Abstract
Nucleoside reverse transcriptase (RT) inhibitors constitute the backbone of current therapies against human immunodeficiency virus type 1 and type 2 (HIV-1 and HIV-2, respectively). However, mutational pathways leading to the development of nucleoside analogue resistance are different in both types of HIV. In HIV-2, resistance to all approved nucleoside analogues is conferred by the combination of RT substitutions K65R, Q151M and M184V. Nucleotide incorporation kinetic analyses of mutant and wild-type (WT) HIV-2 RTs show that the triple-mutant has decreased catalytic efficiency due to the presence of M184V. Although similar effects were previously reported for equivalent mutations in HIV-1 RT, the HIV-2 enzymes were catalytically less efficient. Interestingly, in highly divergent HIV-1 RTs, K65R confers several-fold increased accuracy of DNA synthesis. We have determined the intrinsic fidelity of DNA synthesis of WT HIV-2 RT and mutants K65R and K65R/Q151M/M184V. Our results show that those changes in HIV-2 RT have a relatively small impact on nucleotide selectivity. Furthermore, we found that there were less than two-fold differences in error rates obtained with forward mutation assays using mutant and WT HIV-2 RTs. A different conformation of the β3-β4 hairpin loop in HIV-1 and HIV-2 RTs could probably explain the differential effects of K65R.
Human immunodeficiency viruses type 1 and type 2 (HIV-1 and HIV-2, respectively) share many traits, including similar genetic organizations, major intracellular replication pathways, mode of transmission, and clinical effects leading to the acquired immunodeficiency syndrome (AIDS)1. However, HIV-2 is less pathogenic, undergoes a longer asymptomatic phase and shows lower rates of transmission2,3. Despite the slower progression to AIDS, many HIV-2-infected patients develop clinical features indistinguishable from the syndrome caused by HIV-1, while having fewer treatment options available. Attempts to correlate mutation frequencies in HIV-1 and HIV-2 with the attenuated progression to AIDS have been inconclusive4,5,6. Several factors including dNTP pools, viral proteins and cell types may have an important effect on the viral mutation rate. Next-generation sequencing analysis of amplicons obtained from infected cells showed that HIV-2 had lower mutation rates than HIV-1, although the observed differences were due to the high frequency of G → A transitions found in HIV-17. The higher susceptibility of this virus to APOBEC3-mediated hypermutation could explain these results8. Nucleotide incorporation assays carried out with heteropolymeric template-primers under steady-state conditions revealed only subtle differences in misinsertion and mispair extension fidelity between HIV-1BH10 and HIV-2ROD reverse transcriptases (RTs)9. However, a proper assessment of the intrinsic fidelity of the HIV-2 RT, including estimates of base substitution and frameshift error rates relative to other RTs is still missing.
HIV-1BH10 and HIV-2ROD RTs share similar DNA polymerase catalytic properties10,11. However, the HIV-2ROD enzyme has reduced processivity, particularly in the presence of low dNTP concentrations12. In addition, Lenzi et al. have recently shown that RTs of several HIV-2 and simian immunodeficiency virus (SIV) strains require higher dNTP concentrations for efficient DNA synthesis13. These differences would be most relevant when considering viral replication kinetics in macrophages and non-dividing cells that harbour very low dNTP concentrations. In myeloid cells, expression of the host factor SAMHD1 (sterile α motif and HD domain-containing protein-1) restricts HIV replication by reducing the intracellular dNTP pools, due to its deoxynucleoside triphosphate triphosphohydrolase (dNTPase) activity14,15,16,17. However, viral proteins such as Vpx (in HIV-2 and in SIVsmm- and SIVrcm-related viruses) and certain variants of Vpr (e.g. those found in SIV strains from African green monkeys) counteract the effects of SAMHD1 by targeting the protein for proteasomal degradation18,19. Interestingly, Vpx-mediated degradation of SAMHD1 decreases HIV susceptibility to zidovudine (3′-azido-3′-deoxythymidine (AZT)) and stavudine (2′,3′-didehydro-2′,3′-dideoxythymidine (d4T)) in macrophages and lymphocytes20. Those findings were consistent with data from a previous report showing that purified SAMHD1 has significantly lower dNTPase activity against AZT-triphosphate than against natural dNTPs21. A link between the properties of RTs and Vpx/Vpr proteins has been suggested from phylogenetic analysis of their coding sequences in HIV and SIV genomes22.
HIV-2 shows natural resistance to a number of antiretroviral drugs designed to suppress HIV-1 propagation. Thus, nonnucleoside RT inhibitors (NNRTIs), the fusion inhibitor enfuvirtide and several protease inhibitors are ineffective against HIV-2, while the clinical efficacy of maraviroc is uncertain23. Unlike HIV-1, HIV-2 does not develop resistance to nucleoside RT inhibitors (NRTIs) through the excision pathway (involving amino acid substitutions M41L, D67N, K70R, L210W, T215F/Y and K219E/Q in the viral RT24), but relies exclusively on nucleotide discrimination12. However, clinical and epidemiological studies have shown that HIV-2 has lower genetic barriers than HIV-1 to the development of multidrug resistance25. NRTIs (usually tenofovir/emtricitabine) constitute the backbone of commonly prescribed therapies against HIV-223,26. RT substitutions K65R and Q151M are frequently identified in virus obtained from treated HIV-2-infected patients27, and the presence of both changes together with M184V confers classwide NRTI resistance28. An estimated prevalence of 9% has been reported for the combination of K65R, Q151M and M184V in a European cohort of HIV-2-infected patients treated with antiretroviral drugs29.
Phenotypic studies using recombinant HIV-2 have shown that Q151M alone confers high-level resistance to AZT (zidovudine), and moderate- to low-level resistance to stavudine, abacavir, didanosine and emtricitabine. However, it has a small impact on the viral susceptibility to lamivudine and tenofovir28,29,30,31. As previously shown in HIV-124, K65R arises in HIV-2-infected patients exposed to tenofovir and other NRTIs27,32, while M184V confers high-level resistance to approved cytidine analogues (i.e. lamivudine and emtricitabine)28. K65R can be selected in vitro after HIV-2 exposure to increasing doses of tenofovir33 and produces a 2- to 7-fold decrease in viral susceptibility to the drug29,33. In addition, K65R confers low-level resistance to didanosine and moderate to high-level resistance to lamivudine and emtricitabine in HIV-2 strains28,30. Lys65, Gln151 and Met184 locate at the dNTP binding site of the RT34. By themselves, amino acid substitutions K65R, Q151M or M184V in HIV-1 RT confer reduced catalytic efficiency of nucleotide analogue incorporation (review24). Consistently with those findings, mutant HIV-1 with RT substitutions K65R or M184V show delayed replication kinetics in comparison with the wild-type (WT) virus35,36. In the case of M184V, the fitness defect is more pronounced at low dNTP concentrations and is expected to be more relevant for viruses replicating in non-dividing cells (e.g. macrophages)37. HIV-1 RTs with Arg65 instead of Lys showed decreased nucleotide incorporation rates (kpol) in pre-steady-state kinetic assays38,39,40. Other studies have shown that K65R produces a >8-fold increase in the intrinsic fidelity of the HIV-1 RT, an effect that has been demonstrated with RTs of highly divergent HIV-1 group M (subtype B) and group O strains40,41. In agreement with enzymatic data, K65R produced a 3.3-fold mutant frequency reduction in single round of replication assays, using an HIV-1NL43 vector containing the lacZ gene42.
