Summary
Although a variety of transgenic plants that are tolerant to drought stress have been generated, many of these plants show growth retardation. To improve drought tolerance and plant growth, we applied a gene‐stacking approach using two transcription factor genes: DEHYDRATION‐RESPONSIVE ELEMENT‐BINDING 1A (DREB1A) and rice PHYTOCHROME‐INTERACTING FACTOR‐LIKE 1 (OsPIL1). The overexpression of DREB1A has been reported to improve drought stress tolerance in various crops, although it also causes a severe dwarf phenotype. OsPIL1 is a rice homologue of Arabidopsis PHYTOCHROME‐INTERACTING FACTOR 4 (PIF4), and it enhances cell elongation by activating cell wall‐related gene expression. We found that the OsPIL1 protein was more stable than PIF4 under light conditions in Arabidopsis protoplasts. Transactivation analyses revealed that DREB1A and OsPIL1 did not negatively affect each other's transcriptional activities. The transgenic plants overexpressing both OsPIL1 and DREB1A showed the improved drought stress tolerance similar to that of DREB1A overexpressors. Furthermore, double overexpressors showed the enhanced hypocotyl elongation and floral induction compared with the DREB1A overexpressors. Metabolome analyses indicated that compatible solutes, such as sugars and amino acids, accumulated in the double overexpressors, which was similar to the observations of the DREB1A overexpressors. Transcriptome analyses showed an increased expression of abiotic stress‐inducible DREB1A downstream genes and cell elongation‐related OsPIL1 downstream genes in the double overexpressors, which suggests that these two transcription factors function independently in the transgenic plants despite the trade‐offs required to balance plant growth and stress tolerance. Our study provides a basis for plant genetic engineering designed to overcome growth retardation in drought‐tolerant transgenic plants.
Keywords: Dehydration‐Responsive Element‐Binding protein 1A (DREB1A), Rice Phytochrome‐Interacting factor‐Like 1 (OsPIL1), Drought stress tolerance, Cell elongation, Flowering, Arabidopsis
Introduction
Drought is one of the most serious environmental stresses that affect global agriculture. In recent years, prolonged droughts have caused severe damage to crops in many of the most agriculturally productive areas of the world. At present, approximately 800 million people are undernourished (FAO, IFAD and WFP, 2015). In addition, the world population is expected to experience dramatic grow; thus, the world food crisis appears to be worsening. Therefore, to ensure a stable supply of foods and biomass materials, it is imperative to improve drought stress tolerance in crops.
Plant growth is repressed in response to environmental stresses, indicating that there are trade‐offs between growth and stress tolerance (Claeys and Inze, 2013). Although a variety of transgenic plants that are tolerant to drought stress have been generated, many of these plants show growth retardation (Yamaguchi‐Shinozaki and Shinozaki, 2006). To reduce the negative effects on plant growth, stress‐inducible promoters have been used to drive the expression of transgenes in transgenic plants (Bhatnagar‐Mathur et al., 2007; Kasuga et al., 1999; Pino et al., 2007; Suo et al., 2012). However, growth retardation appears to be unavoidable under the long‐term drought stress conditions because of the prolonged overexpression of transgenes, even when using stress‐inducible promoters. Additionally, it is necessary to select the optimal stress‐inducible promoters for each plant species. For example, an Arabidopsis RESPONSIVE TO DEHYDRATION 29A (RD29A) promoter is useful for overexpression during abiotic stress conditions in both Arabidopsis and tobacco, whereas it functions in the roots, but not the leaves, in rice (Ito et al., 2006; Kasuga, 2004). Therefore, additional approaches are required to improve the growth of stress‐tolerant plants.
Pyramiding (gene‐stacking) breeding is essential for biotechnology applications (Halpin, 2005). For example, plants that produce several Bacillus thuringiensis (Bt) proteins have been shown to improve insect resistance (Carriere et al., 2015). ‘Golden rice’ plants similarly contain three carotenoid biosynthesis genes for accumulating pro‐vitamin A (Ye et al., 2000). The co‐expression of 9‐cis‐epoxycarotenoid dioxygenase (NCED) and D‐arabinono‐1,4‐lactone oxidase (ALO) increases abscisic acid (ABA) and ascorbic acid levels and improves tolerance to drought and chilling in planta (Bao et al., 2016). Many genetically modified (GM) crops cultivated worldwide have been generated by gene stacking (Halpin, 2005). However, few studies have focused on engineering plants to improve both drought stress tolerance and growth using gene‐stacking approaches.
DREB1A is an APETALA2/ethylene‐responsive element‐binding factor (AP2/ERF)‐type transcription factor that specifically binds dehydration‐responsive elements (DREs) and up‐regulates stress‐inducible target gene expression. The overexpression of DREB1A improves the tolerance to drought, salt and freezing stress by enhancing late embryogenesis‐abundant (LEA) protein levels and the compatible solute contents in Arabidopsis (Kasuga et al., 1999; Maruyama et al., 2004, 2009). Overexpression of the Arabidopsis DREB1A gene was reported to enhance abiotic stress tolerance in many crops, such as rice, soybean, peanut and wheat (Bhatnagar‐Mathur et al., 2014; Ito et al., 2006; Pellegrineschi et al., 2004; Suo et al., 2012). Moreover, the mechanism of improved drought stress tolerance was revealed by transcriptome and metabolome analyses as presented above. Therefore, DREB1A appears to be one of the most agriculturally useful genes for improving abiotic stress tolerance in crops. However, DREB1A overexpression causes dwarfism and late flowering in plants, and additional investigations are required before it can be used in efficient agricultural applications.
Arabidopsis phytochrome‐interacting factor (PIF) family proteins, which are basic helix‐loop‐helix (bHLH)‐type transcription factors, were initially isolated through their interaction with phytochrome (Ni et al., 1999). PIFs function to promote seedling skotomorphogenesis, shade avoidance and floral induction and regulate the expression of many downstream genes (Leivar and Quail, 2011). In rice, six PIF‐like genes were identified by in silico analysis (Nakamura et al., 2007). Of these, OsPIL1 was reported to enhance cell elongation through the activation of cell wall synthesis‐related genes in rice (Todaka et al., 2012). Additionally, OsPIL1 expression levels are repressed under drought and low‐temperature conditions; thus, OsPIL1 appears to act as a key regulator of plant growth in response to abiotic stress.
In this study, to overcome the trade‐offs between growth and stress tolerance, we generated transgenic plants overexpressing both OsPIL1 and DREB1A and characterized these double overexpressors by phenotypic analyses, metabolome analyses and genomewide transcriptome analyses. We propose that OsPIL1 partially enhances plant growth and accelerates flowering time, even in the double overexpressors, without negative effects on DREB1A‐mediated drought stress tolerance.
Results
Stability of the rice OsPIL1 protein and transactivation activity of OsPIL1 with DREB1A in Arabidopsis and rice protoplasts
Arabidopsis PIF family proteins have been reported to enhance cell elongation, and their protein levels are regulated by light stimulation (Leivar and Quail, 2011). Light triggers the interaction of PIF family proteins with phytochrome B (PhyB), which results in the degradation of the PIF proteins by the 26S proteasome. However, we previously reported that one of the four amino acid residues important for PhyB binding in Arabidopsis PIF proteins is not conserved in the rice PIF, including protein OsPIL1, which does not interact with OsPhyB in yeast two‐hybrid and bimolecular fluorescence complementation (BiFC) assays (Todaka et al., 2012). Thus, we analysed the OsPIL1 protein stability in Arabidopsis mesophyll protoplasts under light conditions. sGFP‐fused OsPIL1 and PIF4 proteins were transiently expressed in Arabidopsis protoplasts, and the levels of these proteins were then analysed by immunoblotting (Figure S1). In addition, 3 × Flag‐tagged sGFP was co‐expressed in protoplasts as an internal control. To determine whether the OsPIL1 protein is degraded under light conditions, we incubated the transfected protoplasts in the dark and then transferred the protoplasts to light conditions in the absence or presence of the 26S proteasome inhibitor MG132 (Figure S1a). In the absence of MG132, the PIF4 protein was degraded in a light‐ and time‐dependent manner as previously reported (Nozue et al., 2007), whereas the OsPIL1 protein was more stable under the same conditions (Figure S1b). In addition, PIF4 and OsPIL1 were not degraded in the presence of MG132 (Figure S1b). These results demonstrate that OsPIL1 is more stable than PIF4 and show that OsPIL1 does not appear to be degraded by the 26S proteasome in response to light irradiation. Taken together, these results suggest that OsPIL1 may enhance plant growth more effectively than PIF4 in Arabidopsis under light conditions.
OsPIL1 and DREB1A have been reported to recognize the cis‐acting promoter elements G box (CACGTG) and DRE (A/GCCGAC), respectively (Liu et al., 1998; Todaka et al., 2012). To examine whether these transcription factors affect each other's transactivation activity, we performed transactivation assays using Arabidopsis and rice mesophyll protoplasts (Figures 1 and S2). 35SΩ:OsPIL1 and 35SΩ:DREB1A were used as effector constructs, and G box:GUS or DRE:GUS was used as a reporter construct (Liu et al., 1998; Todaka et al., 2012). The expression of OsPIL1 with the G box:GUS reporter construct showed high transactivation activity (Figures 1b and S2b). Similarly, when both OsPIL1 and DREB1A were co‐expressed, high transactivation activity was observed. When DRE:GUS was used as the reporter, the transactivation activity was enhanced by the co‐expression of both OsPIL1 and DREB1A in a manner similar to the expression of DREB1A (Figures 1c and S2c). These results indicate that OsPIL1 and DREB1A act independently and do not interfere with each other's transactivation activities in Arabidopsis and rice protoplasts.
Figure 1.
