Abstract
Background
Despite extensive study on hemoglobins and hemocyanins, little is known about hemerythrin (Hr) evolutionary history. Four subgroups of Hrs have been documented, including: circulating Hr (cHr), myohemerythrin (myoHr), ovohemerythrin (ovoHr), and neurohemerythrin (nHr). Annelids have the greatest diversity of oxygen carrying proteins among animals and are the only phylum in which all Hr subgroups have been documented. To examine Hr diversity in annelids and to further understand evolution of Hrs, we employed approaches to survey annelid transcriptomes in silico.
Results
Sequences of 214 putative Hr genes were identified from 44 annelid species in 40 different families and Bayesian inference revealed two major clades with strong statistical support. Notably, the topology of the Hr gene tree did not mirror the phylogeny of Annelida as presently understood, and we found evidence of extensive Hr gene duplication and loss in annelids. Gene tree topology supported monophyly of cHrs and a myoHr clade that included nHrs sequences, indicating these designations are functional rather than evolutionary.
Conclusions
The presence of several cHrs in early branching taxa suggests that a variety of Hrs were present in the common ancestor of extant annelids. Although our analysis was limited to expressed-coding regions, our findings demonstrate a greater diversity of Hrs among annelids than previously reported.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-017-0933-z) contains supplementary material, which is available to authorized users.
Keywords: Blood pigments, Respiratory proteins, Transcriptome, Annelida
Background
Metabolism in metazoans requires oxidation of organic molecules. Thus natural selection has presumably favored proteins that can reversibly bind and transport oxygen [1]. Such oxygen-binding proteins likely originated from enzymes whose primary function was to protect the body from oxygen toxicity, and, secondarily, these enzymes acquired the ability to carry oxygen molecules [2]. Several different classes of oxygen-carrying proteins, or respiratory pigments, are found across animal life. Although these molecules can reversibly bind oxygen, their binding affinities and evolutionary origins differ. In animals, oxygen-binding proteins are usually divided into two main groups: proteins that use iron to bind oxygen, including hemoglobins and hemerythrins, and hemocyanins that use copper [3]. Although hemoglobins and hemocyanins have been extensively investigated [4–8], knowledge on the evolutionary history of hemerythrins is limited [9]. Interestingly, medical sciences have increasingly been taking advantage of oxygen-binding proteins as blood substitutes [10, 11] or as carrier proteins for synthetic vaccines, (e.g., cancer vaccines; [12, 13]) making further study of oxygen binding protein diversity and evolution appealing.
Hemerythrins (Hrs) are a non-heme oligomeric protein family within the ‘four-helical up-and-down bundle’ fold and ‘all alpha proteins’ class according to the Structural Classification of Proteins database (SCOP) [14]. Oxygen-binding Hr proteins contain approximately 120 amino acid residues in a single domain and transport oxygen with the aid of two iron ions that bind directly to the polypeptide chain. Residues involved in iron binding include histidines (His) in positions 26 56, 75, 79, and 108, glutamic acid residue (Glu) in position 60 and aspartic acid residue (Asp) in position 113 (position numbers from Themiste zostericola in [15]). Presence of these signature residues indicates putative respiratory function for Hrs [16]. Functional Hr subunits usually form a homooctamer, although dimeric, trimeric, or tetrameric Hrs have been observed in some sipunculid species, including Phascolosoma arcuatum, P. agasizii, and Siphonosoma funafuti [17–19]. The crystal structure of Hrs consists of a bundle of four antiparallel α-helices (A, B, C, and D) formed by polypeptides: an A α-helix formed by 19 amino acid residues from position 19 to 38, B α-helix with 23 amino acids residues from position 43 to 65, C α-helix formed by 16 amino acids residues from position 72 to 88, and D α-helix formed by 20 amino acids residues from position 98 to 118, using T. zostericola as the reference sequence [9]. The core of active sites contains two iron atoms bridged by two carboxylate groups from aspartate and glutamate residues and an oxygen-containing ligand [20, 21]. Binding of oxygen apparently requires other currently unknown cellular factors since purified Hr, by itself, usually does not bind oxygen [22, 23]. Observed oxygen binding capacity is about 25% greater in Hrs than heme-based proteins, including hemoglobins [15].
Although Hr-like proteins have also been reported in prokaryotes [24–26] oxygen binding Hrs have only been reported from marine invertebrates belonging to Annelida (which include sipunculids; [27]) Brachiopoda, Priapulida, Bryozoa, and a single species of both Cnidaria (Nematostella vectensis) and Arthropoda (Calanus finmarchicus) [9, 18, 28, 29]. Given this phylogenetic breadth of animals, whether all metazoan Hrs share a common origin is debated [28, 29]. Overall, Hr proteins exhibit variation in their quaternary structure, and four groups have been reported based mainly on their primary structure and location within animal bodies [9]. Specifically, hemerythrins found in vascular tissue, referred to here as circulating hemerythrins (cHrs), and muscle-specific myohemerythrins (myoHr), have been better characterized compared to the other two hemerythrin groups, ovohemerythrins (ovoHr) and neurohemerythrins (nHr) [28, 30]. cHrs are polymeric intracellular proteins that occur inside nucleated cells, hemerythrocytes or pink blood cells located in coelomic fluid or vascular systems of Hr-bearing organisms [15]. In contrast, myoHrs are monomeric cytoplasmic proteins present in muscle cells of annelids [31]. The main difference between these groups is the presence of a five-codon insertion found in myoHr immediately before the D α-helix. Expression of myoHrs seems not to be restricted to cHrs-bearing organisms, considering that some annelid species possess both myoHrs and hemoglobins [32]. The other two groups of Hrs, ovohemerythrin (ovoHr) and neurohemerythrin (nHr), are also intracellular and non-circulating. ovoHr was identified in oocytes of the leech Theromyzon tessulatum and its presence during oogenesis possibly suggests a complex function in iron storage and detoxification [32, 33]. On the other hand, nHr was recently discovered in neural and non-neuronal tissues from the body wall of the leech Hirudo medicinalis, and it exhibits upregulation in response to septic injury [34]. Nevertheless, Vanin et al. suggested that nHr of leech may in fact be a myoHr [9]. Such diversification in Hr function may have involved gene duplications resulting in new proteins via neo- or subfunctionalization [32]. Moreover, ovoHrs and nHrs have only been reported in the literature a few times, and more studies are required to understand their function and evolution.
Annelids have the greatest diversity of oxygen-binding proteins among metazoans [35] and it is the only phylum from which all subtypes of Hr proteins have been documented [28]. While Hrs of annelids have been studied since the middle of the 20th century, until the 1990s, Hrs were recorded only from sipunculids and from a single polychaete family, Magelonidae [18, 36]. Later, Vanin et al. [9] found Hrs in a nereid and a leech and Bailly et al. [28] discovered Hrs genes in seven annelid species, suggesting Hrs are broadly distributed in annelids. Given the diversity of lifestyles among annelids known to have Hrs [29, 37], and the lack of information about Hrs in general [9], the occurrence and diversity of these molecules may be higher than currently recognized. Thus, to examine a wide diversity of annelid taxa for Hrs and to further understand how different forms of Hrs are evolutionarily related to each other, we employed approaches to survey Hrs from a diverse array of annelid transcriptomes in silico. We identified Hrs in 44 taxa and further describe the molecular diversity and evolution of Hrs in the light of annelid phylogeny [27, 38, 39]. Along with this, we assess whether described Hr subtypes consist of evolutionary lineages or result of independent adaptations to different organismal tissues.
