Abstract
We demonstrate the usefulness of utilizing a segmentation step for improving the performance of sparsity based image reconstruction algorithms. In specific, we will focus on retinal optical coherence tomography (OCT) reconstruction and propose a novel segmentation based reconstruction framework with sparse representation, termed segmentation based sparse reconstruction (SSR). The SSR method uses automatically segmented retinal layer information to construct layer-specific structural dictionaries. In addition, the SSR method efficiently exploits patch similarities within each segmented layer to enhance the reconstruction performance. Our experimental results on clinical-grade retinal OCT images demonstrate the effectiveness and efficiency of the proposed SSR method for both denoising and interpolation of OCT images.
Index Terms: Optical coherence tomography, retina, ophthalmic imaging, image reconstruction, sparse representation, denoising, interpolation, layer segmentation
I. Introduction
Optical coherence tomography (OCT) is a non-invasive imaging modality which is employed in diverse medical applications [1, 2], especially for diagnostic ophthalmology. Automated remote analysis of ophthalmologic OCT images is becoming more prevalent for the diagnosis and study of ocular diseases [3]. However, sample-based speckle and detector noise corrupts OCT images. On another front, to accelerate the acquisition process, relatively low spatial sampling rate is often used in capturing clinical OCT images [3–6]. Both the heavy noise and low spatial sampling rate negatively affect automated and even manual OCT image analysis performance, necessitating utilization of effective denoising and interpolation techniques, respectively.
Denoising and interpolation are two well-known reconstruction problems in the image processing [7]. In the past decade, various models have been proposed to reconstruct high quality OCT images for various applications [8–17]. Classical reconstruction approaches often construct a smoothness priori based model (e.g., anisotropic filtering, Tikhonov filtering, and total variation [9]), and reconstruct the image in spatial domain. Some recent approaches transform the input degraded image into another domain (e.g., using the wavelet transformation [18], dual tree complex wavelet transformation [19], and curvelets transformation [20]). While the transform based methods (e.g. wavelet) usually achieve a better reconstruction performance than the spatial domain methods, they are built on a fixed mathematical model and may have limited adaptability [21] for representing structures inocular OCT volumes.
Inspired by the sparse coding mechanism of human vision system [22], the sparse representation is demonstrated to be a powerful tool for many image processing applications [4, 23–30]. The sparse representation decomposes an input image into a linear combination of an over-complete dictionary of basis functions. Basis functions can be chosen from a set of training images similar to the input image [31] and thus can be more adaptive for the representation of specific features. Several recent works have also applied the sparse representation to OCT image reconstruction problems [4, 6,12, 14–16, 24, 32–34]. While different retinal layers have varied pathologic structures [35–37] and even speckle patterns, most of the sparse reconstruction methods only train one general dictionary to represent complex structures and textures in the ocular OCT images.
The above traditional sparse model directly decomposes each local image patch. Both heavy noise in the test patch and high correlations in the dictionary atoms [23] may mislead the sparse decomposition process, thus negatively affecting the final reconstruction. More recently, a nonlocal sparse reconstruction model [38–40] has been introduced, which exploits the nonlocal patch self-similarities to improve the reconstruction. Specifically, for each processed patch, the nonlocal based sparse model searches similar patches from the whole image (or a large searching window) and then jointly decomposes the similar patches on the dictionary [40] to find a more accurate sparse solution. Nonetheless, the nonlocal based sparse representation requires searching the whole image to find the similar patches, which creates very high computational burdens and also will be interfered by the heavy noise in the OCT image. Therefore, how to construct effective spare dictionaries for representing complex structures and efficiently exploiting patch self-similarities for accurate sparse decomposition are the two key issues in the sparse reconstruction model.
In this paper, we utilize the segmentation algorithm for OCT image reconstruction. We particularly focus on the retina, a layered structure where each layer has its own specific features. Noting that since structural elements of each layer, resulting in speckle, have different size, shape, and distribution, the signal and noise model of each layer is expected to be different [3, 35, 36, 41]. Moreover, pathologic structures with distinct features appear in specific layers. For example, drusenoid structures appears immediately below or above the retinal pigment epithelium (RPE) layer [42], while hyper-reflective foci associated with age-related macular degeneration are not expected to appear in the nerve fiber layer (NFL) [43]. Utilizing the structural information in the layers, we propose a segmentation based sparse reconstruction (SSR) model to develop a fast and accurate reconstruction algorithm. Our general approach first utilizes a graph based algorithm [35] to automatically segment the retinal OCT images into multiple layers. Then, for each layer, SSR constructs a dedicated structural dictionary to better represent the anatomic and pathologic structures within this layer. Finally, instead of searching the whole image, SSR efficiently searches for the similar patches within each layer and exploits the patches’ similarities within each layer to improve the sparse decomposition.
Note that, the strategy of segmentation-based denoising has been previously applied to several other image denoising problems [44–46]. In contrast, this paper proposes a segmentation based sparse representation (SSR) model for retinal OCT image reconstruction, which utilizes the segmented layer information to enhance the effectiveness and efficiency of the sparse reconstruction model. Specifically, the main contributions of our paper are detailed as follows.
The SSR method introduces a layer segmentation based structural dictionary construction strategy, which effectively preserves the anatomic and pathologic structures in the OCT image.
The SSR method proposes a layer segmentation based sparse reconstruction strategy, which utilizes the segmented layer information to significantly accelerate the similar patch searching process and exploits the correlations among similar patches to enhance the reconstruction performance.
The remainder of this paper is organized as follows. Section II briefly reviews the sparse reconstruction model and nonlocal means reconstruction model. We introduce the proposed SSR method for the denoising and interpolation of OCT images in Section III. Experimental results on clinical OCT data are presented in Section IV. Section V concludes this paper and suggests for future works.
II. Background: Sparse Reconstruction Model and Nonlocal Means Based Reconstruction Model
A. Sparse Reconstruction Model
Given a degraded image, most sparse reconstruction models first divide the input image into ϒ overlapping patches Xi ∈ ℝn×m, i = 1, 2,…, ϒ. The vector form for each patch Xi is denoted as xi ∈ ℝq×1 (q = n × m), obtained by lexicographic ordering. For a denoising problem, the sparse reconstruction model assumes that the clean retinal OCT signal can be well represented by a weighted linear combination of a few atoms selected from the dictionary D ∈ ℝq×z, q < z), whereas the noise cannot be decomposed on the dictionary. Therefore, the sparse reconstruction model for the denoising problem can be formulated as follows [47]:
| (1) |
where αi ∈ ℝz×1 represents the sparse coefficients vector for the xi and ||αi||0 is the ℓ0 -norm, counting the number of non-zero coefficients in αi. To solve (1), there are two main considerations: 1) dictionary D construction, and 2) sparse coefficients vector αi estimation. To address the first problem, popular algorithms [31, 48] train one dictionary D from a number of relevant sampled patches H = {x1, …, xZ}, where Z is the number of training patches. Concretely, the dictionary D can be trained by the following optimization problem:
| (2) |
where A = {αi, …, αZ} is the sparse coefficient matrix for the H. This equation can be solved by the K-SVD algorithm [31]. To address the second problem, which is known to be a nondeterministic polynomial-time hard (NP-hard) [49], the orthogonal matching pursuit (OMP) [50] algorithms can be utilized to obtain an approximate solution for the sparse coefficient vector α̂i. Then, we can use Dα̂i to reconstruct the related patch and all the reconstructed patches are returned to their original positions to create the final denoised image.
For the image interpolation problem, we first denote an original high resolution image as YH ∈ ℝN×M, the decimation operator as S, and the corresponding low resolution image as YL = SYH ∈ ℝ(N/S)×(M/S). Given the observation degraded image YL, the objective of the image interpolation is to obtain the ŶH such that ŶH ≈ YH. To solve the interpolation problem, recent sparse reconstruction models [51] attempt to infer the relationship of the sparse coefficients and dictionaries between the high resolution space χH and low resolution space χL. In [51], Yang et. al. jointly trained both the low resolution dictionary DL and the high resolution dictionary DH, which ensures that the sparse coefficient in the space χL is the same as the sparse coefficient in the space χH. Inspired by the work of Yang et. al., we utilized a semi-coupled learning algorithm to construct the matched dictionaries DL and DH, and then trained a mapping function M to link the sparse coefficients and [4]. After obtaining the relationship between the spaces χL and χH, the sparse reconstruction model can first find the sparse coefficient of each observation patch , and then restore the latent image patches as well as the corresponding high resolution image ŶH by utilizing the high resolution dictionary DH and mapping function M.
