Abstract
Background
Diabetic cardiomyopathy develops in insulin-dependent diabetic patients who have no hypertension, cardiac hypertrophy or vascular disease. Diabetes increases cardiac fatty acid oxidation, but cardiac hypertrophy limits fatty acid oxidation. Here we examined effects of diabetes on gene expression in rat hearts.
Methods
We used oligonucleotide microarrays to examine effects of insulin-dependent diabetes in the rat heart. RTQ PCR confirmed results of microarrays. Specific antibodies were used to examine changes in the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2).
Results
A surprising result of diabetes was increased mRNA encoding all enzymes of the ketone body synthesis pathway. Increased mRNA expression for these enzymes was confirmed by RTQ PCR. The mRNA encoding HMGCS2, the rate-controlling enzyme, was 27 times greater in diabetic hearts. Total HMGCS2 protein increased 8-fold in diabetic hearts, but no difference was found in HMGCS2 protein in control vs. diabetic liver.
Conclusions
Insulin-dependent diabetes induced the enzymes of ketone body synthesis in the heart, including HMGCS2, as well as increasing enzymes of fatty acid oxidation.
General significance
The mammalian heart does not export ketone bodies to other tissues, but rather is a major consumer of ketone bodies. Induction of HMGCS2, which is normally expressed only in the fetal and newborn heart, may indicate an adaptation by the heart to combat “metabolic inflexibility” by shifting the flux of excess intramitochondrial acetyl-CoA derived from elevated fatty acid oxidation into ketone bodies, liberating free CoA to balance the acetyl-CoA/CoA ratio in favor of increased glucose oxidation through the pyruvate dehydrogenase complex.
Keywords: Heart, Metabolic inflexibility, Carnitine acyltransferases, HMG-CoA synthase 2, Diabetes, Fatty acid, Ketone body
1. Introduction
In non-diabetic individuals, cardiomyopathy is thought to develop following hypertension and cardiac hypertrophy [1]. However, diabetic cardiomyopathy develops in insulin-dependent diabetic patients who have no hypertension, no cardiac hypertrophy and no vascular disease [2]. Heart failure developing from hypertension and hypertrophy is characterized by diminished rates of fatty acid oxidation [3], while insulin-dependent diabetes is clearly associated with increased availability and oxidation of fatty acids [4]. Paradoxically, there is substantial evidence that decreasing fatty acid oxidation and increasing glucose utilization in failing hearts is important for improved heart function [5]. Overall, there is a vital need for a better understanding of the control of both fatty acid and glucose metabolism in the heart, especially with the onset of diabetes.
The major interest of our laboratory has been the regulation of fatty acid metabolism in liver and heart at the level of carnitine palmitoyltransferase I (Cpt1), which is located in the outer membranes of mitochondria in all mammalian cells and represents the predominant site for regulation of mitochondrial fatty acid oxidation [4]. Cpt1 has been shown to respond to the actions of several hormones, especially insulin [6–8]. We have found that both diabetes and fasting increase the level of expression of the Cpt1a isoform in the heart as well as in liver [9]. The regulation of Cpt1a is somewhat different in these two tissues because of the expression of different Cpt1 isoforms [10]. In the heart, Cpt1 isoform switching from the Cpt1a isoform in the fetal and newborn heart to the Cpt1b isoform in the adult. Cpt1a reappears in the heart during fasting and diabetes [9]. Two alternate splicing isoforms of Cpt1b have been reported [11], but their role in regulation of fatty acid metabolism is not understood [12].
Another carnitine acyltransferase important to heart function is the short-chain transferase, carnitine acetyl-transferase (CrAT). That enzyme has been found to be important in heart and skeletal muscles for preventing “metabolic inflexibility,” a condition in which muscles are unable to support high flux of glucose oxidation through the pyruvate dehydrogenase pathway due to an elevated acetyl-CoA/CoA ratio produced by increased mitochondrial fatty acid oxidation. CrAT allows that ratio to be normalized by converting excess acetyl-CoA to acetylcarnitine that can be easily eliminated from the heart mitochondria [13]. Problems arise with this mechanism if the CrAT activity is diminished [14].
The goal of the study reported here was to examine the effects of streptozotocin diabetes (IDDM) on the expression of as many genes as possible in the rat heart in order to ascertain which genes regulating fatty acid and glucose metabolism, as well as any other genes in the heart, would respond to a lack of insulin (and/or a concomitant increase in blood glucose and fatty acids). We were surprised to find reexpression of several genes normally expressed in the perinatal period, including the enzymes of the pathway for ketone body synthesis. This study further suggests that ketone body synthesis (followed by immediate utilization) by the heart during excess mitochondrial fatty acid oxidation might allow another mechanism that would avoid metabolic inflexibility by converting excess intramitochondrial acetyl-CoA to acetyl-carnitine plus CoA.
2. Materials and methods
2.1. Animals
Animals were Sprague-Dawley rats, 150–175 g. All animals were given food (Ralston Purina Co., Richmond, IN) and water ad libitum. Four animals were injected intraperitonally with 150 mg/kg body weight of streptozotocin, known to produce ketotic diabetic animals [8–9]. Urine of these animals was checked at 48 h. Only animals having 80 mg/dl of ketone bodies or greater and urine glucose exceeded 2000 mg/dl as measured using Ames Bili-Labstix (Miles Inc., Elkhart, IN) were used in experiments. All animal were checked to be sure stomachs were filled with food at the time of the experiment.
2.2. Isolation and analysis of RNA
Heart samples were taken from the ventricle and homogenized in RNA-Stat-60 (Tel-Test “B”, Inc., Friendswood, TX). Five ml were used for each 50 mg sample. Following precipitation with ethanol, the RNA extraction procedure was repeated using 4 ml of RNA-Stat-60 to ensure removal of genomic DNA. Analysis of RNA, production of cDNA, labeling and final hybridization and analysis was done by Genome Explorations (Memphis, TN, USA) using Rat Genome U34A Gene Chips (Affymetrix). Means and SEMs of reported fluorescence data were calculated for all expression values that exceeded a two-fold change (either increased or decreased) from normal fed to diabetic rats. Some mRNAs that were not altered by 2-fold were included for comparison.
