Abstract
Gastric cancer is one of the major causes of death due to cancer in the world. It is a multi-factorial disease with epigenetic factors being also involved in its development. FAT4 is a tumor suppressor gene exerting an important role in cell adhesion. This study aimed at analyzing FAT4 expression and promoter methylation in gastric cancer. FAT4 expression was studied in 30 tumoral tissues and their non-tumoral counterparts using Taqman real time PCR method. Promoter methylation was assessed using bisulfite conversion method followed by sequencing. Tumor tissues showed reduced FAT4 expression (P = 0.04). FAT4 downregulation was associated with tumor grade, with higher repression at advanced grades. Significant increase of promoter methylation was observed in tumoral tissues. Reduced expression of FAT4 and increased methylation of its promoter may be one of the effective processes in turning a healthy stomach tissue into a tumor tissue.
Keywords: Gastric cancer, FAT4, Gene expression, Promoter methylation
Introduction
Gastric cancer is one of the most malignant cancer types due to its late diagnosis and low survival rate (Park et al. 2011). According to the world health organization, gastric cancer has the third place among cancer – related deaths (World Health Organization 2014). In Iran, the northern and northwestern regions have the highest incidence of gastric cancer (Malekzadeh et al. 2009). Based on Lauren classification, intestinal and diffuse types are the most frequent forms of gastric cancer (Akhavan-Niaki and Samadani 2014). Notwithstanding the decline in intestinal type incidence, increasing in diffuse type can be observed in some geographic parts of the world (Lee et al. 2014). Remarkably, gastric cancer is a heterogenic disease in which the accumulation of genetic variations can be seen at different levels, including over expression of oncogenes and inactivation of tumor suppressor genes. Ineffective functioning of tumor suppressor genes may have irreversible effects on cells (Jemal et al. 2011; Qu et al. 2013). Relatively, there are many epigenetic factors that are involved in cancer development among which DNA methylation that is divided into hyper and hypo methylation is one of the most important ones (Herman 1999). Considerably, methylation can confirm and assure the appropriate regulation of gene expression and/or silencing (Akhavan-Niaki and Samadani 2013). A growing body of evidence have determined that epigenetic alteration is an important event during carcinogenesis and plays a critical role in transcriptional silencing of tumor suppressor genes (Galm and Esteller 2004; Jones and Baylin 2002). The addition of a methyl group happens consistently in cytosine within CpG dinucleotides in regions which are called CpG islands. Aberrant CpG dinucleotides hyper methylation which can be found in the promoter site of tumor suppressor genes, is a hallmark of epigenetic changes that may lead to expression repression (Worm and Guldberg 2002). Accordingly, there are many main genes that are engaged in carcinogenesis (Samadani and Akhavan-Niaki 2015). FAT4 is one of those genes which is a tumor suppressor and the human ortholog of Drosophila fat gene and belongs to E-cadherin gene family and encodes a huge transmembrane protein of over 5000 amino acids. Moreover, this gene is a member of hippo signaling pathway which has significant action in controlling organ size. The final part of this pathway leads the transcriptional coactivator (YAP/TAZ) to arrange cells for proliferation, evasion from apoptosis, or to promote organ size control through amplification of progenitor/stem cells (Ito et al. 2015; Jung et al. 2015; Zhao et al. 2010). Recent studies showed that FAT4 expression is suppressed in breast and lung cancers (Katoh 2012; Qi et al. 2009). Furthermore, it has been shown that FAT4 is down regulated in some gastric cell lines and gastric cancer tumors belonging to Japanese and Chinese populations (Cai et al. 2015; Yoshida et al. 2016). In this study, we investigated the methylation status of FAT4 promoter alongside its expression level in gastric cancer patients.
Materials and methods
Patients
The study was performed on 30 patients who had been diagnosed for gastric adenocarcinoma. Tissue samples were collected from tumoral and non-tumoral (region surrounding tumor) parts of the stomach during the surgery of patients for whom their relatives have signed an informed consent form, followed by deep freezing of tissue samples in liquid nitrogen. Ultimately, tissue samples were stored at −80 °C for further analyzes. The study was approved by the ethics committee of Babol University of Medical Sciences. 23 patients were men and 7 were women. Clinicopathological characteristics of the gastric cancer patients are illustrated in Table 1.
Table 1.