B. Nonlocal Means Reconstruction Model
For the image denoising and interpolation problems, another very effective reconstruction model is the nonlocal means, which exploits the self-similarities inherent to images [52]. Specifically, for each patch xi extracted from the degraded image, the nonlocal means method first searches the W patches with the highest similarity to xi from the whole image (or a large searching window). The similarity can be measured by the squared ℓ2 -norm of the intensity differences between two patches:
| (3) |
where Λ is a set containing the indexes of all patches in the whole image. Then, the patches that are found to be similar are processed by a weighted average filtering or patch similarity penalty to achieve denoising [52] or interpolation [53], respectively.
In the sparse reconstruction model, the sparse coefficient estimations are affected by the noise in observation image, thus leading to suboptimal reconstruction. To suppress noise interference, recent works including [23, 24, 40] incorporate the nonlocal means into the sparse reconstruction model. Specifically, the nonlocal sparse model first conducts the similar patch search [using (3)] in the whole image and then jointly exploits correlations among similar patches by decomposing them on the same atoms of the dictionary to improve the sparse coefficient solution. The performance improvement is due to, in the sparse solution process, jointly considering the decisions of multiple similar patches is usually more robust to external disturbances [54], similar to the principles of majority voting. However, the nonlocal sparse model requires searching for similar patches across the whole image, which is computational costly. In addition, the heavy noise in the OCT image might still interfere with the similar patch searching process.
III. Proposed SSR Method For OCT Reconstruction
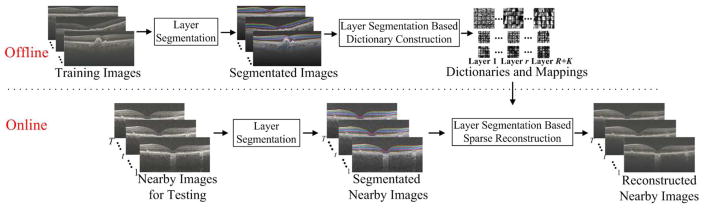
We propose the SSR method, which utilizes the layer specific structural information to enhance the effectiveness and cost efficiency of our previous sparse reconstruction techniques. The SSR method is composed of three main parts: a) layer segmentation; b) layer segmentation based dictionary construction; and c) layer segmentation based sparse reconstruction. The outline of the proposed SSR method is illustrated in Fig. 1.
Fig. 1.
Outline of the proposed SSR algorithm.
A. Layer Segmentation
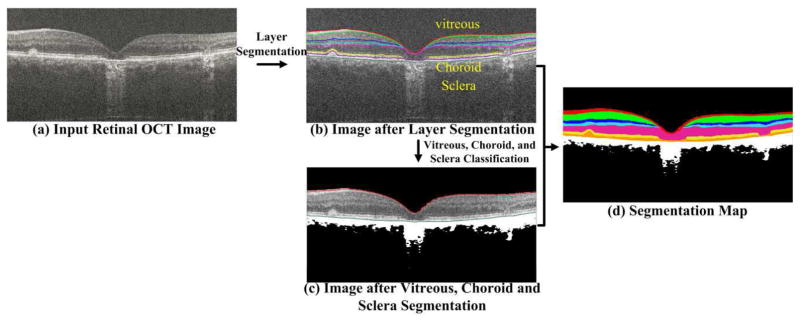
For all the testing and training retinal images, we utilize the popular graph theory and dynamic programming (GTDP) method [35] to automatically segment these images into R layers, as illustrated in Fig. 2(b). The region between two color lines in Fig. 2(b) represents one layer. Note that the intensity of the vitreous and choroid/sclera regions (above and below the segmented layers, respectively) are not uniformly distributed (due to pathology or limited penetration of the 800 nm OCT beam in the choroid layer). Therefore, we further utilize a k-means algorithm to classify the vitreous and choroid/sclera regions into K parts (e.g., see the white and black regions in Fig. 2(c)). To exploit the spatial information for the classification, we extract a patch (of size n×m) for each pixel in the vitreous and choroid/sclera regions, and use its vectorization form for the k-means clustering. We then fuse the results from both the layer segmentation and classification to create a segmentation map, which includes R+K layers (see the Fig. 2(d), each color represents one layer). The whole segmentation process is illustrated in Fig. 2.
Fig. 2.
Outline for the OCT segmentation. In (b), the retinal OCT image is segmented into seven layers using eight color lines, and the region between two color lines represents one layer. Three distinct regions, above and below the retina include vitreous, visible choroid, and sclera. In (c), the vitreous and choroid/sclera regions are further classified into two clusters (one is the black area and one is the white area). In (d), different color regions represent varied segmented layers or classified regions. The dimly visible regions of choroid can be regarded as the background area (delineated in black) with less structural information. The visible regions of choroid can be considered as the foreground area with comparatively rich structural information, delineated in white. Therefore, we set the clustering number K of the X-means algorithm to 2, which further clusters the vitreous, choroid, and sclera regions into two groups, i.e. the black and white areas in (c) and (d).
B. Layer Segmentation Based Dictionary Construction
In the retinal OCT image, different layers contain various types of anatomic and pathologic structures (e.g., vessels, drusen, edema, and fovea), and different thicknesses and speckle structures [3, 35, 36, 41]. Therefore, to well represent complex structures in varied layers, we utilize the segmented layer information to train multiple structural dictionaries (each corresponding to one layer), rather than one general dictionary [40,47].
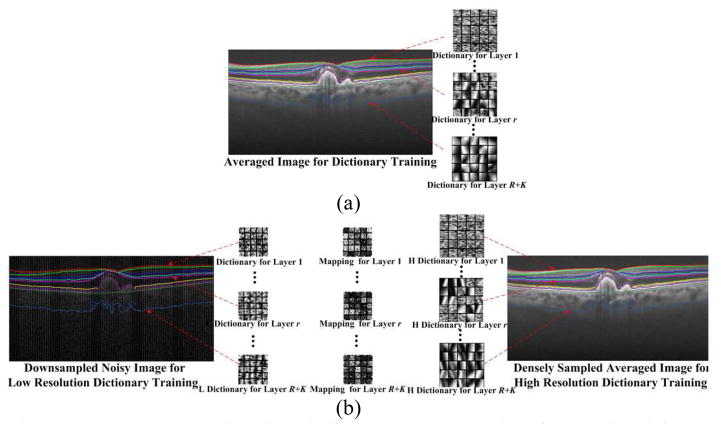
Specifically, for denoising, we first employ segmentation maps of training images to extract patches for each specific layer and construct the related R+K training sets . As in [24], the training images can be considered noiseless, which are obtained by registration and averaging of a number of repeated B-scans acquired from spatially very close positions. Then, we train one structural dictionary Dr (of size q×s) on one training set Hr [see Fig. 3(a)], by modifying (2) as the following optimization problem,
Fig. 3.
Layer segmentation based dictionary construction for (a) denoising problem; (b) interpolation problem.
| (4) |
where Ar is the sparse coefficient matrix for the training set Hr. The optimization problem in (4) is solved by separately performing the K-SVD algorithm [31] on each set Hr. Note that, to construct the structural dictionary for the layer in the black regions (e.g., vitreous and sclera), we randomly select a small number of training patches from that layer as the dictionary atoms. This is because these regions have less structural information and are easy to be represented.
For the image interpolation problem, following [4], we use a set of high SNR and high resolution images, each acquired by averaging a set of repeated densely sampled B-scans, as our high-resolution framing image set . We subsample one frame from each set of the repeated B-Scans, to attain the corresponding low-resolution training image set . Utilizing the segmentation maps of the and , we can extract spatially matched low resolution and high resolution patches for each layer as the matching training sets and . Then, we separately adopt the semi-coupled dictionary learning algorithm [4] on each training set pair and , and obtain R + K matched dictionary pairs and [see Fig. 3(b)]. In addition, as described in Section II. A, we further train R + K mapping functions [see Fig. 3(b)] to relate the sparse coefficients and for each layer,
| (5) |
Following [4], we train these mapping functions using the low resolution sparse matrices and high resolution sparse matrices created from the dictionary learning stage,
| (6) |
where β is the regularization parameter for balancing the objective terms. This optimization problem has a closed form solution [55],
| (7) |
where I is an identity matrix.