Real-time quantitative polymerase chain reaction (RTQ-PCR) measurements of mRNA were carried out for confirming the levels of expression of several genes. For these measurements, 2 µg of RNA from the samples described above were used after treatment with Dnase-I (Ambion, Austin, TX, USA) for reverse transcription. Synthesis of single-stranded cDNA was done using the reverse transcriptase Superscript II-RNase H (Invitrogen, Carlsbad, CA, USA). Total cDNA was diluted with DEPC-water 1:100 times and analyzed by RTQ-PCR on an iCycler (Bio-Rad Laboratories, Hercules, CA, USA). Primers for RTQ-PCR were designed to have a size of about 23 bp and approximately 50%G/C content (Table 1). The target fragment sizes were approximately 100 bp. RTQ-PCR was performed in a total volume of 25 µl of 2 × SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK), 2.5 µl of each primer (100 nM), 2.5 µl of DEPC-water, and 5 µl cDNA diluted 1:100 (~50 pmol). The following protocol was used: 10 min at 95 °C for denaturation, 40 cycles for amplification and temperature annealing at 62 °C and extension at 72 °C. A DNA dissociation curve was run on all samples to insure that all amplicons were consistent with the intended targets and to avoid secondary products. Single-stranded cDNA oligonucleotides of approximately 100 bp were synthesized and used as standards. Correlation coefficients and amplification efficiencies were calculated by the iCycler software. The correlation coefficient was always >0.99 and PCR efficiency was between 95% and 100% in all experiments. For normalization of PCR results, we used 18S RNA as an endogenous control.
Table 1.
Primers for real-time quantitative polymerase chain reaction. Sequences for the forward and reverse primers used for RTQ-PCR are given with the GenBank accession number and name for the gene.
| Gene | Forward primer | Reverse primer | Accession no. |
|---|---|---|---|
| Acetoacetyl-CoA thiolase | agctcaagacagtgttccagaa | gctccgtcgttcagtgtgc | D13921 |
| HMG-CoA synthase 2 | ctgacatcgagggcatagatacc | cagttggcagcgttgaagag | M33648 |
| HMG-CoA lyase | cagaagtttcccggcatcaa | cctgcagctaccgcttcct | AI171090 |
| Pyruvate dehydrogenase kinase, isoenzyme 4 | ggattactgaccgcctctttagtt | gcattccgtgaattgtccatc | AF034577 |
2.3. Western blot analysis
Western blot analysis was performed on samples from the hearts and livers of control and streptozotocin-treated rats. Proteins from these samples were harvested in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (Sigma 11697498001). Protein was quantified by BCA protein assay kit (Pierce 23225). An equal amount of protein was loaded on a 4–12% Bis-Tris acrylamide gel and transferred to a 0.45-µm nitrocellulose membrane (Bio-Rad, Hercules, CA). Membranes were immunoblotted with primary antibodies for HMGCS2 (Abcam 137034) and GAPDH (Cell Signaling Technology 3683) in Tris-buffered saline with Tween 20 containing 5% bovine serum albumin. Membranes were incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibody (Southern Blot 4050-05). Immunoreactive proteins were detected using Supersignal West Femto Chemiluminescent Substrate (Thermo Scientific). The signals were quantified using ImageJ software.
3. Results
3.1. Expression of genes encoding the carnitine palmitoyltransferases
Oligonucleotide microarray data were searched for mRNA representing all the genes encoding the six known members of the carnitine palmitoyltransferase (Cpt family. Every control sample in the U34A rat Gene Chip database was found to contain data indicating the presence of mRNA for all six putative enzymes: peroxisomal carnitine octanoyltransferase (COT), Cpt2, Cpt1α, and Cpt1β with its three splicing isoforms. COT was expressed at the lowest level of all of the CPT family of enzymes, possibly because of the minimal content of peroxisomes in the heart (Fig. 1). The expression of COT was only slightly elevated by diabetes (Table 2). Cpt2 was expressed at a higher level, and it was also slightly elevated by diabetes. Cpt1β was the predominant isoform of the malonyl-CoA inhibitable Cpt's, and there was a small increase (1.4-fold) in its expression in diabetes while Cpt1α was increased by a factor of 2.0. The mRNAs for the proposed alternate splicing isoforms of Cpt1β were expressed at levels greater than that of Cpt1a (Fig. 1). It has not yet been demonstrated that these alternate splicing isoforms of Cpt1β produce a functional enzyme, and in fact one published account of their expression found no activity of the proteins expressed in yeast [15]. The sequences of these alternate splicing isoforms lack some of the functional aspects of the normal Cpt1β including one of the membrane spanning regions [11]. For these reasons, it is not known what the function of these splicing isoforms might be. It is very interesting that the mRNAs for these splicing isoforms were found in such high relative abundance in this study, which would seem to suggest some regulatory function. The expression of these isoforms was not changed by diabetes. They were previously reported to be expressed at only a small percentage of Cpt1b [11], but diabetes produced no change.