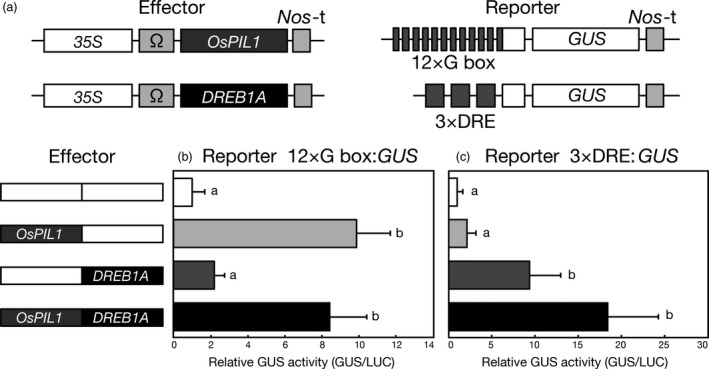
Effect of co‐expressing both OsPIL1 and DREB1A on each transactivation activity in Arabidopsis protoplasts. (a) Schematic diagram of the effector and reporter constructs used in the transactivation analysis of the Arabidopsis protoplasts. The effector construct contains a CaMV 35S promoter and a TMV Ω sequence fused to the coding sequence of OsPIL1 or DREB1A. Nos‐t indicates the polyadenylation signal of the gene for nopaline synthetase. The reporter construct 12 × G box:GUS contains 12 tandem 28‐bp G box‐containing fragments. The reporter construct 3 × DRE:GUS contains three tandem 71‐bp DRE‐containing fragments. (b, c) Transactivation effects of the co‐expression of OsPIL1 and DREB1A. The reporter 12 × G box:GUS (b) or 3 × DRE:GUS (c) and the effectors were co‐transfected into Arabidopsis protoplasts. To normalize for transfection efficiency, an emerald luciferase (ELUC) reporter gene driven by the CaMV 35S promoter was co‐transfected as a control in each experiment. Bars show the standard deviation (SD) of more than four replicates. The letters indicate significant differences among the assays (P < 0.05 according to Games–Howell's multiple range test).
Enhanced cell elongation and improved drought stress tolerance in transgenic plants overexpressing both OsPIL1 and DREB1A (double overexpressors)
Next, we generated transgenic Arabidopsis plants overexpressing both OsPIL1 and DREB1A (double overexpressors) by crossing DREB1A overexpressors with OsPIL1 overexpressors (Figure 2a, Table S1). OsPIL1 or DREB1A was overexpressed under the control of the CaMV 35S promoter and the TMV Ω sequence in transgenic plants. To explore whether the double overexpressors have phenotypes related to the overexpression of each OsPIL1 and DREB1A, we observed the morphology of these transgenic plants. Overexpression of rice OsPIL1 was reported to enhance hypocotyl elongation in Arabidopsis (Nakamura et al., 2007; Todaka et al., 2012). Thus, we measured the hypocotyl length of 7‐day‐old double overexpressors compared with that of 35SΩ:OsPIL1 and 35SΩ:DREB1A plants (Figure 2b, c). Significant differences were not observed in hypocotyl length between the two vector control plants, pGKX and pGHX (Figure 2c). The hypocotyl length of the double overexpressors was intermediate between the 35SΩ:OsPIL1 and the vector control plants. The hypocotyl length of the 35SΩ:DREB1A plants was equivalent to that of the vector control plants. We also observed the lengths of the hypocotyl cells in these transgenic plants and found that hypocotyl length correlated with cell length (Figure S3). In addition, we found that the internode and shoot lengths of 8‐week‐old double overexpressors were longer than those of the 35SΩ:DREB1A plants (Figures S4 and S5). These results indicate that overexpressing OsPIL1 enhances cell elongation of the hypocotyl and stem, even in the double overexpressors, and show that overexpressing DREB1A has a negative effect on OsPIL1‐mediated cell elongation. Furthermore, DREB1A overexpression has been reported to improve drought stress tolerance while causing a severe dwarf phenotype (Liu et al., 1998). Under normal growth conditions, the double overexpressors also showed small rosette leaves and short primary roots, which is similar to the 35SΩ:DREB1A plants (Figure S6a, b, c, d). Next, we weighed the total dry biomass of the transgenic Arabidopsis plants. The double overexpressors had a dry weight similar to the 35SΩ:DREB1A plants (Figure S6e). In addition, we evaluated the seed yield of the plants. Significant differences were not observed in average silique number between the vector control, 35SΩ:DREB1A and double overexpressor plants, but this number was decreased in the 35SΩ:OsPIL1 plants compared with the other plants (Figure S6f). The average seed number per silique was similar in all the transgenic plants (Figure S6g). Therefore, overexpression of OsPIL1 decreased the seed yield of the transgenic plants. This may be due to the early flowering and senescence phenotypes of the 35SΩ:OsPIL1 plants. Overexpression of Arabidopsis PIF4 is also known to induce early senescence in transgenic plants (Sakuraba et al., 2014). However, overexpression of OsPIL1 did not affect the seed yield in the double overexpressors.
Figure 2.
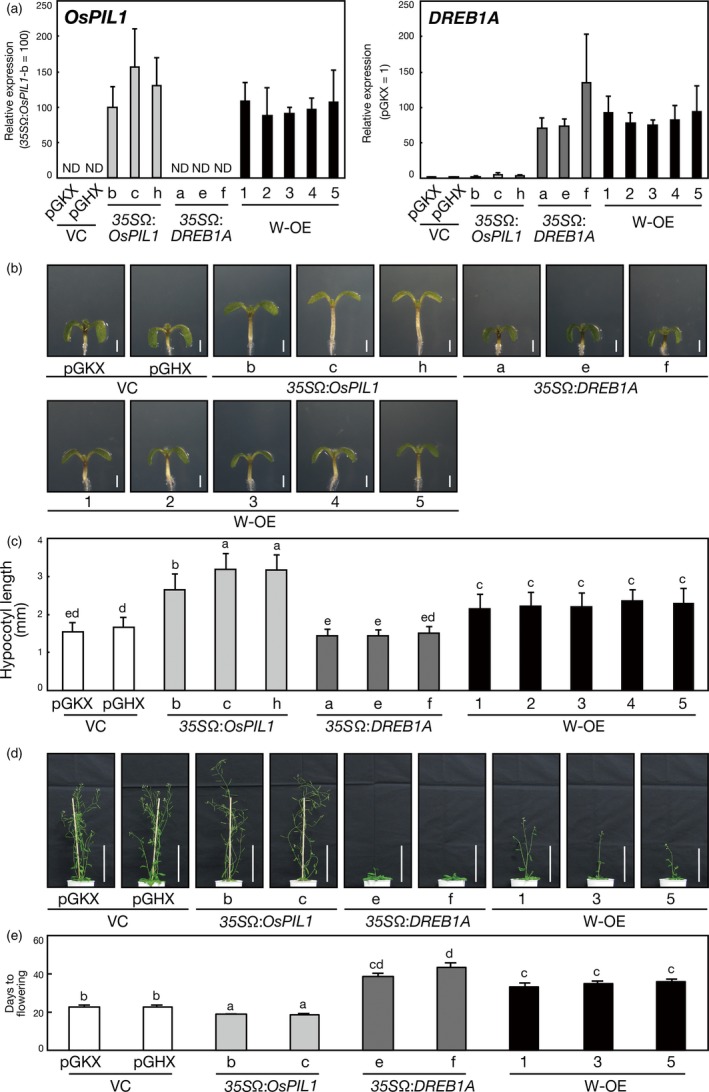
Generation of OsPIL1 and DREB1A double‐overexpressing Arabidopsis plants. (a) Expression levels of the transgenes in the single‐ or double‐overexpressing plants were analysed by quantitative RT‐PCR. The plants were grown on agar medium for 2 weeks. The error bars indicate the SD of more than four samples. ND represents not detected and VC and W‐OE represent vector control and OsPIL1 DREB1A double overexpressor, respectively. (b) Morphology of the transgenic Arabidopsis seedlings. The plants were grown on agar medium for 7 days. Bars = 1 mm. (c) Hypocotyl length calculated from the seedlings grown as in (b). The error bars show the SD of more than 50 seedlings. The letters indicate significant differences among the seedlings (P < 0.01 according to Games–Howell's multiple range test). (d, e) Flowering time of the single‐ or double‐overexpressing Arabidopsis plants. (d) Growth of 6‐week‐old transgenic plants. The plants were grown on agar medium for 2 weeks and then in soil pots. Bars = 10 cm. (e) Days to flowering of the plants grown as in (d). The error bars show the SD of more than four plants. The letters indicate significant differences among the plants (P < 0.05 according to Games–Howell's multiple range test).
In addition to growth retardation, the overexpression of DREB1A causes late‐flowering phenotypes in transgenic Arabidopsis plants (Seo et al., 2009), whereas the overexpression of PIF4, an Arabidopsis homologue of OsPIL1, causes an extremely early flowering phenotype by elevating FLOWERING LOCUS T (FT) expression levels (Kumar et al., 2012). Therefore, we analysed the flowering time of the transgenic plants (Figure 2d, e). Compared with the vector control plants, the 35SΩ:OsPIL1 plants showed early flowering and the 35SΩ:DREB1A plants showed late flowering. The flowering time of the double overexpressors was earlier than that of the 35SΩ:DREB1A plants, but later than that of the vector control and 35SΩ:OsPIL1 plants. These results suggest that OsPIL1 and DREB1A antagonistically regulate flowering time in the double overexpressors. Thus, OsPIL1 is expected to be useful for weakening the late‐flowering phenotypes caused by DREB1A overexpression. Taken together, the double overexpressors present phenotypes caused by the overexpression of each OsPIL1 and DREB1A.
We then evaluated the drought stress tolerance of the double overexpressors by withholding water for 2 weeks. No improvement in drought stress tolerance was observed in the 35SΩ:OsPIL1 plants, whereas the double overexpressors showed the improved drought stress tolerance similar to that of the 35SΩ:DREB1A plants (Figure 3), indicating that overexpressing OsPIL1 has no negative effects on the DREB1A‐mediated improvement of drought stress tolerance in the double overexpressors.
Figure 3.
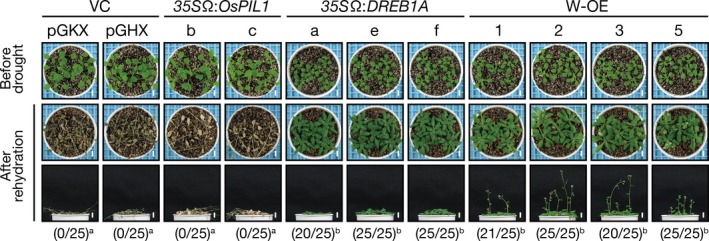
Drought stress tolerance of the single or double overexpressors. Number codes show the number of surviving plants out of the total number. A total of 25 plants were used in the survival test. Bars = 1 cm. The letters indicate significant differences among the plants (P < 0.05 according to Tukey's multiple range test of the ratio).