Note that, since each layer based structural dictionary is designed for one specific layer, it has a significantly smaller number of atoms compared to the universal dictionary in [31, 47], and thus can achieve a more efficient representation. In addition, the dictionary construction step is an off-line process and in practice does not add to the computational cost of denoising or interpolation.
C. Layer Segmentation Based Sparse Reconstruction
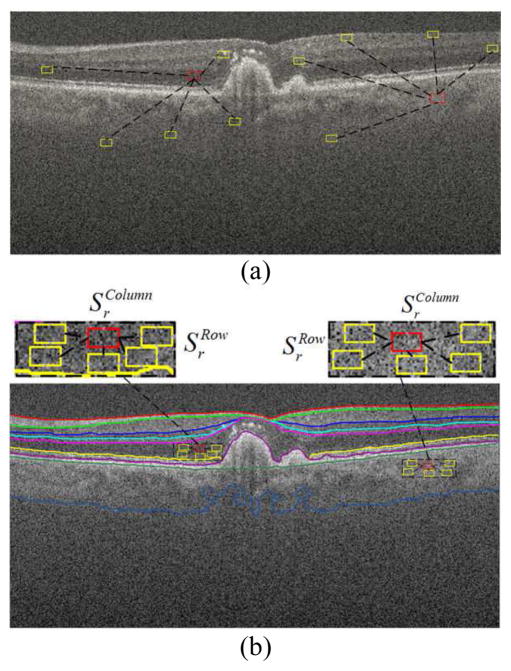
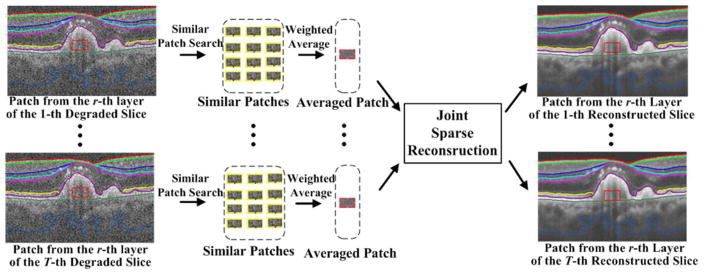
As noted, the anatomic and pathologic structures, intensities, and speckle patterns within each layer are expected to have strong similarities. Therefore, instead of searching the whole image [52], we propose to seek the similar patches in a searching window within each segmented layer, which can greatly reduce the search space [see Fig. 4(b)]. Specifically, for each patch in the r-th layer of the test image, we first define a search window (of size ) within the r-th layer, and then find J patches with the highest similarity to using the similarity measure in (3). Note that if the rectangular search window goes beyond the boundaries of r-th layer, only the window part within the layer is considered as part of the search region. After finding these similar patches, instead of directly conducting the sparse decomposition (as in [23, 24, 40]), we apply a weighted average on these similar patches to obtain an average patch ,
Fig. 4.
Similar patch searching by (a) nonlocal searching; (b) layer segmentation based searching.
| (8) |
where is the weight for the patch , computed as
| (9) |
where W is a predetermined scalar. This weighted averaging scheme is an effective way to reduce noise, which affects direct sparse decomposition of the similar patches. Next, we use the average patches for the sparse reconstruction. Since the 3-D OCT image also has strong correlations among nearby slices [4, 32], we simultaneously process the averaged patches from the same position of nearby slices (called as the nearby patches , where T is number of nearby patches) with a joint sparse decomposition technique. This technique decomposes the nearby patches on the same dictionary atom with different coefficient values, which can enhance the decomposition efficiency. The joint sparse reconstruction for the denoising and interpolation are separately described, as follows (see the Fig. 5).
Fig. 5.
Illustration of nearby patch estimation process in the proposed layer segmentation based sparse reconstruction.
1) Joint Sparse Reconstruction for Denoising
In denoising, simultaneous decomposition of the nearby averaged patches in the r-th layer of nearby images with the joint sparse technique amounts to the following problem,
| (10) |
where F is the maximum number of nonzero coefficients in . This optimization problem is efficiently solved by a simultaneous orthogonal matching pursuit (SOMP) algorithm [56]. Then, we estimate the reconstructed nearby patches as
| (11) |
Note that, for the patches in the layer of the black region (e.g., vitreous and sclera), rather using the SOMP, we use the Euclidean distance to search the most representative atom for each patch. Finally, as in [32], a weighted average operation is conducted on the nearby patches and then these recovered nearby patches are returned to the original positions to reconstruct the denoised nearby images.
2) Joint Sparse Reconstruction for Interpolation
For the interpolation problem, given the averaged patches from nearby low resolution images, we obtain the low resolution sparse matrix by utilizing the SOMP to solve (10) with the dictionary . Then, we estimate the corresponding high resolution sparse matrix as
| (12) |
and the nearby high resolution patch set as
| (13) |
As in [32], we also conduct a weighted average operation on the nearby patches, and return the set of nearby patches to the original positions to reconstruct the high resolution nearby images.
IV. Experimental Results
A. Data Sets
Our experiments used a dataset of retinal OCT volumes from 41 different human subjects with and without nonneovascular age-related macular degeneration (AMD). The images in this dataset were imaged by a Bioptigen SDOCT system (Durham, NC, USA) with an axial resolution of ~4.5 μm per pixel in tissue. This dataset was originally introduced in our previous works [4, 24], and can be freely downloaded online1. This dataset is attained with adherence to the tenets of the Declaration of Helsinki and is a subset of the image captured in the A2A SD-OCT Study, which was registered at ClinicalTrials.gov (Identifier: NCT00734487) and approved by the institutional review boards of the 4 A2A SD-OCT clinics (Devers Eye Institute, Duke Eye Center, Emory Eye Center, and National Eye Institute) [57].
For the denoising problem, images from 28 different human subjects are utilized. Two sets of scans were acquired from each subject. The first scan was focused at the fovea with 40 repeatedly sampled B-scans, each with 900 A-scans. The second scan was a 3D volume, with 900 A-scans and 100 B-scans including the fovea. We registered the first set of repeatedly sampled scans using the StackReg image registration plug-in in ImageJ [58] and then averaged them to obtain an averaged image of the fovea. The averaged image can be regarded as the “noiseless” reference image to train the dictionaries or compute the quantitative metrics. We selected datasets of 18 subjects to test the denoising performance of the proposed SSR method, while the datasets of the other 10 subjects were utilized for training the dictionaries and setting the parameters. Note that, the datasets of subjects used in the dictionary training stage were different from those used in the testing stage and the segmentation labels in the training stage have been manually corrected.
The dataset for interpolation problem included the images used for the denoising experiment. We used the averaged image created from the first scan as the high resolution image and subsampled the central foveal B-scan within the second scan as the corresponding low resolution image. Similar to the denoising problem, the high resolution and low resolution image pairs of 10 different subjects are used for training the dictionaries and mappings, while the remaining image pairs from 18 subjects are used for the testing. In addition, densely sampled and real subsampled datasets from another 13 human subjects are also used for the testing. For each subject, the real subsampled dataset has 450 A-scans per B-scan and 100 B-scans per volume, containing the fovea area.
B. Compared Methods
For the OCT image denoising problem, the proposed SSR method is compared with other five well known denoising approaches: BRFOE [59], K-SVD [47], PGPD [60], BM3D [39], and MSBTD [24]. The BRFOE is a modification of the field-of-experts model, which constructs filters by modeling the noisy image statistics. The K-SVD is a popular sparsity based denoising method, which trains one universal dictionary on the input noisy image. The BM3D is a widely popular benchmark denoising method, which exploits patch self-similarities of the input image by nonlocal searching and 3-D collaborative filtering. The MSBTD is a nonlocal based structural sparse denoising method, which utilizes nonlocal searching to exploit patch self-similarities and uses structural clustering to construct multiple structural dictionaries.