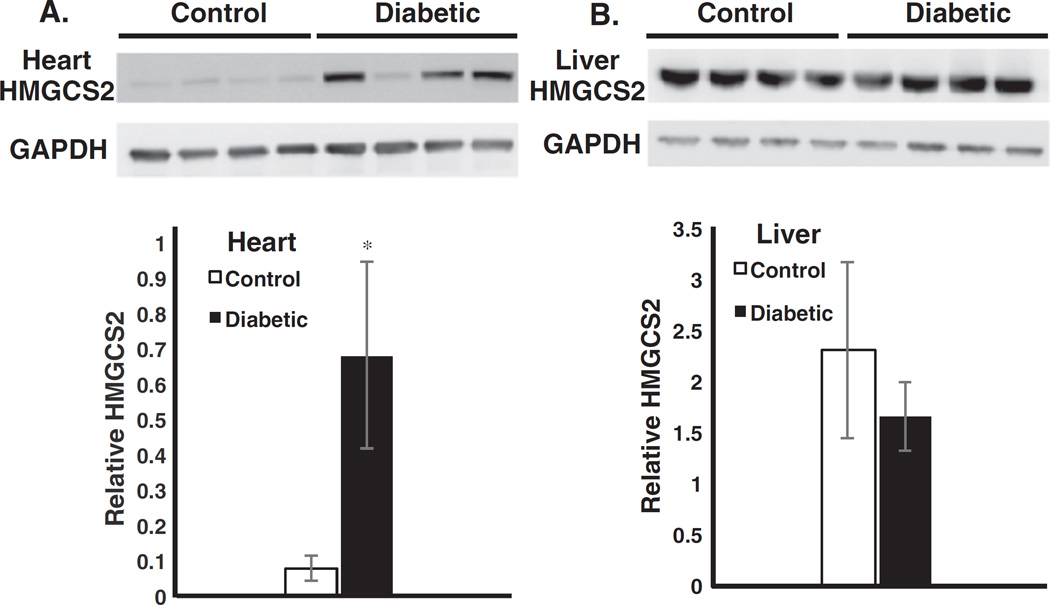
Fig. 1.
Identification of HMGCS2 protein in rat heart and liver. A. At the top of the panel, data from western blots using antibodies to HMGCS2 and GAPDH from four untreated and four diabetic hearts are shown. The data were quantified in the bar graph below. The data are the means±SEM of 4 samples (*p < 0.05). B. Western blot data are shown for the same proteins from rat liver. The data are quantified below and represent the means ± SEM of four samples.
Table 2.
Effects of diabetes on gene expression in the rat heart. Data are from affymetrix gene chip U34A for mRNA of normal and diabetic hearts. Raw data were treated as indicated in Materials and methods and are presented here showing the fold increase or decrease resulting from diabetes.
| Accession number |
Gene name | Fold decrease |
Fold increase |
|---|---|---|---|
| Glucose metabolism | |||
| S67900 | Fructose 6-phosphate,2-kinase:fructose 2,6-bisphosphatase |
3.3 | |
| S56464 | Hexokinase II (HKII) | 3.3 | |
| U25651 | Phosphofructokinase, muscle isoform (PFK-M) | 1.9 | |
| AF034577 | Pyruvate dehydrogenase kinase, isoenzyme 4 (PDK4) |
20.1 | |
| AF062741 | Pyruvate dehydrogenase phosphatase, isoenzyme 2 |
3.3 | |
| Lipid metabolism | |||
| D13921 | Acetyl-coenzyme A acetyltransferase 1, mitochondrial |
1.2 | |
| AB010428 | Acyl-CoA hydrolase, peroxisomal | 8.0 | |
| J02844 | Carnitine octanoyltransferase | 1.2 | |
| L07736 | Carnitine palmitoyltransferase 1a | 2.0 | |
| AF063302 | Carnitine palmitoyltransferase 1b | 1.4 | |
| J0570 | Carnitine palmitoyltransferase 2 | 1.3 | |
| D00569 | Dienoyl-CoA reductase | 2.5 | |
| D00729 | Enoyl-CoA isomerase | 1.8 | |
| U08976 | Enoyl hydratase, peroxisomal | 2.3 | |
| AI171090 | Hydroxymethylglutaryl CoA lyase | 1.9 | |
| M29249 | Hydroxymethylglutaryl-CoA reductase | 1.1 | |
| X52625 | Hydroxymethylglutaryl-CoA synthase 1, cytosolic | 1.4 | |
| M33648 | Hydroxymethylglutaryl-CoA synthase 2, mitochondrial |
27.1 | |
| U40001 | Lipase, hormone sensitive | 3.1 | |
| M15114 | Stearoyl-coenzyme A desaturase 2 | 4.1 | |
| Other metabolism | |||
| J03190 | 5-Aminolevulinate synthase | 3.7 | |
| D89070 | Carbonyl reductase 1 | 3.1 | |
| M11670 | Catalase | 2.2 | |
| AF056333 | Cytochrome P450 2E1 (CYP2E1) | 12.7 | |
| M29853 | Cytochrome P450 isozyme 5 (CYP4B2) | 2.4 | |
| S48325 | Diabetes inducible cytochrome P450RLM6 | 4.9 | |
| M26125 | Epoxide hydrolase 1 | 2.1 | |
| J03752 | Microsomal glutathione S-transferase 1 | 2.9 | |
| Transport/trafficking proteins | |||
| M15883 | Clathrin, light polypeptide (Lcb) | 2.2 | |
| AB005743 | Fatty acid transporter (FAT/CD36) | 2.2 | |
| U89529 | Fatty acid transport protein (FATP) | 1.7 | |
| S68135 | Glucose transporter 1 (GLUT1) | 2.5 | |
| D28561 | Glucose transporter 4 (GLUT4) | 2.1 | |
| AB015433 | Solute carrier family 3, member 2 (activator of
aa transport) |
2.4 | |
| AF054826 | Vesicle-associated membrane protein 5 | 2.1 | |
| Muscle proteins | |||
| L00088 | Fast myosin alkali light chain | 4.8 | |
| X00975 | Myosin, light polypeptide 2 | 2.9 | |
| L24897 | Myosin heavy chain | 2.8 | |
| Transcription & growth regulation | |||
| M63282 | Activating transcription factor 3 | 2.3 | |
| X06769 | c-fos | 3.5 | |
| M65149 | CCAAT/enhancerbinding, protein (C/EBP) delta | 2.4 | |
| D14014 | Cyclin D1 | 2.2 | |
| AF039583 | Decay accelerating factor, GPI-form | 3.8 | |
| M99223 | Fos-related antigen | 3.3 | |
| U17254 | Immediate early gene transcription factor NGFI-B | 4.9 | |
| U75397 | Krox-24 | 1.9 | |
| M18416 | Nerve growth factor-induced A protein (NGFI-A), Egr1 |
2.3 | |
| X86003 | Neuron-derived orphan receptor 2 | 2.9 | |
| AB012235 | Nuclear factor I/X | 2.5 | |
| AB015724 | Nuclear receptor binding factor-1, binds PPARalpha |
2.0 | |
| S74898 | Prostaglandin F2 alpha receptor | 2.3 | |
| X63594 | RL/IF-1 | 2.9 | |
| X60769 | Silencer factor B, (C/EBP) beta | 3.7 | |
| Structural proteins | |||
| V01227 | Alpha-tubulin | 2.1 | |
| X70369 | Collagen, type 3, alpha 1 | 2.6 | |
| X52140 | Integrin alpha 1 | 2.7 | |
| Signalling | |||
| D13376 | Adenylate kinase 1 | 2.2 | |
| AB009636 | Phosphatidylinositol 3-kinase, C2 domain | 3.7 | |
| X51529 | Phospholipase A2, group IIA (platelets, synovial fluid) |
2.6 | |
| D01046 | RAB11B, member RAS oncogene family | 2.1 | |
| AJ012603 | TNF-alpha converting enzyme | 2.3 | |