Clinicopathological characteristics of the gastric cancer patients
| Sex (n = 30) | Tumor stage: n | Metastasis: n | Age (n): |
|---|---|---|---|
| Female =7 Male =23 |
Stage 1: 2 Stage 2: 9 Stage 3: 18 Stage 4: 1 |
Yes: 21 No: 9 |
10 < 60 years 20 ≥ 60 years |
DNA methylation analysis
DNA was obtained using DNA extraction Kit (Bioneer, Korea). The EZ DNA methylation gold kit (Zymo research, USA) was used to carry out bisulfite conversion according to the manufacturer’s instructions. Bisulfite treatment can convert unmethylated cytosine into uracil while leaving methylated cytosine unchanged. A fragment with 10 CpGs of the FAT4 promoter region was amplified using HotStarTaq Plus DNA Polymerase (Qiagen, Germany) and forward 5’GGGTAAGTTYTAAAGTTTYTGAAGA3’ and reverse 5’CGCCARCAAAAATTCARCTRAC3’ primers. PCR amplification included an initial incubation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, 57 °C for 50 s and 72 °C for 50 s, followed by one cycle of 72 °C for 14 min. PCR products were directly sequenced after purification using an ABI 3730xl DNA Analyzer (Applied Biosystems, USA).
Gene expression analysis
RNA was isolated from human tissue specimen according to a previously described method (Samadani et al. 2015) using Trizol (Ambion™, USA). Briefly, 50–100 mg of tissue was crushed to powder by a mortar and pestle in the presence of tiny amounts of liquid nitrogen and 15–20 mg of powdered tissue was used for RNA isolation. RNA integrity was checked on agarose gel according to the presence of ribosomal RNA bands, and the quantity of RNA was measured by a nanodrop instrument (TC100, USA).
Reverse transcription (RT) reactions were carried on RNA samples and using a stem - loop specific primer. Briefly, in this method, RT reactions were performed by an mRNA specific stem-loop primer in the presence of 0.3-1 μg total RNA, 4 μL 5X reaction buffer, 10 mM each of dNTPs, and 1 μL (200 U/μL) RevertAid Reverse Transcriptase (Fermentas, Lithuania ) in a final volume of 20 μL, by 60 min incubation at 42 °C. QPCR was performed on StepOne™ Real-Time (Applied Biosystems, USA) by using universal reverse primer and universal Taqman probe as well as mRNA specific forward primer. The expression levels of FAT4 were normalized against GAPDH RNA. The 20 μL qPCR mixture included 1 μL RT product, 0.25 mM universal probe, 0.5 mM each forward and reverse primers. The PCR reagents were all from Qiagen HotStar-Taq reagent set. The mixtures were incubated at 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s and 60 °C for 1 min. All reactions were performed in triplicate. The universal probe and reverse primer are able to bind one after another with the loop and stem part of the initial stem-loop RT-qPCR primer while the melting temperature of the stem part decreases. The threshold cycle (Ct) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. The sequence of forward and reverse primers along with universal Taqman probe are presented in Table 2.
Table 2.
Sequences of primers and probe used for FAT4 and GAPDH expression analysis
| Forward FAT4 | 5'ACACTGTGATTGCCAGGAGAG3' |
| Reverse-Universal | 5'GTATCCAGTGCTGCGACCGT3' |
| probe-Universal | FAM 5'TGGATGTGTCTGCGGCGTTTTATCAT3' BHQ-1 |
| R-specific FAT 4 | 5' GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTAT-CATGCACTGGATACGAC CAAGAGTCCAGTC3' |
| Forward GAPDH | 5'TGGAGTCCACTGGCGTCTTCAC3' |
| R-specific GAPDH | 5'GTCGTATCCAGTGCTGCGACCGTATGGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGACAGGCATTGCTGA 3' |
Statistical analyzes
Relative gene expression analysis was determined using the 2-ΔΔCT (Livak and Schmittgen 2001) method by StepOne software v2.3. The significant difference was statistically analyzed by paired Student’s t test. A value of P < 0.05 was considered as statistically significant. QUMA (QUantification tool for Methylation Analysis) software was used for gathering information for methylation status of the tumorous tissues and the tissue surrounding the tumors (Kumaki et al. 2008). Analyzes were performed using commercially available statistical software (SPSS, version 19.0) using X 2 test.
