestry between the discovery and replication samples may have contributed to non-replicability for those non-replicated variants, because the discovery association signals might have been driven by the non-European samples. To check this possibility, we analyzed the discovery European samples separately. The results listed in Supplementary Table 5 showed that European samples dominated the overall association signals in the discovery samples. Therefore, the non-replicability was probably attributable to true negative associations, or insufficient replication sample size, rather than to mismatched ancestry between discovery and replication samples.
GREM2 which is expressed in osteoblasts during skeletogenesis [45], was originally identified in embryonic stem cells by gene trapping [46]. The GREM2 gene product contains a domain that is rich in cysteine residues, that is common in regulators of BMP activity, known as acysteine knot domain [47]. Its ability to bind to, and block the activities of, BMP family members has been demonstrated in both in vivo and in vitro studies [48, 49]. Its expression is, in turn, regulated by BMP2 during osteoblast differentiation [50]. Gene ontology (GO) annotations related to this gene include cytokine activity.
NAB1 was originally isolated in yeasts by interactive cloning [38]. It represses the transcriptional activity of EGR1 through a direct interaction, which is essential for transcriptional regulation of BMPR2 [51]. BMPR2, together with other BMP receptors, is involved in the BMP signaling pathway.
In conclusion, by performing a genome-wide association study of Ward's triangle BMD, we have identified 1q43 and 2q32.2 for areal BMD, and have replicated three previously reported loci. Our findings provide useful insights that further enhance our understanding of bone development, osteoporosis, and fracture pathogenesis.
Supplementary Material
Acknowledgments
This study was partially supported by the national natural science foundation of China (31571291 and 31100902 to L.Z., 31501026 to Y.F.P.), the NIH (P50AR055081, R01AG026564, R01AR050496, RC2DE020756, R01AR057049, and R03TW008221 to H.W.D.), the natural science foundation of Jiangsu province of China (BK20150323 to Y.F.P.), the natural science foundation of Inner Mongolia of China (2014MS08130 to R.H.), the Franklin D. Dickson/Missouri Endowment and the Edward G. Schlieder Endowment (to H.W.D.), the startup funding project of Soochow university (Q413900214 to L.Z. and Q413900114 to Y.F.P.) and a project of the priority academic program development of Jiangsu higher education institutions. Computing service was partially provided by the university of Shanghai for science and technology computing cluster. The funders had no role in study design, data collection and analysis, results interpretation or preparation of the manuscript.
The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL-64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. Funding support for the Framingham Whole Body and Regional Dual X-ray Absorptiometry (DXA) dataset was provided by NIH grants R01 AR/AG 41398. The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession phs000342.v14.p10.
The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. This manuscript was not prepared in collaboration with investigators of the WHI, has not been reviewed and/or approved by the Women’s Health Initiative (WHI), and does not necessarily reflect the opinions of the WHI investigators or the NHLBI. Funding for WHI SHARe genotyping was provided by NHLBI Contract N02-HL-64278. The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession phs000200.v10.p3.
Funding support for the Genetic Determinants of Bone Fragility (the Indiana fragility study) was provided through the NIA Division of Geriatrics and Clinical Gerontology. Genetic Determinants of Bone Fragility is a genome-wide association studies funded as part of the NIA Division of Geriatrics and Clinical Gerontology. Support for the collection of datasets and samples were provided by the parent grant, Genetic Determinants of Bone Fragility (P01-AG018397). Funding support for the genotyping which was performed at the Johns Hopkins University Center for Inherited Diseases Research was provided by the NIH NIA. The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession phs000138.v2.p1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All authors declare that they have no conflict of interest.