In contrast to the well-studied HIV-1 RT, studies on the effects of NRTI resistance-associated mutations on the nucleotide specificity and fidelity of the HIV-2 polymerase are limited to a few single- and double-mutants with substitutions at positions 74, 89, 111 and 215 (i.e. L74V, E89G, V111I, S215Y, L74V/S215Y and E89G/S215Y), whose effects were evaluated for a small subset of misincorporations or mispairs using nucleotide discrimination assays29,43. Unfortunately, those studies could not provide reliable mechanistic interpretations of the data because assays were performed under steady-state conditions. None of the studied residues were part of the complex conferring multi-NRTI resistance, although sometimes V111I appears as an accessory mutation that compensates for the lower fitness of classwide NRTI-resistant HIV-229. In our study, we have examined the effects of K65R, Q151M and M184V on nucleotide incorporation, using pre-steady-state kinetics. We show that WT and mutant HIV-2 RTs have reduced nucleotide binding affinities in comparison with the equivalent HIV-1 enzymes, while M184V was found to be responsible for the reduced catalytic efficiency of the triple mutant K65R/Q151M/M184V. Unexpectedly, and unlike in the case of HIV-1 RTs, K65R had a relatively small impact on misinsertion and mispair extension ratios suggesting only subtle effects on the fidelity of HIV-2 RT. Furthermore, the error rates of mutant RTs K65R and K65R/Q151M/M184V were similar to those obtained with the WT HIV-2 RT, using an M13mp2 lacZα forward mutation assay. Although the RT mutation K65R seems to be selected in HIV-1 and HIV-2 after exposure to tenofovir and other NRTIs, our results suggest that there is limited functional equivalence between Lys65 in both HIV RTs.
Results
Nucleotide incorporation kinetics
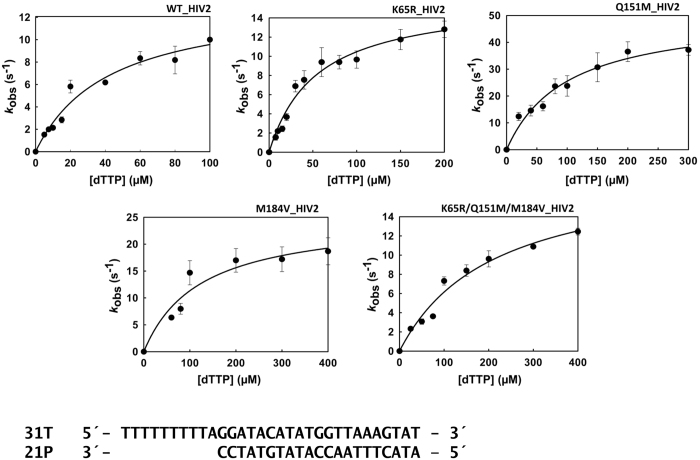
The catalytic efficiency of nucleotide incorporation was determined for the WT HIV-2ROD RT and mutants K65R, Q151M, M184V and K65R/Q151M/M184V under pre-steady-state conditions, using a 31/21mer heteropolymeric template-primer. Catalytic rates and nucleotide affinity constants (kpol and Kd, respectively) were obtained after representing nucleotide incorporation rates (kobs) versus dNTP concentrations and fitting the data to a hyperbola (Fig. 1). In these assays, the WT HIV-2 RT showed 3.5 times higher catalytic efficiency than the triple mutant K65R/Q151M/M184V (Table 1). These differences could be attributed to the higher Kd of the triple mutant. The amino acid substitution M184V alone produced a 3.9-fold reduction of the dNTP binding affinity and was found to be responsible for the lower catalytic efficiency of the K65R/Q151M/M184V HIV-2 RT. In contrast, K65R had minor effects on the catalytic constants determined in these assays, while Q151M produced a modest increase in the catalytic efficiency of the HIV-2 RT, despite reducing the nucleotide binding affinity by two-fold. Interestingly, in comparison with WT HIV-1 RTs of group M-subtype B (BH10 strain)44 or group O (ESP49 strain)40, the HIV-2 enzymes showed 2.6- to 3.9-fold reduced catalytic efficiencies, as a consequence of their lower affinity for incoming dNTPs.
Figure 1. Nucleotide concentration dependence for the incorporation of dTTP into the 31T/21P duplex DNA for wild-type (WT) and mutant HIV-2 RTs.
The continuous line represents the best fit of the data to the Michaelis-Menten equation.
Table 1. Pre-steady-state kinetic constants for the incorporation of dTTP on a heteropolymeric template-primer by WT and mutant HIV-2 RTs.
| Enzyme | kpol (s−1) | Kd (μM) | kpol/Kd (μM−1·s−1) |
|---|---|---|---|
| WT HIV-2 RT | 13.6 ± 1.6 | 42.2 ± 11.1 | 0.32 ± 0.09 |
| K65R HIV-2 RT | 16.0 ± 1.1 | 53.7 ± 9.1 | 0.30 ± 0.06 |
| Q151M HIV-2 RT | 50.8 ± 4.7 | 99.0 ± 21.2 | 0.51 ± 0.12 |
| M184V HIV-2 RT | 27.5 ± 5.9 | 164 ± 77 | 0.168 ± 0.087 |
| K65R/Q151M/M184V HIV-2 RT | 19.1 ± 2.4 | 210 ± 53 | 0.091 ± 0.025 |
| WT BH10 HIV-1 RT | 11.6 ± 0.5 | 13.4 ± 1.9 | 0.86 ± 0.13 |
The template-primer 31T/21P was used as the substrate. Mean values ± standard deviations are given for each enzyme. Assays were performed independently at least three times. Reported values for the WT BH10 HIV-1 RT were taken from Matamoros et al.44.
Pre-steady-state kinetic analysis of fidelity of DNA synthesis of WT and mutant HIV-2 RTs
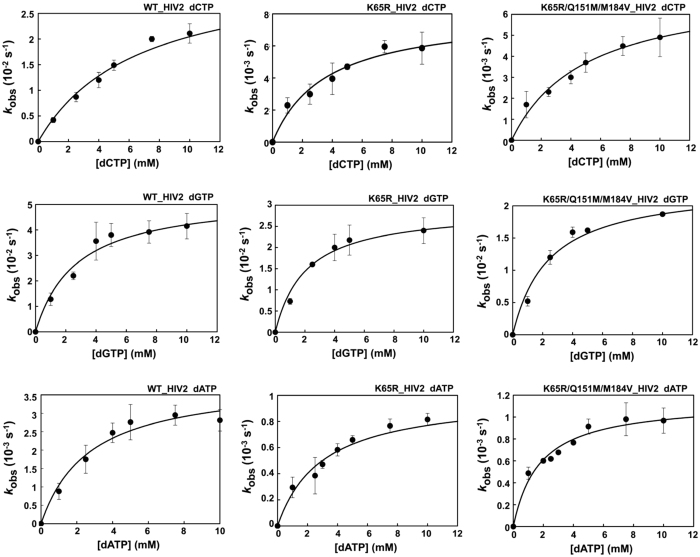
Previous studies with prototypic HIV-1 RTs had shown that K65R produced large increases in the fidelity of DNA synthesis in the absence40,41 and in the presence of the NRTI-associated resistance mutation V75I40. We analysed the nucleotide specificity of HIV-2ROD RT mutants K65R and K65R/Q151M/M184V in misinsertion and mispair extension fidelity assays. The kinetics of misincorporation of dCTP, dGTP and dATP at the 3′ end of the primer into the 31T/21P heteroduplex were determined under single-turnover conditions for mutant and WT RTs. The obtained incorporation rates (kobs) were represented versus dNTP concentrations, and the data fitted to the Michaelis-Menten equation (Fig. 2). The obtained kpol and Kd values, and the catalytic efficiencies (kpol/Kd) of nucleotide misincorporation are given in Table 2. The highest catalytic efficiencies were observed for the incorporation of non-complementary dCTP (1.5 × 10−6–5.2 × 10−6) and dGTP (9.6 × 10−6–2.0 × 10−5), and were about four to five orders of magnitude lower than those obtained for the correct nucleotide. Large reductions in the catalytic efficiencies of incorporation of C, G or A opposite A result from their lower kpol and the large increases in the Kd values obtained in comparison with the corresponding parameters determined for the correct nucleotide (i.e. dTTP). Differences in the misinsertion ratios between the WT HIV-2ROD RT and mutants K65R and K65R/Q151M/M184V were small and non-significant except for the misincorporation of A opposite A which was about 4 times less efficient for the single-mutant K65R than for the WT RT. Nevertheless, dATP incorporation is very inefficient in this sequence context, with kpol/Kd values 6.5 and 44 times lower than those obtained with dCTP or dGTP, respectively, suggesting that this type of errors would be rare during reverse transcription.