OsPIL1 and DREB1A affect sugar and amino acid metabolism in the double overexpressors
To reveal the metabolite profiles of the double overexpressors, we performed gas chromatography–time‐of‐flight mass spectrometry (GC‐TOF‐MS) analyses of 2‐week‐old (vegetative stage) transgenic plants and identified 43 metabolites, including sugars, amino acids and organic acids (Figures 4, S7a). We extracted the soluble metabolites from whole plants harvested at the subjective dawn (Zeitgeber time [ZT] = 0) and the subjective sundown (ZT = 16). Because the metabolite profiles of the double overexpressors harvested at these two sampling points were nearly equivalent, the differences in the metabolite profiles among the tested transgenic plants were independent of their daily changes (Figure S7b, c). According to principal components analysis (PCA), the contribution ratios of the first (PC1) and second principal components (PC2) were 30.1% and 21.8%, respectively (Figure 4a). The 35SΩ:DREB1A and 35SΩ:OsPIL1 plants were separated from the vector control plants by PC1 and PC2. Interestingly, the metabolite profiles of the double overexpressors were clearly different from those of each single overexpressor. The score plot of the double overexpressors was described by the metabolites that contributed to the positive side of the PC1 axis and the negative side of the PC2 axis (Figure 4a). We then compared the loadings of PC1 and PC2 to clarify the metabolites that contributed to the separation of the PCA score (Figure 4b). The top five metabolites contributing to the positive side of PC1 were raffinose, glycerol‐3‐phosphate, β‐alanine, proline and galactinol, and those contributing to the negative side were myo‐inositol, aspartic acid, fumaric acid, maltotriose and dehydroascorbic acid dimer. However, the top five positive contributors to PC2 were glucose, arginine, fructose, ornithine and guanidine, and the top five negative contributors were erythritol, citric acid, glutamic acid, 4‐hydroxyproline and 4‐aminobutyric acid (GABA).
Figure 4.
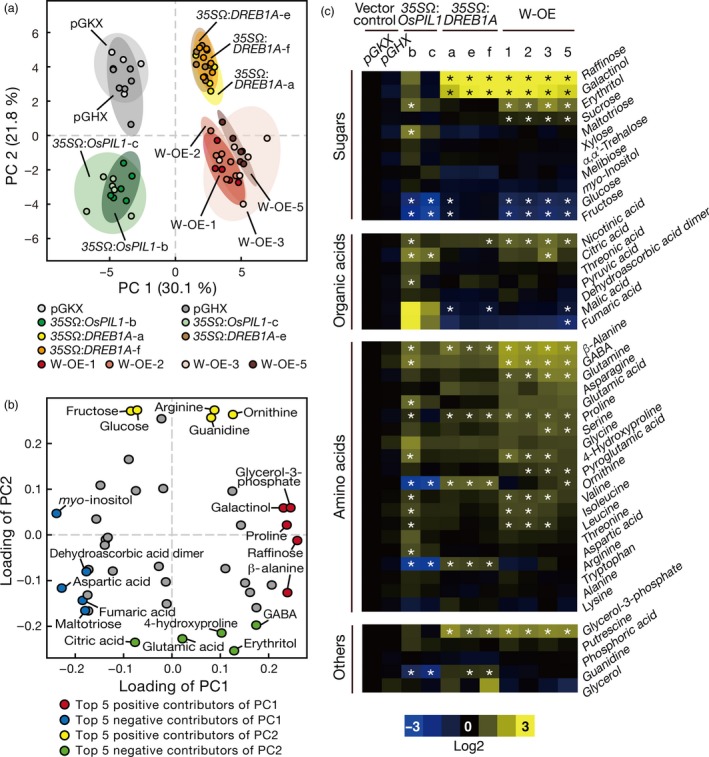
Metabolite profiles of the single‐ or double‐overexpressing plants. (a, b) Principal components analysis (PCA) of metabolite profile in the transgenic plants. Score plot (a) and loading plot (b) of the metabolite profiles in the transgenic plants. The plants were grown on agar medium for 2 weeks and harvested at Zeitgeber time (ZT) = 0. GABA, 4‐aminobutyric acid. (c) Heat maps of the metabolite profile in the transgenic plants. The relative metabolite amounts were normalized with vector control plants (pGKX) and transformed to log2. Yellow and blue colours show increased and decreased levels, respectively. Asterisks indicate significant differences between the vector control plants (both pGKX and pGHX) (P < 0.05 according to Tukey's multiple range test, n = 6).
In the 35SΩ:DREB1A plants, the amounts of raffinose, galactinol, β‐alanine, proline, ornithine, arginine and glycerol‐3‐phosphate were significantly increased relative to the vector control plants (Figure 4c). Galactinol and raffinose have been reported to act as osmoprotectants in drought stress tolerance in plants (Taji et al., 2002). Proline and β‐alanine are precursors of quaternary ammonium compounds (QAC), which also act as osmoprotectants (Hanson et al., 1994). Following the overexpression of OsPIL1, the amount of citric acid was increased, whereas the amounts of glucose, fructose, ornithine, arginine and guanidine were decreased relative to the vector control plants (Figure 4c). The metabolism of ornithine and arginine appeared to be oppositely affected by the overexpression of DREB1A and OsPIL1, and the levels of these metabolites in the double overexpressors were the average of those in the single overexpressors. The amounts of GABA and glutamine were slightly increased in the 35SΩ:DREB1A plants and clearly increased in the double overexpressors (Figure 4c). These results suggest that both OsPIL1 and DREB1A independently regulate sugar and amino acid metabolism in the double overexpressors.
Downstream genes of both OsPIL1 and DREB1A were activated in the double overexpressors
To reveal the transcript profiles, we performed microarray experiments using 2‐week‐old 35SΩ:OsPIL1 plants and double overexpressors harvested at ZT = 0. The overexpression of OsPIL1 alone and OsPIL1 and DREB1A in combination up‐regulated the expression of 30 and 356 genes, respectively (fold change > 2, false discovery rate [FDR] P < 0.05) (Figure 5a, left). We compared these up‐regulated genes with those in the 35SΩ:DREB1A plants (Kidokoro et al., 2015) and found 110 common genes between the 35SΩ:DREB1A plants and the double overexpressors, and these genes included many abiotic stress‐inducible genes (Table S2). Ten genes overlapped between the 35SΩ:OsPIL1 plants and the double overexpressors, including many cell elongation‐related genes (Table S3). No genes overlapped among all three transgenic Arabidopsis plants. The number of down‐regulated genes in the 35SΩ:DREB1A plants, 35SΩ:OsPIL1 plants and double overexpressors was 235, 29 and 437, respectively (fold change < 0.5, FDR P < 0.05) (Figure 5a, right). Fifty‐nine genes were common between the 35SΩ:DREB1A plants and the double overexpressors, including cell wall‐related genes (Table S4). Seven genes overlapped between the 35SΩ:OsPIL1 plants and the double overexpressors, although the functions of these genes were not relevant (Table S5). We observed many up‐regulated or down‐regulated genes in the double overexpressors, but not in the 35SΩ:OsPIL1 or 35SΩ:DREB1A plants alone. However, the majority of these genes were either up‐regulated or down‐regulated at low levels. The fold change values for approximately 70% of the up‐regulated or down‐regulated genes were less than threefold or more than 0.4‐fold (Tables S6 and S7).
Figure 5.
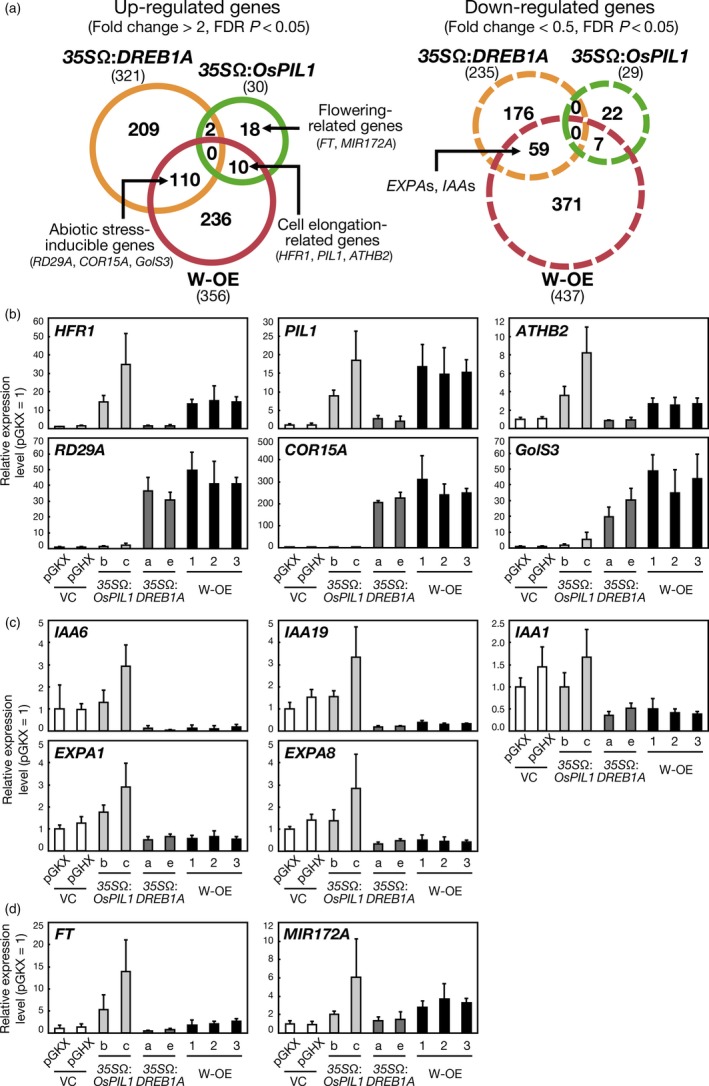
Comparative analyses of the transcriptomes of single‐ or double‐overexpressing plants. (a) Venn diagrams comparing up‐regulated and down‐regulated genes among the 35SΩ:DREB1A plants (orange circles), the 35SΩ:OsPIL1 plants (green circles) and the double overexpressors (red circles). The plants were grown on agar medium for 2 weeks and harvested at ZT = 0. Up‐regulated and down‐regulated genes are in solid circles and dashed circles, respectively. The total numbers of up‐regulated and down‐regulated genes are shown in parentheses. (b, c, d) Expression levels of up‐regulated genes (b), down‐regulated genes (c) and flowering regulator genes (d) in the transgenic Arabidopsis plants were analysed by quantitative RT‐PCR. The plants were grown on agar medium for 2 weeks and harvested at ZT = 0. The error bars indicate the SD of more than four samples.