| Others | |||
| AB000507 | Aquaporin 7 | 2.2 | |
| M31178 | Calbindin 1 | 4.3 | |
| E00775 | Cardionatrin precursor | 2.4 | |
| X60767 | Cell division cycle 2 homolog A | 5.4 | |
| M88469 | F-spondin | 2.7 | |
| L13619 | Growth response protein (CL-6) | 2.8 | |
| X76985 | Latexin | 6.6 | |
| X01118 | Natriuretic peptide precursor type A | 2.6 | |
| M60921 | Nerve growth factor-inducible protein PC3 | 3.5 | |
| M24067 | Plasminogen activator inhibitor-1 (PAI-1) | 3.9 | |
| S49491 | Proenkephalin | 4.9 | |
| AF022774 | Rabphilin 3A-like (without C2 domains) | 2.1 | |
| D10233 | Renin-binding protein | 2.9 | |
| M27902 | Sodium channel, voltage-gated, type V, alpha polypeptide |
3.4 | |
| M58040 | Transferrin receptor | 10.5 | |
| AF037272 | WAP four-disulfide core domain protein (ps20) | 2.8 | |
3.2. Increased expression of genes involved in fatty acid transport and metabolism in diabetes
The mRNA expression of several genes regulating metabolism in the heart were increased by 2-fold or more (Table 2). Both of the fatty acid transporters (CD36 and FATP)were elevated by approximately 2-fold in the diabetic heart, suggesting an overall increase in the transport of fatty acids into the heart. Both Cpt1α and Cpt1β were increased, further suggesting increased fatty acid oxidation.
The largest change found in this study was the 27-fold increase in 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2)mRNA (Table 2). This increase was unexpected since the HMGCS2 enzyme is normally found in the heart only during the early stages of perinatal development [16]. HMGCS2 is the well-known regulatory enzyme in the pathway of ketone body synthesis in the liver during states of high fatty acid availability and oxidation [17]. Although unexpected, this particular diabetic effect in the heart may fit in well with the current hypothesis that cardiomyopathy is a condition resulting from an increased fetal/neonatal program of expression [18]. HMG-CoA lyase and acetoacetyl-CoA thiolase, the other two enzymes of the ketone body synthesis pathway were also induced, but to a smaller extent, by diabetes. Using real-time quantitative PCR (RTQ-PCR) of normal and diabetic heart samples, we confirmed that HMGCS2 was induced in the heart by diabetes (Table 3). We also confirmed the microarray results for HMG-CoA lyase and acetoacetyl-CoA thiolase. These results indicate that the ketone body synthesis pathway is induced in insulin-dependent diabetes.
Table 3.
Confirmation by real-time quantitative PCR of microarray results for the effect of diabetes on mRNA expression in the rat heart. Adult rats were made diabetic as indicated in Materials and methods. Values are presented as molar ratios of each specific mRNA to 18 s RNA. Values are means ± S.D. (n= 4).
| Genes | Control hearts | Diabetic hearts | Fold induction |
|---|---|---|---|
| Cpt1a | 4.22 ± 0.48 × 10−5 | 1.65 ± 0.10 × 10−5 | 3.90 |
| Cpt1b | 1.17 ± 0.44 × 10−4 | 2.18 ± 0.36 × 10−4 | 1.86 |
| PDK4 | 2.40 ± 0.77 × 10−6 | 6.06 ± 0.68 × 10−5 | 25.25 |
| HMG-CoA lyase | 1.23 ± 0.13 × 10−6 | 3.19 ± 0.35 × 10−6 | 2.59 |
| HMG-CoA synthase | 1.47 ± 0.11 × 10−6 | 4.83 ± 0.48 × 10−5 | 32.85 |
| AA-CoA thiolase | 3.32 ± 0.44 × 10−5 | 3.39 ± 0.13 × 10−5 | 1.02 |
Two additional major inductions of expression by diabetes were found in acyl-CoA thioesterase (8-fold) and cytochrome P-450 CYP2E1 (13-fold), whose functions are not clearly elucidated in the heart but act to regulate or eliminate concentrations of fatty acids and ketone bodies in other tissues [19–21]. The cytochrome P-4502E1 (CYP 2E1), is known to metabolize ethanol, ketone bodies and fatty acids and can be induced by diabetes [22]. The gene for this enzyme is listed three times in Table 2 under three different accession numbers (AF056333, M29853 and S48325). Using the nucleotide sequence for S48325, a partial cDNA sequence, we did a Blast search of the Rat Genome and found that these three sequences all have identical nucleotide sequences and all represent the same rat gene (the CYP2E1 gene) which can be targeted to the mitochondria as well as microsomes [42].
The gene for hormone sensitive lipase was also upregulated by 3.1-fold in diabetes. Hormone sensitive lipase has been reported in rat heart [23] and plays a regulatory role in the release of free fatty acids from triglycerides within the heart muscle [24].
