Abstract
Optogenetic and chemogenetic control of proteins has revealed otherwise inaccessible facets of signaling dynamics. Here we use light or ligand-sensitive domains to modulate the structural disorder of diverse proteins, thereby generating robust allosteric switches. Sensory domains were inserted into non-conserved, surface exposed loops that were tight and identified computationally as allosterically coupled to active sites. Allosteric switches introduced into motility signaling proteins (kinases, GTPases, guanine exchange factors) controlled conversion between conformations closely resembling natural active and inactive states, and modulated the morphodynamics of living cells. Our results illustrate a broadly applicable approach to design physiological protein switches.
Proteins have been engineered to respond to light or small molecules in living cells through a number of mechanisms, including oligomerization (1–4), control of targeting sequences (1, 5, 6), split proteins (2), sterically blocking the active site (1, 7, 8), and engineered allosteric control (9–13). Engineering allostery has the potential to be especially versatile and valuable, as sensory domains can be inserted where they do not interfere with normal protein interactions, more readily producing fully functional analogs to replace endogenous protein. Allosteric sites have been identified using both screening approaches (10, 14) and rational analysis of sequence conservation (10, 11). A number of domains, including the light-sensitive LOV2 domain (11) and drug-responsive uniRapR domain (12, 13) used here, have been applied. Nonetheless, it remains challenging to identify allosteric sites via rational analysis, and to produce reversible, robust allosteric switches whose on and off states replicate natural levels of protein activity. Here we present a generalizable approach to identify surface loops where the disorder of the attached domain can be effectively transferred to the active site in a controlled fashion (harnessing extrinsic disorder (9, 10, 12–15)), to produce on and off states that mimic naturally occurring protein states.
In previous work, we identified an allosteric site in kinases where insertion of an engineered domain (uniRapR) conferred rapamycin-induced kinase activation (12, 13). Insertion of the uniRapR domain rendered kinases catalytically inactive until the domain bound rapamycin (12, 13, 16). Molecular dynamics simulations indicated that the protein loop containing the insertion site was mechanically coupled to the ATP binding site (13). We hypothesized that insertion of a light-responsive domain at the same site could lead to optogenetic control of kinases (Fig. 1A). We chose the LOV2 domain because the 10 Å spacing between its N and C termini enabled us to insert the domain with minimal perturbation into a loop (Fig. 1B) that connects parts of a tightly folded structural unit (two interacting anti-parallel β-strands). The spacing between LOV2’s termini becomes much more flexible upon irradiation, due to disordering of terminal helices (17, 18); this could perturb the conformation of the loop and the β-strands. Introducing disorder into the loop would cause protein inhibition, rather than the activation caused by rapamycin-induced folding of the uniRapR domain (fig. S1). The light-induced conformational changes in LOV2 (3.8 kcal/mol of free energy (19)) should be more than sufficient to disorder portions of host proteins (fig. S2), whose overall stabilities are on the order of 5–15 kcal/mol (20).
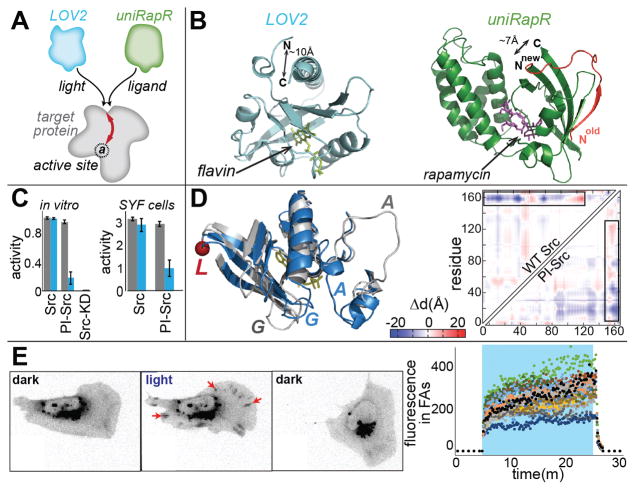
Fig. 1. Design concept and PI-Src.