REFERENCES
- 1.Reginster JY, Burlet N. Osteoporosis: a still increasing prevalence. Bone. 2006;38:S4–S9. doi: 10.1016/j.bone.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Cummings SR, Melton LJ. Epidemiology and outcomes of osteoporotic fractures. Lancet. 2002;359:1761–1767. doi: 10.1016/S0140-6736(02)08657-9. [DOI] [PubMed] [Google Scholar]
- 3.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23:303–326. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 4.Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, Wilson SG, Andrew T, Falchi M, Gwilliam R, Ahmadi KR, Valdes AM, Arp P, Whittaker P, Verlaan DJ, Jhamai M, Kumanduri V, Moorhouse M, van Meurs JB, Hofman A, Pols HA, Hart D, Zhai G, Kato BS, Mullin BH, Zhang F, Deloukas P, Uitterlinden AG, Spector TD. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Center JR, Nguyen TV, Bagger Y, Gulcher JR, Eisman JA, Christiansen C, Sigurdsson G, Kong A, Thorsteinsdottir U, Stefansson K. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 6.Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, Richards JB, Zillikens MC, Kavvoura FK, Amin N, Aulchenko YS, Cupples LA, Deloukas P, Demissie S, Grundberg E, Hofman A, Kong A, Karasik D, van Meurs JB, Oostra B, Pastinen T, Pols HA, Sigurdsson G, Soranzo N, Thorleifsson G, Thorsteinsdottir U, Williams FM, Wilson SG, Zhou Y, Ralston SH, van Duijn CM, Spector T, Kiel DP, Stefansson K, Ioannidis JP, Uitterlinden AG. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong DH, Liu XG, Guo YF, Tan LJ, Wang L, Sha BY, Tang ZH, Pan F, Yang TL, Chen XD, Lei SF, Yerges LM, Zhu XZ, Wheeler VW, Patrick AL, Bunker CH, Guo Y, Yan H, Pei YF, Zhang YP, Levy S, Papasian CJ, Xiao P, Lundberg YW, Recker RR, Liu YZ, Liu YJ, Zmuda JM, Deng HW. Genome-wide association and follow-up replication studies identified ADAMTS18 and TGFBR3 as bone mass candidate genes in different ethnic groups. Am J Hum Genet. 2009;84:388–398. doi: 10.1016/j.ajhg.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Y, Tan LJ, Lei SF, Yang TL, Chen XD, Zhang F, Chen Y, Pan F, Yan H, Liu X, Tian Q, Zhang ZX, Zhou Q, Qiu C, Dong SS, Xu XH, Guo YF, Zhu XZ, Liu SL, Wang XL, Li X, Luo Y, Zhang LS, Li M, Wang JT, Wen T, Drees B, Hamilton J, Papasian CJ, Recker RR, Song XP, Cheng J, Deng HW. Genome-wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis. PLoS Genet. 2010;6:e1000806. doi: 10.1371/journal.pgen.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koller DL, Ichikawa S, Lai D, Padgett LR, Doheny KF, Pugh E, Paschall J, Hui SL, Edenberg HJ, Xuei X, Peacock M, Econs MJ, Foroud T. Genome-wide association study of bone mineral density in premenopausal European-American women and replication in African-American women. J Clin Endocrinol Metab. 2010;95:1802–1809. doi: 10.1210/jc.2009-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan EL, Danoy P, Kemp JP, Leo PJ, McCloskey E, Nicholson GC, Eastell R, Prince RL, Eisman JA, Jones G, Sambrook PN, Reid IR, Dennison EM, Wark J, Richards JB, Uitterlinden AG, Spector TD, Esapa C, Cox RD, Brown SD, Thakker RV, Addison KA, Bradbury LA, Center JR, Cooper C, Cremin C, Estrada K, Felsenberg D, Gluer CC, Hadler J, Henry MJ, Hofman A, Kotowicz MA, Makovey J, Nguyen SC, Nguyen TV, Pasco JA, Pryce K, Reid DM, Rivadeneira F, Roux C, Stefansson K, Styrkarsdottir U, Thorleifsson G, Tichawangana R, Evans DM, Brown MA. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011;7:e1001372. doi: 10.1371/journal.pgen.1001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, Koller DL, Li G, Liu CT, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao SM, Yerges-Armstrong LM, Zheng HF, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, Garcia-Giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim GS, Kooperberg C, Koromila T, Kruk M, Laaksonen M, Lacroix AZ, Lee SH, Leung PC, Lewis JR, Masi L, Mencej-Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi ML, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, Gonzalez-Macias J, Kahonen M, Karlsson M, Khusnutdinova E, Koh JM, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren O, Lorenc RS, Marc J, Mellstrom D, Obermayer-Pietsch B, Olmos JM, Pettersson-Kymmer U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Lagboom PE, Tang NL, Urreizti R, Van Hul W, Viikari J, Zarrabeitia MT, Aulchenko YS, Castano-Betancourt M, Grundberg E, Herrera L, Ingvarsson T, Johannsdottir H, Kwan T, Li R, Luben R, Medina-Gomez C, Th Palsson S, Reppe S, Rotter JI, Sigurdsson G, van Meurs JB, Verlaan D, Williams FM, Wood AR, Zhou Y, Gautvik KM, Pastinen T, Raychaudhuri S, Cauley JA, Chasman DI, Clark GR, Cummings SR, Danoy P, Dennison EM, Eastell R, Eisman JA, Gudnason V, Hofman A, Jackson RD, Jones G, Jukema JW, Khaw KT, Lehtimaki T, Liu Y, Lorentzon M, McCloskey E, Mitchell BD, Nandakumar K, Nicholson GC, Oostra BA, Peacock M, Pols HA, Prince RL, Raitakari O, Reid IR, Robbins J, Sambrook PN, Sham PC, Shuldiner AR, Tylavsky FA, van Duijn CM, Wareham NJ, Cupples LA, Econs MJ, Evans DM, Harris TB, Kung AW, Psaty BM, Reeve J, Spector TD, Streeten EA, Zillikens MC, Thorsteinsdottir U, Ohlsson C, Karasik D, Richards JB, Brown MA, Stefansson K, Uitterlinden AG, Ralston SH, Ioannidis JP, Kiel DP, Rivadeneira F. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012 doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng HF, Forgetta V, Hsu YH, Estrada K, Rosello-Diez A, Leo PJ, Dahia CL, Park-Min KH, Tobias JH, Kooperberg C, Kleinman A, Styrkarsdottir U, Liu CT, Uggla C, Evans DS, Nielson CM, Walter K, Pettersson-Kymmer U, McCarthy S, Eriksson J, Kwan T, Jhamai M, Trajanoska K, Memari Y, Min J, Huang J, Danecek P, Wilmot B, Li R, Chou WC, Mokry LE, Moayyeri A, Claussnitzer M, Cheng CH, Cheung W, Medina-Gomez C, Ge B, Chen SH, Choi K, Oei L, Fraser J, Kraaij R, Hibbs MA, Gregson CL, Paquette D, Hofman A, Wibom C, Tranah GJ, Marshall M, Gardiner BB, Cremin K, Auer P, Hsu L, Ring S, Tung JY, Thorleifsson G, Enneman AW, van Schoor NM, de Groot LC, van der Velde N, Melin B, Kemp JP, Christiansen C, Sayers A, Zhou Y, Calderari S, van Rooij J, Carlson C, Peters U, Berlivet S, Dostie J, Uitterlinden AG, Williams SR, Farber C, Grinberg D, LaCroix AZ, Haessler J, Chasman DI, Giulianini F, Rose LM, Ridker PM, Eisman JA, Nguyen TV, Center JR, Nogues X, Garcia-Giralt N, Launer LL, Gudnason V, Mellstrom D, Vandenput L, Amin N, van Duijn CM, Karlsson MK, Ljunggren O, Svensson O, Hallmans G, Rousseau F, Giroux S, Bussiere J, Arp PP, Koromani F, Prince RL, Lewis JR, Langdahl BL, Hermann AP, Jensen JE, Kaptoge S, Khaw KT, Reeve J, Formosa MM, Xuereb-Anastasi A, Akesson K, McGuigan FE, Garg G, Olmos JM, Zarrabeitia MT, Riancho JA, Ralston SH, Alonso N, Jiang X, Goltzman D, Pastinen T, Grundberg E, Gauguier D, Orwoll ES, Karasik D, Davey-Smith G, Smith AV, Siggeirsdottir K, Harris TB, Zillikens MC, van Meurs JB, Thorsteinsdottir U, Maurano MT, Timpson NJ, Soranzo N, Durbin R, Wilson SG, Ntzani EE, Brown MA, Stefansson K, Hinds DA, Spector T, Cupples LA, Ohlsson C, Greenwood CM, Jackson RD, Rowe DW, Loomis CA, Evans DM, Ackert-Bicknell CL, Joyner AL, Duncan EL, Kiel DP, Rivadeneira F, Richards JB. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Choi HJ, Estrada K, Leo PJ, Li J, Pei YF, Zhang Y, Lin Y, Shen H, Liu YZ, Liu Y, Zhao Y, Zhang JG, Tian Q, Wang YP, Han Y, Ran S, Hai R, Zhu XZ, Wu S, Yan H, Liu X, Yang TL, Guo Y, Zhang F, Guo YF, Chen Y, Chen X, Tan L, Deng FY, Deng H, Rivadeneira F, Duncan EL, Lee JY, Han BG, Cho NH, Nicholson GC, McCloskey E, Eastell R, Prince RL, Eisman JA, Jones G, Reid IR, Sambrook PN, Dennison EM, Danoy P, Yerges-Armstrong LM, Streeten EA, Hu T, Xiang S, Papasian CJ, Brown MA, Shin CS, Uitterlinden AG, Deng HW. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet. 2014;23:1923–1933. doi: 10.1093/hmg/ddt575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang TL, Chen XD, Guo Y, Lei SF, Wang JT, Zhou Q, Pan F, Chen Y, Zhang ZX, Dong SS, Xu XH, Yan H, Liu X, Qiu C, Zhu XZ, Chen T, Li M, Zhang H, Zhang L, Drees BM, Hamilton JJ, Papasian CJ, Recker RR, Song XP, Cheng J, Deng HW. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am J Hum Genet. 2008;83:663–674. doi: 10.1016/j.ajhg.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshihashi AK, Drake AJ, 3rd, Shakir KM. Ward's triangle bone mineral density determined by dual-energy x-ray absorptiometry is a sensitive indicator of osteoporosis. Endocr Pract. 1998;4:69–72. doi: 10.4158/EP.4.2.69. [DOI] [PubMed] [Google Scholar]
- 16.The Women's Health Initiative Study Group. Design of the Women's Health Initiative clinical trial and observational study. The Women's Health Initiative Study Group. Control. Clin. Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 17.Mahmood SS, Levy D, Vasan RS, Wang TJ. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. Lancet. 2014;383:999–1008. doi: 10.1016/S0140-6736(13)61752-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Pei YF, Fu X, Lin Y, Wang YP, Deng HW. FISH: fast and accurate diploid genotype imputation via segmental hidden Markov model. Bioinformatics. 2014;30:1876–1883. doi: 10.1093/bioinformatics/btu143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Li J, Pei YF, Liu Y, Deng HW. Tests of association for quantitative traits in nuclear families using principal components to correct for population stratification. Ann. Hum. Genet. 2009;73:601–613. doi: 10.1111/j.1469-1809.2009.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 26.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 32.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van Iterson M, Almlof J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HP, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Montgomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Lehrach H, Schreiber S, Sudbrak R, Carracedo A, Antonarakis SE, Hasler R, Syvanen AC, van Ommen GJ, Brazma A, Meitinger T, Rosenstiel P, Guigo R, Gut IG, Estivill X, Dermitzakis ET. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leslie R, O'Donnell CJ, Johnson AD. GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics. 2014;30:i185–i194. doi: 10.1093/bioinformatics/btu273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuniga E, Rippen M, Alexander C, Schilling TF, Crump JG. Gremlin 2 regulates distinct roles of BMP and Endothelin 1 signaling in dorsoventral patterning of the facial skeleton. Development. 2011;138:5147–5156. doi: 10.1242/dev.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russo MW, Sevetson BR, Milbrandt J. Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci U S A. 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bradley EW, Ruan MM, Oursler MJ. Novel pro-survival functions of the Kruppel-like transcription factor Egr2 in promotion of macrophage colony-stimulating factor-mediated osteoclast survival downstream of the MEK/ERK pathway. J Biol Chem. 2008;283:8055–8064. doi: 10.1074/jbc.M709500200. [DOI] [PubMed] [Google Scholar]