For the OCT image interpolation problem, we compared the proposed SSR method with five well-known interpolation approaches: Bicubic, Tikhonov [61], BM3D [39]+Bicubic, ScSR [51], and SBSDI [4]. The BM3D+Bicubic method is a combination of the BM3D denoising approach and the Bicubic interpolation approach. The ScSR is a popular sparsity based interpolation method, which utilizes the joint dictionary learning strategy to train a pair of low resolution and high resolution dictionaries. The SBSDI uses structural clustering to train multiple low resolution and high resolution dictionary pairs and employs a joint sparse operation to exploit the correlations among nearby slices for interpolation.
C. Algorithm Parameters
The parameters of the proposed SSR algorithm were empirically selected based on our experiments on the training data, and kept unchanged for all the test images. As in [35], the number of segmented layers R was set to seven. Since the vitreous, choroid, and sclera regions can be generally classified into background areas with less structural information and foreground areas with comparatively rich structural information, the cluster number K was selected to two. We chose the number of nearby slices T to be 5, based on the azimuthal resolution of the OCT volume, as slices from farther distances might have large differences, decreasing the effectiveness of the joint sparse reconstruction. The parameter W in (9) was set to 80. Using larger W values increases the averaging property in (8) and thus will create a stronger smoothing effect. The size of the search window was set to 6×40. Increasing the size of the search window might further enhance the reconstruction performance, but create more computational cost. In each search window, the target number of similar patches J was set to 60. Further increasing in the number of similar patches number might result in over-smoothing. The sparsity level F for both the dictionary construction and the sparse reconstruction stages was set to 3. The above parameters were similarly set for both denoising and interpolation problems. For the denoising problem, the number of dictionary atoms is separately set to 30 and 80 for the layer of black region (e.g., vitreous and sclera) and other layers, respectively. For the interpolation problem, the number of dictionary atoms is set to 80 for all the layers. Increasing the number of dictionary atoms will improve the performances, but results in higher computational burden. Since most of the structures in the OCT image lay in the horizontal direction, the patch size n×m was set to 5×10 for the denoising problem. For the interpolation problem, the low resolution image patch size was set to 4×4, while the high resolution image patch sizes were chosen to be 4×8 and 4×16, when 50% and 75% data are missing, respectively. In the Section IV. G, we will further discuss the effect of the patch size on the performance of the proposed SSR method. The parameters of the compared methods: BRFOE [59], K-SVD [47], PGPD [60], BM3D [39], and MSBTD [24], BM3D [39]+Bicubic, ScSR [51], and SBSDI [4] are set to the default values as in their references [4, 24, 39, 47, 51, 59, 60]. The parameters for the Tikhonov method were tuned to reach the best results.
D. Quantitative Metrics
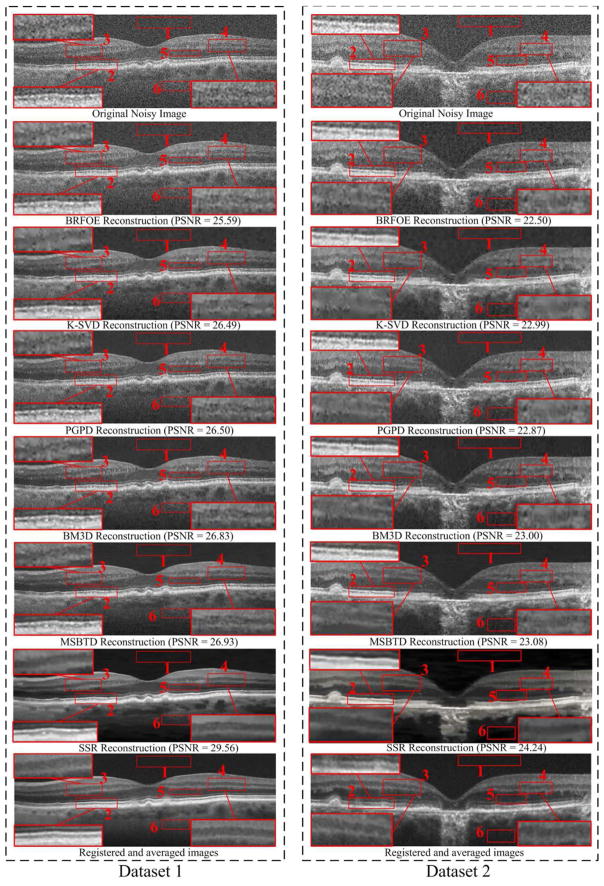
We adopted the peak signal-to-noise-ratio (PSNR), mean-to-standard-deviation ratio (MSR) [62], contrast-to-noise ratio (CNR) [63] and Wilcoxon signed-rank test [64] to quantitatively evaluate the results of the reconstruction methods. PSNR is a global quality metric and its calculation needs a high-quality reference image. Since all the datasets used in our experiments are acquired in clinic (not simulated) with heavy noise, we utilized the registered and averaged images created from the repeatedly sampled scans as the “noiseless” reference image. We used StackReg in [58] to register the reference image and reconstructed images in order to reduce the motion between them. After the registration, both the reference and recovered images might be slightly resized to compute the PSNR. MSR and CNR are two non-reference quality metrics and their calculations focus on some selected local regions. That is, MSR and CNR compute the means and standard deviations of background regions (e.g., red box #1 in Fig. 6) and foreground regions (e.g., red box #2–#6 in Fig. 6) in the reconstructed images. The Wilcoxon signed-rank test was used to test for statistical significance between the SSR method and each compared method on the evaluated PSNR, MSR and CNR.
Fig. 6.
Two retinal OCT datasets and their Denoising results using the BRFOE [59], K-SVD [47], PGPD [60], BM3D [39], MSBTD [24] and SSR methods.
E. Results for OCT Image Denoising
Fig. 6 provides visual comparison of denoising results obtained from the BRFOE [59], K-SVD [47], PGPD [60], BM3D [39], MSBTD [24], and SSR methods on two real retinal OCT images. As can be observed, results from the BPFOE exhibit rather noisy appearance. The K-SVD, PGPD, and BM3D methods significantly remove noise, but appear over-smoothed with visible artifacts (see the zoomed layer areas in red boxes #2, #3, and #4 in Fig. 6). Though the MSBTD method can further reduce the artifacts, it still has indistinct layer boundaries (see the zoomed layer areas in red boxes #2, #3, and #4 in Fig. 6). By contrast, utilization of the proposed SSR method can achieve noticeably improved noise suppression while preserving the layer structural details as compared to other methods. This demonstrates that the proposed layer-segmentation based dictionary construction and sparse reconstruction strategies can significantly suppress the noise while still achieving better layer structure representation.
Average quantitative results (over 18 foveal images) of all the test methods are tabulated m the Table I. The proposed SSR method consistently outperforms the compared methods in terms of the three quantitative metrics (i.e., PSNR, MSR, and CNR). The average running time (over 18 foveal images) of all the test methods are reported in Table I. All the compared methods as well as the segmentation and sparse reconstruction parts of the proposed SSR method (in both the denoising and interpolation applications) are executed on a desktop PC with an 17-5930 CPU at 3.5 GHz and 64 GB of RAM. As can be seen, the reconstruction part of the SSR method needs much less running time than the non-local based denoising approaches: PGPD, BM3D, and MSBTD. This shows that searching for similar patches within each specific layer can greatly reduce the computational cost in comparison with the non-local similar patch searching strategy. Furthermore, we observe that the reconstruction part of the SSR method is faster than that of the original sparse denoising method (K-SVD). This is because the proposed SSR utilizes much smaller size layer based dictionaries and can reduce the computational cost for the sparse solution. Note that, the main part of the proposed SSR algorithm (except the SOMP code2) is coded with MATLAB, and can be further greatly accelerated with more efficient coding and a general purposed graphics processing unit (GPU).
TABLE I.