(A) Domains conferring either photo-inhibition (LOV2) or activation induced by small molecules (uniRapR) function at the same allosteric site. (B) The termini of LOV2 and uniRapR domains are closely spaced for insertion. (C) Paxillin phosphorylation assays show that PI-Src’s catalytic activity is inhibited upon irradiation. SYF cells expressing PI-Src(YF) show reduced phosphoylation of cell lysates blotted with anti-pTyr antibody. Blue denotes irradiation. Error bars show s.e.m (n=3). (D) Inactive (blue) and active (gray) conformations of WT Src. The red circle (L) is the insertion site. Conformational changes were quantified by displaying the pairwise distance changes (Δd) between all residues as a heat map. The upper left triangle shows distances for WT Src, computed from published crystal structures. The lower triangle shows distances for PI-Src, determined using molecular dynamics simulations of the dark and lit states. Blue = decreased distance, red = increased distance. (E) In SYF cells, irradiation causes PI-Src(WT) to translocate to focal adhesions (FA, red arrows), edge movements to increase, and cells to polarize and translocate. When cells are returned to the dark, FA translocation is reversed but effects on morphodynamics persist. (blue = irradiation, n= 18 cells, quantitation in fig. S7)
We focused on Src kinase because of evidence that its activity is controlled by spatio-temporal dynamics in vivo (16, 21). The LOV2 domain was inserted at position G296 of constitutively active Src mutant Y535F (YF). We named the new Src analog PI-Src, for photo-inhibitable Src. In vitro kinase activity assays revealed that this LOV2-Src fusion was indeed strongly inhibited upon exposure to blue light (Fig. 1C). Light had no effect on the activity of kinase dead (KD) or constitutively active (YF) Src. Inhibition was not sensitive to small variations in linker length (fig. S3). To test PI-Src in living cells, we generated SYF cells (cells lacking the Src family members Src, Yes and Fyn expressing PI-Src(YF). Phosphotyrosine blots of cell lysates showed that irradiation inhibited phosphorylation of multiple Src substrates (Fig. 1C and fig. S4). Consistent with Src’s reported roles in motility (13, 16, 21), irradiation reduced migration rates, and caused reversible collapse of lamellipodia (fig. S5 and movie S1). Similar effects were produced by the Src inhibitor PP2 (fig. S5 and movie S2).
To investigate how distortion of the insertion loop led to a reduction in kinase activity, we built a structural model of PI-Src, and examined its light-induced conformational changes using discrete molecular dynamics (22, 23). While we expected to observe randomly distorted conformations of PI-Src in the lit state, we saw instead that the active and inactive states of PI-Src were strikingly similar to the crystal structures of active and inactive wild type (WT) Src (Fig. 1D, correlation coefficient = 0.82). This suggested that irradiation shifts PI-Src from its physiological active state to a conformation strongly resembling Src’s native inactive state (fig. S6).
The physiologically relevant conformations of PI-Src raised the possibility of examining how Src controls the motility of living cells. We generated SYF cells expressing PI-Src(WT). In the dark, PI-Src(WT) showed the perinuclear distribution (Fig. 1E) typical of endogenous, inactive Src (16). Upon irradiation, the PI-Src translocated to focal adhesions at the cell periphery and induced changes in cell morphodynamics (increased protrusion and retraction, polarization and in some cases polarized movement, movie S3). Returning the cells to the dark reversed focal adhesion localization (Fig. 1E) but not effects on cell morphodynamics (fig. S7). It is thought that Src localization to focal adhesions is mediated by Src SH2/SH3 domains, and is not dependent on kinase activity (24, 25). The light-induced distortion of the catalytic domain that inhibits the kinase may also perturb autoinhibitory interactions, exposing the SH2 and SH3 domains; this likely generated the reversible focal adhesion localization we observed, and suggested that focal adhesion localization was sufficient to affect cell morphodynamics even without catalytic activity. Supporting this hypothesis, we found that PI-Src(KD) also translocated reversibly to adhesions and induced changes in morphodynamics upon irradiation, and that elimination of the SH2 and SH3 domains from PI-Src(KD) prevented both focal adhesion localization and effects on cell morphodynamics (fig. S7). Movement away from focal adhesions, which occurred 30–40 seconds after irradiation was halted, indicated that the conformational change of PI-Src was reversible in cells (fig. S7) and was consistent with LOV2’s rate of return to the dark conformation (t1/2 = 18.5 sec at 37°C in cells (26)). In contrast, effects on cell morphodynamics were retained for >20 minutes after irradiation was halted, potentially due to feedback kinetics and/or inactivation pathways not involving Src.