Next, we analysed the expression patterns of up‐regulated and down‐regulated genes in each transgenic Arabidopsis plant using the microarray database at Genevestigator (https://genevestigator.com/gv/). The downstream genes in the double overexpressors were responsive to ABA, cold, drought, osmotic and salt stress, which is similar to the effects in the 35SΩ:DREB1A plants (Figure S8). Certain up‐regulated genes in the 35SΩ:OsPIL1 plants were responsive to ABA, indole‐3‐acetic acid (IAA) and far‐red (FR) light (Figure S8). To obtain additional information on the molecular functions of the downstream genes, we categorized these genes according to overrepresentation analyses using PageMan software (http://mapman.mpimp-golm.mpg.de/). Compared with the categorization of the complete Arabidopsis genome, the up‐regulated genes in the 35SΩ:DREB1A plants showed higher ratios of the categories secondary metabolism (12.7%), stress (11.7%), hormone metabolism (6.4%) and sugar metabolism (4.5%) (Figure S9). The ratios of the genes in the categories RNA (27.1%), hormone metabolism (12.9%) and signalling (8.6%) were increased in the 35SΩ:OsPIL1 plants (Figure S9). The ratios of the genes in the categories hormone metabolism (13.6%) and cell wall (5.0%) were higher among the down‐regulated genes in the 35SΩ:DREB1A plants (Figure S10). Genes in the categories secondary metabolism (11.5%), sugar metabolism (6.4%) and cell wall (11.5%) were overrepresented among the down‐regulated genes in the 35SΩ:OsPIL1 plants (Figure S10). The ratios of each category of up‐regulated and down‐regulated genes in the double overexpressors were similar to the average of the values in the 35SΩ:OsPIL1 and 35SΩ:DREB1A plants (Figures S9 and S10). Furthermore, because raffinose and galactinol were accumulated in the double overexpressors, sugar metabolism in the transgenic plants was analysed using the MapMan database (http://mapman.gabipd.org/). In the sugar metabolism pathway, genes for galactinol synthases and raffinose synthase were up‐regulated in 35SΩ:DREB1A and the double overexpressors (Figure S11). However, a gene for sucrose invertase was down‐regulated in 35SΩ:OsPIL1 (Figure S11).
We confirmed the expression levels of the downstream genes in the transgenic plants by using quantitative RT‐PCR (Figure 5b, c, d). LONG HYPOCOTYL IN FAR‐RED (HFR1), PHYTOCHROME‐INTERACTING FACTOR 3‐LIKE 1 (PIL1) and ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2 (ATHB2) are cell elongation‐related target genes of PIF4 in Arabidopsis (Hornitschek et al., 2012; Kunihiro et al., 2011). The expression levels of these three genes increased in the 35SΩ:OsPIL1 plants and the double overexpressors (Figure 5b). RD29A, COLD‐REGULATED 15A (COR15A) and GALACTINOL SYNTHASE 3 (GolS3) are major abiotic stress‐inducible targets of DREB1A (Maruyama et al., 2009), and the expression of these genes was elevated in the 35SΩ:DREB1A plants and the double overexpressors (Figure 5b). These up‐regulated genes may be important for cell elongation and drought stress tolerance in the double overexpressors. However, INDOLE‐3‐ACETIC ACID‐INDUCIBLE (IAA) genes and EXPANSIN A (EXPA) genes were down‐regulated in the 35SΩ:DREB1A plants and the doble overexpressors (Figure 5c). IAA genes are auxin inducible and act as repressors in auxin signalling (Reed, 2001). EXPAs are known to enhance cell wall enlargement by cell wall loosening (Cosgrove, 2000). In addition, we analysed the expression levels of flowering‐related genes, such as FT and MICRORNA172A (MIR172A), in the transgenic plants (Figure 5d). FT encodes florigen, whose overexpression causes early flowering (Turck et al., 2008). Overexpression of MIR172A also causes early flowering by down‐regulating AP2‐like floral repressors (Aukerman and Sakai, 2003). The expression levels of FT and MIR172A were elevated in the 35SΩ:OsPIL1 plants (Figure 5d). However, the expression levels of FT in the 35SΩ:DREB1A plants were approximately half of those in the vector control plants. In the double overexpressors, the expression levels of FT and MIR172A were higher than those in the control plants, which suggests that OsPIL1 enhances floral induction by regulating FT and MIR172A expression in the transgenic plants.
Furthermore, we analysed the expression levels of other PIF4 downstream genes, namely XYLOGLUCAN ENDOTRANSGLYCOSYLASE 7 (XTR7), PACLOBUTRAZOL RESISTANCE1 (PRE1) and IAA29 (Hornitschek et al., 2012; de Lucas et al., 2008; Nomoto et al., 2012; Oh et al., 2012); however, significant differences were not observed in the 35SΩ:OsPIL1 plants and double overexpressors compared with the vector control plants (Figure S12a). These results suggest that certain OsPIL1 target genes differ from those of PIF4. We then analysed the expression levels of SMALL AUXIN‐UPREGULATED RNA (SAUR) genes (Figure S12b). SAURs positively regulate cell elongation to promote hypocotyl growth, and PIF4 has been shown to up‐regulate the expression of SAUR19‐24 at high temperatures (Franklin et al., 2011; Ren and Gray, 2015). The expression levels of SAUR19, SAUR22 and SAUR23 were elevated in the 35SΩ:OsPIL1 plants and the double overexpressors (Figure S12b), indicating that OsPIL1 also enhances cell elongation through the up‐regulation of SAUR genes, which is similar to the action of PIF4 in the transgenic Arabidopsis plants. These results suggest that OsPIL1 acts as a positive regulator of auxin signalling.
To characterize the promoters of the up‐regulated and down‐regulated genes in the double overexpressors, we assessed the enrichment of all hexamer motifs (from AAAAAA to TTTTTT, 46 = 4096) in the promoters of the genes in each transgenic plant (Figure 6). We compared the frequency of each hexamer sequence in the promoters of the genes with their normalized frequencies in Arabidopsis promoters (Maruyama et al., 2012). In the 35SΩ:OsPIL1 plants, G box (CACGTG) and ABRE sequences (ACGTGG and CGTGGG) ranked among the top 10 up‐regulated genes (Figure 6a, top). For the 35SΩ:DREB1A plants, we analysed microarray data from a previous study (Figure 6a, middle; Kidokoro et al., 2015). Consistent with previous work, the most overrepresented sequence in the 35SΩ:DREB1A plants was DRE (Maruyama et al., 2012). In the double overexpressors, DRE sequences occupied the top 10, and G box or ABRE sequences were ranked within the top 15 among the up‐regulated genes (Figure 6a, bottom). The double overexpressor‐specific elements were not found in the promoter regions of the up‐regulated genes. These results indicate that OsPIL1 and DREB1A activate the expression of each downstream gene independently via each cis‐acting element in the double overexpressors. However, known cis elements were not identified among the enriched hexamer motifs in the promoters of the down‐regulated genes in the 35SΩ:OsPIL1 plants and the double overexpressors (Figure 6b). In the 35SΩ:DREB1A plants, known hexamer motifs were also not observed, suggesting that IAA and EXPA gene expression is indirectly repressed by DREB1A.
Figure 6.
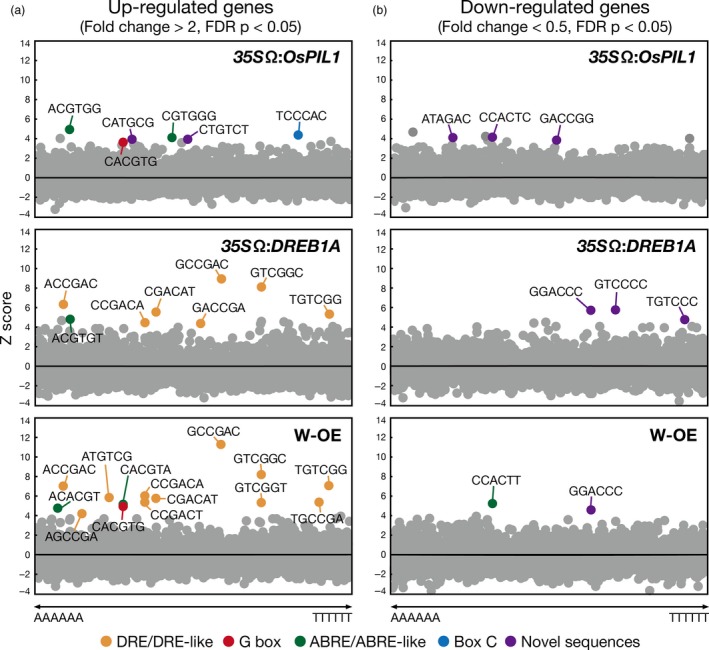
Overrepresentation analyses of hexamer sequences in the promoters of up‐regulated and down‐regulated genes in the single or double overexpressors. Scatter plots showing Z scores (y‐axes) for the observed frequencies of all hexamer sequences (x‐axes) in the 1‐kb promoters of up‐regulated (a) and down‐regulated genes (b) in each transgenic plant. DRE/DRE‐like (orange), G box (red), ABRE/ABRE‐like (green), Box C (blue) and novel sequences (purple) are shown.
Discussion
Considerable effort has been devoted to producing GM plants with improved tolerance to abiotic stress, such as drought and high salinity, by using molecular breeding strategies. The overexpression of DREB1A induces many stress‐inducible target genes and enhances drought stress tolerance in various crops, although it also causes the growth retardation (Liu et al., 1998). To improve drought stress tolerance and plant growth, we generated transgenic plants overexpressing two transcription factors: DREB1A and OsPIL1. OsPIL1 is reported to be a homologue of Arabidopsis PIF4 and acts as a key regulator of internode elongation in rice by up‐regulating many cell elongation‐related genes, including cell wall‐related genes (Todaka et al., 2012). We observed that the overexpression of OsPIL1 also up‐regulated several cell elongation‐related genes and increased hypocotyl and stem lengths in Arabidopsis plants (Figures 2b, c, 5b, S3, S4 and S5). The double overexpressors showed improved drought stress tolerance and enhanced hypocotyl elongation compared with the vector control plants (Figures 2b, c and 3), suggesting that plant genetic engineering using gene‐stacking approaches will be useful for generating transgenic plants that exhibit improved drought stress tolerance and enhanced growth.