Results
Loss of FAT4 expression in gastric cancer
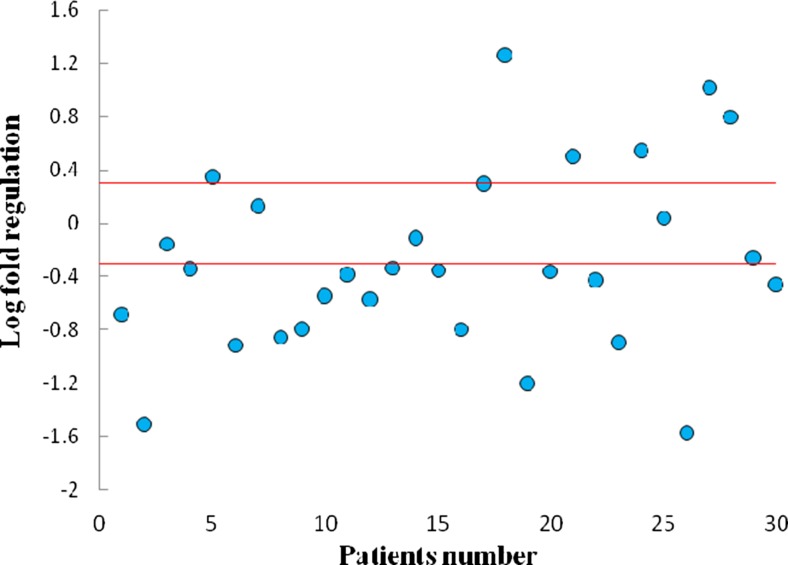
Gene expression analysis was performed by specific Taqman probe using real time PCR. The data were analyzed with Relative Expression Software Tool (REST) (Pfaffl et al. 2002) and Fig. 1 represents the scatter plot analysis of relative expression of FAT4 in gastric cancer patients. About 60 % (18 out of 30) of analyzed samples showed decreased FAT4 expression.
Fig. 1.
Scatter plot of FAT 4 gene expression in tumoral and non-tumoral tissues. Horizontal red lines represent cut-off values log2 fold changes in expression. The upper part of the graphs indicates up-regulation in the tumoral compared to the non-tumoral tissue; the lower part of the graph indicates down-regulation in the tumoral compared to the non-tumoral tissue (differences in expression ≥2; P < 0.05)
FAT4 methylation status in the tumor margin and tumor samples
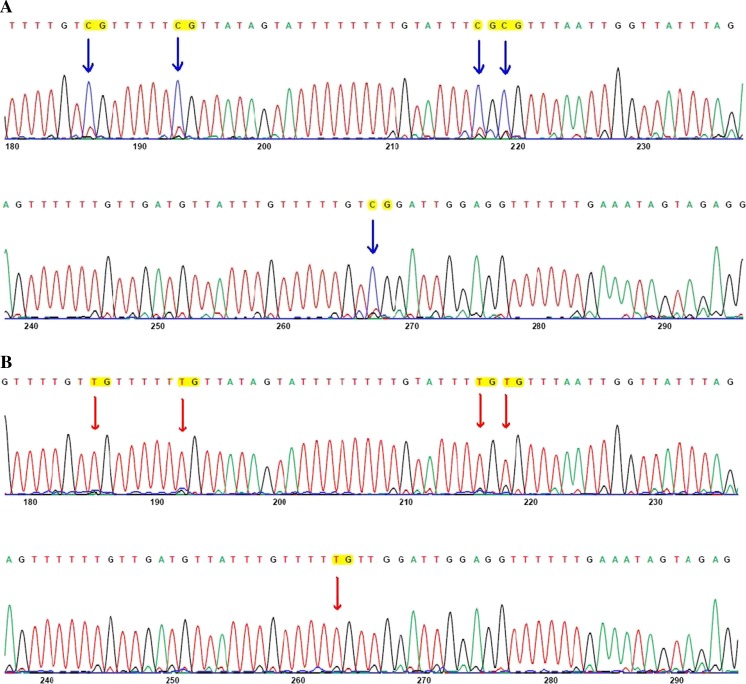
Using the QUMA software, the sequence of the promoter region of tumoral tissues and their margin were examined. 10 CpGs in the promoter area of FAT4 gene were informative. The percentage of methylation at each position was calculated by the software and the corresponding graphs were plotted (Fig. 2). Figure 3 is a representative electrophoregram of FAT4 promoter methylation pattern. Among 10 analyzed CpG sites, 6 showed significant increase of methylation (Table 3).
Fig. 2.