Controlling protein activity via LOV2 insertion would be much more valuable if it could be readily applied to multiple proteins families. We tested a more generalizable approach by identifying an allosteric site for LOV2 insertion in the Rho family GTPase Rac1. We focused on surface-exposed and evolutionarily non-conserved loops to avoid possible structural perturbations, and asked whether these loops were mechanically coupled to the active site. For the kinase analogs above, we found that both dynamic coupling analysis (12) and a static contact map analysis (fig. S8 and S9) were effective in identifying mechanically-coupled loops. (The latter offers a simpler approach accessible to many laboratories). Dynamic coupling and static contact map analyses indicated several loops in Rac1 (Fig. 2A and fig. S10), but much of the surface of Rac1 is used to interact with other proteins, limiting the loops appropriate for insertion. Considering all these criteria we selected loop L1, which connects β-strands between the S1 and S2 regions (Fig. 2A). L1 was similar to the insertion loop of Src in that it connected interacting secondary structures, enabling efficient propagation of structural changes from the inserted domain to the host protein. All the examples that follow indicate that such ‘tight loops’ are useful insertion sites (fig. S9 and S11).
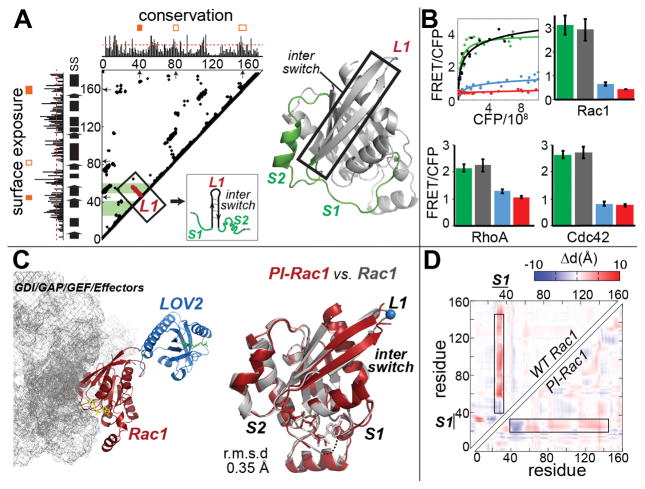
Fig. 2. Designing PI-GTPases.
(A) Sequence conservation, surface exposure, loop “tightness”, and contact maps were used to select insertion loops (fig. S9). Orange filled boxes indicate loops fulfilling selection criteria (red dashed lines = thresholds, SS = secondary structure). Lines extending perpendicular to the diagonal indicate loops (L) that connect tightly interacting elements of secondary structure. When these lines reached the active site (green bands) the loop was selected for testing. For PI-Rac1, we selected L1, which connects strands of the β-pleated sheet in the interswitch region. (B) GTPase activity in HEK293T cells reported using biosensors fused to PI-GTPases in a high throughput assay. gray = dark state mutant, blue = lit state mutant, red = T17N (Rac1 and Cdc42) and T19N (RhoA) dominant negative mutants, green = wild type GTPase positive control. CFP x axis indicates expression level of biosensor-GTPase fusion. Error bars show s.e.m (n=3). (C) (Left) Crystal structure of PI-Rac1 with interacting proteins (gray mesh); (right) structures of wild type Rac1 (gray) and PI-Rac1 (red) are in excellent agreement. L1 loop is the LOV2 insertion loop. (D) Map showing inter-residue distances for WT Rac1 versus PI-Rac1, suggesting that these molecules undergo similar conformational changes.