- 40.Zheng HF, Duncan EL, Yerges-Armstrong LM, Eriksson J, Bergstrom U, Leo PJ, Leslie WD, Goltzman D, Blangero J, Hanley DA, Carless MA, Streeten EA, Lorentzon M, Brown MA, Spector TD, Pettersson-Kymmer U, Ohlsson C, Mitchell BD, Richards JB. Meta-analysis of genome-wide studies identifies MEF2C SNPs associated with bone mineral density at forearm. J Med Genet. 2013;50:473–478. doi: 10.1136/jmedgenet-2012-101287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 42.Kramer I, Baertschi S, Halleux C, Keller H, Kneissel M. Mef2c deletion in osteocytes results in increased bone mass. J Bone Miner Res. 2012;27:360–373. doi: 10.1002/jbmr.1492. [DOI] [PubMed] [Google Scholar]
- 43.Khalid AB, Krum SA. Estrogen receptors alpha and beta in bone. Bone. 2016;87:130–135. doi: 10.1016/j.bone.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhattacharjee S, Rajaraman P, Jacobs KB, Wheeler WA, Melin BS, Hartge P, Yeager M, Chung CC, Chanock SJ, Chatterjee N. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet. 2012;90:821–835. doi: 10.1016/j.ajhg.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ideno H, Takanabe R, Shimada A, Imaizumi K, Araki R, Abe M, Nifuji A. Protein related to DAN and cerberus (PRDC) inhibits osteoblastic differentiation and its suppression promotes osteogenesis in vitro. Exp Cell Res. 2009;315:474–484. doi: 10.1016/j.yexcr.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Minabe-Saegusa C, Saegusa H, Tsukahara M, Noguchi S. Sequence and expression of a novel mouse gene PRDC (protein related to DAN and cerberus) identified by a gene trap approach. Dev Growth Differ. 1998;40:343–353. doi: 10.1046/j.1440-169x.1998.t01-1-00010.x. [DOI] [PubMed] [Google Scholar]
- 47.Pearce JJ, Penny G, Rossant J. A mouse cerberus/Dan-related gene family. Dev Biol. 1999;209:98–110. doi: 10.1006/dbio.1999.9240. [DOI] [PubMed] [Google Scholar]
- 48.Merino R, Rodriguez-Leon J, Macias D, Ganan Y, Economides AN, Hurle JM. The BMP antagonist Gremlin regulates outgrowth, chondrogenesis and programmed cell death in the developing limb. Development. 1999;126:5515–5522. doi: 10.1242/dev.126.23.5515. [DOI] [PubMed] [Google Scholar]
- 49.Sudo S, Avsian-Kretchmer O, Wang LS, Hsueh AJ. Protein related to DAN and cerberus is a bone morphogenetic protein antagonist that participates in ovarian paracrine regulation. J Biol Chem. 2004;279:23134–23141. doi: 10.1074/jbc.M402376200. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki D, Yamada A, Aizawa R, Funato S, Matsumoto T, Suzuki W, Takami M, Miyamoto Y, Suzawa T, Yamamoto M, Baba K, Kamijo R. BMP2 differentially regulates the expression of Gremlin1 and Gremlin2, the negative regulators of BMP function, during osteoblast differentiation. Calcif Tissue Int. 2012;91:88–96. doi: 10.1007/s00223-012-9614-5. [DOI] [PubMed] [Google Scholar]
- 51.Gaddipati R, West JD, Loyd JE, Blackwell T, Lane KA, Lane NM, Lane KB. EGR1 is essential for transcriptional regulation of BMPR2. American Journal of Molecular Biology. 2011;1:131–139. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.