3.3. Decreased expression of genes involved in glucose transport and metabolism in diabetes
Concomitant with increased expression of fatty acid transport and metabolism in the diabetic rat, we found decreased expression of mRNA encoding enzymes of glucose transport and metabolism. Both GLUT1 and GLUT4 mRNAs were diminished in the diabetic state by a factor of 2, indicating decreased glucose transport into heart cells (Table 2). The muscle form [25] of hexokinase (HK2) was downregulated by a factor of 3.3. The muscle form of phosphofructokinase was downregulated by a factor of 2 and the bifunctional enzyme fructose 6-phosphate,2-kinase:fructose 2,6-bisphosphatase was downregulated by a factor of 3.3, all of which suggests a strong inhibition of glycolysis [26,27]. The very strong induction of pyruvate dehydrogenase kinase, isoenzyme 4 (PDK4) (20-fold) coupled with the repression of pyruvate dehydrogenase phosphatase 2, indicate that the oxidation of glucose (through inhibition of pyruvate oxidation) was also minimized in the diabetic animals. The change in PDK4 was expected in diabetes and indicates that most likely the increased availability of fatty acids in the diabetic state produced a dramatic increase in expression of this mRNA through the actions of PPARα [28].
3.4. Other changes in expression of genes involved in metabolic processes
Catalase was induced 3.3-fold in diabetic mice, which most likely occurred through PPARα [29]. Catalase induction in heart cells has been reported to provide protection against oxidative cell injury [30]. Although it has been suggested that catalase in the heart may be located to some extent in the mitochondrial matrix its location is mostly in peroxisomes [31]. Increased levels of catalase have been reported in failing human hearts where it may provide some protection, also [32]. Catalase overexpression in transgenic mice was shown to ameliorate the reduction in contractile force and heart rate caused by hypoxiareoxygenation, and overexpression eliminated reoxygenation-induced arrhythmia [33]. Some other related enzymes were also induced in the heart during diabetes, including carbonyl-reductase-1 (3.1-fold), epoxide hydrolase 1 (2.1-fold) and glutathione S-transferase 1 (2.9-fold).
A relatively large number of transcription factors and other growth regulators were affected by diabetes, as well as genes regulating synthesis of muscle proteins and proteins involved in several signalling pathways. These are all presented in Table 2.
3.5. Induction of HMGCS2
Our next experiments were designed to ascertain whether HMGCS2 protein was increased by diabetes in the heart and also whether that enzyme was altered in the liver. Using an HMGCS2 antibody, we were able to detect HMGCS2 in the heart of control rats, and this expression was increased by eight-fold in the diabetic hearts (Fig 1A). All the streptozocin-treated rats had elevated urine glucose, and all except one (lane 6) had highly elevated ketone bodies in their urine (80 mg/dl). Expression of HMGCS2 was higher in liver than in heart, but surprisingly HMGCS2was elevated in hearts but not in livers by the diabetic state (Fig 1B).
4. Discussion
A previous study of streptozotocin-induced diabetes has been carried out using the Affymetrix oligonucleotide microarray system to examine the effects of diabetes on skeletal muscle metabolism in mice [34]. In that study, only about 20 genes were up- or down-regulated by a factor of 2. In the mouse study, GLUT4, HK2 and pyruvate dehydrogenase were all decreased, and many enzymes of lipid metabolism, including acyl-CoA thioesterase, hormone sensitive lipase, carnitine acetyltransferase, enoyl-CoA hydratase, enoyl-CoA isomerase and FATP were upregulated [34].
The major unexpected discovery of this study using rat hearts was the very large upregulation of the mitochondrial isoform of HMG-CoA synthase that regulates the pathway of ketone body synthesis. This induction of expression by the diabetic state of HMGCS2was the largest of any we found, and it was confirmed by RTQ-PCR. We also found mRNAs for the other two enzymes of the ketone body synthesis pathway by both oligonucleotide microarray and by RTQ-PCR. Our finding is not the first report of these three enzymes in the heart. The mRNAs for all three enzymes were previously observed in hearts of 11-day and 28-day old rats in experiments using ribonuclease protection assays [16]. The mRNA for HMGCS2 has also been found in human hearts but at lower levels than in liver [35]. In a study of hormone sensitive lipase transgenic mice, the mRNA for HMGCS2 was detected in the hearts of control mice by oligonucleotide microarray analysis [24]. None of these studies examined the effects of diabetes, but it seems clear that the enzymes of the ketone body synthesis pathway exist in the mammalian heart. Proof of the functioning of the ketone body synthesis pathway in the rat heart has come from the laboratory of Dr. Lionel Opie from experiments in which they analyzed oxidation products from rat hearts perfused with fatty acids [36]. Opie's laboratory reported that while the heart was capable of utilizing both acetoacetate and 3-hydroxybutyrate as oxidizable substrates, it was also capable of synthesizing ketone bodies when fatty acids were supplied as substrates. They reported that the Langendorf perfused heart produced ketone bodies at 4 times the rate of the working heart [36]. We have been unable to find any additional reports in which ketone body synthesis in the heart has been studied, and we did not find that heart cells exported ketone bodies (unpublished).
There has been a recent report of ketone body synthesis in the retinal pigment epithelial cells indicating these cells oxidize fatty acids to ketone bodies that are then utilized as energy substrates by the retina [37]. It appears that expression of enzymes of the ketone body synthesis pathway is more widespread than previously believed.
A possible beneficial effect for the heart from induction of HMGCS2 is that this could represent a mechanism by which the heart could continue to utilize excess free fatty acids in the diabetic state but avoid any additional increase the intramitochondrial acetyl-CoA/CoA that is so unfavorable to pyruvate oxidation. Such a mechanism would allow increased exchange of mitochondrial acetoacetate with cytosolic pyruvate via the mitochondrial monocarboxylate carrier to increase mitochondrial pyruvate and stimulate pyruvate oxidation [38]. It is possible that removal of excess free fatty acids could help to prevent overproduction of triglycerides from fatty acids. One study in hepatectomized dogs has discouraged investigation in this area [39]. That study presented the concept of “pseudoketogenesis” in which changes in the radiolabelling patterns of ketone bodies that were first thought to represent net ketone body production by skeletal muscle were shown to be due to an artifact of radiolabelling [39]. This has led to the current dogma that no muscle cells, including the heart, can make ketone bodies. None of the pseudoketogenesis studies were conducted in starved or diabetic animals. The stimulation of pyruvate uptake by the mitochondria would be expected to prevent decreased contractile function of rat hearts by stimulating pyruvate carboxylation [40].