Figure 2. Nucleotide concentration dependence on the first-order rates for dCTP, dGTP and dATP incorporation into the DNA/DNA 31/21-mer 31T/21P, obtained with WT and the mutant HIV-2 RTs K65R and K65R/Q151M/M184V.
The first-order rates of the nucleotide incorporation reaction (kobs) were plotted against the corresponding nucleotide concentration. The data were fitted to the Michaelis-Menten equation to obtain Kd and kpol values (best fit represented with a solid line).
Table 2. Pre-steady-state kinetic parameters for misincorporation.
| Enzyme | Nucleotide | kpol (s−1) | Kd (μM) | kpol/Kd (μM−1·s−1) | Misinsertion ratio (fins)a |
|---|---|---|---|---|---|
| WT HIV-2 RT | dCTP | (3.4 ± 0.4) × 10−2 | 6597 ± 1508 | (5.2 ± 1.3) × 10−6 | (1.6 ± 0.6) × 10−5 |
| dGTP | (5.3 ± 0.4) × 10−2 | 2702 ± 654 | (2.0 ± 0.5) × 10−5 | (6.1 ± 2.4) × 10−5 | |
| dATP | (4.0 ± 0.4) × 10−3 | 2715 ± 753 | (1.5 ± 0.4) × 10−6 | (4.6 ± 1.9) × 10−6 | |
| K65R HIV-2 RT | dCTP | (8.0 ± 0.6) × 10−3 | 3562 ± 783 | (2.2 ± 0.5) × 10−6 | (7.5 ± 2.2) × 10−6 (2.1) |
| dGTP | (2.8 ± 0.2) × 10−2 | 1928 ± 542 | (1.5 ± 0.4) × 10−5 | (4.8 ± 1.7) × 10−5 (1.3) | |
| dATP | (1.0 ± 0.1) × 10−3 | 2980 ± 699 | (3.4 ± 0.8) × 10−7 | (1.1 ± 0.3) × 10−6 (4.1) | |
| K65R/Q151M/M184V HIV-2 RT | dCTP | (7.3 ± 0.6) × 10−3 | 4985 ± 927 | (1.5 ± 0.3) × 10−6 | (1.6 ± 0.5) × 10−5 (1.0) |
| dGTP | (2.3 ± 0.2) × 10−2 | 2400 ± 511 | (9.6 ± 2.1) × 10−6 | (1.1 ± 0.4) × 10−4 (0.6) | |
| dATP | (1.1 ± 0.1) × 10−3 | 1754 ± 277 | (6.5 ± 1.1) × 10−7 | (7.1 ± 2.3) × 10−6 (0.6) |
The template-primer 31T/21P was used as the substrate. Data shown are the mean values ± standard deviation. Each of the assays was performed independently at least three times.
afins = [kpol(incorrect)/Kd(incorrect)]/[kpol(correct)/Kd(correct)], where incorrect nucleotides were dCTP, dGTP or dATP, and the correct nucleotide was dTTP. Numbers between parentheses represent the relative increase of fidelity, as determined for each incorrect nucleotide as the ratio: fins (WT_HIV-2 RT)/fins (mutant RT).
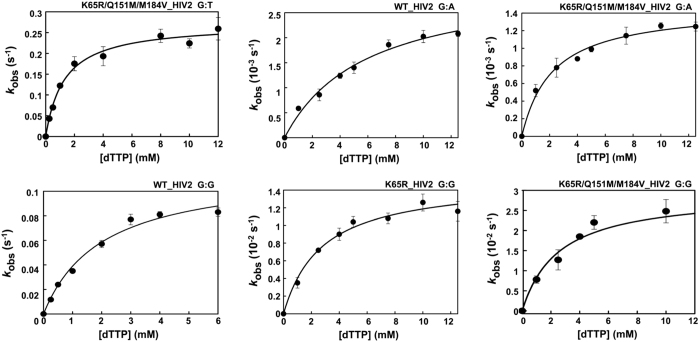
Previous studies with WT and mutant HIV-1 RTs have shown that mispair extension fidelity assays are more reliable for detecting differences in the accuracy of those enzymes40,44. We determined kinetic parameters for the incorporation of dTTP using template-primers 31T/21PT, 31T/21PG, and 31T/21PA. These complexes were identical to 31T/21P but contained mismatched 3′ ends (i.e. G:T, G:G and G:A in duplexes 31T/21PT, 31T/21PG, and 31T/21PA, respectively). The kinetic constants obtained for mispair extension reactions catalysed by WT and mutant HIV-2 RTs are shown in Table 3. As expected, G:T mismatches were extended more efficiently than G:G or G:A mispairs. However, the kpol of the nucleotide incorporation reaction on G:T mismatches could not be reliably estimated for the WT and the mutant K65R RT, because both enzymes showed extremely low dTTP binding affinity in this sequence context. The same difficulties were also found for the K65R HIV-2 RT in G:A mispair extension assays. Plots of nucleotide incorporation rates versus dTTP concentrations are given in Fig. 3 for those mispairs providing valid estimates of kpol and Kd. Nucleotide incorporation rates (kpol) for the extension of G:T, G:G and G:A mispairs were very small except for reactions catalysed by the WT RT using G:T mismatched template-primers. However, where kinetic parameters were reliably determined, the differences between WT HIV-2 RT and the classwide NRTI-resistant K65R/Q151M/M184V RT were not significant. The K65R mutant enzyme showed increased fidelity in comparison with the WT RT in extension reactions with template-primers having a G:G mismatch at their 3′ ends. However, in this case the catalytic efficiency of nucleotide incorporation was very low (5.4 × 10−6 μM−1 s−1).
Table 3. Pre-steady-state kinetic parameters for mispair extension.
| Enzyme | Base pair at the 3′ enda | kpol (s−1) | Kd (μM) | kpol/Kd (μM−1•s−1) | Mismatch extensión ratio (fext)b |
|---|---|---|---|---|---|
| WT HIV-2 RT | G:T | 2.41c | >8000 | <2.4 × 10−4 | ndd |
| G:G | 0.12 ± 0.01 | 2023 ± 381 | (5.8 ± 1.2) × 10−5 | (1.8 ± 0.6) × 10−4 | |
| G:A | (3.1 ± 0.2) × 10−3 | 5885 ± 990 | (5.3 ± 1.0) × 10−7 | (1.6 ± 0.6) × 10−6 | |
| K65R HIV-2 RT | G:T | 0.27c | >8000 | <2.7 × 10−5 | nd |
| G:G | (1.5 ± 0.1) × 10−2 | 2758 ± 492 | (5.4 ± 1.0) × 10−6 | (1.8 ± 0.5) × 10−5 (9.8) | |
| G:A | <4.1 × 10−4 | >8000 | nd | nd | |
| K65R/Q151M/M184V HIV-2 RT | G:T | 0.27 ± 0.01 | 1299 ± 175 | (2.1 ± 0.3) × 10−4 | (2.3 ± 0.7) × 10−3 (0.3) |
| G:G | (3.0 ± 0.3) × 10−2 | 2542 ± 773 | (1.2 ± 0.4) × 10−5 | (1.3 ± 0.5) × 10−4 (1.4) | |
| G:A | (1.5 ± 0.1) × 10−3 | 2213 ± 302 | (6.8 ± 1.0) × 10−7 | (7.4 ± 2.3) × 10−6 (0.3) |
The template-primer 31T/21P was used as the substrate. Data shown are the mean values ± standard deviation. Each of the assays was performed independently at least three times.
aThe first base corresponds to the template and the second to the primer.
bfext = [kpol(mismatched)/Kd(mismatched)]/[kpol(matched)/Kd(matched)]. Numbers between parentheses represent the relative increase of fidelity, as determined for each mispair as the ratio: fext (WT_HIV-2 RT)/fext (mutant RT).
cIndicated values are observed catalytic rates (kobs) at 10 mM concentration of dTTP.
dnd, not determined.
Figure 3. Nucleotide concentration dependence on the first-order rates for dTTP incorporation into DNA/DNA 31/21-mers with G:T, G:A and G:G mismatches at their 3′ end.