Results
Our bioinformatic analyses (Additional file 1) recovered a total of 415 unique nucleotide sequences of hemerythrin-like genes. Following translation Pfam domain evaluation and manual removal of sequences with less than 100 amino acid residues, 214 putative novel Hr genes were retained from all taxa examined in this study, representing 44 annelid species in 40 different families (Table 1). Novel Hr genes accession numbers for each species is available in Additional file 2. The number of expressed Hrs in a given species ranged from one in Alciopa sp., Cossura longocirrata, Enchytraeus albidus, Schizobranchia insignis, and Syllis cf. hyaline to 11 in Magelona berkeleyi and Phascolosoma agassizii.
Table 1.
List of taxa, including collection site, total number of base pairs sequenced, total number of contigs after assembly, number of putative Hr genes, and GenBank accession numbers. GenBank accession numbers for each Hr copy is indicated in Additional file 2
| Species | Collection site | Total bp | Total contigs number | Hr genes number | Accession number |
|---|---|---|---|---|---|
| ALCIOPIDAE | |||||
| Alciopa sp. | N 33° 07.’ W 076° 06.4’ | 157,869,560 | 233,051 | 1 | KY007275 |
| ALVINELLIDAE | |||||
| Paralvinella palmiformis Desbruyères & Laubier, 1986 | N 47° 56.9’ W 129° 05.9’ | 59,602,987 | 85,363 | 1 | KY007423 |
| AMPHINOMIDAE | |||||
| Paramphinome jeffreysii (McIntosh, 1868) | N 63° 30.8’ E 10° 25.0’ | 104,449,511 | 165,337 | 8 | KY007424 to KY007431 |
| APHRODITIDAE | |||||
| Aphrodita japonica Marenzeller, 1879 | N 48° 28.6’ W 122° 58.7’ | 84,662,357 | 120,025 | 9 | KY007279 to KY007287 |
| ASPIDOSIPHONIDAE | |||||
| Aspidosiphon laevis de Quatrefages, 1865 | N 09° 22.6’ W 82° 18.1’ | 120,601,137 | 168,072 | 4 | KY007297 to KY007300 |
| CAPITELLIDAE | |||||
| Heteromastus filiformis (Claparede, 1864) | N 41° 41.5’ W 070° 37.6’ | 94,824,555 | 148,196 | 8 | KY007364 to KY007371 |
| CHAETOPTERIDAE | |||||
| Chaetopterus variopedatus (Renier, 1804) | N 41° 41.5’ W 070° 43.5’ | 166,610,386 | 147,132 | 2 | KY007307 and KY007309 |
| Mesochaetopterus taylori Potts, 1914 | N 48° 29.0’ W 123° 04.3’ | 86,521,966 | 83,209 | 3 | KY007383 to KY007386 |
| CHRYSOPETALIDAE | |||||
| Arichlidon gathofi Watson Russell, 2000 | N 33° 59.6’ W 76° 42.1’ | 105,115,966 | 140,980 | 8 | KY007288 to KY007296 |
| CIRRATULIDAE | |||||
| Chaetozone sp. | N 66° 33.2’ E 33° 06.7’ | 85,730,053 | 143,587 | 4 | KY007310 to KY007313 |
| COSSURIDAE | |||||
| Cossura longocirrata Webster & Benedict, 1887 | N 33° 29.4’ W 074° 48.0’ | 45,732,505 | 75,079 | 1 | KY007319 |
| ENCHYTRAEIDAE | |||||
| Enchytraeus albidus Henle, 1837 | N 59o 5.2' E 16o 03.3' | 13,345,974 | 22,776 | 1 | KY007331 |
| EUNICIDAE | |||||
| Eunice pennata (Müller, 1776) | N 39° 47.2’ W 70° 46.3’ | 59,429,144 | 93,814 | 5 | KY007332 to KY007336 |
| EUPHROSINIDAE | |||||
| Euphrosine capensis Kinberg, 1857 | S 34° 09.9’ E 18° 26.0’ | 27,221,777 | 72,220 | 3 | KY007337 to KY007339 |
| FLABELLIGERIDAE | |||||
| Poeobius meseres Heath, 1930 | N 36° 41.2’ W 122° 02.0’ | 25,964,726 | 70,078 | 2 | KY007441 and KY007442 |
| GLYCERIDAE | |||||
| Glycera dibranchiata Ehlers, 1868 | N 41° 54.1’ W 070° 00.4’ | 51,282,233 | 101,455 | 3 | KY007350 to KY007352 |
| GOLFINGIIDAE | |||||
| Thysanocardia nigra (Ikeda, 1904) | N 48° 28.6’ W 122° 58.7’ | 57,399,340 | 58,011 | 6 | KY007480 to KY007485 |
| GONIADIDAE | |||||
| Glycinde armigera Moore, 1911 | N 36° 23.0’ W 121° 57.9’ | 32,178,692 | 79,528 | 4 | KY007353 to KY007356 |
| HAPLOTAXIDAE | |||||
| Delaya leruthi (Hrabe, 1958) | N 43° 0.8’ E 01° 2.5’ | 93,863,431 | 118,020 | 7 | KY007320 to KY007326 |
| HESIONIDAE | |||||
| Oxydromus pugettensis (Johnson, 1901) | N 48° 34.3’ W 123° 10.1’ | 45,242,396 | 92,341 | 2 | KY007421 and KY007422 |
| LUMBRINERIDAE | |||||
| Ninoe sp. | N 35° 29.4’ W 074° 48.0’ | 120,256,564 | 151,183 | 5 | KY007409 to KY007413 |
| MAGELONIDAE | |||||
| Magelona berkeleyi Jones, 1971 | N 36° 22.8’ W 121° 58.1’ | 16,339,407 | 50,123 | 10 | KY007372 to KY007382 |
| MALDANIDAE | |||||
| Clymenella torquata (Leidy, 1855) | N 41° 42.7’ W 070° 19.7’ | 62,661,529 | 111,567 | 5 | KY007314 to KY007314 |
| Nicomache venticola Blake & Hilbig, 1990 | N 47° 57.0’ W 129° 05.9’ | 64,130,139 | 124,708 | 4 | KY007405 to KY007408 |
| NEPHTYIDAE | |||||
| Nephtys incisa Malmgren, 1865 | N 40° 53.0’ W 070° 25.0’ | 126,720,409 | 188,338 | 6 | KY007396 to KY007401 |