Some of the effects of diabetes reported here have been reported previously to be controlled by PPARα or PPARβ/δ including decreased expression of GLUT4 and HK2 and upregulation of Cpt1β [41]. The control of HMGCS2 is also recognized to be upregulated in different tissues through PPARα [17]. Specific cardiac overexpression of PPARα in mice has also been shown to produce several of the effects seen here, including upregulation of Cpt1β, FATP and FAT/CD36 [41]. Interestingly, the latter studies produced not only changes in gene expression found in diabetes, but also many of the pathological effects of diabetes in heart such as increased lipotoxicity, ventricular hypertrophy and alteration in systolic ventricular dysfunction [42].
The data presented here strongly support the role of insulin and elevated fatty acid levels in regulating all aspects fatty acid transport and metabolism in the heart and suggest that the lack of insulin and/or the elevation of fatty acids and glucose results in re-expression of many fetal/neonatal isoforms of metabolic enzymes including the ketone body synthesis pathway in the diabetic heart. Most of the mRNA's encoding the carnitine acyl-transferases (Cpt1a, Cpt1b, COT) were elevated in diabetes, but carnitine acetyl-transferase (CrAT) was not significantly affected. CrAT has been suggested to alleviate “metabolic inflexibility9201D; in muscles by lowering the intramitochondrial acetyl-CoA/CoA ratio that can limit the oxidation of glucose in states of abnormally elevated fatty acid oxidation. While it is known that the mammalian heart usually does not export ketone bodies to other tissues, but rather is a major consumer of ketone bodies, these results may indicate an adaptation by the heart in response to the diabetic state to combat “metabolic inflexibility” by shifting the flux of excess intramitochondrial acetyl-CoA derived from highly elevated fatty acid oxidation into ketone bodies, decreasing acetyl-CoA and liberating free CoA to balance the acetyl-CoA/CoA ratio in favor of increased glucose oxidation through the pyruvate dehydrogenase complex. It should be emphasized that mRNA for PDK4, which phosphorylates and inhibits the PDH, was elevated 20-fold in our study and is probably playing the major role in decreasing acetyl-CoA production. As pointed out in the introduction, carnitine acetyl-transferase is able to decrease mitochondrial acetyl-CoA content to diminish the problem of metabolic inflexibility [13] and it has been shown that decreased activity of this enzyme in obesity and diabetes can lead to decreased glucose utilization [14]. Decreased activity of this enzyme in the diabetic state may further increase the need for a mechanism to overcome metabolic inflexibility such as increased production of ketone bodies. It has been shown that treatment of rats with diets containing ketone body esters that increase the content of 3-hydroxybutyrate in heart is beneficial to heart function and, in fact, have effects similar to insulin treatment [43]. Effects of ketone body diets in diabetes and other pathological states have been reviewed [44]. It is not known whether production of ketone bodies in the heart could have similar effects.
Our data suggest that the lack of insulin and/or the elevation of fatty acids and glucose results in re-expression of many fetal/neonatal isoforms of metabolic enzymes including the ketone body synthesis pathway in the diabetic heart. They also suggest a need to re-evaluate the role of ketone body synthesis and elimination in the heart.
Acknowledgments
This work was supported by grants (to G.A.C.) from the National Institutes of Health (HL66924) and the American Heart Association (00-50224N) and a grant (to EAP) from the Veterans Administration (1I01BX002408).
Footnotes
Uncited reference
[25]
References
- 1.de las Fuentes L, Herrero P, Peterson LR, Kelly DP, Gropler RJ, Davila-Roman VG. Myocardial fatty acid metabolism: independent predictor of left ventricular mass in hypertensive heart disease. Hypertension. 2003;41:83–87. doi: 10.1161/01.hyp.0000047668.48494.39. [DOI] [PubMed] [Google Scholar]
- 2.Kannel WB, Hjortland M, Castelli WP. Role of diabetes in congestive heart failure: the Framingham study. Am. J. Cardiol. 1974;34:29–34. doi: 10.1016/0002-9149(74)90089-7. [DOI] [PubMed] [Google Scholar]
- 3.Barger PM, Kelly DP. Fatty acid utilization in the hypertrophied and failing heart: molecular regulatory mechanisms. Am. J. Med. Sci. 1999;318:36–42. doi: 10.1097/00000441-199907000-00006. [DOI] [PubMed] [Google Scholar]
- 4.McGarry JD, Foster DW. Regulation of hepatic fatty acid oxidation and ketone body production. Annu. Rev. Biochem. 1980;49:395–420. doi: 10.1146/annurev.bi.49.070180.002143. [DOI] [PubMed] [Google Scholar]
- 5.Chandler MP, Chavez PN, McElfresh TA, Huang H, Harmon CS, Stanley WC. Partial inhibition of fatty acid oxidation increases regional contractile power and efficiency during demand-induced ischemia. Cardiovasc. Res. 2003;59:143–151. doi: 10.1016/s0008-6363(03)00327-4. [DOI] [PubMed] [Google Scholar]
- 6.Cook GA, Gamble MS. Regulation of carnitine palmitoyltransferase by insulin results in decreased activity and decreased apparent Ki values for malonyl-CoA. J. Biol. Chem. 1987;262:2050–2055. [PubMed] [Google Scholar]