The first-order rates of the incorporation reaction (kobs), obtained with WT and mutant HIV-2 RTs (K65R and K65R/Q151M/M184V), were plotted against the corresponding nucleotide concentration. The data were fitted to the Michaelis-Menten equation to obtain Kd and kpol values (best fit represented with a solid line).
M13mp2 lacZα forward mutation assays
Nucleotide incorporation assays detected only small differences of fidelity between WT and mutant HIV-2 RTs. However, they provided only limited information on nucleotide specificity since the analyses were restricted to a reduced number of substrates and sequence contexts. M13-based forward mutation assays using lacZα as a reporter gene provide a better fidelity assessment, based on a relatively large number of mutational sites, although silent mutations could not be detected using this method45. These assays were used to determine mutation frequencies and error rates for WT HIV-1BH10 and HIV-2ROD RTs, as well as for mutant HIV-2 RTs K65R and K65R/Q151M/M184V. A double-stranded DNA derived from phage M13mp2 and lacking the lacZα gene sequence in one of the two DNA chains (gapped DNA) was used as substrate. Mutations introduced when the RT copied the gapped region of the lacZ gene changed the colour phenotype of M13mp2 plaques, detected when phages were grown on an appropriate indicator strain, in the presence of isopropyl β-D-1-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal). Mutant frequencies were calculated as the ratio of mutant (pale blue or colourless) to total plaques. In these assays we found that the WT RT of the HIV-1BH10 strain had lower accuracy than the WT HIV-2ROD RT, although differences were relatively small with mutation frequencies of 1.99% and 1.24%, respectively (Table 4). Mutations K65R and K65R/Q151M/M184V had almost no effect on the intrinsic accuracy of the HIV-2ROD RT, with mutant frequencies being only slightly lower in comparison with that of the WT enzyme.
Table 4. Accuracy of WT and mutant RTs in M13mp2 lacZα forward mutation assays.
| RT | Total plaques | Mutant plaques | Mutant frequencya |
|---|---|---|---|
| WT HIV-2 | 9660 | 120 | 0.01242 |
| K65R HIV-2 | 4750 | 56 | 0.01179 (1.1) |
| K65R/Q151M/M184V HIV-2 | 5142 | 53 | 0.01031 (1.2) |
| BH10 HIV-1 | 2915 | 58 | 0.01990 (0.6) |
aBackground frequencies in these assays were estimated to be less than one in 20,000 plaques40. Numbers between parentheses indicate the fold-increase in fidelity relative to the WT HIV-2 RT.
The mutational specificity of the studied RTs was determined after sequencing the lacZα mutants generated in the forward mutation assays. Analysis of the obtained mutational spectra revealed small differences between WT and mutant HIV-2 RTs (Supplementary Figs S1–S4). Nevertheless, a few general trends were common to those three enzymes. For example, more than one third of all errors introduced by HIV-2 RT variants (mutant and WT enzymes) were frameshifts, and in most cases consisted of deletions of one nucleotide. These deletions occurred predominantly at heteropolymeric sequences. Base substitutions were frequently observed in all spectra, with transversions appearing at least three times more frequently than transitions in the analysed lacZα sequence. A predominance of G-to-T substitutions was detected in all mutational spectra. The analysis of error rates (Table 5) demonstrated that all HIV-2 RTs introduced frameshifts with a similar frequency, while differences in fidelity were observed when looking at base substitution error rates. Thus, the frequency of this type of mutations was reduced by less than two-fold in the case of classwide-resistant K65R/Q151M/M184V RT relative to the WT enzyme, while K65R alone produced a mere 1.5-fold increase in nucleotide substitution fidelity. The comparison of WT HIV-2ROD and HIV-1BH10 RTs demonstrated that both enzymes introduced similar types of errors (i.e. frequent one-nucleotide deletions, and more transversions than transitions). However, frameshifts were more abundant in the mutational spectrum of the HIV-2ROD RT (Supplementary Fig. S4). Compared with the WT HIV-1BH10 RT, the HIV-2 enzymes were less prone to introduce single nucleotide substitutions, but showed higher overall frameshift error rates due to their strong tendency to introduce deletions at heteropolymeric sequences.
Table 5. Summary of error rates for WT and mutant RTs, for various classes of mutations, based on M13mp2 DNA-dependent DNA synthesis reactions.
| Mutation type | WT HIV-2 (ROD) | K65R HIV-2 RT | K65R/Q151M/M184V HIV-2 RT | WT HIV-1 (BH10) | ||||
|---|---|---|---|---|---|---|---|---|
| Number of errors | Error rate | Number of errors | Error rate | Number of errors | Error rate | Number of errors | Error rate | |
| All classes | 137 | 1/10027 | 57 | 1/11850 | 54 | 1/13541 | 63 | 1/6580 |
| Base substitutions | 88 | 1/8233 | 28 | 1/12723 | 24 | 1/16069 | 54 | 1/4049 |
| Transitions | 21 (24%) | 1 (4%) | 5 (21%) | 14 (26%) | ||||
| Transversions | 67 (76%) | 27 (96%) | 19 (79%) | 40 (74%) | ||||
| Frameshifts | 49 | 1/17506 | 29 | 1/14545 | 30 | 1/15220 | 9 | 1/28761 |
| Insertions | 0 (0%) | 0 (0%) | 1 (3%) | 0 (0%) | ||||
| Deletions | 49 (100%) | 29 (100%) | 29 (97%) | 9 (100%) | ||||
| At runsa | 5 | 1/91577 | 2 | 1/112575 | 1 | 1/243731 | 2 | 1/69086 |
| At non-runs | 44 | 1/9089 | 27 | 1/7283 | 29 | 1/7341 | 7 | 1/17240 |
aA run is here defined as a row of three or more identical nucleotides.
Discussion
Classwide NRTI resistance in HIV-2 is conferred by mutations K65R, Q151M and M184V28. In agreement with previous reports13,46, we found that HIV-2ROD RT exhibits lower catalytic efficiency than HIV-1 RTs. Previously reported dNTP incorporation rates (kpol) for HIV-1 group M subtype B RTs (e.g. HIV-1BH10 and HIV-1NL43 RTs) were in the range of 11.6–17.4 s−1, with Kd values between 7.1 and 17 μM44,47,48, as determined in assays carried out with the same template-primer used in our study (i.e. 31T/21P). Those kinetic parameters were similar to those previously obtained for WT HIV-1ESP49 group O RT (kpol = 13.2 s−1 and Kd = 17.3 μM)49. Under the same conditions, the HIV-2 RT shows similar incorporation rates but reduced nucleotide affinity, thereby suggesting a possible mechanistic explanation for the lower replication capacity of HIV-2 in different cell types50,51.
Mutations selected under drug pressure frequently result in a loss of viral fitness. Our analysis on the effects of K65R, Q151M and M184V on nucleotide incorporation kinetics revealed that M184V was responsible for the reduced catalytic efficiency of classwide-resistant HIV-2 RT. M184V alone decreases the catalytic efficiency by reducing the nucleotide binding affinity of the RT. Although K65R, Q151M and M184V might be selected under NRTI pressure in HIV-1 as well as in HIV-2, the classwide-resistant triple mutant is characteristic of the HIV-2 RT since Q151M is the preferred mutation selected during treatment with AZT23,24. Interestingly, the effects of those mutations on nucleotide incorporation by HIV-1 RTs had been previously assessed with the 31T/21P duplex, using pre-steady-state kinetics38,39. The results of those studies were consistent with the kinetic data reported here and showed that M184V produced a significant reduction of the catalytic efficiency of the HIV-1 polymerase due to a decreased dNTP binding affinity. In contrast, the substitution of Met for Gln151 (Q151M) enhanced the catalytic efficiency of the HIV-1 RT due to an increased kpol38, an effect that we also observed with this amino acid substitution in the HIV-2 RT. On the other hand, the results with K65R were slightly different in the HIV-1BH10 RT as compared with the HIV-2ROD enzyme. Deval et al.38 showed that K65R reduced the catalytic efficiency of HIV-1BH10 RT by decreasing its nucleotide incorporation rates (kpol) for different dNTPs, while in our assays with HIV-2 RT, the substitution had almost no effect on the catalytic parameters of the enzyme. As in HIV-1BH10 RT, mutant HIV-1 group O RT bearing the K65R change showed reduced kpol, although in this case the enzyme had increased affinity for the incoming dNTP40.