To test the efficacy of LOV2 insertion into the L1 loop, we incorporated the Rac1 analog into a previously characterized fluorescence resonance energy transfer (FRET) biosensor of Rac1 activity (27) (donor fluorescent protein fused to PI-Rac1 and FRET acceptor fused to a fragment from p21-activated kinase that specifically binds activated Rac1). PI-Rac1 containing a dark state LOV2 mutation was activated by the upstream guanine exchange factor (GEF) Vav2, while activation of a lit state mutant was substantially reduced (Fig. 2B). Even when an activating mutation (Q61L) was introduced in Rac1, the lit state LOV2 mutant showed strong inhibition (fig S12). For physiologically relevant studies, we used PI-Rac1 to replace endogenous Rac1 in Rac1-depleted fibroblasts. Irradiation produced reversible cell edge retraction, initially as indentations closely spaced along the edge, and then broad retraction of entire lamellae (fig. S12 and movies S4 and S5). We also generated switches from the Rho family GTPases RhoA and Cdc42, using insertion in the same loop. Both showed effective light-induced inhibition in live cells (Fig. 2B, fig. S13, movies S6 and S7).
To probe the mechanism of photoinhibition, we determined the crystal structure of dark state PI-Rac1 and examined lit/dark conformational changes using molecular dynamics. The crystal structure showed that LOV2 was placed where it did not interfere with Rac1 binding to regulatory or effector proteins (Fig. 2C, fig. S14 and Table S1). Notably, the dark state activated structure was almost identical to that of activated WT Rac1, with a root mean square deviation (RMSD) of only 0.35 Å (Fig. 2C). Molecular dynamics showed that the conformational change between dark and irradiated PI-Rac1 manifested mainly in the S1 and S2 switch regions, and that the conformational changes of PI-Rac1 and WT Rac1 were remarkably similar (Fig. 2D and fig. S15). Together these studies suggested that opto-allosteric perturbation of Rac1, like Src, caused switching between naturally occurring active and inactive states.
We next targeted another protein family, GEFs, beginning with the Rac1 activator Vav2. GEFs present a different challenge in that they induce a conformational change through GTPase binding to a broad area involving many relatively weak interactions. Computational analysis of Vav2’s GTPase-binding DH domain revealed three potential insertion loops (Fig. 3A and fig. S16). We used loop L4 for LOV2 insertion because it produced the most effective switch. PI-Vav2 was tested in living cells by examining its effects on a Rac1 biosensor (Fig. 3B). The activities of the PI-Vav2 dark and lit state mutants resembled those of different activated and inactivated Vav2 mutants (Fig. 3B and figs. S17 and S18). Pull-down assays confirmed that PI-Vav2 was inhibited upon irradiation (fig. S17), and irradiation of PI-Vav2 in HeLa cells produced rapid and reversible retraction (Fig. 3C and movie S8). Insertion of LOV2 into L1 or L2 also produced effective PI-Vav2 constructs, but insertion of LOV2 into a loop that was not “tight” produced no switch (fig. S18). The same analysis was used to create two other photo-inhibitable GEFs: GEF-H1, a Rho GEF, and Intersectin1(ITSN), a Cdc42 GEF (Fig. 3D and figs. S19–S21).
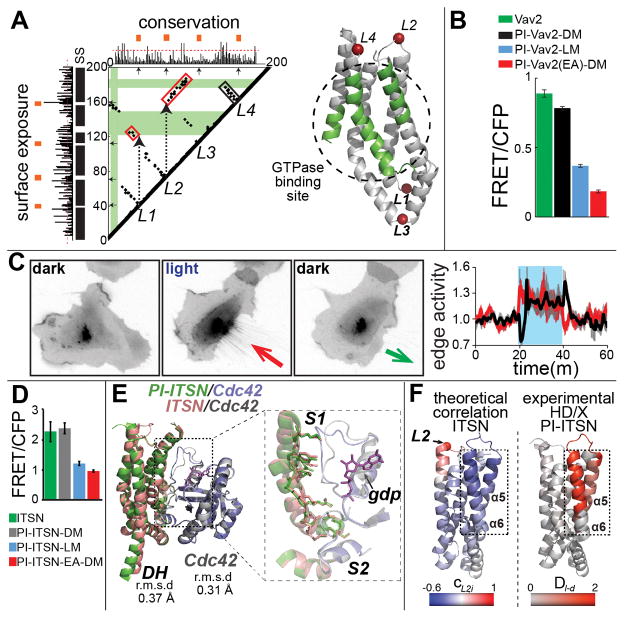
Fig. 3. Designing PI-GEFs.