Mean and Standard Deviation of the PSNR, MSR, CNR Together With Running Time (in Seconds) for 18 Foveal Images From 18 Different Subjects Reconstructed by BRFOE [59], K-SVD [47], PGPD [60], BM3D [39], MSBTD [24], And SSR Methods. “Seg” and “Rec” Denotes the Required Running Time For the Layer Segmentation Part (R Layers Segmentation) And Sparse Reconstruction Part (Including vitreous and choroid/sclera regions Classification), Respectively. When p<0.05, The Metrics (I.E. PSNR, MSR, CNR) For Each Test Method are Considered Statistically Significant and were Marked By “*”. Best Results in the Mean Values are Labeled in Bold.
| Metric\Method | BRFOE | K-SVD | PGPD | BM3D | MSBTD | SSR | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| PSNR | Mean | 25.32 | 27.03 | 27.01 | 27.04 | 27.08 | 28.10 | |
| Stand deviation | 1.72 | 2.47 | 2.51 | 2.49 | 2.47 | 2.63 | ||
| p value | 1.96E-04* | 1.96E-04* | 1.96E-04* | 1.96E-04* | 1.96E-04* | |||
|
| ||||||||
| MSR | Mean | 7.05 | 9.79 | 9.57 | 9.27 | 10.96 | 13.04 | |
| Stand deviation | 0.84 | 2.34 | 2.29 | 2.19 | 3.37 | 3.51 | ||
| p value | 1.96E-04* | 1.96E-04* | 1.96E-04* | 1.96E-04* | 2.33E-04* | |||
|
| ||||||||
| CNR | Mean | 3.32 | 4.41 | 4.12 | 4.06 | 4.58 | 5.34 | |
| Stand deviation | 1.05 | 1.34 | 1.19 | 1.17 | 1.32 | 1.31 | ||
| p value | 1.96E-04* | 1.96E-04* | 1.96E-04* | 1.96E-04* | 1.96E-04* | |||
|
| ||||||||
| Running Time | 63.53 | 2.90 | 69.35 | 4.45 | 1646.35 | Seg | Rec | |
|
| ||||||||
| 0.49 | 1.01 | |||||||
F. Results for OCT Image Interpolation
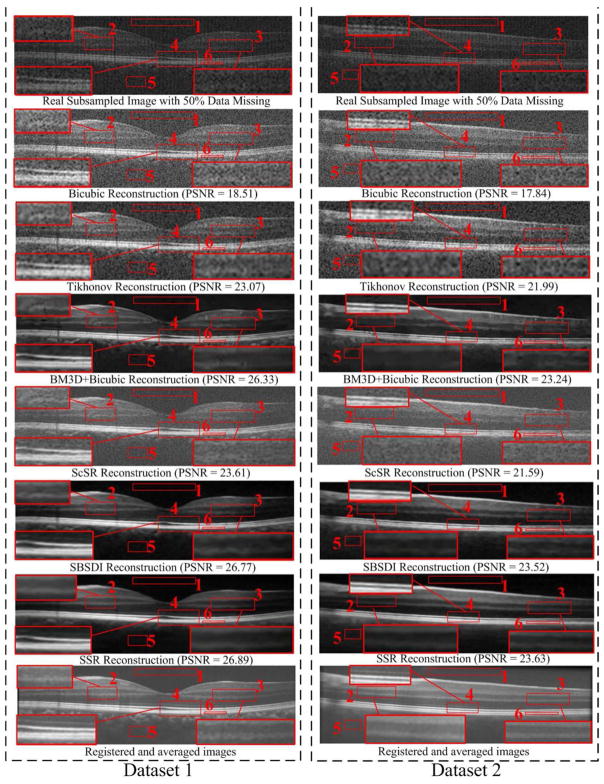
For interpolation, we conducted the experiments on two sampling conditions (with 50% data missing and with 75% data missing). For the condition with 50% missing, we first tested the Bicubic, Tikhonov [61], BM3D [39]+Bicubic, ScSR [51], SBSDI [4], and SSR methods on the real subsampled datasets from 13 different subjects. For each dataset, we adopted the central foveal B-scan and another two B-scans located approximately 1.5 mm below and above the fovea. Therefore, there are totally 3 × 13=39 images for this real subsampled experiment. Fig. 7 shows two real subsampled images and their visually reconstructed results obtained from all the test methods. As can be observed, the Bicubic, Tikhonov, and ScSR methods result in noisy reconstructions. The BM3D+Bicubic can greatly reduce the noise, but blurs the layer boundaries (see the zoomed layer areas in red boxes #2, #3, and #4 in Fig. 7). Although the layer boundaries can be preserved in the SBSDI method, it still introduces some artifacts. In contrast, the proposed SSR method can well preserve the structural details while significantly suppressing noise. In addition, we also adopted a regular sampling pattern to remove the 50% data information of the 18 foveal images used in the above denoising problem and used them for the interpolation testing. Table II reports the average quantitative results (over 39 real subsampled + 18 synthetic subsampled images) of all the test methods. As can be observed, the proposed SSR method delivers the best quantitative results. Furthermore, the reconstruction part of the SSR method requires less running time than the Tikhonov, BM3D+Bicubic, ScSR, and SBSDI methods, but has a higher computational cost than the simple Bicubic method. Note that, for the interpolation problem, the SBSDI and SSR methods adopts similar reconstruction framework, except for the proposed layer segmentation based dictionary construction and similar patch searching strategies.
Fig. 7.
Two real subsampled retinal OCT datasets (with 50% data missing) and their reconstruction results by using the Bicubic, Tikhonov [61], BM3D [39]+Bicubic, ScSR [51], SBSDI [4], and proposed SSR methods.
TABLE II.
Mean and Standard Deviation of the PSNR, MSR, CNR Together With Running Time (in Seconds) for 18 Foveal Images (With 50% Data Missing) From 18 Different Subjects and 39 Real Subsampled Images (Including Foveal and Nonfoveal Images With 50% Data Missing) From 13 Different Subjects Reconstructed by Bicubic, Ttkhonov [61], BM3D [39]+Bicubic, ScSR [51], SBSDI [4], and SSR Methods. “Seg” and “Rec” Denotes the Required Running Time For the Layer Segmentation Part (R Layers Segmentation) And Sparse Reconstruction Part (Including vitreous and choroid/sclera regions Classification), Respectively. When p<0.05, The Metrics (I.E. PSNR, MSR, CNR) For Each Test Method are Considered Statistically Significant and were Marked By “*”. Best Results in the Mean Values are Labeled in Bold.
| Metric\Method | Bicubic | Tikhonov | BM3D+Bicubic | ScSR | SBSDI | SSR | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| PSNR | Mean | 18.24 | 22.47 | 25.55 | 22.74 | 26.07 | 26.20 | |
| Stand deviation | 0.77 | 1.58 | 2.44 | 3.09 | 2.73 | 2.70 | ||
| p value | 5.14E-11* | 5.14E-11* | 5.14E-11* | 5.14E-11* | 2.08E-08* | |||
|
| ||||||||
| MSR | Mean | 3.53 | 5.87 | 9.39 | 6.76 | 13.04 | 14.12 | |
| Stand deviation | 0.37 | 0.70 | 1.83 | 0.92 | 3.18 | 3.73 | ||
| p value | 5.14E-11* | 5.14E-11* | 5.14E-11* | 5.14E-11* | 1.99E-07* | |||
|
| ||||||||
| CNR | Mean | 1.62 | 3.49 | 4.58 | 3.21 | 5.44 | 5.83 | |
| Stand deviation | 0.45 | 0.89 | 1.06 | 1.02 | 1.51 | 1.62 | ||
| p value | 5.14E-11* | 6.71E-11* | 5.14E-11* | 5.14E-11* | 1.44E-06* | |||
|
| ||||||||
| Running Time | 0.01 | 2.24 | 2.34 | 96.29 | 4.61 | Seg | Rec | |
|
| ||||||||
| 0.43 | 0.53 | |||||||
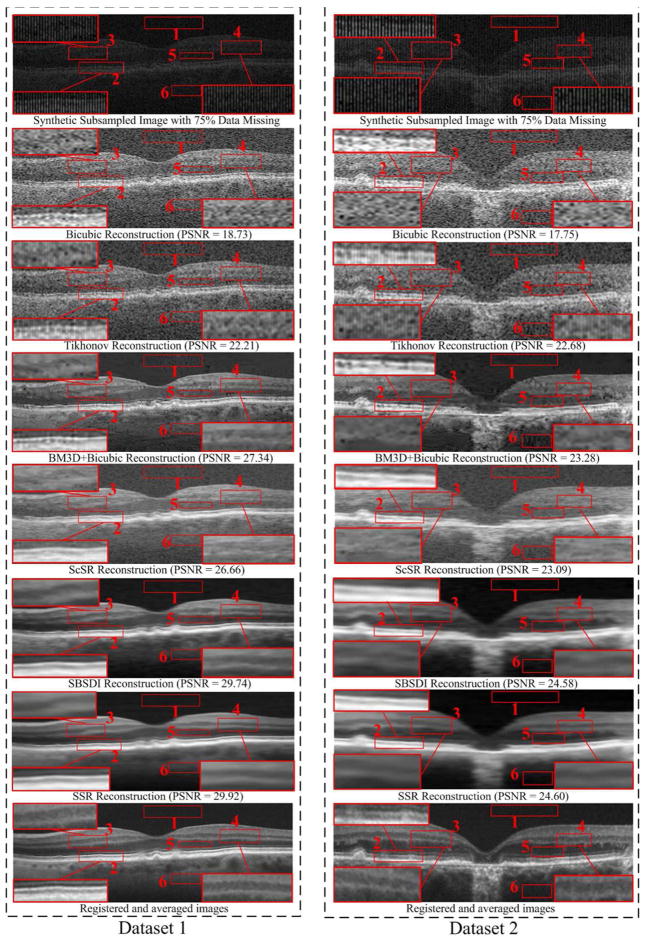
For the condition with 75% data missing, we also used the regular sampling pattern to remove the 75% data information of the 18 foveal images in the denoismg problem. Fig. 8 and Table III show the qualitative and quantitative reconstruction results from all the test methods on datasets with 75% data missing. As can be observed, the proposed SSR method provides the best performance.