The results of our transcriptome and metabolome analyses of the transgenic plants supported the idea that the two transcription factors function additively in the double overexpressors (Figures 4, 5 and 6). Regarding the improvement of drought stress tolerance, we observed an accumulation of compatible solutes, such as galactinol and raffinose, and the up‐regulation of the corresponding biosynthesis genes in the double overexpressors (Figures 4c and S11, Table S2). Because these data were also reported in the DREB1A overexpressors (Maruyama et al., 2009), the drought stress tolerance of the double overexpressors appears to be increased via the same mechanisms in the DREB1A overexpressors. OsPIL1 overexpression increased the amount of citric acid and decreased the amount of glucose, fructose, ornithine, arginine and guanidine, and similar alterations were observed in the amounts of these metabolites in the double overexpressors (Figure 4c). Transcriptome analyses indicated that the expression levels of many downstream genes of OsPIL1 and DREB1A were also up‐regulated in the double overexpressors (Figures 5b and S12b). The major downstream genes of the two transcription factors (RD29A, COR15A, GolS3 and PIL1) were up‐regulated by more than 20‐fold (Tables S2 and S3), while the expression levels of the up‐regulated and down‐regulated genes only in the double overexpressors were low (Tables S6 and S7). Our data suggest that cross‐regulation among the downstream genes may not be critical in the double overexpressors. In addition, we found that both the G box and DRE (which target the cis‐acting elements of OsPIL1 and DREB1A, respectively) were enriched in the promoters of the up‐regulated genes (Figure 6a). These results indicate that OsPIL1 and DREB1A act additively on the promoters of each target gene and suggest that these two transcription factors are unlikely to negatively affect each other in the transgenic plants. Thus, the co‐expression of OsPIL1 does not affect the DREB1A‐mediated improvement of drought stress tolerance in the double overexpressors, indicating that OsPIL1 has the potential to improve the growth of transgenic plants tolerant to abiotic stress.
A late‐flowering phenotype was one of the negative effects of DREB1A overexpression in Arabidopsis (Figure 2d, e), and it has also been reported in other DREB1‐overexpressing plants, such as soybean and barley (Gilmour et al., 2004; Morran et al., 2011; Seo et al., 2009; Suo et al., 2012), indicating that DREB1 overexpression results in a prolonged vegetative stage and delays the harvest time of crops. In the present study, we demonstrated that the delayed flowering time of DREB1A‐overexpressing plants was improved by OsPIL1 co‐expression through the up‐regulation of the floral inducers FT and MIR172A (Figures 2d, e and 5d; Aukerman and Sakai, 2003; Turck et al., 2008). Thus, the appropriate expression of these genes is important for controlling flowering time in agricultural applications, and OsPIL1 overexpression can be used to improve the delayed flowering times caused by transgenic DREB1A overexpression.
Arabidopsis PIF4 is known to promote shade avoidance syndrome (SAS), which consists of hypocotyl elongation, petiole elongation, stem elongation and hyponasty (Franklin et al., 2011; Hornitschek et al., 2012; Koini et al., 2009; Kumar et al., 2012), through the up‐regulation of auxin‐related genes (IAAs and SAURs), shade‐inducible genes (ATHB2, PIL1 and HFR1) and cell wall‐related genes (XTR7) (Casal, 2013). We showed that the overexpression of rice OsPIL1 enhanced the hypocotyl and stem elongation in Arabidopsis (Figures 2b, c, S3 and S4). In the double overexpressors, HFR1, PIL1, ATHB2 and SAUR mRNA levels were increased (Figures 5b and S12b). These results indicate that cell elongation in the 35SΩ:OsPIL1 plants is mainly caused by the elevated expression of SAS‐related genes similar to PIF4. Despite the similar expression levels of SAS‐related genes in the double overexpressors (Figure 5b), the hypocotyl length was shorter than that of the 35SΩ:OsPIL1 plants, suggesting that DREB1A overexpression negatively affects the hypocotyl cell elongation by regulating the expression of genes other than SAS‐related genes.
The function of PIF family proteins in plant metabolic pathways has not been reported. In this study, we observed higher levels of citric acid and lower levels of glucose, fructose, ornithine and arginine in the 35SΩ:OsPIL1 plants compared with the vector control plants (Figure 4c). Higher or lower levels of these primary metabolites have also been reported in the pseudo response regulator (prr) 9‐11 prr7‐10 prr5‐10 triple mutant (d975) of Arabidopsis (Fukushima et al., 2009). PRR9, 7 and 5 are known to function as transcriptional repressors in the circadian clock and repress the expression of PIF genes (Nakamichi et al., 2012). The d975 mutant shows an accumulation of PIF transcripts and hypocotyl elongation (Nakamichi et al., 2009). Therefore, PIF family proteins may act to maintain primary plant metabolism under the regulation of circadian clock components. According to our transcriptome data, the gene expression level of CELL WALL INVERTASE 5 (CWINV5), an enzyme that degrades sucrose into glucose and fructose, was decreased in the 35SΩ:OsPIL1 plants (Figure S11). This reduction in CWINV5 expression partly accounted for the decrease in fructose and glucose. Our results suggest that PIF family proteins function in the regulation of primary metabolism.
Compared with the hypocotyl and stem, the rosette leaf size and dry weight in the double overexpressors were not increased and remained similar to those in the 35SΩ:DREB1A plants (Figure S6a, b, e). The overexpression of OsPIL1 has been shown to enhance the internode elongation, but not flag leaf length, in rice (Todaka et al., 2012). Therefore, the overexpression of OsPIL1 might not affect the phenotypes of rosette leaves in the double overexpressors. We found that the EXPA gene expression levels decreased in the 35SΩ:DREB1A plants and double overexpressors (Figure 5c). The negative regulation of EXPA genes may be a cause of the dwarfed rosette leaves in the 35SΩ:DREB1A plants and double overexpressors.
We demonstrated that OsPIL1 overexpression partially enhances the plant growth in transgenic plants that overexpress OsPIL1 and DREB1A. The co‐overexpression of two transcription factors is considered an effective strategy for plant genetic engineering designed to improve drought stress tolerance and plant growth. In addition, gene‐stacking approaches appear to be able to tilt the balance in favour of growth in abiotic stress‐tolerant transgenic plants. We expect that the growth of stress‐tolerant plants will be further enhanced by the use of stress‐specific promoters that control transgene expression. Moreover, as the rosette leaf size in the double overexpressors was not increased, the utilization of genes encoding other growth regulators for leaf expansion may be an effective strategy for increasing the biomass and yield of stress‐tolerant plants. Our study provides a basis for overcoming growth retardation in stress‐tolerant plants through genetic engineering by gene stacking.
Experimental procedures
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia plants were grown on Murashige–Skoog (MS) agar medium at 22°C under a 16‐h/8‐h light/dark cycle at a photon flux density of 40 ± 10 μmol/m2/s as previously described (Yamaguchi‐Shinozaki and Shinozaki, 1994).
Protein stability analysis in protoplasts
A protein stability analysis using the protoplasts derived from Arabidopsis mesophyll cells was performed according to Mizoi et al. (2013) with minor modifications. OsPIL1‐sGFP and PIF4‐sGFP were expressed under the control of the CaMV 35S promoter and TMV Ω sequence. Approximately 1.6 × 105 protoplasts were transformed with 33.25 μg of OsPIL1‐sGFP or PIF4‐sGFP plasmid and 1.75 μg of 3 × FLAG‐sGFP. A 14‐h incubation at 22°C in the dark was followed by incubation in a solution containing 50 μm MG132 or 0.5% (v/v) dimethyl sulphoxide (DMSO, as a solvent control) under white‐light conditions (50 ± 5 μmol/m2/s). After 2 and 4 h, the protoplasts were precipitated by centrifugation and dissolved in 40 μL of sample buffer. The extracts were denatured at 95°C for 3 min and then analysed by SDS–PAGE and immunoblotting. The sGFP‐fused proteins were detected with a polyclonal antibody against GFP (Tanaka et al., 2012). Horseradish peroxidase‐conjugated anti‐rabbit IgG antibodies (Pierce, Rockford, IL), ECL Plus (GE Healthcare, Chalfont, UK) and an ImageQuant LAS 4000 system (GE Healthcare) were used to visualize the signals.
Transient reporter assays with Arabidopsis and rice protoplasts
Transient expression assays using the protoplasts derived from Arabidopsis and rice mesophyll cells were performed as previously described (Kidokoro et al., 2009; Matsukura et al., 2010; Mizoi et al., 2013). The effector plasmids were constructed as described in Figures 1 and S2. Plasmids containing 12 tandem repeats of a 28‐bp G box‐containing fragment from the rice 1‐AMINOCYCLOPROPANE‐1‐CARBOXYLATE OXIDASE1 (OsACO1) promoter (G box:GUS) or three tandem repeats of a 71‐bp DRE‐containing fragment from the RD29A promoter (DRE:GUS) were used as reporters (Liu et al., 1998; Todaka et al., 2012). An emerald luciferase (ELUC) reporter gene driven by the CaMV 35S promoter (35SΩ:ELUC) and a luciferase (LUC) reporter gene driven by the maize ubiquitin promoter (Ubi:LUC) were used as internal controls in the Arabidopsis and rice protoplasts, respectively (Matsukura et al., 2010; Mizoi et al., 2013).
Construction of plasmids and generation of OsPIL1 and DREB1A double‐overexpressing plants
We used pGKX and pGHX vectors as binary vectors (Fujita et al., 2012; Qin et al., 2008). The pGKX and pGHX vectors include genes that confer antibiotic resistance to kanamycin and hygromycin, respectively. To generate OsPIL1 or DREB1A single overexpressors, the coding sequences were inserted into the backbone binary vectors to construct pGKX‐35SΩ:OsPIL1 and pGHX‐35SΩ:DREB1A plasmids. Arabidopsis plants were transformed using the floral dip method (Clough and Bent, 1998). OsPIL1 and DREB1A double‐overexpressing Arabidopsis plants were generated by crossing 35SΩ:DREB1A plants with 35SΩ:OsPIL1 plants (Table S1).