(A) Computational analysis of the Vav2 catalytic DH domain. (Left) black and red boxes indicate local and non-local interactions (fig. S9) that mediate coupling between loops and the active site. (Right) structural model of the Vav2 DH domain showing insertion loops and the active site (green). (B) In living cells, PI-Vav2 was inhibited in the lit state. DM and LM = dark and lit mutants. EA = E200A/K333A dominant negative mutant. Error bars show s.e.m (n=3). (C) (Left) Reversible retraction induced by irradiation of PI-Vav2 in HeLa cells (n=9); retraction = red arrow, protrusion = black arrow. (D) High content live cell imaging showed that PI-ITSN was inhibited in the lit state. EA = E1244A dominant negative mutant. Error bars show s.e.m (n=3). (E) (Left) Crystal structure of PI-ITSN (L2) in complex with Cdc42 superimposed on the wild type ITSN:Cdc42 complex. (F) Comparison of deuterium exchange (HD/X) results and dynamic coupling computed using molecular dynamics simulations. CL2,I corresponds to the correlation coefficient between the motion of the L2 loop and the motions of each residue. DI-d corresponds to the differences in relative deuteration levels in the dark and light.
Unlike kinase and GTPase catalytic domains, the GEF catalytic domains do not undergo major intra-domain conformational changes. Instead, they are regulated sterically by autoinhibitory domains. To investigate the light-mediated structural changes of PI-ITSN (L2), we determined the crystal structures of PI-ITSN alone and in complex with Cdc42 (Fig. 3E, fig. S22 and Table S2). PI-ITSN and WT ITSN were structurally similar, with a backbone RMSD of 0.37 Å. The structure of Cdc42 in the ITSN:Cdc42 complex, and in the PI-ITSN:Cdc42 complex differed with an RMSD of only 0.31 Å. The interface between ITSN and Cdc42 was also in excellent agreement in the two structures, with an RMSD of 0.36 Å (Fig. 3E). The crystal structure of the PI-ITSN:Cdc42 complex was consistent with the shapes of the PI-ITSN:Cdc42 complex and PI-ITSN in solution, constructed ab initio by small angle X-ray scattering (fig. S24).
Molecular dynamics simulations of ITSN and PI-ITSN revealed that the motions of helices α5 and α6, part of the active site, correlate with the motions of the insertion loop (Fig. 3F), and hydrogen/deuterium exchange coupled to mass spectrometry showed light-induced destabilization of these helices (Fig. 3F and figs. S24–S26). These results suggest that native ITSN and Cdc42 interactions are maintained in PI-ITSN, and support an allosteric connection between the insertion loop and important active site residues.
We also inserted the uniRapR domain in GEFs, to activate GEFs with rapamycin. Vav2, ITSN, Asef, p115, and Tiam1 all showed rapamycin-induced GTPase binding in pulldown assays (fig. S27). Titrating uniRapR-Vav2 with saturating rapamycin in HEK293T cells produced dose-dependent activation of a Rac1 biosensor (fig. S27), and fibroblasts stably expressing uniRapR-Vav2 or uniRapR-ITSN produced protrusions and ruffles upon rapamycin addition (fig. S27 and movie S9).
Successful photo-inhibition suggested that we might also use allosteric control to activate proteins with light. GEFs, and many other proteins, are regulated by intramolecular interactions of autoinhibitory domains (AID), which might be turned on and off allosterically. Analyzing the CH portion of Vav2’s AID showed two potential insertion loops (Fig. 4A and fig. S28). We inserted LOV2 in these and in three additional loops not predicted to control activity (fig. S29). In live cell assays using Rac1 biosensor activity as a readout, only the computed insertion sites produced robust switches (Fig. 4B and fig. S29). In fibroblasts, Vav2 photo-activation induced rounding and lamellae formation rather than the retractions produced by photo-inhibition. Upon removal of light, PA-Vav2 cells underwent marked contractions (panels 1 and 2 in Fig. 4C; fig. S30; movie S10). These results demonstrated that a protein can be allosterically activated by light through control of its AID.
Fig. 4. Designing PA-Vav2 and multiplexed control in living cells.