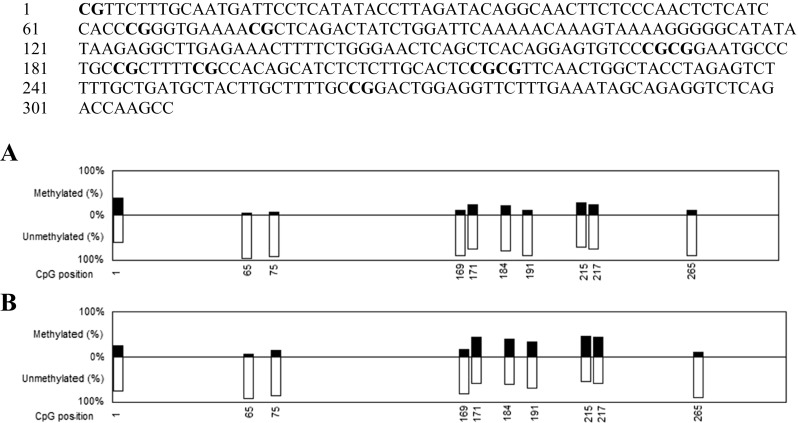
Methylation status of FAT4 promoter. FAT4 promoter sequence corresponding to position-436 to −80 (NCBI Reference Sequence: NG_033865.1) is presented and CpG dinucleotides are in bold. The graphs represent the methylation status of the 10 CpG present in the promoter region of FAT4 in A: tumor margin and B: tumor samples
Fig. 3.
Representative electrophoregrams of FAT4 methylation pattern. These electrophoregrams represent the sequence of nucleotides −210 to −94 of FAT4 corresponding to A: a tumoral tissue and B: the non-tumoral counterpart tissue. Genomic DNA was examined after bisulfite conversion and sequencing. All cytosines were converted to thymine while methylated cytosines remained unchanged and were read as cytosine. Red arrows indicate unmethylated CpG and blue arrows indicate methylated CpG
Table 3.
P-value calculation for each CpG
| CpG number | P value |
|---|---|
| 1 | 0.032317 |
| 2 | 0.194366 |
| 3 | 0.106388 |
| 4 | 0.147487 |
| 5 | 0.010273 |
| 6 | 0.008392 |
| 7 | 0.000235 |
| 8 | 0.012529 |
| 9 | 0.010273 |
| 10 | 1 |
Among 10 CpG sites, 6 showed significant increase of methylation status in tumoral tissues (P < 0.05)
Association of FAT4 expression with clinicopathological variables
To evaluate the clinicopathological consequences of FAT4 expression in gastric cancer, we analyzed the association of various clinicopathological variables with FAT4 mRNA level (Table 4). All patients presenting grade 1 or 2 tumors (11 cases) showed unchanged or decreased expression of FAT4 (mean ± SD: 0.363 ± 0.1), while 13 out of 19 tumor samples that were in grade 3 or 4 had a decreased expression of FAT4 (mean ± SD: 0.150 ± 0.113). Our data show an association of FAT4 expression with tumor grade (P < 0.05).
Table 4.
Correlation of FAT4 gene expression with clinicopathological features of gastric cancer patients
| Factor |
|
P value | |
|---|---|---|---|
| Tumor stage | |||
| 1-2 | 13 | 4 | 0.46 |
| 3-4 | 6 | 13 | |
| Tumor Grade | |||
| 1-2 | 11 | 0 | |
| 3-4 | 6 | 13 | 0.045 |
| Lymph node metastasis | |||
| Yes | 17 | 3 | 0.3 |
| No | 3 | 7 | |
| Distant metastasis | |||
| Yes | 3 | 18 | 0.23 |
| No | 3 | 6 | |
 : decrease or no change of gene expression;
: decrease or no change of gene expression;  : increase of gene expression
: increase of gene expression
Discussion
Gastric cancer is one of the prevalent cancers in the world that causes many deaths every year. Considering its silent development and consequent delayed diagnosis, this type of cancer is also one of the most resistant cancers to chemotherapy and sometimes surgery is the only choice of treatment, which is not sufficient with regard to disease development (Marin et al. 2016; Park et al. 2011; Zhang and Fan 2010).
With due consideration of the complications and unknown processes of creation and development of this type of cancer, a better comprehension of molecular mechanisms involved in its development can lead to early diagnosis and proper treatment of this malignant disease.
Besides mutations that dispose the cell to cancer, it has been found that epigenetic changes may also deactivate tumor suppressor genes and have an important role in tumor development (Bird 2002; Feinberg and Tycko 2004; Sebova and Fridrichova 2010). Accordingly, epigenetic treatment have been added as a new item to the treatment process along with others in various cancer cases (Egger et al. 2004; Mani and Herceg 2010, Yoo and Jones 2006). Regarding their very critical role in preventing various types of cancer, tumor suppressor genes are mostly affected by epigenetic phenomenon, and making an identification list of suppressor genes that undergo epigenetic changes in various types of cancer has been very much headed to in cancer epigenetic field and is currently under process to be completed by certain epigenetics researchers (Egger et al. 2004; Fukushige and Horii 2013).