- 7.Mynatt RL, Park EA, Thorngate FE, Das HK, Cook GA. Changes in carnitine palmitoyltransferase-I mRNA abundance produced by hyperthyroidism and hypothyroidism parallel changes in activity. Biochem. Biophys. Res. Commun. 1994;201:932–937. doi: 10.1006/bbrc.1994.1791. [DOI] [PubMed] [Google Scholar]
- 8.Park EA, Mynatt RL, Cook GA, Kashfi K. Insulin regulates enzyme activity, malonyl-CoA sensitivity and mRNA abundance of hepatic carnitine palmitoyltransferase-I. Biochem. J. 1995;310:853–858. doi: 10.1042/bj3100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook GA, Edwards TL, Jansen MS, Bahouth SW, Wilcox HG, Park EA. Differential regulation of carnitine palmitoyltransferase-I gene isoforms (Cpt1α and Cpt1β) in the rat heart. J. Mol. Cell. Cardiol. 2001;33:317–329. doi: 10.1006/jmcc.2000.1304. [DOI] [PubMed] [Google Scholar]
- 10.Brown NF, Weis BC, Husti JE, Foster DW, McGarry JD. Mitochondrial carnitine palmitoyltransferase I isoform switching in the developing rat heart. J. Biol. Chem. 1995;270:8952–8957. doi: 10.1074/jbc.270.15.8952. [DOI] [PubMed] [Google Scholar]
- 11.van der Leij FR, Cox KB, Jackson VN, Huijkman NC, Bartelds B, Kuipers JR, Dijkhuizen T, Terpstra P, Wood PA, Zammit VA, Price NT. Structural and functional genomics of the CPT1β gene for muscle-type carnitine palmitoyltransferase I in mammals. J. Biol. Chem. 2002;277:26994–27005. doi: 10.1074/jbc.M203189200. [DOI] [PubMed] [Google Scholar]
- 12.Yu GS, Lu YC, Gulick T. Rat carnitine palmitoyltransferase Ibeta mRNA splicing isoforms. Biochim. Biophys. Acta. 1998;1393:166–172. doi: 10.1016/s0005-2760(98)00075-7. [DOI] [PubMed] [Google Scholar]
- 13.Muoio DM, Noland RC, Kovalik J-P, Seiler SE, Davies MN, DeBalsi KL, Ilkayeva OR, Stevens RD, Kheterpal I, Zhang J, Covington JD, Bajpeyi S, Ravussin E, Kraus W, Koves TR, Mynatt RL. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab. 2012;15:764–777. doi: 10.1016/j.cmet.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seiler SE, Martin OJ, Noland RC, Slentz DH, DeBalsi KL, Ilkayeva OR, An J, Newgard CB, Koves TR, Muoio DM. Obesity and lipid stress inhibit carnitine acetyltransferase activity. J. Lipid Res. 2014;55:635–644. doi: 10.1194/jlr.M043448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JY, Koves TR, Yu GS, Gulick T, Cortright RN, Dohm GL, Muoio DM. Evidence of a malonyl-CoA-insensitive carnitine palmitoyltransferase I activity in red skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2002;282:E1014–E1022. doi: 10.1152/ajpendo.00233.2001. [DOI] [PubMed] [Google Scholar]
- 16.Cullingford TE, Dolphin CT, Bhakoo KK, Peuchen S, Canevari L, Clark JB. Molecular cloning of rat mitochondrial 3-hydroxy-3-methylglutaryl-CoA lyase and detection of the corresponding mRNA and of those encoding the remaining enzymes comprising the ketogenic 3-hydroxy-3-methylglutaryl-CoA cycle in central nervous system of suckling rat. Biochem. J. 1998;329:373–381. doi: 10.1042/bj3290373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegardt FG. Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: a control enzyme in ketogenesis. Biochem. J. 1999;338:569–582. [PMC free article] [PubMed] [Google Scholar]
- 18.Lehman JJ, Kelly DP. Transcriptional activation of energymetabolic switches in the developing and hypertrophied heart. Clin. Exp. Pharmacol. Physiol. 2002;29:339–345. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- 19.Hunt MC, Alexson SE. The role Acyl-CoA thioesterases play in mediating intracellular lipid metabolism. Prog. Lipid Res. 2002;41:99–130. doi: 10.1016/s0163-7827(01)00017-0. [DOI] [PubMed] [Google Scholar]
- 20.Hunt MC, Lindquist PJ, Nousiainen S, Huttunen M, Orii K, Svensson TL, Aoyama T, Hashimoto T, Diczfalusy U, Alexson SE. Acyl-CoA thioesterases belong to a novel gene family of peroxisome proliferator-regulated enzymes involved in lipid metabolism. Cell Biochem. Biophys. 2000;32:317–324. doi: 10.1385/cbb:32:1-3:317. [DOI] [PubMed] [Google Scholar]
- 21.Zangar RC, Novak RF. Effects of fatty acids and ketone bodies on cytochromes P450 2B,4A, and 2E1 expression in primary cultured rat hepatocytes. Arch. Biochem. Biophys. 1997;337:217–224. doi: 10.1006/abbi.1996.9785. [DOI] [PubMed] [Google Scholar]
- 22.Woodcroft KJ, Hafner MS, Novak RF. Insulin signaling in the transcriptional and posttranscriptional regulation of CYP2E1 expression. Hepatology. 2002;35:263–273. doi: 10.1053/jhep.2002.30691. [DOI] [PubMed] [Google Scholar]
- 23.Kraemer FB, Patel S, Saedi MS, Sztalryd C. Detection of hormone-sensitive lipase in various tissues. Expression of an HSL/bacterial fusion protein and generation of anti-HSL antibodies. J. Lipid Res. 1993;34:663–671. [PubMed] [Google Scholar]
- 24.Suzuki J, Shen WJ, Nelson BD, Patel S, Veerkamp JH, Selwood SP, Murphy GM, Reaven E, Kraemer FB. Absence of cardiac lipid accumulation in transgenic mice with heart-specific HSL overexpression. Am. J. Physiol. Endocrinol. Metab. 2001;281:E857–E866. doi: 10.1152/ajpendo.2001.281.4.E857. [DOI] [PubMed] [Google Scholar]
- 25.Printz RL, Koch S, Potter LR, O'Doherty RM, Tiesinga JJ, Moritz S, Granner DK. Hexokinase II mRNA and gene structure, regulation by insulin, and evolution. J. Biol. Chem. 1993;268:5209–5219. [PubMed] [Google Scholar]