(A) (Left) Computational analysis of Vav2’s AID indicated that loops L1 and L2 are coupled to the active site (green) through non-local (red box) and local (black box) interactions. CH and AC denote calponin-homology and acidic motifs. (Right) A structural model of the AID showing the connection of L1 and L2 to the active site. (B) PA-Vav2 is activated in the lit state, assayed as in Fig. 2B. Error bars show s.e.m (n=3). (C) Effects of irradiation and cessation of irradiation on cells expressing PA-Vav2 alone, PI-Rac1 alone, or both in the same cell. Blue box denotes irradiation, ea = edge activity, and envelopes show s.e.m (n= 15 for PA-Vav2, n= 18 for PI-Rac1, n= 17 for PA-Vav2+PI-Rac1).
With optogenetics, effects of protein manipulation can be assayed rapidly, before the cell has a chance to compensate as it does with genetic manipulations. We tested whether photoactivation and photoinhibition could be combined, to assay how photoactivation of one protein is affected by downstream inhibition of another. Unlike the rounding and large lamellae produced by irradiating PA-Vav2 (panel 1 in Fig. 4C and movie S10), irradiating PI-Rac1 cells (produced via knockdown/rescue of Rac1) caused retraction (panels 4 and 5 in Fig. 4C and movie S5), with formation of a complex perimeter and increased velocity of both retractions and protrusions (panel 6 in Fig. 4C). When PA-Vav2 and PI-Rac1 were irradiated in the same cell, area and roundness responded similarly to PA-Vav2 alone, but the cell edge showed a phenotype clearly different from that produced by the individual proteins, with a strong reduction in edge dynamics during irradiation (panels 7–9 in Fig. 4C, movie S11, fig. S30). Thus PI-Rac1 was being activated and affecting results of Vav2 activation. Interpretation will require further study, but these results are consistent with recent work showing that Vav2 can act independent of Rac1 (28–30).
In summary, we have controlled proteins with light or small molecules in living cells by harnessing order/disorder transitions. Sensory domains were inserted into ‘tight’, non-conserved, surface loops that are allosterically coupled to active sites. Engineered proteins switched between naturally occurring, physiologically relevant active and inactive states, generating effective tools to manipulate living cells.
Supplementary Material
An SYF cell expressing PI-Src(YF). PI-Src was localized to focal adhesions before, during, and after irradiation. Irradiation led to retraction and a reduced migration rate. Cells were visualized using PI-Src(YF)-mCherry. Blue dot indicates irradiation.
An SYF cells expressing PI-Src(YF) retracted and showed reduced edge dynamics when treated with 10 μM PP2 inhibitor. Cells were visualized using differential interference contrast.
SYF cells expressing PI-Src(WT). Irradiation caused the PI-Src(WT) to reversibly translocate to focal adhesions. This was accompanied to varying degrees by cell polarization, cell translation, and increased edge dynamics. Blue dot indicates irradiation. Cells were visualized using PI-Src(WT)-mCherry.
A fibroblast expressing PI-Rac1(Q61L). Irradiation produced reversible cell edge retraction, initially as indentations closely spaced along the edge, and then broader retraction of lamellae. Blue dot indicates irradiation. Cells were visualized using PI-Rac1(Q61L)-mCherry.
A fibroblast expressing PI-Rac1(WT). Irradiation produced reversible cell edge retraction. Blue dot indicates irradiation. Cells were visualized using PI-Rac1(WT)-mCherry.
A fibroblast expressing PI-RhoA(Q63L) visualized using Lifeact-mCherry fluorescence. Irradiation produced reversible cell protrusion. Blue dot indicates irradiation.
A fibroblast expressing PI-Cdc42(Q61L) visualized using Lifeact-mCherry fluorescence. Irradiation produced partially reversible cell retractions and accumulation of actin at the cell periphery. Blue dot indicates irradiation.
HeLa cells expressing PI-Vav2 shrank in the light, and rapidly recovered in the dark. Blue dot indicates irradiation. White arrows point to regions undergoing retraction. Cells were visualized using Lifeact-mCherry.
A fibroblast expressing uniRapR-Vav2 produced protrusions upon addition of rapamycin. Cells were visualized using mCherry-uniRapR-Vav2.