Drought stress tolerance test
A dehydration treatment was performed as previously described (Liu et al., 1998) with minor modifications. The plants were grown on agar medium for 2 weeks and then in pots with soil for a week. Watering was withheld from 3‐week‐old plants for 2 weeks, and the survival rates were determined after 4 days of recovery following rehydration.
Metabolome analysis
Metabolites were extracted from the whole plants grown on agar medium for 2 weeks. Extraction, derivatization and GC‐TOF‐MS analyses were performed as previously described (Lisec et al., 2006). The data were processed using TagFinder (Luedemann et al., 2008) and Xcalibur software (Thermo Scientific, Waltham, MA), and the relative amounts were normalized using ribitol as an internal standard and the fresh weight of the transgenic plants.
Expression analysis
Total RNA was isolated from 2‐week‐old plants using RNAiso plus (TaKaRa Bio, Otsu, Japan) according to the manufacturer's instructions. cDNA synthesis and quantitative RT‐PCR were performed as previously described (Sato et al., 2014). The relative expression levels were calculated using the delta Ct method, and the obtained values were normalized to the 18S rRNA. The primer sequences are shown in Table S8.
Microarray experiment and data processing
Genome‐wide expression analyses using the Arabidopsis 3 Oligo Microarray were performed as previously described (Mizoi et al., 2013) with minor modifications. Normalization and statistical analyses of the data were performed using Subio Platform software (https://www.subio.jp/). The signal intensities were normalized by the Lowess method, and the significance of the expression changes was evaluated by one‐sample t‐tests. The p values were corrected by the Benjamini–Hochberg false discovery rate (FDR) method, and probes with an FDR of less than 0.05 were used for further analyses. All microarray data are available at Array Express under accession numbers E‐MTAB‐4073 and E‐MTAB‐4739.
Supporting information
Figure S1. Stability of the OsPIL1 protein in Arabidopsis protoplasts under light conditions. (a) Schematic diagram of the treatments used in the assay. Transfected protoplast suspensions were treated with 0.5% dimethyl sulfoxide (DMSO) or 50 μM MG132 under white light (50 ± 5 μmol photons/m2/s). (b) Stability of the OsPIL1 and PIF4 proteins. OsPIL1 and PIF4 proteins were expressed as sGFP fusion proteins under the control of the CaMV 35S promoter and the TMV Ω sequence. To normalize for transfection efficiency, 3 × Flag‐tag fused‐sGFP driven by the CaMV 35S promoter was co‐transfected into the protoplasts as an internal control. The levels of the fusion proteins were analyzed by immunoblotting using an antibody against GFP.
Figure S2. Effect of co‐expressing both OsPIL1 and DREB1A on their individual transactivation activity in rice protoplasts. (a) Schematic diagram of the effector and reporter constructs used in the transactivation analysis of the rice protoplasts. The effector construct contains the maize ubiquitin promoter fused to the coding sequence of OsPIL1 or DREB1A. (b, c) Transactivation effects of DREB1A and OsPIL1 co‐expression. The reporter 12 × G box:GUS (b) or 3 × DRE:GUS (c) and the effectors were co‐transfected into rice protoplasts. To normalize for transfection efficiency, the luciferase (LUC) reporter gene driven by the maize ubiquitin promoter was co‐transfected as a control in each experiment. Bars show the SD of 4 replicates. The letters indicate significant differences among the assays (P < 0.01 according to Games‐Howell's multiple range test).
Figure S3. Hypocotyl cell size of the single‐ or double‐overexpressing plants. (a) Cell surface in the hypocotyls of transgenic plants. The plants were grown on agar medium for 7 days. The cells were stained with toluidine blue. Bars = 50 μm. (b) Cell length calculated from the plants grown as in (a). The error bars show the SD of more than 50 cells in more than 4 seedlings (n > 200). The letters indicate significant differences among the seedlings (P < 0.01 according to Games‐Howell's multiple range test). VC and W‐OE represent vector control and OsPIL1 DREB1A double overexpressor, respectively.
Figure S4. Internode length of the single‐ or double‐overexpressing plants. (a) Stem morphology in 8‐week‐old transgenic plants. The plants were grown on agar medium for 2 weeks and then in soil pots for 6 weeks. Bars = 1 cm. (b) Internode length of the transgenic plants grown as in (a). The error bars show the SD of 10 internodes in 6 plants (n = 60). The letters indicate significant differences among the plants (P < 0.05 according to Games‐Howell's multiple range test).
Figure S5. Shoot length of the single‐ or double‐overexpressing plants. (a) Morphology of 8‐week‐old transgenic plants. The plants were grown on agar medium for 2 weeks and then in soil pots for 6 weeks. Bars = 10 cm. (b) Shoot length of the transgenic plants grown as in (a). The error bars show the SD of more than 4 plants. The letters indicate significant differences among the plants (P < 0.05 according to Games‐Howell's multiple range test).
Figure S6. Growth of the single‐ or double‐overexpressing plants. (a) Growth of 3‐week‐old transgenic Arabidopsis plants. The plants were grown on agar medium. Bars = 1 cm. (b) Average radius of the rosette calculated from the plants grown as in (a). The error bars show the SD of 14 plants. The letters indicate significant differences among the plants (P < 0.01 according to Tukey's multiple range test). (c) Growth of 12‐day‐old transgenic Arabidopsis plants on vertically oriented agar medium. Black bars show the root apex of the transgenic plants. White bar = 1 cm. (d) Average primary root length of the transgenic plants grown as shown in (c). The error bars show the SD of more than 4 plants. The letters indicate significant differences among the plants (P < 0.05 according to Tukey's multiple range test). (e) Average dry weight of 6‐week‐old transgenic Arabidopsis plants. The error bars show the SD of 6 plants. The letters indicate significant differences among the plants (P < 0.01 according to Games‐Howell's multiple range test). (f) Silique number in the transgenic Arabidopsis plants. The plants were grown on agar medium for 2 weeks and then in soil pots for more than 10 weeks. The error bars show the SD of more than 4 plants. The letters indicate significant differences among the plants (P < 0.05 according to Tukey's multiple range test). (g) Seed number per silique for the transgenic plants grown as shown in (f). The error bars show the SD of 5 siliques in more than 4 plants (n > 20).
Figure S7. Comparison of the metabolite profiles in the transgenic plants at different sampling time points. (a) Growth of 2‐week‐old transgenic Arabidopsis plants. The plants were grown on agar medium. Bars = 1 cm. Metabolite profiles of the transgenic plants harvested at the start of the day (ZT = 0) (b) and the end of the day (ZT = 16) (c). The relative amounts of the metabolites are displayed as heat maps. Yellow and blue colors show increased and decreased levels, respectively.
Figure S8. Hormone metabolism, abiotic stress, and light responses of the up‐regulated and down‐regulated genes in the single or double overexpressors. The expression ratios of the up‐regulated and down‐regulated genes in each plant (x axis, from left to right) in response to various hormones, abiotic stress, or light treatments (y axis) are displayed as heat maps by Genevestigator software. ABA, abscisic acid; GA, gibberellin; IAA, indole‐3‐acetic acid; R/FR, red/far‐red.
Figure S9. Functional categorization of the up‐regulated genes in each transgenic Arabidopsis plant. Up‐regulated genes (fold change > 2, FDR P < 0.05) in the transgenic plants were annotated by PageMan software. The numbers in each pie chart indicate the ratio against the total number of up‐regulated genes in each transgenic plant.
Figure S10. Functional categorization of the down‐regulated genes in each transgenic Arabidopsis plant. Down‐regulated genes (fold change < 0.5, FDR P < 0.05) in the transgenic plants were annotated by PageMan software. The numbers in each pie chart indicate the ratio against the total number of down‐regulated genes in each transgenic plant.
Figure S11. Map of sugar metabolism. The map was constructed based on the microarray data of the single‐ or double‐overexpressing plants analyzed by MapMan software. Squares and circles indicate the amounts of metabolites and transcripts, respectively. The expression level of the gene shows the highest or the lowest expression in each gene family. Yellow and blue colors show increased and decreased levels, respectively.
Figure S12. Expression analysis of the downstream genes of Arabidopsis PIF4 and auxin‐inducible genes in the single or double overexpressors. Expression levels of PIF4 downstream genes (a) and auxin‐inducible SAUR genes (b) in the transgenic plants analyzed by quantitative RT‐PCR. The plants were grown on agar medium for 2 weeks and harvested at ZT = 0. The error bars indicate the SD of more than 4 samples.
Table S1. Generation of OsPIL1 and DREB1A double‐overexpressing Arabidopsis plants.
Table S2. Overlapping up‐regulated genes between the 35SΩ:DREB1A plants and the double overexpressors.
Table S3. Overlapping up‐regulated genes between the 35SΩ:OsPIL1 plants and the double overexpressors.
Table S4. Overlapping down‐regulated genes between the 35SΩ:DREB1A plants and the double overexpressors.
Table S5. Overlapping down‐regulated genes between the 35SΩ:OsPIL1 plants and the double overexpressors.
Table S6. Genes up‐regulated only in the double overexpressors.
Table S7. Genes down‐regulated only in the double overexpressors.
Table S8. Primer sequences used in this study.
Acknowledgements
We thank Y. Tanaka and S. Alseekh for providing technical assistance, E. Toma for providing skilful editorial assistance, K. Kodaira for providing material support and T. Tohge for providing fruitful discussions. This work was supported by a Grant‐in‐Aid for JSPS Fellows (No. 16J01053 to M.K.), for Scientific Research on Innovative Areas (No. 15H05960 to K.Y.‐S.) and for Scientific Research (A) (No. 25251031 to K.Y.‐S.), JSPS ‘Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation’ (No. S2503) from the Japan Society for the Promotion of Science, and Technology of Japan and by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (BRAIN) of Japan (to K.Y.‐S.). The authors declare no conflict of interest.