- 26.Ma Z, Ramanadham S, Kempe K, Hu Z, Ladenson J, Turk J. Characterization of expression of phosphofructokinase isoforms in isolated rat pancreatic islets and purified beta cells and cloning and expression of the rat phosphofructokinase-A isoform. Biochim. Biophys. Acta. 1996;1308:151–163. doi: 10.1016/0167-4781(96)00088-7. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe F, Sakai A, Furuya E, Uyeda K. Molecular cloning and tissue specific expression of fructose 6-phosphate,2-kinase:fructose 2,6-bisphosphatase of rat brain. Biochem. Biophys. Res. Commun. 1994;198:335–340. doi: 10.1006/bbrc.1994.1047. [DOI] [PubMed] [Google Scholar]
- 28.Huang B, Wu P, Bowker-Kinley MM, Harris RA. Regulation of pyruvate dehydrogenase kinase expression by peroxisome proliferator-activated receptor-alpha ligands, glucocorticoids, and insulin. Diabetes. 2002;51:276–283. doi: 10.2337/diabetes.51.2.276. [DOI] [PubMed] [Google Scholar]
- 29.Chen C, Hennig GE, Whiteley HE, Manautou JE. Protection against acetaminophen hepatotoxicity by clofibrate pretreatment: role of catalase induction. J. Biochem. Mol. Toxicol. 2002;16:227–234. doi: 10.1002/jbt.10043. [DOI] [PubMed] [Google Scholar]
- 30.Peng X, Li Y. Induction of cellular glutathione-linked enzymes and catalase by the unique chemoprotective agent,3H-1,2-dithiole-3-thione in rat cardiomyocytes affords protection against oxidative cell injury. Pharmacol. Res. 2002;45:491–497. doi: 10.1006/phrs.2002.0991. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Z, Kang YJ. Cellular and subcellular localization of catalase in the heart of transgenic mice. J. Histochem. Cytochem. 2000;48:585–594. doi: 10.1177/002215540004800502. [DOI] [PubMed] [Google Scholar]
- 32.Dieterich S, Bieligk U, Beulich K, Hasenfuss G, Prestle J. Gene expression of antioxidative enzymes in the human heart: increased expression of catalase in the end-stage failing heart. Circulation. 2000;101:33–39. doi: 10.1161/01.cir.101.1.33. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Yu A, Saari JT, Kang YJ. Repression of hypoxia-reoxygenation injury in the catalase-overexpressing heart of transgenic mice. Proc. Soc. Exp. Biol. Med. 1997;216:112–116. doi: 10.3181/00379727-216-44162b. [DOI] [PubMed] [Google Scholar]
- 34.Yechoor VK, Patti ME, Saccone R, Kahn CR. Coordinated patterns of gene expression for substrate and energy metabolism in skeletal muscle of diabetic mice. Proc Nat Acad Sci, USA. 99:10587–10592. doi: 10.1073/pnas.142301999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mascaro C, Buesa C, Ortiz JA, Haro D, Hegardt FG. Molecular cloning and tissue expression of human mitochondrial 3-hydroxy-3-methyl-glutatryl-CoA synthase. Arch. Biochem. Biophys. 1995;317:385–390. doi: 10.1006/abbi.1995.1178. [DOI] [PubMed] [Google Scholar]
- 36.Opie LH, Owen P. Effects of increased mechanical work by isolated perfused rat heart during production or uptake of ketone bodies. Biochem. J. 1975;148:403–415. doi: 10.1042/bj1480403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adijanto J, Du J, Moffat C, Seifert EL, Hurle JB, Philp NJ. The retinal pigment epithelium utilizes fatty acids for ketogenesis, implications for metabolic coupling with the outer retina. J. Biol. Chem. 2014;289:20570–20582. doi: 10.1074/jbc.M114.565457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel TB, Barron LL, Olson MS. The stimulation of hepatic gluconeogenesis by acetoacetoacetate precursors. A role for the monocaroxylate translocator. J. Biol. Chem. 1984;259:7525–7531. [PubMed] [Google Scholar]
- 39.Des Rosiers C, Montgomery JA, Garneau M, David F, Mamer OA, Daloze P, Toffolo G, Cobelli C, Landau BR, Brunengraber H. Pseudoketogenesis in hepatectomized dogs. Am. J. Phys. 1990;258:E519–E528. doi: 10.1152/ajpendo.1990.258.3.E519. [DOI] [PubMed] [Google Scholar]
- 40.Russell RR, Taegtmeyer H. Pyruvate carboxylation prevents the decline in contractile function of rat hearts. Am. J. Phys. 1991;261:H1756–H1762. doi: 10.1152/ajpheart.1991.261.6.H1756. [DOI] [PubMed] [Google Scholar]
- 41.Gilde AJ, van der Lee KA, Willemsen PH, Chinetti G, van der Leij FR, van der Vusse GJ, Staels B, van Bilsen M. Peroxisome proliferator-activated receptor (PPAR)alpha and PPARbeta/delta, but not PPARgamma, modulate the expression of genes involved in cardiac lipid metabolism. Circ. Res. 2003;92:518–524. doi: 10.1161/01.RES.0000060700.55247.7C. [DOI] [PubMed] [Google Scholar]
- 42.Finck BN, Lehman JJ, Leone TC, Welch MJ, Bennett MJ, Kovacs A, Han X, Gross RW, Kozak R, Lopaschuk GD, Kelly DP. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J. Clin. Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kashiwaya Y, King MT, Veech RL. Substrate signaling by insulin: a ketone bodies ratio mimics insulin action in heart. Am. J. Cardiol. 1997;80:50A–64A. doi: 10.1016/s0002-9149(97)00458-x. [DOI] [PubMed] [Google Scholar]
- 44.Veech RL. The therapeutic implications of ketone bodies: the effects of ketone bodies in pathological conditions: ketosis, ketogenic diet, redox states, insulin resistance, and mitochondrial metabolism. Prostaglandins Leukot. Essent. Fat. Acids. 2004;70:309–319. doi: 10.1016/j.plefa.2003.09.007. [DOI] [PubMed] [Google Scholar]