Fibroblasts expressing PA-Vav2 rounded when irradiated, and underwent contraction when returned to the dark. Blue dot denotes irradiation. Cells were visualized using mCherry-PA-Vav2.
PA-Vav2 and PI-Rac1 in the same cell. Upon irradiation, area and roundness changed as they did in response to PA-Vav2 alone, but edge dynamics were clearly reduced, unlike response to either PA-Vav2 alone or PI-Rac1 alone. Blue dot denotes irradiation. Cells were visualized using mVenus-PI-Rac1.
Acknowledgments
This work was supported by NIH grants P01-GM103723 and P41-EB002025 (K.M.H.), R01GM080742 (N.V.D.), and by DFG grant FOR1279 (I.S.). O.D. is a Howard Hughes Medical Institute International Student Research Fellow. X-ray data were collected at the Swiss Light Source, beamline X10SA and X12SA, Paul Scherrer Institute, Villigen, Switzerland. We thank Andreas Menzel and Thomas Barends for collecting the SAXS data. Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes: 5HZJ (PI-ITSN1-WT), 5HZI (PI-ITSN1-C450M), 5HZK (PI-ITSN1-WT:Cdc42), 5HZH (PI-Rac1-C450A). O.D., N.V.D. and K.M.H. designed the research; M.T. performed the crystallographic work and conducted hydrogen/deuterium exchange coupled to mass spectrometry assays under the directions of I.S. who collected the X-ray data.; P.C. helped the characterization of PI-Src; D.S. conducted the simulations of PI-Src; O.D. performed all experiments and computations not listed above; O.D. and K.M.H. wrote the manuscript with input from all authors.
Footnotes
Materials and Methods
References (31–63)
References and Notes
- 1.Weitzman M, Hahn KM. Optogenetic approaches to cell migration and beyond. Curr Opin Cell Biol. 2014;30:112–120. doi: 10.1016/j.ceb.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banaszynski LA, Wandless TJ. Conditional control of protein function. Chem Biol. 2006;13:11–21. doi: 10.1016/j.chembiol.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennedy MJ, et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nature Methods. 2010;7:973–U948. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yumerefendi H, et al. Light-induced nuclear export reveals rapid dynamics of epigenetic modifications. Nature chemical biology. 2016;12:399–401. doi: 10.1038/nchembio.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niopek D, et al. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat Commun. 2014;5:4404. doi: 10.1038/ncomms5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–U111. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical Control of Protein Activity by Fluorescent Protein Domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi JH, Laurent AH, Hilser VJ, Ostermeier M. Design of protein switches based on an ensemble model of allostery. Nat Commun. 2015;6:6968. doi: 10.1038/ncomms7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostermeier M. Designing switchable enzymes. Current Opinion in Structural Biology. 2009;19:442–448. doi: 10.1016/j.sbi.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J, et al. Surface sites for engineering allosteric control in proteins. Science. 2008;322:438–442. doi: 10.1126/science.1159052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM. Engineered allosteric activation of kinases in living cells. Nat Biotechnol. 2010;28:743–U1756. doi: 10.1038/nbt.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dagliyan O, et al. Rational design of a ligand-controlled protein conformational switch. Proc Natl Acad Sci U S A. 2013;110:6800–6804. doi: 10.1073/pnas.1218319110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oakes BL, et al. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat Biotechnol. 2016;34:646–651. doi: 10.1038/nbt.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tucker CL, Fields S. A yeast sensor of ligand binding. Nat Biotechnol. 2001;19:1042–1046. doi: 10.1038/nbt1101-1042. [DOI] [PubMed] [Google Scholar]
- 16.Chu PH, et al. Engineered kinase activation reveals unique morphodynamic phenotypes and associated trafficking for Src family isoforms. Proc Natl Acad Sci U S A. 2014;111:12420–12425. doi: 10.1073/pnas.1404487111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 18.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 19.Yao X, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-J alpha photoswitch. Nature chemical biology. 2008;4:491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fersht AR, Serrano L. Principles of protein stability derived from protein engineering experiments. Current Opinion in Structural Biology. 1993;3:75–83. [Google Scholar]