Previous studies have shown reduced FAT4 gene expressions in other cancer samples, like breast cancer and lung cancer (Katoh 2012; Qi et al. 2009; Mizuno et al. 2009). Recently FAT4 expression reduction was also shown in gastric cancer (Yoshida et al. 2016) but in the present study we found a meaningful gene expression reduction in gastric tumor tissues in comparison with tumor margin tissues. Since FAT4 is considered as a preventive factor for activation of YAP/TAZ, reduced expression of this gene may result in activation of this pathway and will be an assisting factor in transformation of healthy cells to cancerous cells.
While the previous studies indicated an increase of methylation in FAT4 promoter in some cancer types like breast, and lung (Qi et al. 2009; Rauch et al. 2012), we examined FAT4 promoter methylation status in gastric cancer samples in 10 CpG sites and found a meaningful methylation increase in 6 CpGs in tumor tissues in comparison with tumor margin tissues. It is noteworthy that investigation of melanoma in a large cohort revealed the presence of missense and nonsense somatic mutations in FAT4 (Nikolaev et al. 2012), suggesting the possibility of point mutations and deletions occurrence, leading to non-functional FAT4 protein production in gastric cancer. Our findings suggest that FAT4 reduced expression and increase of methylation may be one of the effective processes in turning a healthy tissue into a tumoral one. Remarkably, the clinicopathological evaluation revealed that tumor stage which refers to the size and spreading of the tumor to neighboring tissues/organs, and consequently disease advance, did not show a significant association with FAT4 expression. However, we observed that reduction of FAT4 expression was grade specific. Tumor grade, refers to the organization and the extent of differentiation of the tumor. Low grade tumors are better differentiated and tend to grow and spread more slowly than high grade tumors which are poorly differentiated or undifferentiated. With increase of tumoral grade, a higher FAT4 down regulation was observed, suggesting the implication of FAT4 in tumor growth rate control.
These findings together with previous observations highlight the importance of FAT4 repression in gastric cancer development.
Acknowledgments
This work was supported by the Research Deputy of Babol University of Medical Sciences. Authors would like to express their gratitude to the director and staffs of Razi Pathobiology laboratory, Babol, Iran, and North Research Center of Pasteur Institute, Amol, Iran for providing necessary instruments for accomplishing some experimental parts of this study. We appreciate the helpful comments from the staff members of the said university, research center and laboratory.
Compliance with ethical standards
Conflict of interest
Authors declare no conflict of interest.
References
- Akhavan-Niaki H, Samadani AA. DNA methylation and cancer development: molecular mechanism. Cell Biochem Biophys. 2013;67:501–513. doi: 10.1007/s12013-013-9555-2. [DOI] [PubMed] [Google Scholar]
- Akhavan-Niaki H, Samadani AA. Molecular insight in gastric cancer induction: an overview of cancer stemness genes. Cell Biochem Biophys. 2014;68:463–473. doi: 10.1007/s12013-013-9749-7. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Cai J, Feng D, Hu L, Chen H, Yang G, Cai Q, Gao C, Wei D. FAT4 functions as a tumour suppressor in gastric cancer by modulating Wnt/β-catenin signalling. Br J Cancer. 2015;113:1720–1729. doi: 10.1038/bjc.2015.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Fukushige S, Horii A. DNA methylation in cancer: a gene silencing mechanism and the clinical potential of its biomarkers. Tohoku J Exp Med. 2013;229:173–185. doi: 10.1620/tjem.229.173. [DOI] [PubMed] [Google Scholar]
- Galm O, Esteller M. Beyond genetics--the emerging role of epigenetic changes in hematopoietic malignancies. Int J Hematol. 2004;80:120–127. doi: 10.1532/IJH97.04075. [DOI] [PubMed] [Google Scholar]
- Herman JG. Hypermethylation of tumor suppressor genes in cancer. Semin Cancer Biol. 1999;9:359–367. doi: 10.1006/scbi.1999.0138. [DOI] [PubMed] [Google Scholar]