In studies carried out with HIV-1 group O RT, K65R was found to have a large impact on nucleotide selectivity. Introduction of K65R in the sequence of HIV-1ESP49 RT decreased misinsertion and mispair extension ratios for all tested nucleotides and mismatched template-primers40. Furthermore, studies with the group O enzyme also showed the increased nucleotide selectivity of the double-mutant K65R/V75I in comparison with the RT bearing the single amino acid substitution V75I. In general, the results of our misinsertion and mispair extension fidelity assays using the 31/21-mer template-primer did not reveal significant differences between HIV-2 RTs bearing mutation K65R and the WT enzyme, except for the extension of G:G mispairs which was less efficient in the case of the mutant enzyme. Unfortunately, data were inconclusive for most mispairs, including the ones that were extended more efficiently, due to the low efficiency of dNTP incorporation observed with the used template-primers.
To overcome those limitations, we obtained more reliable estimates of the intrinsic fidelity of HIV-2 RTs by using forward mutation assays. It is well-known that lentiviral RTs (e.g. those of HIV-1, SIV or feline immunodeficiency virus) are less accurate than oncoretroviral RTs, such as those of murine leukaemia virus (MLV) or avian myeloblastosis virus52,53. Our study shows that the WT HIV-2ROD RT is only 1.6 times more faithful than the reference HIV-1BH10 RT. These differences are similar to those reported for other primate lentiviral RTs when compared with HIV-1 group M subtype B polymerases, such as the RTs of HIV-1 strains BH10 or HXB2, using the M13-based forward mutation assay. Thus, the HIV-1BH10 RT showed about two-fold decreased fidelity in comparison with a prototypic WT HIV-1 group O RT54,55, while SIVmne and SIVagm RTs were only 1.3 and 1.8 times more accurate, respectively, than HIV-1 group M subtype B RTs56,57. Despite the relatively small differences in mutation rates, HIV-1 group M subtype B RTs emerge as the less faithful polymerases among those of primate lentiviruses. The higher intrinsic fidelity of HIV-2 RT in comparison with HIV-1BH10 RT results from its lower nucleotide substitution error rate, a trend that was also observed with the SIVagm RT56. In contrast, our analysis shows that WT HIV-2ROD RT has a relatively high frameshift error rate (5.7 × 10−5), ranking among the highest reported for RTs of primate lentiviruses.
There were relatively small differences in fidelity between the classwide NRTI-resistant K65R/Q151M/M184V RT and the WT HIV-2ROD enzyme, despite the presence of K65R in the mutational complex. These effects were unexpected considering that K65R alone produced large increases in fidelity in forward mutation assays carried out with HIV-1 RTs from phylogenetically distant strains such as HXB2 (group M subtype B)41 or ESP49 (group O)40. Furthermore, estimates of HIV-1 mutant frequencies obtained in cell culture after one round of replication were consistent with the enzymatic data and showed a 3.3-fold reduction for virus containing the K65R mutation in their RT42. In contrast, our results show that K65R in HIV-2ROD RT had almost no effect on the overall fidelity of the enzyme, although it produced a modest decrease of its base substitution error rate. The effects of K65R on the evolutionary rate of HIV-2 have not been assessed. However, a recent work has shown the quick reversion of the mutation when present in SIVmac239 infecting pigtailed macaques58. Interestingly, WT and mutant SIVmac239 showed similar mutation rates in cell-based assays although K65R-containing viruses showed slightly reduced variability. The differences in mutation rates were estimated to be less than 1.3-fold, but considered statistically significant58. Interestingly, mutant MLV carrying the equivalent substitution in its RT (i.e. K103R) was found to be less infectious than the WT. However, no differences were found in assays measuring the frequency of lacZ inactivation after one round of replication in cells infected with WT and mutant viruses59, thereby suggesting that the mutation had no effect on the intrinsic fidelity of the MLV RT.
Although several studies carried out with HIV-1 RT variants have shown that K65R and other mutations conferring resistance to nucleoside analogues (e.g. M184V) increase their DNA polymerase fidelity, there are examples of NRTI resistance mutations having little effect on the accuracy of HIV-1 RT (e.g. L74V, Q151M)52,53. Our results show that in HIV-2, classwide NRTI resistance may not be associated with significant changes in the intrinsic fidelity of its RT, although a lower fitness is expected for drug-resistant viruses as a result of the lower catalytic efficiency of their RT. Therefore, the relationship between drug resistance and increased fidelity cannot be established, particularly for the major mutations found in HIV-2 strains resistant to NRTIs.
The increased intrinsic fidelity conferred by K65R appears to be a specific characteristic of HIV-1 RT and not shared by RTs of the HIV-2/SIV lineage. Although a structural justification of these differences is very difficult due to the lack of structural information for HIV-2 RT, a number of amino acid sequence differences found in the β3-β4 hairpin loops (residues 63–75) of RTs of HIV-1 and HIV-2 could have a significant impact on its conformation and mobility, thereby affecting neighbouring contacts between the side-chains of Lys or Arg65. In support of this notion, crystal structures of ternary complexes composed of mutant K65R HIV-1BH10 RT, double-stranded DNA and incoming nucleotides (i.e. dATP or tenofovir-diphosphate) have shown stacking interactions between guanidinium groups of Arg65 and Arg72 that restrict the conformational adaptability of both residues60. The amino acid sequences of the β3-β4 hairpin loops of HIV-1 group M subtype B and group O RTs are identical (IKKKDSTKWRKLV), but different from those found in HIV-2ROD and SIVmac239 RTs (IKKKDKNKWRMLI). The distribution of charged residues along the hairpin loop sequence is rather different in both pairs of RTs and it is tempting to speculate that substitutions of basic residues in these structures could have an impact on fidelity. Nevertheless, how interactions between hairpin loop residues may influence the intrinsic fidelity of the enzyme is a question open for future research.
Experimental Procedures
Expression and purification of RTs
WT heterodimeric (p66/p51) HIV-1BH10 RT was obtained as previously described61,62. Histidine-tagged WT and mutant HIV-2ROD RTs were obtained as p68/p54 heterodimers with constructs derived from plasmid pT5m that were used to transform Escherichia coli strain BL21(DE3)(pLysS). The pT5m vector containing the p68-coding region of the WT HIV-2ROD RT and the HIV-2D194 protease has been previously described63 and was kindly provided by Amnon Hizi (Tel Aviv University, Israel). A modified version of the method described by Boyer et al.12 was used to purify HIV-2 RTs. Briefly, bacteria (in one-liter Luria-Broth cultures, containing 2 mM glucose) were grown at 30 °C and RT expression was induced for 3 h with 1 mM IPTG. Cells were collected by centrifugation and the obtained pellets were re-suspended in 6 ml of 50 mM sodium phosphate pH 8.0 buffer, containing 50 mM NaCl, 0.75 mg/ml hen egg-white lysozyme and 1.5 mM phenylmethylsulfonyl fluoride (PMSF), and kept on ice for 25 min. After addition of 0.43 ml of 4 M NaCl, the solution was subjected to brief periods of sonication, and then centrifuged at 23,000 rpm for 60 min at 4 °C using a Beckman JA 25.50 rotor. The obtained supernatants were diluted with one volume of 66 mM sodium phosphate pH 7.0 buffer, containing 0.3 M NaCl, and applied on a 3 ml Ni2+-nitriloacetic acid-agarose column (Invitrogen, Carlsbad, CA, USA), previously equilibrated with 50 mM sodium phosphate pH 7.0 buffer containing 0.3 M NaCl. The column was then washed with 50 mM sodium phosphate pH 6.0 buffer containing 0.3 M NaCl, 20 mM imidazole and 10% glycerol. The RT was eluted with a 20 mM–0.5 M imidazole gradient in the same buffer. The fractions containing the heterodimer were analysed by SDS-PAGE, pooled and dialyzed against 50 mM sodium phosphate pH 6.8 buffer, and then applied on a 5 ml P11 phosphocellulose column (Whatman, Maidstone, Kent, England), previously equilibrated with the same buffer. After washing the column with 50 mM sodium phosphate pH 6.8 buffer containing 0.3 M NaCl, the RT was eluted with a 30-ml gradient from 0.3 to 1 M NaCl. Fractions eluted from the salt gradient were analysed by SDS-PAGE, and those containing the heterodimer were pooled, dialyzed against 50 mM Tris-HCl pH 7.0, containing 25 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT) and 10% glycerol, concentrated in Centriprep-30 and Centricon-30 to <0.5 ml, and stored at −20 °C. Enzymes were quantified by active site titration64,65 before biochemical studies.