Fig. 8.
Two synthetic Subsampled Retinal OCT datasets (with 75% data missing) and their reconstruction results by using the Bicubic, Tikhonov [61], BM3D [39]+Bicubic, ScSR [51], SBSDI [4], and proposed SSR methods.
TABLE III.
Mean and Standard Deviation of the PSNR, MSR, CNR Together With Running Time (in Seconds) for 18 Foveal Images (With 75% Data Missing) From 18 Different Subjects by Bicubic, Ttkhonov [61], BM3D [39]+Bicubic, ScSR [51], SBSDI [4], and SSR Methods. “Seg” and “Rec” Denotes the Required Running Time For Layer Segmentation Part (R Layers Segmentation) And Sparse Reconstruction Part (Including vitreous and choroid/sclera regions Classification), Respectively. When p<0.05, The Metrics (I.E. PSNR, MSR, CNR) For Each Test Method are Considered Statistically Significant and were Marked By “*”. Best Results in the Mean Values are Labeled in Bold.
| Metric\Method | Bicubic | Tikhonov | BM3D+Bicubic | ScSR | SBSDI | SSR | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| PSNR | Mean | 18.52 | 22.52 | 27.26 | 25.76 | 28.30 | 28.31 | |
| Stand deviation | 0.49 | 0.95 | 2.50 | 1.63 | 2.54 | 2.50 | ||
| p value | 1.96E-04* | 1.96E-04* | 1.96E-04* | 1.96E-04* | 0.31 | |||
|
| ||||||||
| MSR | Mean | 3.63 | 5.35 | 10.72 | 9.00 | 14.53 | 15.26 | |
| Stand deviation | 0.47 | 0.55 | 0.27 | 1.05 | 4.57 | 4.48 | ||
| p value | 1.96E-04* | 1.96E-04* | 1.96E-04* | 1.96E-04* | 0.01* | |||
|
| ||||||||
| CNR | Mean | 1.68 | 2.53 | 4.62 | 3.82 | 5.89 | 6.25 | |
| Stand deviation | 0.56 | 0.80 | 1.31 | 1.26 | 1.43 | 1.64 | ||
| p value | 1.96E-04* | 1.96E-04* | 1.96E-04* | 1.96E-04* | 1.96E-04* | |||
|
| ||||||||
| Running Time | 0.01 | 1.48 | 1.14 | 48.88 | 2.32 | Seg | Rec | |
|
| ||||||||
| 0.51 | 0.31 | |||||||
G. Effects of Patch Size
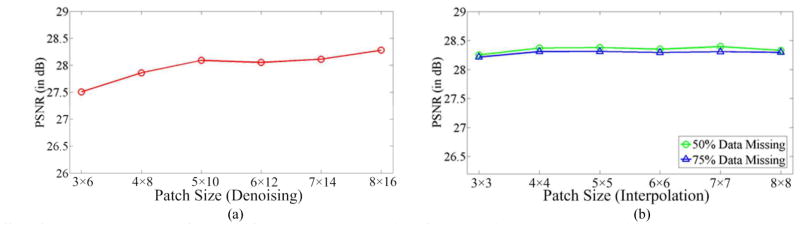
In this section, we analyze the effect of the patch size on the performance (in the PSNR sense) of the proposed SSR method. In the analysis of each specific parameter, we vary this parameter in a certain range and set the other parameters to the fixed values described in Section IV. C. Note that, the reported values are the averaged results over all 18 synthetic test images described in Section IV. E and F. For the denoismg problem, the patch size was varied from 3 × 6 to 8 × 16. The performance of the proposed SSR algorithm under varied patch sizes is shown in Fig. 9(a). For the interpolation problem, we changed the low resolution patch size from 3×3 to 8×8. The corresponding high resolution patch was enlarged in the horizontal direction according to the interpolation scale number (e.g., two times for 50% data missing and four times for 75% data missing). The performance of the proposed SSR algorithm under different patch sizes for two interpolation cases (e.g., 50% and 75% data missing) is illustrated in Fig. 9(b). As can be observed in Fig. 9, small patch sizes negatively affect the performance of our SSR algorithm. This is due to the fact that small patch size contains very limited spatial information for the reconstruction. The SSR algorithm generally performs the best when the patch size is increased to 5 × 10 and 4 × 4 for the denoising and interpolation problems, respectively. The performance of the proposed SSR algorithm, when using larger patch sizes, might become stable or even slightly worse based on qualitative evaluation. This is because using too large patches might create an over-smoothing effect, which blurs the final reconstruction results. In addition, using larger patches has higher computational costs.
Fig. 9.
Effect of the patch size on the performance of the proposed SSR algorithm for (a) Denoising and (b) Interpolation.
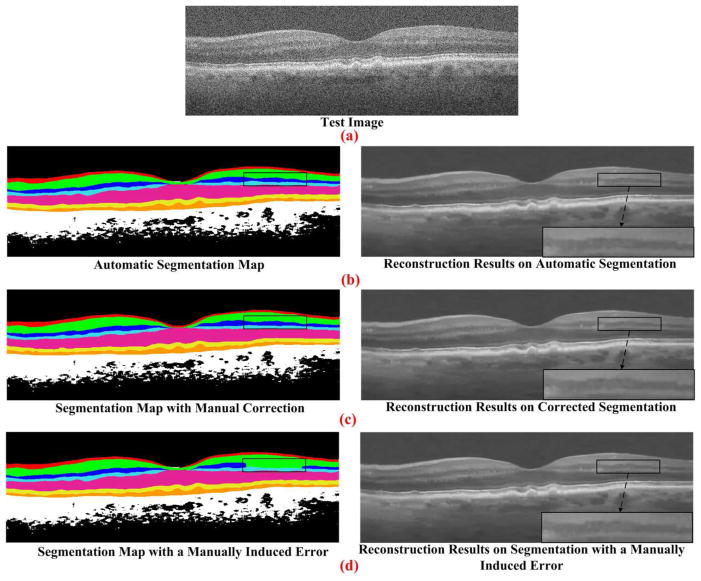
H. Effect of Segmentation Errors
Our next experiment assessed the sensitivity of our denoising algorithm to small errors in segmentation. We segmented a B-Scan using the automatic GTDP technique [35]. Next, we modified this B-Scan to correct segmentation errors (corrected segmentation). Since automatic and corrected segmentation results were very close, we manually modified the automatic segmentation, intentionally introducing errors in segmentation (manually induced error). As shown in Fig. 10, the corrected segmentation case might slightly perform better than the automatic and manually induced error segmentation cases. However, these differences are negligible.
Fig. 10.