References
- Aukerman, M.J. and Sakai, H. (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2‐like target genes. Plant Cell, 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, G. , Zhuo, C. , Qian, C. , Xiao, T. , Guo, Z. and Lu, S. (2016) Co‐expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol. J. 14, 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar‐Mathur, P. , Devi, M.J. , Reddy, D.S. , Lavanya, M. , Vadez, V. , Serraj, R. , Yamaguchi‐Shinozaki, K. et al (2007) Stress‐inducible expression of At DREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water‐limiting conditions. Plant Cell Rep. 26, 2071–2082. [DOI] [PubMed] [Google Scholar]
- Bhatnagar‐Mathur, P. , Rao, J.S. , Vadez, V. , Dumbala, S.R. , Rathore, A. , Yamaguchi‐Shinozaki, K. and Sharma, K.K. (2014) Transgenic peanut overexpressing the DREB1A transcription factor has higher yields under drought stress. Mol. Breed. 33, 327–340. [Google Scholar]
- Carriere, Y. , Crickmore, N. and Tabashnik, B.E. (2015) Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat. Biotechnol. 33, 161–168. [DOI] [PubMed] [Google Scholar]
- Casal, J.J. (2013) Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64, 403–427. [DOI] [PubMed] [Google Scholar]
- Claeys, H. and Inze, D. (2013) The agony of choice: how plants balance growth and survival under water‐limiting conditions. Plant Physiol. 162, 1768–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2000) Loosening of plant cell walls by expansins. Nature, 407, 321–326. [DOI] [PubMed] [Google Scholar]
- FAO, IFAD and WFP (2015) The State of Food Insecurity in the World 2015. Meeting the 2015 International Hunger Targets: Taking Stock of Uneven Progress. Rome: Food and Agriculture Organization of the United Nations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, K.A. , Lee, S.H. , Patel, D. , Kumar, S.V. , Spartz, A.K. , Gu, C. , Ye, S. et al (2011) Phytochrome‐interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl Acad. Sci. USA, 108, 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, M. , Fujita, Y. , Iuchi, S. , Yamada, K. , Kobayashi, Y. , Urano, K. , Kobayashi, M. et al (2012) Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proc. Natl Acad. Sci. USA, 109, 6343–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima, A. , Kusano, M. , Nakamichi, N. , Kobayashi, M. , Hayashi, N. , Sakakibara, H. , Mizuno, T. et al (2009) Impact of clock‐associated Arabidopsis pseudo‐response regulators in metabolic coordination. Proc. Natl Acad. Sci. USA, 106, 7251–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J. , Fowler, S.G. and Thomashow, M.F. (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54, 767–781. [DOI] [PubMed] [Google Scholar]
- Halpin, C. (2005) Gene stacking in transgenic plants – the challenge for 21st century plant biotechnology. Plant Biotechnol. J. 3, 141–155. [DOI] [PubMed] [Google Scholar]
- Hanson, A.D. , Rathinasabapathi, B. , Rivoal, J. , Burnet, M. , Dillon, M.O. and Gage, D.A. (1994) Osmoprotective compounds in the Plumbaginaceae: a natural experiment in metabolic engineering of stress tolerance. Proc. Natl Acad. Sci. USA, 91, 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek, P. , Kohnen, M.V. , Lorrain, S. , Rougemont, J. , Ljung, K. , Lopez‐Vidriero, I. , Franco‐Zorrilla, J.M. et al (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711. [DOI] [PubMed] [Google Scholar]
- Ito, Y. , Katsura, K. , Maruyama, K. , Taji, T. , Kobayashi, M. , Seki, M. , Shinozaki, K. et al (2006) Functional analysis of rice DREB1/CBF‐type transcription factors involved in cold‐responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153. [DOI] [PubMed] [Google Scholar]
- Kasuga, M. (2004) A combination of the Arabidopsis DREB1A gene and stress‐inducible rd29A promoter improved drought‐ and low‐temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45, 346–350. [DOI] [PubMed] [Google Scholar]
- Kasuga, M. , Liu, Q. , Miura, S. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress‐inducible transcription factor. Nat. Biotechnol. 17, 287–291. [DOI] [PubMed] [Google Scholar]
- Kidokoro, S. , Maruyama, K. , Nakashima, K. , Imura, Y. , Narusaka, Y. , Shinwari, Z.K. , Osakabe, Y. et al (2009) The phytochrome‐interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 151, 2046–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro, S. , Watanabe, K. , Ohori, T. , Moriwaki, T. , Maruyama, K. , Mizoi, J. , Myint Phyu Sin Htwe, N. et al (2015) Soybean DREB1/CBF‐type transcription factors function in heat and drought as well as cold stress‐responsive gene expression. Plant J., 81, 505–518. [DOI] [PubMed] [Google Scholar]
- Koini, M.A. , Alvey, L. , Allen, T. , Tilley, C.A. , Harberd, N.P. , Whitelam, G.C. and Franklin, K.A. (2009) High temperature‐mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408–413. [DOI] [PubMed] [Google Scholar]
- Kumar, S.V. , Lucyshyn, D. , Jaeger, K.E. , Alos, E. , Alvey, E. , Harberd, N.P. and Wigge, P.A. (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature, 484, 242–U127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunihiro, A. , Yamashino, T. , Nakamichi, N. , Niwa, Y. , Nakanishi, H. and Mizuno, T. (2011) Phytochrome‐interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin‐inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol. 52, 1315–1329. [DOI] [PubMed] [Google Scholar]
- Leivar, P. and Quail, P.H. (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec, J. , Schauer, N. , Kopka, J. , Willmitzer, L. and Fernie, A.R. (2006) Gas chromatography mass spectrometry‐based metabolite profiling in plants. Nat. Protoc. 1, 387–396. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Kasuga, M. , Sakuma, Y. , Abe, H. , Miura, S. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought‐ and low‐temperature‐responsive gene expression, respectively, in Arabidopsis. Plant Cell, 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas, M. , Daviere, J.‐M. , Rodriguez‐Falcon, M. , Pontin, M. , Iglesias‐Pedraz, J.M. , Lorrain, S. , Fankhauser, C. et al (2008) A molecular framework for light and gibberellin control of cell elongation. Nature, 451, 480–U411. [DOI] [PubMed] [Google Scholar]
- Luedemann, A. , Strassburg, K. , Erban, A. and Kopka, J. (2008) TagFinder for the quantitative analysis of gas chromatography ‐ mass spectrometry (GC‐MS)‐based metabolite profiling experiments. Bioinformatics, 24, 732–737. [DOI] [PubMed] [Google Scholar]
- Maruyama, K. , Sakuma, Y. , Kasuga, M. , Ito, Y. , Seki, M. , Goda, H. , Shimada, Y. et al (2004) Identification of cold‐inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38, 982–993. [DOI] [PubMed] [Google Scholar]
- Maruyama, K. , Takeda, M. , Kidokoro, S. , Yamada, K. , Sakuma, Y. , Urano, K. , Fujita, M. et al (2009) Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150, 1972–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, K. , Todaka, D. , Mizoi, J. , Yoshida, T. , Kidokoro, S. , Matsukura, S. , Takasaki, H. et al (2012) Identification of cis‐acting promoter elements in cold‐ and dehydration‐induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura, S. , Mizoi, J. , Yoshida, T. , Todaka, D. , Ito, Y. , Maruyama, K. , Shinozaki, K. et al (2010) Comprehensive analysis of rice DREB2‐type genes that encode transcription factors involved in the expression of abiotic stress‐responsive genes. Mol. Genet. Genomics, 283, 185–196. [DOI] [PubMed] [Google Scholar]
- Mizoi, J. , Ohori, T. , Moriwaki, T. , Kidokoro, S. , Todaka, D. , Maruyama, K. , Kusakabe, K. et al (2013) GmDREB2A;2, a canonical DEHYDRATION‐RESPONSIVE ELEMENT‐BINDING PROTEIN2‐type transcription factor in soybean, is posttranslationally regulated and mediates dehydration‐responsive element‐dependent gene expression. Plant Physiol. 161, 346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran, S. , Eini, O. , Pyvovarenko, T. , Parent, B. , Singh, R. , Ismagul, A. , Eliby, S. et al (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9, 230–249. [DOI] [PubMed] [Google Scholar]
- Nakamichi, N. , Kusano, M. , Fukushima, A. , Kita, M. , Ito, S. , Yamashino, T. , Saito, K. et al (2009) Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50, 447–462. [DOI] [PubMed] [Google Scholar]
- Nakamichi, N. , Kiba, T. , Kamioka, M. , Suzuki, T. , Yamashino, T. , Higashiyama, T. , Sakakibara, H. et al (2012) Transcriptional repressor PRR5 directly regulates clock‐output pathways. Proc. Natl Acad. Sci. USA, 109, 17123–17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y. , Kato, T. , Yamashino, T. , Murakami, M. and Mizuno, T. (2007) Characterization of a set of phytochrome‐interacting factor‐like bHLH proteins in Oryza sativa. Biosci. Biotechnol. Biochem. 71, 1183–1191. [DOI] [PubMed] [Google Scholar]
- Ni, M. , Tepperman, J.M. and Quail, P.H. (1999) Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature, 400, 781–784. [DOI] [PubMed] [Google Scholar]
- Nomoto, Y. , Kubozono, S. , Yamashino, T. , Nakamichi, N. and Mizuno, T. (2012) Circadian clock‐ and PIF4‐controlled plant growth: a coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 53, 1950–1964. [DOI] [PubMed] [Google Scholar]
- Nozue, K. , Covington, M.F. , Duek, P.D. , Lorrain, S. , Fankhauser, C. , Harmer, S.L. and Maloof, J.N. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature, 448, 358–U311. [DOI] [PubMed] [Google Scholar]
- Oh, E. , Zhu, J.‐Y. and Wang, Z.‐Y. (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–U864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrineschi, A. , Reynolds, M. , Pacheco, M. , Brito, R.M. , Almeraya, R. , Yamaguchi‐Shinozaki, K. and Hoisington, D. (2004) Stress‐induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome, 47, 493–500. [DOI] [PubMed] [Google Scholar]
- Pino, M.‐T. , Skinner, J.S. , Park, E.‐J. , Jeknic, Z. , Hayes, P.M. , Thornashow, M.F. and Chen, T.H.H. (2007) Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol. J. 5, 591–604. [DOI] [PubMed] [Google Scholar]
- Qin, F. , Sakuma, Y. , Tran, L.‐S.P. , Maruyama, K. , Kidokoro, S. , Fujita, Y. , Fujita, M. et al (2008) Arabidopsis DREB2A‐interacting proteins function as RING E3 ligases and negatively regulate plant drought stress‐responsive gene expression. Plant Cell, 20, 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W. (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425. [DOI] [PubMed] [Google Scholar]