| NEREIDIDAE | |||||
| Alitta succinea (Leuckart, 1847) | N 41° 54.0’ W 070° 00.3’ | 105,821,565 | 153,011 | 3 | KY007402 to KY007404 |
| OENONIDAE | |||||
| Drilonereis sp. | N 39° 54.1’ W 070° 35.1’ | 3,490,940 | 12,598 | 4 | KY007327 to KY007330 |
| Oenone fulgida (Savigny in Lamarck, 1818) | S 34° 37.0’ E 19° 21.4’ | 92973167 | 144,726 | 3 | KY007414 to KY007416 |
| ORBINIIDAE | |||||
| Naineris laevigata (Grube, 1855) | S 34° 35.0’ E 19° 20.9’ | 123,970,343 | 218,272 | 4 | KY007392 to KY007395 |
| OWENIIDAE | |||||
| Galathowenia oculata (Zachs, 1923) | N 66° 33.2’ E 33° 6.7’ | 128,195,375 | 179,612 | 10 | KY007340 to KY007349 |
| PECTINARIIDAE | |||||
| Pectinaria gouldii (Verrill, 1874) | N 41° 37.9’ W 070° 53.3’ | 63,132,019 | 92,091 | 9 | KY007432 to KY007440 |
| PHASCOLOSOMATIDAE | |||||
| Phascolosoma agassizii Keferstein, 1866 | N 48° 31.2’ W 123° 01.0’ | 78,749,017 | 87,403 | 11 | KY007443 to KY007453 |
| PILARGIDAE | |||||
| Ancistrosyllis groenlandica McIntosh, 1879 | N 40° 27.3’ W 070° 47.6’ | 39,327,753 | 94,924 | 3 | KY007276 to KY007278 |
| POLYNOIDAE | |||||
| Halosydna brevisetosa Kinberg, 1856 | N 48° 29.5’ W 123° 01.1’ | 61,671,140 | 118,418 | 7 | KY007357 to KY007363 |
| RANDIELLIDAE | |||||
| Randiella sp. | S 14o 40.0' E 145o 27.1' | 139,189,396 | 151,934 | 6 | KY007454 to KY007459 |
| SABELLIDAE | |||||
| Myxicola infundibulum (Montagu, 1808) | N 48° 28.6’ W 122° 58.7’ | 156,042,620 | 217,996 | 5 | KY007387 to KY007391 |
| Schizobranchia insignis Bush, 1905 | N 48° 33.3’ W 122° 56.5’ | 55,085,979 | 102,002 | 1 | KY007460 |
| SPARGANOPHILIDAE | |||||
| Sparganophilus sp. | N 40° 50.3’ W 92° 5.3’ | 117,343,038 | 123,905 | 8 | KY007461 to KY007468 |
| SPIONIDAE | |||||
| Boccardia proboscidea Hartman, 1940 | N 48° 29.2’ W 123° 04.1’ | 78,374,988 | 117,570 | 7 | KY007301 to KY007307 |
| STERNASPIDAE | |||||
| Sternaspis scutata Ranzani, 1817 | N 48° 29.1’ W 123° 04.3’ | 81,147,455 | 115,096 | 3 | KY007469 to KY007471 |
| SYLLIDAE | |||||
| Syllis cf. hyalina Grube, 1863 | S 34° 37.0’ E 19° 21.4’ | 76,801,405 | 106,283 | 1 | KY007472 |
| THEMISTIDAE | |||||
| Themiste pyroides (Chamberlin, 1919) | N 48° 21.4’ W 123° 43.4’ | 75,495,745 | 88,157 | 7 | KY007473 to KY007479 |
| TOMOPTERIDAE | |||||
| Tomopteris sp. | N 36° 41.2’ W 122° 02.0’ | 30,525,410 | 66,655 | 3 | KY007486 to KY007488 |
| Oligochaeta gen. sp. (unidentified Crassiclitellata - Place Kabary 2) | S 12° 59.0’ E 49° 17.4’ | 107,638,847 | 146,018 | 4 | KY007417 to KY007420 |
Following trimming alignment of translated transcripts possessed 132 residue positions, with nearly all sequences, the exception being Aphrodita japonica, starting with a methionine residue. We decided to keep the apparently incomplete sequence from A. japonica due to its high similarity with the remaining sequences. All sequences in the alignment contained signature residues involved in iron binding, indicating putative respiratory function for these putative Hrs [16]. For the 214 sequences, 100 were unique and 114 identical for at least two species at the amino acid level.
Sequences were assembled into a final dataset containing 225 sequences being 214 new, two Hr sequences from Lingula reevii, a brachiopod, and nine annelid sequences previous used as “queries”, with 396 aligned nucleotides positions. Of these, 209 sequences contained a five-codon insertion between the C and D α-helices described for myoHr, but not cHrs (Fig. 1) [9]. Datasets supporting conclusions of this article are available in the Figshare repository, under DOIs: https://doi.org/10.6084/m9.figshare.3505883.v1 and https://doi.org/10.6084/m9.figshare.3505886.v1.
Fig. 1.
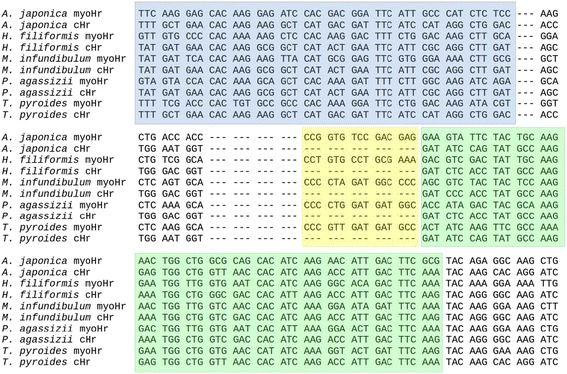
Alignment of nucleotide dataset of few species. Showing the five-codon insertion (yellow background) between C α-helix (blue background) and D α-helix (green background)
Every species analyzed possessed at least one copy of myoHr gene. Bayesian inference rooted using two brachiopod Hr sequences revealed two major clades with one corresponding to cHrs clade (p.p. = 0.93; Fig. 2 blue clade) and the other supported monophyly for a myoHr clade (p.p. = 0.95; Fig. 2 black clade). The leech nHr sequence included in the dataset was found inside the myoHr clade (Fig. 2; orange). Within the myoHr clade low nodal support values and polytomies were found. However, such results are not uncommon for single protein or protein family trees [40].
Fig. 2.