SSR reconstruction results using three different layer segmentations. The image in (a) is a raw OCT B-Scan. Automatic segmentation of retinal layers using GTDP algorithm (delineated in different colors in the left column) is used to produce the denoised image of the right column in (b). We carefully manually corrected the automatic segmentation of layer boundaries, resulting in the slightly modified layer boundaries shown in the left column of the (c). Corresponding denoised image is shown in the right column of the (c). To artificially create more severe segmentation errors, we intentionally introduced errors in segmentation of the inner nuclear layer in the black box region (manually induced error), resulting in the images of the (d).
In general, the segmentation information has been utilized in three stages of the proposed SSR algorithm: dictionary construction, similar patch searching, and sparse representation. Errors in segmentation can potentially affect each of these stages. Since manual corrections can be applied to the automatically segmented images in the offline dictionary construction stage, the segmentation accuracy in this stage can be as high as needed. In the similar patch searching stage, since the similarity metrics are tested for each patch, the search process significantly reduces the effects of segmentation errors on reconstruction. The sparse representation stage is relatively more sensitive to segmentation accuracy, as segmentation error can result in the selection of a dictionary from an incorrect layer to represent the patch being processed. However, even with a suboptimally selected dictionary, the sparse solution process (e.g. the orthogonal matching pursuit algorithm [50]) can find well-suited atoms to represent the processed patch and the final reconstruction result is expected to be stable, as shown in Fig. 10. As indicated in Section IV E and F, only with the classic GTDP automatic segmentation algorithm [35], the proposed SSR algorithm’s performance is superior to several state-of-the-art non-segmentation based reconstruction methods.
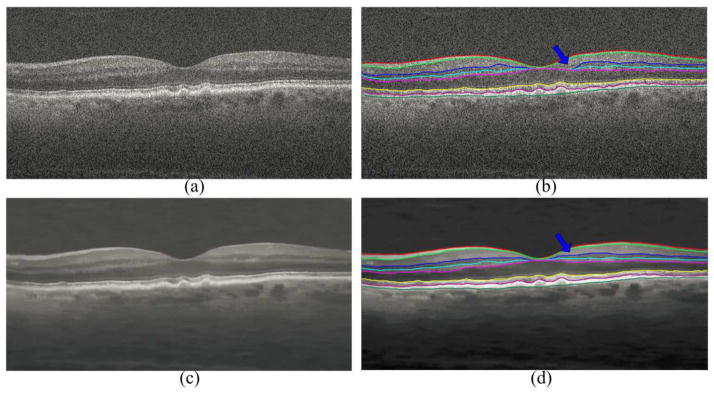
I. Effect of Image Reconstruction to the Layer Segmentation
The experiment in Fig. 10 showed that our denoising algorithm is not sensitive to small errors in automated segmentation. In a recursive process, once the SSR denoised images are in hand, they can be segmented again using the GTDP method, which is expected to result in equal or better segmentation. The experiment in Fig. 11 shows such a case. In this example, there is a small error in the segmentation of the inner nuclear layer boundaries delineated by blue and cyan colors near the fovea, when the GTDP technique is applied on the noisy images. When GTDP is applied on the SSR denoised image, these small segmentation errors are mostly eliminated.
Fig. 11.
Impact of SSR reconstruction on automatic GTDP layer segmentation performance. The raw B-Scan in (a) is segmented using automated GTDP technique in (b). The blue arrow points to a small region with errors in automatic segmentation of the inner nuclear layer boundaries (delineated in blue and cyan colors). SSR denoised image using the segmentation results in (b) is shown in (c). Automatic segmentation of the image in (c) using GTDP technique is shown in (d), where the accuracy of segmentation is improved for the previous erroneously segmented region.
V. CONCLUSIONS
In this paper, we presented a novel layer-segmentation based sparse reconstruction method named SSR for efficient denoising and interpolation of 3D SDOCT images. Unlike our previous sparse denoising [24] and interpolation [4] methods, the proposed SSR method utilizes segmented layer information to construct the structural dictionaries and conduct similar-patch searching. Both the layer-segmentation based dictionaries and similar-patches searching can aid sparse reconstruction in order to achieve better representation, while greatly reducing the computational cost. Our experiments demonstrated the high efficiency and effectiveness of the proposed SSR method over several current state-of-the-art denoising and interpolation approaches.
Following our previous work, the denoising and interpolation software developed for this project publically will be available at https://sites.google.com/site/levuanfang/home.
There is space for improving the performance_of our SSR algorithm. One limiting factor in our proposed technique is the utilization of fixed-size rectangular patches. Today, OCT technology is used for imaging a variety of pathologic structures in eyes and other human and animal organs, the shape of which might not be optimally represented by the fixed-size rectangular patches. Therefore, in our future work, we will incorporate shape adaptive patches (e.g. utilizing superpixel [65] or adaptive kernels [66]) into the proposed sparse model, which we expect to improve the reconstruction performance. In addition to the fixed-size patch, several parameters need to be tuned for the proposed method. So, a future line of work will also be on designing efficient automatic parameter selection algorithms.
There are two potential utilities for our segmentation-based reconstruction technique. The first application is the OCT image quality improvement for visual inspection of complex anatomic and pathologic structures (e.g., vessels, drusen, hyperreflective foci, edema, etc.). The second important utility of our reconstruction approach is improving the performance of automated segmentation algorithms. Note that, our algorithm utilizes automated layer segmentation (a problem for which several solutions are available in the literature) and results in denoised images from which other features (e.g. hyperreflective foci, edema, vessel, etc.) can be automatically segmented. The proposed SSR method based on the layer segmentation can even recursively improve the accuracy of segmentation. While our automatic GTDP retinal layer segmentation algorithm is accurate, it is not perfect and occasionally results in erroneous segmentation, especially for noisy images. Utilizing our SSR reconstruction algorithm can improve the GTDP segmentation accuracy as shown in the experiment of Fig. 12. A complete quantitative assessment of the proposed method impact for enhancing the performance of various automated segmentation methods is a part of our ongoing works.
In this paper, we utilized the segmented retinal layer information for enhancing volumetric OCT images. However, the proposed mathematical framework is general and applicable to virtually any other tissue. Also, for segmentation, we utilized the GTDP method of [35], which can be replaced by any other layer or feature segmentation technique [24, 67–69]. Therefore, in our future publications, we will investigate the applicability of the proposed segmentation based reconstruction model for enhancing the quality of a wide variety of images from different tissues (e.g. dermatology, Gastroenterology, and cardiology) captured via OCT or other imaging modalities such as MRI or ultrasound.
Finally, despite the fact that our algorithm utilized the classic GTDP automatic segmentation algorithm [35], the final reconstructed results demonstrated the superiority of the proposed SSR algorithm over several state-of-the-art non-segmentations based reconstruction methods. More recent and advanced 3-D segmentation methods such as [3, 70, 71], which are more robust to imaging and pathologic artifacts, can also be utilized to improve segmentation accuracy, if needed. However, more complex 3-D segmentation algorithms often increase the computational load of the overall algorithm.
Acknowledgments
This work was supported in part by grants from NIH R01 EY022691 and P30 EY005722, the National Natural Science Foundation for Distinguished Young Scholars of China under Grant No. 61325007, and the National Natural Science Foundation for Young Scientist of China under Grant No. 61501180.
We thank the A2A Ancillary SDOCT Study group, especially Dr. Cynthia A. Toth, for sharing their dataset of OCT images.
Footnotes
Datasets were downloaded at: http://people.duke.edu/~sf59/Fang_TMI_2013.htm.
Downloaded from: http://spams-devel.gforge.inria.fr/.
Contributor Information
Leyuan Fang, College of Electrical and Information Engineering, Hunan University, Changsha, 410082, China, and also with the Department of Biomedical Engineering, Duke University, Durham, NC 27708 USA.
Shutao Li, College of Electrical and Information Engineering, Hunan University, Changsha, 410082, China.
David Cunefare, Department of Biomedical Engineering, Duke University, Durham, NC 27708 USA.
Sina Farsiu, Department of Biomedical Engineering, Duke University, Durham, NC 27708 USA.