Mutagenesis
Site-directed mutagenesis was carried out by following the standard QuikChangeTM (Stratagene) protocol. The pT5m plasmid encoding the large subunit of the HIV-2ROD RT was used as template63. Complementary mutagenic primers were used to amplify the entire pT5m(RODRT) plasmid in a thermocycling reaction carried out with high-fidelity Pfu DNA polymerase. The mutagenic primers used were: 5′-CATTTGCAATCAAGAGAAAGGACAAAAAC-3′ and 5′-GTTTTTGTCCTTTCTCTTGATTGCAAAT-3′ for K65R, 5′-TAAAGTCTTGCCAATGGGATGGAAGGGA-3′ and 5′-TCCCTTCCATCCCATTGGCAAGACTTTA-3′ for Q151M, and 5′-ATTATCATTCAGTACGTGGATGATATCT-3′ and 5′-AGATATCATCCACGTACTGAATGATAAT-3′ for M184V. Mutations found in the triple mutant K65R/Q151M/M184V were orderly added starting from mutant K65R, and then introducing mutation Q151M, and finally M184V. All constructs were verified by DNA sequencing.
Pre-steady-state kinetic assays
Transient kinetics experiments were used to determine the catalytic constants kpol and Kd for the incorporation of correct and incorrect nucleotides. Assays were carried out as previously described44,66,67, in 50 mM Tris/HCl buffer (pH 8.0), containing 50 mM KCl and 12 mM MgCl2 (for correct dNTPs) or 15 mM (for incorrect dNTPs), and varying concentrations of dNTP, using the heteropolymeric template-primer 31T/21P at 100 nM (Fig. 1). An excess of Mg2+ concentration over dNTP was used in all assays. The experiments were performed manually or with a rapid quench flow instrument (model QFM-400, Bio-Logic Science Instruments, Claix, France), upgraded with a mixer cross and a special mixer (Bio-Logic) that facilitated the determination of catalytic rate constants greater than 20 s−1. Oligonucleotides 21P and 31T (sequences provided in Fig. 1) were used as primer and template, respectively. Three additional primers (21PT, 21PG and 21PA) were used in mispair extension fidelity assays. These oligonucleotides differed from 21P in having T, G or A (instead of C) at their 3′-end. Before annealing to the template, primers were labelled at their 5′ terminus with [γ-32P]ATP (Perkin Elmer, Boston, MA, USA) and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA).
In misincorporation fidelity assays, dTTP incorporation reactions were performed under single-turnover conditions with 75–125 nM (active sites) RT and 100 nM template-primer. At appropriate time-points, the reactions were quenched with EDTA (0.3 M final concentration). The products were analysed by denaturing PAGE and quantified by phosphorimaging. Reactions involving the incorporation of incorrect nucleotides or mispair extension kinetics (i.e. dCTP, dGTP or dATP incorporation on 31T/21P, or the extension of G:T, G:G and G:A mispairs) were carried out with 120 nM RT and 100 nM template-primer. These conditions were chosen to eliminate the influence of the enzyme turnover rate (kss), which interferes with the determination of low incorporation rates. Due to their lower catalytic rates, incorrect nucleotide incorporation assays and G:G and G:A mispair extension assays were carried out manually, as well as the longer incubation times needed for the determination of kinetic parameters in G:T mispair extensions. The kinetic constants Kd and kpol were determined as described67, from curve-fitting using SigmaPlot, after calculating catalytic rates (kobs) at different dNTP concentrations using the burst equation: [P] = A × [1 − exp(−kobs × t)] + kss × t, where [P] is the product concentration, A the amplitude of the burst, and kobs the apparent kinetic constant of formation of the phosphodiester bond.
M13mp2 lacZα forward mutation assays
The fidelity of DNA-dependent DNA synthesis was determined using the forward mutation assay45, under the previously described conditions40,68,69. Gap filling synthesis reactions were carried out at 37 °C during 60 min, in 25 μl of 25 mM Tris-HCl (pH 8.0) buffer containing 100 mM KCl, 2 mM DTT, 4 mM MgCl2, 250 μM of each dNTP and 100 nM RT. Each reaction contained 100–150 ng of gapped DNA. The gapped DNA was preincubated at 60 °C during 5 min to reduce secondary structure in the single-stranded DNA and avoid undesirable hybridizations. Nine or ten gap filling reactions were performed for each tested RT and the obtained products were analysed on a 0.8% agarose gel to check if the reactions had been completed. Reaction products were electroporated into E. coli MC1061 cells, and the transformed cells were plated onto M9 medium-containing plates with X-Gal and IPTG with CSH50 lawn cells45. Mutant frequencies were calculated as the ratio of mutant (light blue or colourless) plaques to the total number of plaques screened. Mutant phenotypes were confirmed by nucleotide sequencing of the phage replicative-form DNA using primer 5′-GCTTGCTGCAACTCTCTCAG-3′69. It should be noted that silent mutations and mutations that do not affect the β-galactosidase activity may not be detected in these assays. Error rates were calculated as described by Bebenek and Kunkel45. It has been estimated that this limitation results in a 2- to 3-fold underestimation of the actual mutation rates70.
Additional Information
How to cite this article: Álvarez, M. et al. Fidelity of classwide-resistant HIV-2 reverse transcriptase and differential contribution of K65R to the accuracy of HIV-1 and HIV-2 reverse transcriptases. Sci. Rep. 7, 44834; doi: 10.1038/srep44834 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported in part by a grant of the Spanish Ministry of Economy and Competitiveness (BIO2013-48788-C2-1-R). A.S.-M. is a recipient of a predoctoral fellowship of the Spanish Ministry of Education, Culture and Sport (FPU2013-00693). An institutional grant of Fundación Ramón Areces (Madrid, Spain) to the Centro de Biología Molecular “Severo Ochoa” is also acknowledged. The funders had no role in study design, data collection and interpretation, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no competing financial interests.
Author Contributions This study was conceived and designed by L.M.-A. M.A., A.S.-M. and G.G.-M. performed experiments and analysed data. L.M.-A. also analysed data and wrote the manuscript, which was critically revised by M.A. and A.S.-M. All authors read and approved the manuscript.