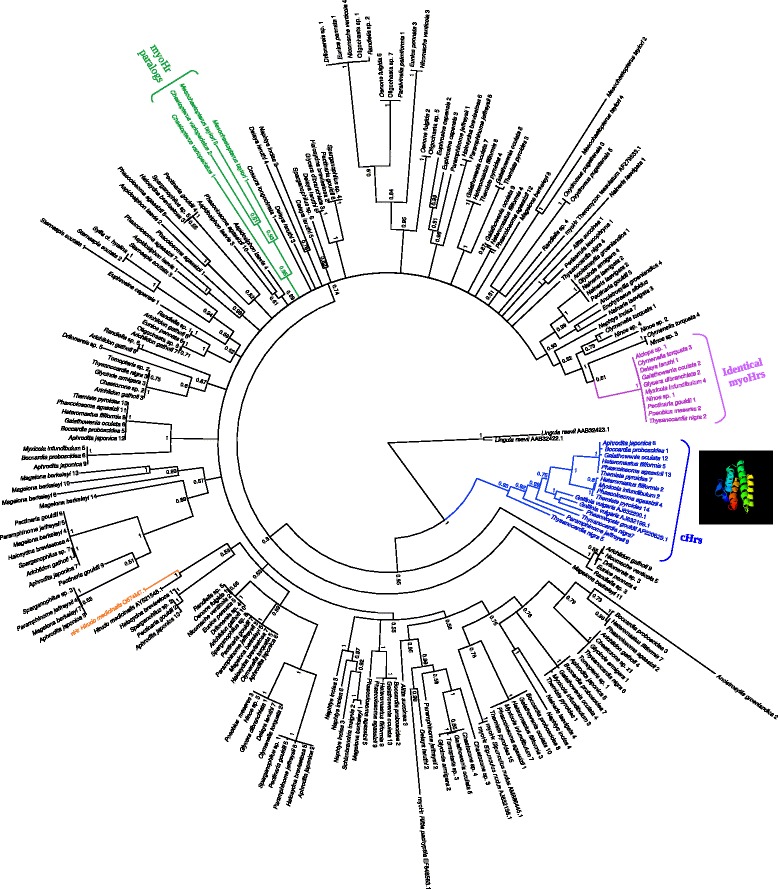
Bayesian tree using MrBayes 3.2.1 [51] rooted with two Hr sequences from Lingula reevii, a brachiopod. The blue clade represents the cHrs othologs sequences with a prediction of the consensus cHr sequence quaternary structure using I-TASSER 4.4 [61]. Black, purple, and green sequences represent the myoHr sequences and orange sequence is the only nHr from Hirudo medicinalis. The green clade shows the paralogs sequences from C. variopedatus and M. taylori and the purple clade represents the identical orthologs myoHr sequences. The number after the name of each sequence indicates the GenBank accession numbers for each Hr gene and it is indicated in Additional file 2: Table S1
Our analysis found 13 putative cHr sequences lacking the characteristic five-codon insertion before the D α-helix that define myoHrs, distributed across nine different families. Besides well-known records of cHrs in sipunculids, such as Themiste pyroides and Phascolosoma agassizii, we discovered cHrs in the sipunculid Thysanocardia nigra as well as six annelid families; Amphinomidae Aproditidae, Capitellidae, Oweniidae, Sabellidae, and Spionidae (Fig. 2; blue clade).
The topology of the Hr gene tree did not mirror recent phylogenies of Annelida based on phylogenomic datasets [27, 38, 39]. For example, we found 10 Hr sequences identical at the nucleotide level (Fig. 2, purple clade) belonging to distant annelid families indicating a strong conservation among those orthologs. Several of these sequences were prepared and sequenced at different times, making cross contamination unlikely. Those 10 identical sequences differed 28.54% (nucleotide level) from the consensus of all others myoHr sequences and the majority of nonsynonymous substitutions are concentrated in A and B α-helices.
Regarding paralogs multiple copies of Hr genes were found for several species, including two paralogs from both Chaetopterus variopedatus and Mesochaetopterus taylori, with these paralogs forming a monophyletic clade (p.p. = 0.85; Fig. 2; green clade). Both species were from Chaetopteridae suggesting a recent paralogous duplication [26]. Given the presence of multiple Hrs, apparently early annelids already contained several copies of Hr genes, with some paralogs arising later (as in C. variopedatus and M. taylori).
Differences in evolutionary rates between cHrs and myoHrs sequences was accessed and relative rates of change in different positions were calculated using two different approaches. Sites with high variation were found not to have significantly different rates among inter-helices sites using DIVERGE [41]. For helix regions A and C α-helices had one site each with high evolutionary rate, while D α-helix did not have any sites with a high evolutionary rate, indicating a highly conservative region. At the same time, B α-helix had five sites, suggesting that this helix is evolving faster than others (Fig. 3). RELAX [42] was also used to assess differences in selection on cHrs relative to myoHrs while accounting for lineage-specific rate differences. Similar to the DIVERGE analyses, no significant differences (P = 0.218) were found between the two sets of genes. Thus, the gene duplication event leading to cHrs versus myoHrs does not seem to have been accompanied by a significant change in substitution rate or selection differences on the two gene lineages. Alternatively, any evidence of such a rate or selection difference could have been lost during the 500+ MY since these genes diverged.
Fig. 3.
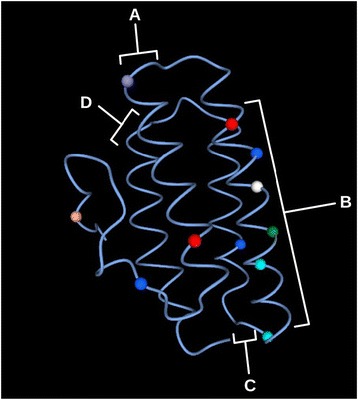
Hr showing differences in evolutionary rates between cHrs and myoHrs sequences calculated with DIVERGE [41] using a 0.7 cutoff. A and C α-helices presented one site each above the cutoff value, D α-helix did not present any sites, and B α-helix presented five sites. Colored dots represent sites above cutoff value, and different colors are only to aid illustration
Discussion
As demonstrated here expression of Hr genes among annelids is much more common than previously reported, revealing unrecognized diversity of these genes in this phylum. All 44 of the examined species possessed actively transcribed myoHrs, while cHrs were less frequently recovered from these transcriptomes. Although this diversity and wide distribution of Hrs in annelids could, in part, be explained by the need to carry oxygen (Hr have approximately 25% greater oxygen affinity than hemoglobins; [15]) secondary functional specializations could also be important for driving diversification. For example, Hrs participate in iron storage, metal detoxification, and immunity in some annelids (e.g., Theromyzon tessulatum, Hirudo medicinalis and Neanthes diversicolor) [33, 34, 43]. Our findings build on recent publications demonstrating that sequence diversity among Hr-bearing species was larger than traditionally suspected [9, 28, 29]. However, those studies were based predominately on genomic data. In this case, use of transcriptomes shows that Hrs genes were present and expressed.
Bayesian phylogenetic reconstruction recovered monophyletic clades for annelid cHrs sequences as well as myoHrs sequences with strong support corroborating Vanin et al.’s [9] previous findings. The presence of multiple copies of myoHr genes across the annelid phylogeny implies these proteins have undergone several instances of gene duplication during their evolution, as previously reported for other bacterial, archeal, and eukaryotic taxa [26]. Moreover, the unexpected diversity of myoHrs could be associated with functional diversification of this gene, as observed for myoglobins [44] and also for Hrs involved in heavy metal detoxification and aspects of innate immunity [45].