- Ren, H. and Gray, W.M. (2015) SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant, 8, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba, Y. , Jeong, J. , Kang, M.‐Y. , Kim, J. , Paek, N.‐C. and Choi, G. (2014) Phytochrome‐interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5, 4636. [DOI] [PubMed] [Google Scholar]
- Sato, H. , Mizoi, J. , Tanaka, H. , Maruyama, K. , Qin, F. , Osakabe, Y. , Morimoto, K. et al (2014) Arabidopsis DPB3‐1, a DREB2A interactor, specifically enhances heat stress‐induced gene expression by forming a heat stress‐specific transcriptional complex with NF‐Y subunits. Plant Cell, 26, 4954–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, E. , Lee, H. , Jeon, J. , Park, H. , Kim, J. , Noh, Y.‐S. and Lee, I. (2009) Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering‐time gene SOC1 and its upstream negative regulator FLC. Plant Cell, 21, 3185–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo, H. , Ma, Q. , Ye, K. , Yang, C. , Tang, Y. , Hao, J. , Zhang, Z.J. et al (2012) Overexpression of AtDREB1A causes a severe dwarf phenotype by decreasing endogenous gibberellin levels in soybean [Glycine max (L.) Merr]. PLoS ONE, 7, e45568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji, T. , Ohsumi, C. , Iuchi, S. , Seki, M. , Kasuga, M. , Kobayashi, M. , Yamaguchi‐Shinozaki, K. et al (2002) Important roles of drought‐ and cold‐inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 29, 417–426. [DOI] [PubMed] [Google Scholar]
- Tanaka, H. , Osakabe, Y. , Katsura, S. , Mizuno, S. , Maruyama, K. , Kusakabe, K. , Mizoi, J. et al (2012) Abiotic stress‐inducible receptor‐like kinases negatively control ABA signaling in Arabidopsis. Plant J. 70, 599–613. [DOI] [PubMed] [Google Scholar]
- Todaka, D. , Nakashima, K. , Maruyama, K. , Kidokoro, S. , Osakabe, Y. , Ito, Y. , Matsukura, S. et al (2012) Rice phytochrome‐interacting factor‐like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc. Natl Acad. Sci. USA, 109, 15947–15952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck, F. , Fornara, F. and Coupland, G. (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. [DOI] [PubMed] [Google Scholar]
- Yamaguchi‐Shinozaki, K. and Shinozaki, K. (1994) A novel cis‐acting element in an Arabidopsis gene is involved in responsiveness to drought, low‐temperature, or high‐salt stress. Plant Cell, 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. [DOI] [PubMed] [Google Scholar]
- Ye, X.D. , Al‐Babili, S. , Kloti, A. , Zhang, J. , Lucca, P. , Beyer, P. and Potrykus, I. (2000) Engineering the provitamin A (beta‐carotene) biosynthetic pathway into (carotenoid‐free) rice endosperm. Science, 287, 303–305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Stability of the OsPIL1 protein in Arabidopsis protoplasts under light conditions. (a) Schematic diagram of the treatments used in the assay. Transfected protoplast suspensions were treated with 0.5% dimethyl sulfoxide (DMSO) or 50 μM MG132 under white light (50 ± 5 μmol photons/m2/s). (b) Stability of the OsPIL1 and PIF4 proteins. OsPIL1 and PIF4 proteins were expressed as sGFP fusion proteins under the control of the CaMV 35S promoter and the TMV Ω sequence. To normalize for transfection efficiency, 3 × Flag‐tag fused‐sGFP driven by the CaMV 35S promoter was co‐transfected into the protoplasts as an internal control. The levels of the fusion proteins were analyzed by immunoblotting using an antibody against GFP.
Figure S2. Effect of co‐expressing both OsPIL1 and DREB1A on their individual transactivation activity in rice protoplasts. (a) Schematic diagram of the effector and reporter constructs used in the transactivation analysis of the rice protoplasts. The effector construct contains the maize ubiquitin promoter fused to the coding sequence of OsPIL1 or DREB1A. (b, c) Transactivation effects of DREB1A and OsPIL1 co‐expression. The reporter 12 × G box:GUS (b) or 3 × DRE:GUS (c) and the effectors were co‐transfected into rice protoplasts. To normalize for transfection efficiency, the luciferase (LUC) reporter gene driven by the maize ubiquitin promoter was co‐transfected as a control in each experiment. Bars show the SD of 4 replicates. The letters indicate significant differences among the assays (P < 0.01 according to Games‐Howell's multiple range test).
Figure S3. Hypocotyl cell size of the single‐ or double‐overexpressing plants. (a) Cell surface in the hypocotyls of transgenic plants. The plants were grown on agar medium for 7 days. The cells were stained with toluidine blue. Bars = 50 μm. (b) Cell length calculated from the plants grown as in (a). The error bars show the SD of more than 50 cells in more than 4 seedlings (n > 200). The letters indicate significant differences among the seedlings (P < 0.01 according to Games‐Howell's multiple range test). VC and W‐OE represent vector control and OsPIL1 DREB1A double overexpressor, respectively.
Figure S4. Internode length of the single‐ or double‐overexpressing plants. (a) Stem morphology in 8‐week‐old transgenic plants. The plants were grown on agar medium for 2 weeks and then in soil pots for 6 weeks. Bars = 1 cm. (b) Internode length of the transgenic plants grown as in (a). The error bars show the SD of 10 internodes in 6 plants (n = 60). The letters indicate significant differences among the plants (P < 0.05 according to Games‐Howell's multiple range test).
Figure S5. Shoot length of the single‐ or double‐overexpressing plants. (a) Morphology of 8‐week‐old transgenic plants. The plants were grown on agar medium for 2 weeks and then in soil pots for 6 weeks. Bars = 10 cm. (b) Shoot length of the transgenic plants grown as in (a). The error bars show the SD of more than 4 plants. The letters indicate significant differences among the plants (P < 0.05 according to Games‐Howell's multiple range test).
Figure S6. Growth of the single‐ or double‐overexpressing plants. (a) Growth of 3‐week‐old transgenic Arabidopsis plants. The plants were grown on agar medium. Bars = 1 cm. (b) Average radius of the rosette calculated from the plants grown as in (a). The error bars show the SD of 14 plants. The letters indicate significant differences among the plants (P < 0.01 according to Tukey's multiple range test). (c) Growth of 12‐day‐old transgenic Arabidopsis plants on vertically oriented agar medium. Black bars show the root apex of the transgenic plants. White bar = 1 cm. (d) Average primary root length of the transgenic plants grown as shown in (c). The error bars show the SD of more than 4 plants. The letters indicate significant differences among the plants (P < 0.05 according to Tukey's multiple range test). (e) Average dry weight of 6‐week‐old transgenic Arabidopsis plants. The error bars show the SD of 6 plants. The letters indicate significant differences among the plants (P < 0.01 according to Games‐Howell's multiple range test). (f) Silique number in the transgenic Arabidopsis plants. The plants were grown on agar medium for 2 weeks and then in soil pots for more than 10 weeks. The error bars show the SD of more than 4 plants. The letters indicate significant differences among the plants (P < 0.05 according to Tukey's multiple range test). (g) Seed number per silique for the transgenic plants grown as shown in (f). The error bars show the SD of 5 siliques in more than 4 plants (n > 20).
Figure S7. Comparison of the metabolite profiles in the transgenic plants at different sampling time points. (a) Growth of 2‐week‐old transgenic Arabidopsis plants. The plants were grown on agar medium. Bars = 1 cm. Metabolite profiles of the transgenic plants harvested at the start of the day (ZT = 0) (b) and the end of the day (ZT = 16) (c). The relative amounts of the metabolites are displayed as heat maps. Yellow and blue colors show increased and decreased levels, respectively.
Figure S8. Hormone metabolism, abiotic stress, and light responses of the up‐regulated and down‐regulated genes in the single or double overexpressors. The expression ratios of the up‐regulated and down‐regulated genes in each plant (x axis, from left to right) in response to various hormones, abiotic stress, or light treatments (y axis) are displayed as heat maps by Genevestigator software. ABA, abscisic acid; GA, gibberellin; IAA, indole‐3‐acetic acid; R/FR, red/far‐red.
Figure S9. Functional categorization of the up‐regulated genes in each transgenic Arabidopsis plant. Up‐regulated genes (fold change > 2, FDR P < 0.05) in the transgenic plants were annotated by PageMan software. The numbers in each pie chart indicate the ratio against the total number of up‐regulated genes in each transgenic plant.
Figure S10. Functional categorization of the down‐regulated genes in each transgenic Arabidopsis plant. Down‐regulated genes (fold change < 0.5, FDR P < 0.05) in the transgenic plants were annotated by PageMan software. The numbers in each pie chart indicate the ratio against the total number of down‐regulated genes in each transgenic plant.
Figure S11. Map of sugar metabolism. The map was constructed based on the microarray data of the single‐ or double‐overexpressing plants analyzed by MapMan software. Squares and circles indicate the amounts of metabolites and transcripts, respectively. The expression level of the gene shows the highest or the lowest expression in each gene family. Yellow and blue colors show increased and decreased levels, respectively.
Figure S12. Expression analysis of the downstream genes of Arabidopsis PIF4 and auxin‐inducible genes in the single or double overexpressors. Expression levels of PIF4 downstream genes (a) and auxin‐inducible SAUR genes (b) in the transgenic plants analyzed by quantitative RT‐PCR. The plants were grown on agar medium for 2 weeks and harvested at ZT = 0. The error bars indicate the SD of more than 4 samples.
Table S1. Generation of OsPIL1 and DREB1A double‐overexpressing Arabidopsis plants.
Table S2. Overlapping up‐regulated genes between the 35SΩ:DREB1A plants and the double overexpressors.
Table S3. Overlapping up‐regulated genes between the 35SΩ:OsPIL1 plants and the double overexpressors.
Table S4. Overlapping down‐regulated genes between the 35SΩ:DREB1A plants and the double overexpressors.
Table S5. Overlapping down‐regulated genes between the 35SΩ:OsPIL1 plants and the double overexpressors.
Table S6. Genes up‐regulated only in the double overexpressors.
Table S7. Genes down‐regulated only in the double overexpressors.
Table S8. Primer sequences used in this study.