- Ito T, Taniguchi H, Fukagai K, Okamuro S, Kobayashi A. Inhibitory mechanism of FAT4 gene expression in response to actin dynamics during Src-induced carcinogenesis. PLoS One. 2015;10:e0118336. doi: 10.1371/journal.pone.0118336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg962. [DOI] [PubMed] [Google Scholar]
- Jung HY, Cho H, MH O, Lee JH, Lee HJ, Jang SH, Lee MS. Loss of FAT atypical cadherin 4 expression is associated with high pathologic T stage in radically resected gastric cancer. J Gastric Cancer. 2015;15:39–45. doi: 10.5230/jgc.2015.15.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Function and cancer genomics of FAT family genes (review. Int J Oncol. 2012;41:1913–1918. doi: 10.3892/ijo.2012.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Cho YS, Lee GK, Lee S, Kim YW, Jho S, Kim HM, Hong SH, Hwang JA, Kim SY, Hong D, Choi IJ, Kim BC, Kim BC, Kim CH, Choi H, Kim Y, Kim KW, Kong G, Kim HL, Bhak J, Lee SH, Lee JS. Genomic profile analysis of diffuse-type gastric cancers. Genome biol. 2014;15:R55. doi: 10.1186/gb-2014-15-4-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Malekzadeh R, Derakhshan MH, Malekzadeh Z. Gastric cancer in Iran: epidemiology and risk factors. Arch Iran Med. 2009;12:576–583. [PubMed] [Google Scholar]
- Mani S, Herceg Z. DNA demethylating agents and epigenetic therapy of cancer. Adv Genet. 2010;70:327–340. doi: 10.1016/B978-0-12-380866-0.60012-5. [DOI] [PubMed] [Google Scholar]
- Marin JJ, Al-Abdulla R, Lozano E, Briz O, Bujanda L, Banales JM, Macias RI. Mechanisms of resistance to chemotherapy in gastric cancer. Anti Cancer Agents Med Chem. 2016;16:318–334. doi: 10.2174/1871520615666150803125121. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genet. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2012;44:133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- Park JH, Park J, Choi JK, Lyu J, Bae MG, Lee YG, Bae JB, Park DY, Yang HK, Kim TY, Kim YJ. Identification of DNA methylation changes associated with human gastric cancer. BMC Med Genet. 2011;4:82. doi: 10.1186/1755-8794-4-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C, Zhu YT, Hu L, Zhu Y-J. Identification of Fat4 as a candidate tumor suppressor gene in breast cancers. Int J Cancer. 2009;124:793–798. doi: 10.1002/ijc.23775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Rauch TA, Wang Z, Wu X, Kernstine KH, Riggs AD, Pfeifer GP. DNA methylation biomarkers for lung cancer. Tumour Biol. 2012;33:287–296. doi: 10.1007/s13277-011-0282-2. [DOI] [PubMed] [Google Scholar]
- Samadani A, Akhavan-Niaki H. Interaction of sonic hedgehog (SHH) pathway with cancer stem cell genes in gastric cancer. Med Oncol. 2015;32:1–7. doi: 10.1007/s12032-015-0492-3. [DOI] [PubMed] [Google Scholar]
- Samadani AA, Nikbakhsh N, Fattahi S, Pourbagher R, Aghajanpour Mir SM, Mousavi Kani N, Abedian Z, Akhavan-Niaki H. RNA extraction from animal and Human’s cancerous tissues: does tissue matter? Int J Mol Cell Med. 2015;4:54–59. [PMC free article] [PubMed] [Google Scholar]
- Sebova K, Fridrichova I. Epigenetic tools in potential anticancer therapy. Anti-cancer. Drugs. 2010;21:565–577. doi: 10.1097/CAD.0b013e32833a4352. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2014) (February 2014. Retrieved 10 June 2014) Cancer Fact sheet N°297Available from: http://www.who.int/mediacentre/factsheets/fs297/en/.
- Worm J, Guldberg P. DNA methylation: an epigenetic pathway to cancer and a promising target for anticancer therapy. J Oral Pathol Med. 2002;31:443–449. doi: 10.1034/j.1600-0714.2002.00034.x. [DOI] [PubMed] [Google Scholar]
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Yamashita S, Niwa T, Mori A, Ito S, Ichinose M, Ushijima T. Epigenetic inactivation of FAT4 contributes to gastric field cancerization. Gastric cancer. 2016 doi: 10.1007/s10120-016-0593-5. [DOI] [PubMed] [Google Scholar]
- Zhang D, Fan D. New insights into the mechanisms of gastric cancer multidrug resistance and future perspectives. Future Oncol. 2010;6:527–537. doi: 10.2217/fon.10.21. [DOI] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. The hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]