Classification of specific Hr subtypes [28, 32–34] was not validated by the gene genealogy. Although our analyses used whole organisms (including reproductive and nerve tissues), our results failed to recover Hr proteins that corresponded to ovoHrs or nHrs. These categories, however, were described based on limited differences in amino acid sequence and do not reflect distinct monophyletic subgroups within the Hr gene family. Given this, recognizing only two primary types of Hrs, circulating Hrs (cHr) and non-circulating Hrs (ncHrs), is perhaps more appropriate. Although tentative, this reinterpretation deserves further consideration.
Incongruence between our gene genealogy relative to current knowledge of annelid evolutionary history indicates that Hrs have a complex history which possibly involved events of gene losses and duplications, paralogs replacements and lateral gene transfer [26]. Although previous work supported the idea of a monomeric protein as an ancestral myoHr within annelids [9, 15], the presence of cHrs in Paramphinome jeffreysii (Amphinomidae), Galathowenia oculata (Oweniidae), Phascolosoma agassizii, Themiste pyroides, and Thysanocardia nigra (the last three belonging to Sipuncula), all members of lineages near the base of the annelid tree [27, 38, 39] (Fig. 4), indicates that both cHrs and myoHrs were likely present in the ancestor of annelids, which date back to the Cambrian [46]. Interestingly, Hrs from multiple species, representing hundreds of millions of years of evolution, possessed identical amino acid, and in some cases nucleotide, sequences (Fig. 2, purple clade), suggesting a level of conservation and selection orders of magnitude greater than most of the genome. Additional studies across metazoan together with studies of the gene structure of Hr proteins and physiological aspects of organisms are the next important steps toward a better understanding of the evolutionary patterns involved in this family of oxygen carrying proteins.
Fig. 4.
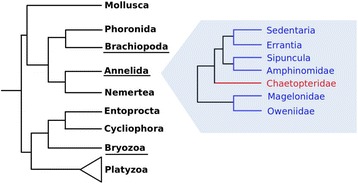
Lophotrochozoa and annelid relationships based on current knowledge [39, 62]. Underlined phyla represent the Hr-bearing representants. Annelid taxa in blue possess both cHr and myoHr genes and taxa in red possess just myoHr genes
Conclusions
Our findings demonstrate that sequence diversity among Hr-bearing annelid species is much greater than traditionally suspected and that many of these Hrs are actively expressed. There are two primary types of Hrs circulating Hrs (cHr) and non-circulating Hrs (ncHrs), instead of the four subtypes reported in the literature. Incongruence between our gene genealogy relative to current knowledge of annelid evolutionary history indicates that Hrs have a complex history. Our findings indicate that both cHrs and myoHrs were likely present in the ancestor of annelids, as both subtypes occur in all lineages near the base of the annelid tree.
Methods
Sample collection
Information on species employed herein is provided in Table 1. Transcriptomes of these species were collected as part of the WormNet II project to resolve annelid phylogeny and were collected with a variety of techniques, including intertidal sampling, dredge and box cores. Upon collection, all samples were either preserved in RNALater or frozen at −80 °C.
Data collection & sequence assembly
RNA extraction, cDNA preparation and high-throughput sequencing generally followed Kocot et al. [47] and Whelan et al. [48]. Briefly, total RNA was extracted from either whole animals (for small specimens) or the body wall and coelomic region (for larger specimens). After extraction, RNAs were purified using TRIzol (Invitrogen) or the RNeasy kit (Qiagen) with on-column DNase digestion, respectively. The SMART cDNA Library Construction Kit (Clonetech) was utilized to reverse transcribe single stranded RNA template. Double stranded cDNA synthesis was completed with The Advantage 2 PCR system (Clontech). Libraries were barcoded and sequenced with Illumina technology by The Genomic Services Lab at the Hudson Alpha Institute (Huntsville, Alabama, USA). Because transcriptomic sequencing was conducted from 2012–2015, Paired End (PE) runs were of 100 bp or 125 bp lengths, utilizing either v3 or v4 chemistry on Illumina HiSeq 2000 or 2500 platforms (San Diego, California). To facilitate sequence assembly, paired-end transcriptome data were digitally normalized to an average k-mer coverage of 30 using normalize-by-median.py [49] and assembled using Trinity r2013-02-25 with default settings [50].
Data mining and gene identification
Two approaches were utilized in searching transcriptomic data for putative Hr genes in silico. The first approach employed BLASTX [51] at an e-value cutoff of 10−6 to compare each assembled transcriptome contig (‘queries’) to a protein database composed of 21 protein sequence from the National Center for Biotechnology (NCBI) database (Additional file 3: Table S2) of at least 110 amino acid residues and previously identified as annelid Hrs (n = 11), myoHrs (n = 9), or nHr (n = 1). Sequences of ovoHrs were not included since there were only two relatively short (18 amino acid residues each) sequences available from NCBI. The BLASTX approach, rather than a tBLASTn, assured that any transcriptome contig with a significant ‘hit’ to an Hr would be further evaluated in the pipeline. Contigs recovered from initial BLAST searches were then utilized in BLASTX searches against the NCBI protein database (minimum e-value of 10−10) and only top hits longer than 300 nucleotides retained. These were considered putative cHr, myoHr or nHr genes, as appropriate.
The second approach processed the transcriptomic data through the Trinotate annotation pipeline (http://trinotate.github.io/) [50], which utilizes a BLAST-based approach to provide, among others, GO (The Gene Ontology Consortium) annotation [52]. Recovered sequences were verified by the functional annotation they received. Transcripts annotated as hemerythrins, using the 10−6 e-value cutoff obtained by using BLASTX, were also considered putative hemerythrin-like gene orthologs.
Contigs putatively identified as Hr genes by the two approaches were subsequently translated into amino acids using TransDecoder with default settings [53]. Since TransDecoder can produce multiple open reading frames (ORFs), all translations were additionally subject to a Pfam Domain evaluation using the EMBL-EBI database with an e-value cutoff of 10−5. Translations returning an Hr-like Pfam domain and that were longer than 100 amino acids residues were retained for subsequent analyses. Moreover, we manually evaluated the presence of residues involved in iron binding, which are: histidine residues (His) in positions 26, 56, 75, 79, and 108; glutamic acid residue (Glu) in position 60; and aspartic acid residue (Asp) in position 113, numbered by reference sequence T. zostericola. Presence of these signature residues indicates putative respiratory function for Hrs. Transcripts passing the criteria described above were considered Hr genes (Table 1). ovoHr has been just reported once for a single species and there is no complete sequence available at GenBank, so we are not able to investigate if ovoHr is present in our dataset.