- 21.Playford MP, Schaller MD. The interplay between Src and integrins in normal and tumor biology. Oncogene. 2004;23:7928–7946. doi: 10.1038/sj.onc.1208080. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y, Karplus M. Interpreting the folding kinetics of helical proteins. Nature. 1999;401:400–403. doi: 10.1038/43937. [DOI] [PubMed] [Google Scholar]
- 23.Ding F, Tsao D, Nie H, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fincham VJ, Frame MC. The catalytic activity of Src is dispensable for translocation to focal adhesions but controls the turnover of these structures during cell motility. EMBO J. 1998;17:81–92. doi: 10.1093/emboj/17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunton VG, et al. Identification of Src-specific phosphorylation site on focal adhesion kinase: dissection of the role of Src SH2 and catalytic functions and their consequences for tumor cell behavior. Cancer Res. 2005;65:1335–1342. doi: 10.1158/0008-5472.CAN-04-1949. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, et al. LOVTRAP: an optogenetic system for photoinduced protein dissociation. Nat Methods. 2016;13:755–758. doi: 10.1038/nmeth.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Razidlo GL, Schroeder B, Chen J, Billadeau DD, McNiven MA. Vav1 as a central regulator of invadopodia assembly. Curr Biol. 2014;24:86–93. doi: 10.1016/j.cub.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe K, et al. Vav2 is an activator of Cdc42, Rac1, and RhoA. J Biol Chem. 2000;275:10141–10149. doi: 10.1074/jbc.275.14.10141. [DOI] [PubMed] [Google Scholar]
- 30.Samson T, Welch C, Monaghan-Benson E, Hahn KM, Burridge K. Endogenous RhoG is rapidly activated after epidermal growth factor stimulation through multiple guanine-nucleotide exchange factors. Mol Biol Cell. 2010;21:1629–1642. doi: 10.1091/mbc.E09-09-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An SYF cell expressing PI-Src(YF). PI-Src was localized to focal adhesions before, during, and after irradiation. Irradiation led to retraction and a reduced migration rate. Cells were visualized using PI-Src(YF)-mCherry. Blue dot indicates irradiation.
An SYF cells expressing PI-Src(YF) retracted and showed reduced edge dynamics when treated with 10 μM PP2 inhibitor. Cells were visualized using differential interference contrast.
SYF cells expressing PI-Src(WT). Irradiation caused the PI-Src(WT) to reversibly translocate to focal adhesions. This was accompanied to varying degrees by cell polarization, cell translation, and increased edge dynamics. Blue dot indicates irradiation. Cells were visualized using PI-Src(WT)-mCherry.
A fibroblast expressing PI-Rac1(Q61L). Irradiation produced reversible cell edge retraction, initially as indentations closely spaced along the edge, and then broader retraction of lamellae. Blue dot indicates irradiation. Cells were visualized using PI-Rac1(Q61L)-mCherry.
A fibroblast expressing PI-Rac1(WT). Irradiation produced reversible cell edge retraction. Blue dot indicates irradiation. Cells were visualized using PI-Rac1(WT)-mCherry.
A fibroblast expressing PI-RhoA(Q63L) visualized using Lifeact-mCherry fluorescence. Irradiation produced reversible cell protrusion. Blue dot indicates irradiation.
A fibroblast expressing PI-Cdc42(Q61L) visualized using Lifeact-mCherry fluorescence. Irradiation produced partially reversible cell retractions and accumulation of actin at the cell periphery. Blue dot indicates irradiation.
HeLa cells expressing PI-Vav2 shrank in the light, and rapidly recovered in the dark. Blue dot indicates irradiation. White arrows point to regions undergoing retraction. Cells were visualized using Lifeact-mCherry.
A fibroblast expressing uniRapR-Vav2 produced protrusions upon addition of rapamycin. Cells were visualized using mCherry-uniRapR-Vav2.
Fibroblasts expressing PA-Vav2 rounded when irradiated, and underwent contraction when returned to the dark. Blue dot denotes irradiation. Cells were visualized using mCherry-PA-Vav2.
PA-Vav2 and PI-Rac1 in the same cell. Upon irradiation, area and roundness changed as they did in response to PA-Vav2 alone, but edge dynamics were clearly reduced, unlike response to either PA-Vav2 alone or PI-Rac1 alone. Blue dot denotes irradiation. Cells were visualized using mVenus-PI-Rac1.