References
- Nyamweya S. et al. Comparing HIV-1 and HIV-2 infection: Lessons for viral immunopathogenesis. Rev. Med. Virol. 23, 221–240, doi: 10.1002/rmv.1739 (2013). [DOI] [PubMed] [Google Scholar]
- Rowland-Jones S. L. & Whittle H. C. Out of Africa: what can we learn from HIV-2 about protective immunity to HIV-1? Nat. Immunol. 8, 329–331, doi: 10.1038/ni0407-329 (2007). [DOI] [PubMed] [Google Scholar]
- Campbell-Yesufu O. T. & Gandhi R. T. Update on human immunodeficiency virus (HIV)-2 infection. Clin. Infect. Dis. 52, 780–787, doi: 10.1093/cid/ciq248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P. et al. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput. Biol. 3, e29, doi: 10.1371/journal.pcbi.0030029 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil A. et al. Long-term intrapatient viral evolution during HIV-2 infection. J. Infect. Dis. 195, 726–733, doi: 10.1086/511308 (2007). [DOI] [PubMed] [Google Scholar]
- Skar H. et al. HIV-2 genetic evolution in patients with advanced disease is faster than that in matched HIV-1 patients. J. Virol. 84, 7412–7415, doi: 10.1128/JVI.02548-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson J. M., Landman S. R., Reilly C. S. & Mansky L. M. HIV-1 and HIV-2 exhibit similar mutation frequencies and spectra in the absence of G-to-A hypermutation. Retrovirology 12, 60, doi: 10.1186/s12977-015-0180-6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro A. C. et al. Functional analysis of Vif protein shows less restriction of human immunodeficiency virus type 2 by APOBEC3G. J. Virol. 79, 823–833, doi: 10.1128/JVI.79.2.823-833 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhanashvili M. & Hizi A. The fidelity of the reverse transcriptases of human immunodeficiency viruses and murine leukemia virus, exhibited by the mispair extension frequencies, is sequence dependent and enzyme related. FEBS Lett. 319, 201–205, doi: 10.1016/0014-5793(93)80067-5 (1993). [DOI] [PubMed] [Google Scholar]
- Hizi A., Tal R., Shaharabany M. & Loya S. Catalytic properties of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2. J. Biol. Chem. 266, 6230–6239 (1991). [PubMed] [Google Scholar]
- Hizi A. & Herschhorn A. Retroviral reverse transcriptases (other than those of HIV-1 and murine leukemia virus): a comparison of their molecular and biochemical properties. Virus Res. 134, 203–220, doi: 10.1016/j.virusres.2007.12.008 (2008). [DOI] [PubMed] [Google Scholar]
- Boyer P. L., Clark P. K. & Hughes S. H. HIV-1 and HIV-2 reverse transcriptases: different mechanisms of resistance to nucleoside reverse transcriptase inhibitors. J. Virol. 86, 5885–5894, doi: 10.1128/JVI.06597-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi G. M., Domaoal R. A., Kim D.-H., Schinazi R. F. & Kim B. Kinetic variations between reverse transcriptases of viral protein X coding and noncoding lentiviruses. Retrovirology 11, 111, doi: 10.1186/s12977-014-0111-y (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N. et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657, doi: 10.1038/nature10117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K. et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661, doi: 10.1038/nature10195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H. et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228, doi: 10.1038/ni.2236 (2012). Erratum in: Nat. Immunol. 14, 877 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauveau L. et al. HIV-2 infects resting CD4+ T cells but not monocyte-derived dendritic cells. Retrovirology 12, 2, doi: 10.1186/s12977-014-0131-7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E. S. et al. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204, doi: 10.1016/j.chom.2012.01.004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N. et al. Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11, 205–217, doi: 10.1016/j.chom.2012.01.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballana E. et al. SAMHD1 specifically affects the antiviral potency of thymidine analog HIV reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 58, 4804–4813, doi: 10.1128/AAC.03145-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amie S. M. et al. Anti-HIV host factor SAMHD1 regulates viral sensitivity to nucleoside reverse transcriptase inhibitors via modulation of cellular deoxyribonucleoside triphosphate (dNTP) levels. J. Biol. Chem. 288, 20683–20691, doi: 10.1074/jbc.M113.472159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y., Doi N., Miyazaki Y., Adachi A. & Nomaguchi M. Phylogenetic insights into the functional relationship between primate lentiviral reverse transcriptase and accessory proteins Vpx/Vpr. Front. Microbiol. 7, 1655, doi: 10.3389/fmicb.2016.01655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez-Arias L. & Álvarez M. Antiretroviral therapy and drug resistance in human immunodeficiency virus type 2 infection. Antiviral Res. 102, 70–86, doi: 10.1016/j.antiviral.2013.12.001 (2014). [DOI] [PubMed] [Google Scholar]
- Menéndez-Arias L. Molecular basis of human immunodeficiency virus type 1 drug resistance: Overview and recent developments. Antiviral Res. 98, 93–120, doi: 10.1016/j.antiviral.2013.01.007 (2013). [DOI] [PubMed] [Google Scholar]
- Gottlieb G. S. et al. Emergence of multiclass drug-resistance in HIV-2 in antiretroviral-treated individuals in Senegal: implications for HIV-2 treatment in resource-limited West Africa. Clin. Infect. Dis. 48, 476–483 (2009). Erratum in: Clin. Infect. Dis. 48, 848, doi: 10.1086/596504 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthard H. F. et al. Antiretroviral Drugs for Treatment and Prevention of HIV Infection in Adults: 2016 Recommendations of the International Antiviral Society-USA Panel. JAMA 316 , 191–210, doi: 10.1001/jama.2016.8900 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntemgwa M. L., Toni d. T., Brenner B. G., Camacho R. J. & Wainberg M. A. Antiretroviral drug resistance in human immunodeficiency virus type 2. Antimicrob. Agents Chemother. 53, 3611–3619, doi: 10.1128/AAC.00154-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. A., Anderson D. J., Pyrak C. L., Preston B. D. & Gottlieb G. S. Antiretroviral drug resistance in HIV-2: three amino acid changes are sufficient for classwide nucleoside analogue resistance. J. Infect. Dis. 199, 1323–1326, doi: 10.1086/597802 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuzing I. P. et al. Mutation V111I in HIV-2 reverse transcriptase increases the fitness of the nucleoside analogue-resistant K65R and Q151M viruses. J. Virol. 89, 833–843, doi: 10.1128/JVI.02259-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damond F. et al. In vitro phenotypic susceptibility to nucleoside reverse transcriptase inhibitors of HIV-2 isolates with the Q151M mutation in the reverse transcriptase gene. Antivir. Ther. 10, 861–865 (2005). [PubMed] [Google Scholar]
- Bennett M. et al. Characterization of the Q151M and V111I mutations of HIV-2 reverse transcriptase. Antivir. Ther. 12, S120 (2007). [Google Scholar]
- Peterson K., Jallow S., Rowland-Jones S. L. & de Silva T. I. Antiretroviral therapy for HIV-2 infection: recommendations for management in low-resource settings. AIDS Res. Treat. 2011, 463704, doi: 10.1155/2011/463704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta K., Miller M. D. & White K. L. HIV-2 antiviral potency and selection of drug resistance mutations by the integrase strand transfer inhibitor elvitegravir and NRTIs emtricitabine and tenofovir in vitro. J. Acquir. Immune Defic. Syndr. 62, 367–374, doi: 10.1097/QAI.0b013e31827b55f1 (2013). [DOI] [PubMed] [Google Scholar]
- Huang H., Chopra R., Verdine G. L. & Harrison S. C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: Implications for drug resistance. Science 282, 1669–1675, doi: 10.1126/science.282.5394.1669 (1998). [DOI] [PubMed] [Google Scholar]
- Back N. K. T. et al. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15, 4040–4049 (1996). [PMC free article] [PubMed] [Google Scholar]
- Cong M., Heneine W. & García-Lerma J. G. The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J. Virol. 81, 3037–3041, doi: 10.1128/JVI.02712-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keulen W., Back N. K., van Wijk A., Boucher C. A. & Berkhout B. Initial appearance of the 184Ile variant in lamivudine-treated patients is caused by the mutational bias of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 71, 3346–3350 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval J. et al. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J. Biol. Chem. 279, 509–516, doi: 10.1074/jbc.M308806200 (2004). [DOI] [PubMed] [Google Scholar]