Sequence alignment
The protein dataset consisted of 225 sequences, including two Hr sequences from Lingula reevii, a brachiopod, nine annelid sequences previous used as “queries” (Additional file 3: Table S2), and a remaining 214 sequences from translated transcripts (Additional file 4). All sequences were initially aligned with MAFFT using the “accurate E-INS-i” algorithm [54], followed by visual inspection and manual curation in order to remove spuriously aligned sequences based on similarity to the protein alignment as a whole. Subsequently, ends of aligned sequences were manually trimmed in Geneious 8.1.6 [55] to exclude 5’residues leading to the putative start codon and 3’ residues following the first seven amino acids subsequent to the end of the D α-helix.
In order to employ nucleotide sequences in the phylogenetic analysis, an alignment from corresponding aligned protein sequences (Additional file 4) was performed using PAL2NAL [56]. The resulting nucleotide alignment was used for all subsequent analyses (Additional file 5).
Phylogenetic analysis
JModelTest2 was applied to carry out statistical selection of best-fit models of nucleotide substitution for the dataset using the Akaike and Bayesian Information Criteria (AIC and BIC, respectively) methods [57]. Bayesian phylogenetic inference was performed with MrBayes 3.2.1 [58] using the GTR + G substitution model. Two independent runs with four Metropolis-coupled chains were run for 107 generations, sampling the posterior distribution every 500 generations. In order to confirm if chains achieved stationary and determine an appropriate burn-in, we evaluated trace plots of all MrBayes parameter output in Tracer v1.6 [59]. The first 25% of samples were discarded as burn-in and a majority rule consensus tree generated using MrBayes. Bayesian posterior probabilities were used for assessing statistical support of each bipartition.
Two alternative approaches were used to root the Hr gene genealogy. Firstly, the tree inferred in MrBayes was rooted using two Hr sequences from Lingula reevii (AAB32422.1 and AAB32423.1) as outgroup. We also inferred the root of the tree using using BEAST 1.8.3 [60] to infer a rooted tree of Hrs under the strict molecular clock. This was done because we were unable to decisively rule out the possibility that Hr sequences from Lingula reevii were closely evolutionarily related to one of the annelid Hr lineages. The strategy using BEAST is similar to midpoint rooting, although the Bayesian implementation in BEAST allows for a more flexible treatment of the evolutionary rate via a normal prior with mean and standard deviation equal to 1. Moreover, tree topology was jointly estimated. In BEAST, we adopted the same substitution model settings used in MrBayes and the MCMC algorithm was run for 50,000,000 generations and sampled every 1,000th generation, with 50% of the run discarded as burn-in. Trees was summarized in TreeAnnotator 1.8.3 [60] and Markov chain stationarity was assessed in Tracer by ESS values > 1,000. Since the inferred root node separated Lingula reevii sequences from annelid sequences with 0.86 posterior probability, the results are reported using the gene genealogy rooted with outgroup.
Evolutionary rate analyses
The protein alignment (Additional file 4) was used in DIVERGE [41] to examine site-specific shifted evolutionary rates and assesses whether there has been a significant change in evolution rate after duplication or speciation events by calculating the coefficient of divergence (θD) and determining if the null hypothesis of no functional divergence between myoHrs and cHrs could be statistically rejected. We employed a cutoff of 0.7 for detection of site-specific shifted evolutionary rates.
Additionally, we used RELAX [42] to detect changes in selection intensity between cHRs and myoHrs based on aligned nucleotide data (Additional file 5). Rather than just looking in dN/dS ratios averaged across branches using a ‘branch-site’ model approach, RELAX examines dN/dS ratios along each branch at a given site by drawing values from a discrete distribution of dN/dS ratios independent of other branches. In addition to assessing values in a branch independent manner, RELAX results are analyzed within a phylogenetic framework.
Additional files
Flow chart of bioinformatics pipeline. Rounded purple rectangles represent input/output files, orange ovals represent software or scripts, and the green hexagon represents a step which involving manual evaluation. Nine annelid Hrs sequences previous used as query and two Lingula (Brachipoda) sequences from Genbank (Additional file 3) were also included in the dataset. (DOC 1870 kb)
Hr genes with their respective accession numbers. Novel Hr genes accession numbers for each species. From the manuscript of Costa-Paiva et al. BMC Evolutionary Biology. (DOC 217 kb)
Outgroup and query sequences used to search assembled translated transcriptomes. Query seqeunces used to search transcriptomes. The Hr sequence for H. medicinalis possesses the myoHr five codon insertion between the C and D α-helix, so in this work we considered it a myoHr. Sequences in bold was also included in the dataset previous to the alignment. From the manuscript of Costa-Paiva et al. BMC Evolutionary Biology (DOC 53 kb)
The amino acid alignment used in analyses. The nucleotide alignment used for all subsequent analyses. (TXT 31 kb)
The nucleotide alignment used for analyses. (TXT 89 kb)
Acknowledgments
We thank Fernando Avila Queiroz for valuable help with the figures and Antonio Sole-Cava, Joana Zanol, and Francisco Prosdocimi for critically reading this manuscript. We are grateful to Christer Erséus and Sam James for oligochaete samples used in transcriptomes. A Fellowship for the first author was provided by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil). The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the U.S. Fish and Wildlife Service. Use of SkyNet computational resources at Auburn University is acknowledged. This paper is part of the D. Sc. Requirements of Elisa Maria Costa-Paiva at Biodiversity and Evolutionary Biology Graduate Program of the Federal University of Rio de Janeiro. This is Molette Biology Laboratory contribution 61 and Auburn University Marine Biology Program contribution 153.
Funding
This work was funded by National Science Foundation grant DEB-1036537 to K. M. Halanych and Scott R. Santos and OCE-1155188 to K. M. Halanych.
Availability of data and material
The alignment datasets supporting the conclusions of this article are available in the Figshare repository, under DOI: https://doi.org/10.6084/m9.figshare.3505883.v1 and https://doi.org/10.6084/m9.figshare.3505886.v1. Individual sequences have been deposited to GenBank under accessions KY007275-KY007488 and are listed in Additional file 2: Table S1.
Authors’ contributions
EMC and KMH conceived this study. EMC performed the in silico analysis helped by DSW. EMC, NVW and CGS participate in phylogenetic analysis. EMC, CGS and KMH interpreted the data. EMC and KMH drafted the manuscript and all others revised critically. All the authors read and approved the version to be published.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable
Ethics approval and consent to participate
This work used non vertebrate animals only and as such there were no ethic approval requirements.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- cHr
Circulating hemerythrin
- Hr
Hemerythrin
- myoHr
Myohemerythrin
- ncHr
Non-circulating hemerythrin
- nHr
Neurohemerythrin
- ovoHr
Ovohemerythrin
Contributor Information
Elisa M. Costa-Paiva, Email: elisapolychaeta@gmail.com
Nathan V. Whelan, Email: nathan_whelan@fws.gov
Damien S. Waits, Email: dsw0002@tigermail.auburn.edu
Scott R. Santos, Email: halocaridina@gmail.com
Carlos G. Schrago, Email: carlos.schrago@gmail.com
Kenneth M. Halanych, Email: ken@auburn.edu
References
- 1.Schmidt-Rhaesa A. The Evolution of Organs Systems. New York: Oxford University Press; 2007.
- 2.Terwilliger NB. Functional adaptations of oxygen-transport proteins. J Exp Biol. 1998;201:1085–1098. doi: 10.1242/jeb.201.8.1085. [DOI] [PubMed] [Google Scholar]
- 3.Terwilliger RC, Terwilliger NB, Schabtach E. Comparison of chlorocruorin and annelid hemoglobin quaternary structures. Comp Biochem Phys A. 1976;55(1):51–55. [DOI] [PubMed]
- 4.Burmester T. Origin and evolution of arthropod hemocyanins and related proteins. J Comp Physiol B. 2002;172(2):95–107. doi: 10.1007/s00360-001-0247-7. [DOI] [PubMed] [Google Scholar]
- 5.Burmester T. Evolution of respiratory proteins across the Pancrustacea. Integr Comp Biol. 2015;55(5):765–770. doi: 10.1093/icb/icv079. [DOI] [PubMed] [Google Scholar]
- 6.Lecomte JT, Vuletich DA, Lesk AM. Structural divergence and distant relationships in proteins: evolution of the globins. Curr Opin Struc Biol. 2005;15(3):290–301. doi: 10.1016/j.sbi.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Decker H, Hellmann N, Jaenicke E, Lieb B, Meissner U, Markl J. Minireview: Recent progress in hemocyanin research. Integr Comp Biol. 2007;47(4):631–644. doi: 10.1093/icb/icm063. [DOI] [PubMed] [Google Scholar]
- 8.Vinogradov SN, Hoogewijs D, Bailly X, Arredondo-Peter R, Gough J, Dewilde S, Moens L, Vanfleteren JR. A phylogenomic profile of globins. BMC Evol Biol. 2006;6(1):31. doi: 10.1186/1471-2148-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanin S, Negrisolo E, Bailly X, Bubacco L, Beltramini M, Salvato B. Molecular evolution and phylogeny of sipunculan hemerythrins. J Mol Evol. 2006;62:32–41. doi: 10.1007/s00239-004-0296-0. [DOI] [PubMed] [Google Scholar]
- 10.Rousselot M, Delpy E, Rochelle CD, Lagente V, Pirow R, Rees JF, Hagege A, Guen D, Hourdez S, Zal F. Arenicola marina extracellular hemoglobin: A new promising blood substitute. Biotechnol J. 2006;1(3):333–345. doi: 10.1002/biot.200500049. [DOI] [PubMed] [Google Scholar]
- 11.Zal F, Rousselot M. Use of a haemoglobin for the preparation of dressings and resulting dressings. U.S. Patent 9220929B2, December 28, 2015.
- 12.Jones L. Recent advances in the molecular design of synthetic vaccines. Nat Chem. 2015;7(12):952–960. doi: 10.1038/nchem.2396. [DOI] [PubMed] [Google Scholar]
- 13.Zal, F. Use of haemoglobin of annelids for treating cancer. U.S. Patent 0374796A1, December 31, 2015.
- 14.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:226–229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangum CP. Physiological function of the hemerythrins. In: Mangum CP, editor. Advances in Comparative & Environmental Physiology Vol 13 – Blood and Tissue Oxygen Carriers. Berlin: Springer; 1992. pp. 173–192. [Google Scholar]
- 16.Thompson JW, Salahudeen AA, Chollangi S, Ruiz JC, Brautigam CA, Makris TM, Lipscomb JD, Tomchick DR, Bruick RK. Structural and molecular characterization of iron-sensing hemerythrin-like domain within F-box and leucine-rich repeat protein 5 (FBXL5) J Biol Chem. 2012;287(10):7357–7365. doi: 10.1074/jbc.M111.308684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Addison AW, Bruce RE. Chemistry of Phascolosoma lurco hemerythrin. Arch Biochem Biophys. 1977;183:328–332. doi: 10.1016/0003-9861(77)90446-5. [DOI] [PubMed] [Google Scholar]
- 18.Klippenstein GL. Structural aspects of hemerythrin and myohemerythrin. Am Zool. 1980;20:39–51. doi: 10.1093/icb/20.1.39. [DOI] [Google Scholar]
- 19.Klotz IM, Kurtz DM., Jr Binuclear oxygen carriers: hemerythrin. Accounts Chem Res. 1984;17(9):16–22. doi: 10.1021/ar00097a003. [DOI] [Google Scholar]
- 20.Stenkamp RE. Dioxygen and hemerythrin. Chem Rev. 1994;94:715–726. doi: 10.1021/cr00027a008. [DOI] [Google Scholar]
- 21.Wirstam M, Lippard SJ, Friesner R. Reversible dioxygen binding to hemerythrin. J Am Chem Soc. 2003;125(13):3980–7. doi: 10.1021/ja017692r. [DOI] [PubMed] [Google Scholar]
- 22.Wells RMG. Respiratory characteristics of the blood pigments of three worms from an intertidal mudflat. New Zeal J Zool. 1982;9:243–248. doi: 10.1080/03014223.1982.10423853. [DOI] [Google Scholar]
- 23.Kurtz DM., Jr . Molecular structure/function of the hemerythrins. In: Mangum CP, editor. Advances in Comparative & Environmental Physiology Vol 13 – Blood and Tissue Oxygen Carriers. Berlin: Springer; 1992. pp. 151–171. [Google Scholar]
- 24.French CE, Bell JML, Ward FB. Diversity and distribution of hemerythrin-like proteins in prokaryotes. FEMS Microbiol Lett. 2008;279(2):131–145. doi: 10.1111/j.1574-6968.2007.01011.x. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Tao J, Hu X, Chan J, Xiao J, Mi K. A bacterial hemerythrin-like protein MsmHr inhibits the SigF-dependent hydrogen peroxide response in mycobacteria. Front Microbiol. 2015;5(Jan):1–11. doi: 10.3389/fmicb.2014.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez-Carreño C, Becerra A, Lazcano A. Molecular evolution of the oxygen-binding hemerythrin domain. PLoS ONE. 2016;11(6):e0157904. doi: 10.1371/journal.pone.0157904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weigert A, Helm C, Meyer M, Nickel B, Arendt D, Hausdorf B, Santos SR, Halanych KM, Purschke G, Bleidorn C, Struck TH. Illuminating the base of the annelid tree using transcriptomics. Mol Biol Evol. 2014;31(6):1391–1401. doi: 10.1093/molbev/msu080. [DOI] [PubMed] [Google Scholar]
- 28.Bailly X, Vanin S, Chabasse C, Mizuguchi K, Vinogradov SN. A phylogenomic profile of hemerythrins, the nonheme diiron binding respiratory proteins. BMC Evol Biol. 2008;8:244. doi: 10.1186/1471-2148-8-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martín-Durán JM, De Mendoza A, Sebé-Pedrós A, Ruiz-Trillo I, Hejnol A. A broad genomic survey reveals multiple origins and frequent losses in the evolution of respiratory hemerythrins and hemocyanins. Genome Biol Evol. 2013;5:1435–1442. doi: 10.1093/gbe/evt102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klippenstein GL, Cote JL, Ludlam SE. The primary structure of myohemerythrin. Biochemistry. 1966;15(5):1128–1136. doi: 10.1021/bi00650a027. [DOI] [PubMed] [Google Scholar]