- Deval J. et al. Mechanistic insights into the suppression of drug resistance by human immunodeficiency virus type 1 reverse transcriptase using α-boranophosphate nucleoside analogs. J. Biol. Chem. 280, 3838–3846, doi: 10.1074/jbc.M411559200 (2005). [DOI] [PubMed] [Google Scholar]
- Barrioluengo V., Álvarez M., Barbieri D. & Menéndez-Arias L. Thermostable HIV-1 group O reverse transcriptase variants with the same fidelity as murine leukaemia virus reverse transcriptase. Biochem. J. 436, 599–607, doi: 10.1042/BJ20101852 (2011). [DOI] [PubMed] [Google Scholar]
- Shah F. S. et al. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 275, 27037–27044, doi: 10.1074/jbc.M002881200 (2000). [DOI] [PubMed] [Google Scholar]
- Mansky L. M., Le Rouzic E., Benichou S. & Gajary L. C. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J. Virol. 77, 2071–2080, doi: (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube R., Avidan O. & Hizi A. The fidelity of misinsertion and mispair extension throughout DNA synthesis exhibited by mutants of the reverse transcriptase of human immunodeficiency virus type 2 resistant to nucleoside analogs. Eur. J. Biochem. 250, 106–114, doi: 10.1111/j.1432-1033.1997.00106.x (1997). [DOI] [PubMed] [Google Scholar]
- Matamoros T., Kim B. & Menéndez-Arias L. Mechanistic insights into the role of Val75 of HIV-1 reverse transcriptase in misinsertion and mispair extension fidelity of DNA synthesis. J. Mol. Biol. 375, 1234–1248, doi: 10.1016/j.jmb.2007.11.021 (2008). [DOI] [PubMed] [Google Scholar]
- Bebenek K. & Kunkel T. A. Analyzing fidelity of DNA polymerases. Methods Enzymol. 262, 217–232, doi: 10.1016/0076-6879(95)62020-6 (1995). [DOI] [PubMed] [Google Scholar]
- Lenzi G. M., Domaoal R. A., Kim D.-H., Schinazi R. F. & Kim B. Mechanistic and kinetic differences between reverse transcriptases of Vpx coding and non-coding lentiviruses. J. Biol. Chem. 290, 30078–30086, doi: 10.1074/jbc.M115.691576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval J. et al. The molecular mechanism of multidrug resistance by the Q151M human immunodeficiency virus type 1 reverse transcriptase and its suppression using α-boranophosphate nucleotide analogues. J. Biol. Chem. 277, 42097–42104, doi: 10.1074/jbc.M206725200 (2002). [DOI] [PubMed] [Google Scholar]
- Kisic M. et al. Mechanistic basis of zidovudine hypersusceptibility and lamivudine resistance conferred by the deletion of codon 69 in the HIV-1 reverse transcriptase coding region. J. Mol. Biol. 382, 327–341, doi: 10.1016/j.jmb.2008.07.030 (2008). [DOI] [PubMed] [Google Scholar]
- Matamoros T., Nevot M., Martínez M. A. & Menéndez-Arias L. Thymidine analogue resistance suppression by V75I of HIV-1 reverse transcriptase: Effects of substituting Valine 75 on stavudine excision and discrimination. J. Biol. Chem. 284, 32792–32802, doi: 10.1074/jbc.M109.038885 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaak H., van der Ende M. E., Boers P. H. M., Schuitemaker H. & Osterhaus A. D. M. E. In vitro replication capacity of HIV-2 variants from long-term aviremic individuals. Virology. 353, 144–154, doi: 10.1016/j.virol.2006.05.029 (2006). [DOI] [PubMed] [Google Scholar]
- Mörner A. et al. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73, 2343–2349 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez-Arias L. Mutation rates and intrinsic fidelity of retroviral reverse transcriptases. Viruses 1, 1137–1165, doi: 10.3390/v1031137 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez-Arias L. HIV reverse transcriptase fidelity, clade diversity, and acquisition of drug resistance In Human Immunodeficiency Virus Reverse Transcriptase: A Bench-to-Bedside Success (ed. Le Grice S. F. J. & Götte M.) 225–252 (Springer Science+Business Media, 2013). [Google Scholar]
- Álvarez M., Matamoros T. & Menéndez-Arias L. Increased thermostability and fidelity of DNA synthesis of wild-type and mutant HIV-1 group O reverse transcriptases. J. Mol. Biol. 392, 872–884, doi: 10.1016/j.jmb.2009.07.081 (2009). [DOI] [PubMed] [Google Scholar]
- Álvarez M., Barrioluengo V., Afonso-Lehmann R. N. & Menéndez-Arias L. Altered error specificity of RNase H-deficient HIV-1 reverse transcriptases during DNA-dependent DNA synthesis. Nucleic Acids Res. 41, 4601–4612, doi: 10.1093/nar/gkt109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuke A. W., Ahmad-Omar O., Hoefer K., Hunsmann G. & Jentsch K. D. Mutations in the SIV env and the M13 lacZα gene generated in vitro by reverse transcriptases and DNA polymerases. Arch. Virol. 142, 1139–1154 (1997). [DOI] [PubMed] [Google Scholar]
- Diamond T. L. et al. Mechanistic understanding of an altered fidelity simian immunodeficiency virus reverse transcriptase mutation, V148I, identified in a pig-tailed macaque. J. Biol. Chem. 278, 29913–29924, doi: 10.1074/jbc.M211754200 (2003). [DOI] [PubMed] [Google Scholar]
- Lloyd S. B. et al. High fidelity simian immunodeficiency virus reverse transcriptase mutants have impaired replication in vitro and in vivo. Virology 492, 1–10, doi: 10.1016/j.virol.2016.02.008 (2016). [DOI] [PubMed] [Google Scholar]
- Halvas E. K., Svarovskaia E. S. & Pathak V. K. Role of murine leukemia virus reverse transcriptase deoxyribonucleoside triphosphate-binding site in retroviral replication and in vivo fidelity. J. Virol. 74, 10349–10358, doi: 10.1128/JVI.74.22.10349-10358.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K. et al. Structural basis for the role of the K65R mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance. J. Biol. Chem. 284, 35092–35100, doi: 10.1074/jbc.M109.022525 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros T. et al. Suppression of multidrug-resistant HIV-1 reverse transcriptase primer unblocking activity by α-phosphate-modified thymidine analogues. J. Mol. Biol. 349, 451–463, doi: 10.1016/j.jmb.2005.04.004 (2005). [DOI] [PubMed] [Google Scholar]
- Boretto J. et al. An integrated system to study multiply substituted human immunodeficiency virus type 1 reverse transcriptase. Anal. Biochem. 292, 139–147, doi: 10.1006/abio.2001.5045 (2001). [DOI] [PubMed] [Google Scholar]
- Sevilya Z., Loya S., Hughes S. H. & Hizi A. The ribonuclease H activity of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2 is affected by the thumb subdomain of the small protein subunits. J. Mol. Biol. 311, 957–971, doi: 10.1006/jmbi.2001.4904 (2001). [DOI] [PubMed] [Google Scholar]
- Kati W. M., Johnson K. A., Jerva L. F. & Anderson K. S. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267, 25988–25997 (1992). [PubMed] [Google Scholar]
- Menéndez-Arias L. Studies on the effects of truncating α-helix E’ of p66 human immunodeficiency virus type 1 reverse transcriptase on template-primer binding and fidelity of DNA synthesis. Biochemistry 37, 16636–16644, doi: 10.1021/bi981830g (1998). [DOI] [PubMed] [Google Scholar]
- Mendieta J., Cases-González C. E., Matamoros T., Ramírez G. & Menéndez-Arias L. A Mg2+-induced conformational switch rendering a competent DNA polymerase catalytic complex. Proteins 71, 565–574, doi: 10.1002/prot.21711 (2008). [DOI] [PubMed] [Google Scholar]
- Matamoros T., Barrioluengo V., Abia D. & Menéndez-Arias L. Major groove binding track residues of the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase enhance cDNA synthesis at high temperatures. Biochemistry 52, 9318–9328, doi: 10.1021/bi401390x (2013). [DOI] [PubMed] [Google Scholar]
- Barrioluengo V., Wang Y., Le Grice S. F. J. & Menéndez-Arias L. Intrinsic DNA synthesis fidelity of xenotropic murine leukemia virus-related virus reverse transcriptase. FEBS J. 279, 1433–1444, doi: 10.1111/j.1742-4658.2012.08532.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez M. & Menéndez-Arias L. Temperature effects on the fidelity of a thermostable HIV-1 reverse transcriptase. FEBS J. 281, 342–351, doi: 10.1111/febs.12605 (2014). [DOI] [PubMed] [Google Scholar]
- Abram M. E., Ferris A. L., Shao W., Alvord W. G. & Hughes S. H. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J. Virol. 84, 9864–9878, doi: 10.1128/JVI.00915-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.