- 31.Ward KB, Hendrickson WA, Klippenstein GL. Quaternary and tertiary structure of hemerythrin. Nature. 1975;257:818–821. doi: 10.1038/257818a0. [DOI] [PubMed] [Google Scholar]
- 32.Coutte L, Slomianny MC, Malecha J, Baert JL. Cloning and expression analysis of a cDNA that encodes a leech hemerythrin. Biochim Biophys Acta. 2001;1518(3):282–286. doi: 10.1016/S0167-4781(00)00312-2. [DOI] [PubMed] [Google Scholar]
- 33.Baert JL, Britel M, Sautière P, Malécha J. Ovohemerythrin, a major 14-kDa yolk protein distinct from vitellogenin in leech. Eur J Biochem. 1992;209:563–569. doi: 10.1111/j.1432-1033.1992.tb17321.x. [DOI] [PubMed] [Google Scholar]
- 34.Vergote D, Sautière PE, Vandenbulcke F, Vieau D, Mitta G, Macagno ER, Salzet M. Up-regulation of neurohemerythrin expression in the central nervous system of the medicinal leech, Hirudo medicinalis, following septic injury. J Biol Chem. 2004;279(42):43828–37. doi: 10.1074/jbc.M403073200. [DOI] [PubMed] [Google Scholar]
- 35.Mangum CP. Major events in the evolution of the oxygen carriers. Amer Zool. 1998;38(1):1–13. doi: 10.1093/icb/38.1.1. [DOI] [Google Scholar]
- 36.Manwell C, Baker CMA. Magelona haemerythrin: tissue specificity, molecular weights and oxygen equilibria. Comp Biochem Phys B. 1988;89(3):453–463. [Google Scholar]
- 37.Rouse G, Pleijel F. Polychaetes. New York: Oxford University Press; 2001.
- 38.Struck TH, Golombek A, Weigert A, Franke FA, Westheide W, Purschke G, Bleidorn C, Halanych KM. The evolution of annelids reveals two adaptive routes to the interstitial realm. Curr Biol. 2015;25(15):1993–1999. doi: 10.1016/j.cub.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Weigert A, Bleidorn C. Current status of annelid phylogeny. Org Divers Evol. 2016;16:1–18. doi: 10.1007/s13127-016-0265-7. [DOI] [Google Scholar]
- 40.DeSalle R. Can single protein and protein family phylogenies be resolved better? J Phylogenetics Evol Biol. 2015;3:e116. doi: 10.4172/2329-9002.1000e116. [DOI] [Google Scholar]
- 41.Gu X, Zou Y, Su Z, Huang W, Zhou Z, Arendsee A, Zeng Y. An update of DIVERGE software for functional divergence analysis of protein family. Mol Biol Evol. 2013;30:1713–1719. doi: 10.1093/molbev/mst069. [DOI] [PubMed] [Google Scholar]
- 42.Wertheim JO, Murrell R, Smith MD, Kosakovsky Pond SL, Scheffler K. RELAX: Detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 2014;32:820–832. doi: 10.1093/molbev/msu400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Demuynck S, Li KW, Van der Schors R, Dhainaut-Courtois N. Amino acid sequence of the small cadmium-binding protein (MP-II) from Nereis diversicolor (Annelida, Polychaeta) – evidence for a myohemerythrin structure. Eur J Biochem. 1993;217:151–156. doi: 10.1111/j.1432-1033.1993.tb18230.x. [DOI] [PubMed] [Google Scholar]
- 44.Koch J, Lüdemann J, Spies R, Last M, Amemiya CT, Burmester T. Unusual diversity of myoglobin genes in the lungfish. Mol Biol Evol. 2016;33(12):3033–3041. doi: 10.1093/molbev/msw159. [DOI] [PubMed] [Google Scholar]
- 45.Coates CJ, Decker H. Immunological properties of oxygen-transport proteins: hemoglobin, hemocyanin and hemerythrin. Cell Mol Life Sci. 2017;74:293. doi: 10.1007/s00018-016-2326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Ou Q, Han J, Li J, Wu Y, Jiao G, He T. Lower Cambrian polychaete from China sheds light on early annelid evolution. Naturwissenschaften. 2015;102(5):1–7. doi: 10.1007/s00114-015-1285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kocot KM, Cannon JT, Todt C, Citarella MR, Kohn AB, Meyer A, Santos SR, Schander C, Moroz LL, Lieb B, Halanych KM. Phylogenomics reveals deep molluscan relationships. Nature. 2011;477:452–456. doi: 10.1038/nature10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whelan NV, Kocot KM, Moroz LL, Halanych KM. Error, signal, and the placement of Ctenophora sister to all other animals. Proc Natl Acad Sci U S A. 2015;112:5773–5778. doi: 10.1073/pnas.1503453112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown CT, Howe A, Zhang Q, Pyrkosz AB, Brom TH. A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv:1203.4802 [q-bio.GN]. 2012.
- 50.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Gene Ontology Consortium The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(1):D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thiever T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(suppl 2):609–612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 59.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6. Available from http://beast.bio.ed.ac.uk/Tracer. 2014. Accessed 20 July 2016.
- 60.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: Protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kocot KM, Struck TH, Merkel J, Waits DS, Todt C, Brannock PM, Weese DA, Cannon JT, Moroz LL, Lieb B, Halanych KM. Phylogenomics of Lophotrochozoa with consideration of systematic error. Syst Biol. 2016. doi:10.1093/sysbio/syw079. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow chart of bioinformatics pipeline. Rounded purple rectangles represent input/output files, orange ovals represent software or scripts, and the green hexagon represents a step which involving manual evaluation. Nine annelid Hrs sequences previous used as query and two Lingula (Brachipoda) sequences from Genbank (Additional file 3) were also included in the dataset. (DOC 1870 kb)
Hr genes with their respective accession numbers. Novel Hr genes accession numbers for each species. From the manuscript of Costa-Paiva et al. BMC Evolutionary Biology. (DOC 217 kb)
Outgroup and query sequences used to search assembled translated transcriptomes. Query seqeunces used to search transcriptomes. The Hr sequence for H. medicinalis possesses the myoHr five codon insertion between the C and D α-helix, so in this work we considered it a myoHr. Sequences in bold was also included in the dataset previous to the alignment. From the manuscript of Costa-Paiva et al. BMC Evolutionary Biology (DOC 53 kb)
The amino acid alignment used in analyses. The nucleotide alignment used for all subsequent analyses. (TXT 31 kb)
The nucleotide alignment used for analyses. (TXT 89 kb)