References
- 1.Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, Hee MR, Flotte T, Gregory K, Puliafito CA, Fujimoto JG. Optical coherence tomography. Science. 1991 Nov;254(5035):1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhat S, Larina IV, Larin KV, Dickinson ME, Liebling M. 4D reconstruction of the beating embryonic heart from two orthogonal sets of parallel optical coherence tomography slice-sequences. IEEE Trans Med Imag. 2013 Mar;32(3):578–588. doi: 10.1109/TMI.2012.2231692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi F, Chen X, Zhao H, Zhu W, Xiang D, Gao E, Sonka M, Chen H. Automated 3-D retinal layer segmentation of macular optical coherence tomography images with serous pigment epithelial detachments. IEEE Trans Med Imag. 2015 Feb;34(2):441–452. doi: 10.1109/TMI.2014.2359980. [DOI] [PubMed] [Google Scholar]
- 4.Fang L, Li S, McNabb R, Nie Q, Kuo A, Toth C, Izatt JA, Farsiu S. Fast Acquisition and Reconstruction of Optical Coherence Tomography Images via Sparse Representation. IEEE Trans Med Imag. 2013 Nov;32(11):2034–2049. doi: 10.1109/TMI.2013.2271904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson MD, Chiu SJ, Toth CA, Izatt J, Lo JY, Farsiu S. Novel applications of super-resolution in medical imaging. In: Milanfar P, editor. Super-Resolution Imaging. Boca Raton: CRC Press; 2010. pp. 383–412. [Google Scholar]
- 6.Kafieh R, Rabbani H, Selesnick I. Three dimensional data-driven multi scale atomic tepresentation of optical coherence tomography. IEEE Trans Med Imag. 2015 May;34(5):1042–1062. doi: 10.1109/TMI.2014.2374354. [DOI] [PubMed] [Google Scholar]
- 7.Milanfar P. A tour of modern image filtering: New insights and methods, both practical and theoretical. IEEE Signal Process Mag. 2013 Jan;30(1):106–128. [Google Scholar]
- 8.Salinas HM, Fernández DC. Comparison of PDE-based nonlinear diffusion approaches for image enhancement and denoising in optical coherence tomography. IEEE Trans Med Imag. 2007 Jun;26(6):761–771. doi: 10.1109/TMI.2006.887375. [DOI] [PubMed] [Google Scholar]
- 9.Ozcan A, Bilenca A, Desjardins AE, Bouma BE, Tearney GJ. Speckle reduction in optical coherence tomography images using digital filtering. J Opt Soc Am. 2007 Jul;24(7):1901–1910. doi: 10.1364/josaa.24.001901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong A, Mishra A, Bizheva K, Clausi DA. General Bayesian estimation for speckle noise reduction in optical coherence tomography retinal imagery. Opt Exp. 2010 Apr;18(8):8338–8352. doi: 10.1364/OE.18.008338. [DOI] [PubMed] [Google Scholar]
- 11.Boroomand A, Wong A, Li E, Cho DS, Ni B, Bizheva K. Multi-penalty conditional random field approach to super-resolved reconstruction of optical coherence tomography images. Biomed Opt Exp. 2013 Apr;4(10):2032–2050. doi: 10.1364/BOE.4.002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Kang JU. Compressive SD-OCT: the application of compressed sensing in spectral domain optical coherence tomography. Opt Exp. 2010 Oct;18(21):22010–22019. doi: 10.1364/OE.18.022010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu D, Vaswani N, Huang Y, Kang JU. Modified compressive sensing optical coherence tomography with noise reduction. Opt Lett. 2012 Aug;37(20):4209–4211. doi: 10.1364/OL.37.004209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu D, Huang Y, Kang JU. Real-time compressive sensing spectral domain optical coherence tomography. Opt lett. 2014 Jan;39(1):76–79. doi: 10.1364/OL.39.000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebed E, Mackenzie PJ, Sarunic MV, Beg FM. Rapid volumetric OCT image acquisition using compressive sampling. Opt Exp. 2010 Aug;18(20):21003–21012. doi: 10.1364/OE.18.021003. [DOI] [PubMed] [Google Scholar]
- 16.Wu AB, Lebed E, Sarunic MV, Beg MF. Quantitative evaluation of transform domains for compressive sampling-based recovery of sparsely sampled volumetric OCT images. IEEE Trans Biomed Eng. 2013 Feb;60(2):470–478. doi: 10.1109/TBME.2012.2199489. [DOI] [PubMed] [Google Scholar]
- 17.Lurie KL, Angst R, Ellerbee AK. Automated mosaicing of feature-poor optical coherence tomography volumes with an integrated white light imaging system. IEEE Trans Biomed Eng. 2014;61(7):2141–2153. doi: 10.1109/TBME.2014.2316535. [DOI] [PubMed] [Google Scholar]
- 18.Rabbani H, Nezafat R, Gazor S. Wavelet-domain medical image denoising using bivariate laplacian mixture model. IEEE Trans Biomed Eng. 2009 Dec;56(12):2826. doi: 10.1109/TBME.2009.2028876. [DOI] [PubMed] [Google Scholar]
- 19.Chitchian S, Fiddy MA, Fried NM. Denoising during optical coherence tomography of the prostate nerves via wavelet shrinkage using dual-tree complex wavelet transform. J Biomed Opt. 2009 Jan;14(1):014031-014031–6. doi: 10.1117/1.3081543. [DOI] [PubMed] [Google Scholar]
- 20.Jian Z, Yu L, Rao B, Tromberg BJ, Chen Z. Three-dimensional speckle suppression in optical coherence tomography based on the curvelet transform. Opt Exp. 2010 Jan;18(2):1024–1032. doi: 10.1364/OE.18.001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubinstein R, Bruckstein AM, Elad M. Dictionaries for sparse representation modeling. Proc IEEE. 2010 Jun;98(6):1045–1057. [Google Scholar]
- 22.Olshausen BA, Field DJ. Emergence of simple-cell receptive field properties by learning a sparse code for natural images. Nature. 1996 Jun;381(6583):607–609. doi: 10.1038/381607a0. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Fang L, Yin H. An efficient dictionary learning algorithm and its application to 3-D medical image denoising. IEEE Trans Biomed Eng. 2012 Feb;59(2):417–427. doi: 10.1109/TBME.2011.2173935. [DOI] [PubMed] [Google Scholar]
- 24.Fang L, Li S, Nie Q, Izatt JA, Toth CA, Farsiu S. Sparsity based denoising of spectral domain optical coherence tomography images. Biomed Opt Express. 2012 May;3(5):927–942. doi: 10.1364/BOE.3.000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong A, Mishra A, Fieguth P, Clausi DA. Sparse reconstruction of breast mri using homotopic minimization in a regional sparsified domain. IEEE Trans Biomed Eng. 2013 Mar;60(3):743–752. doi: 10.1109/TBME.2010.2089456. [DOI] [PubMed] [Google Scholar]
- 26.Li S, Yin H, Fang L. Group-sparse representation with dictionary learning for medical image denoising and fusion. IEEE Trans Biomed Eng. 2012 Dec;59(12):3450–3459. doi: 10.1109/TBME.2012.2217493. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Wu G, Yap P, Feng Q, Lian J, Chen W, Shen D. Hierarchical patch-based sparse representation-A new approach for resolution enhancement of 4D-CT lung data. IEEE Trans Med Imag. 2012 Nov;31(11):1993–2005. doi: 10.1109/TMI.2012.2202245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Q, Yu H, Mou X, Zhang L, Hsieh J, Wang G. Low-dose X-ray CT reconstruction via dictionary learning. IEEE Trans Med Imag. 2012 Sep;31(9):1682–1697. doi: 10.1109/TMI.2012.2195669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang L, Li S, Kang X, Benediktsson JA. Spectral–spatial hyperspectral image classification via multiscale adaptive sparse representation. IEEE Trans Geosci Remote Sens. 2014 Dec;52(12):7738–7749. [Google Scholar]
- 30.Fang L, Li S, Kang X, Benediktsson JA. Spectral–spatial classification of hyperspectral images with a superpixel-based discriminative sparse model. IEEE Trans Geosci Remote Sens. 2015 Feb;53(8):4186–4201. [Google Scholar]
- 31.Aharon M, Elad M, Brackstein AM. The K-SVD: An algorithm for designing of overcomplete dictionaries for sparse representation. IEEE Trans Signal Process. 2006 Nov;54(11):4311–4322. [Google Scholar]
- 32.Fang L, Li S, Kang X, Izatt JA, Farsiu S. 3-D Adaptive Sparsity Based Image Compression with Applications to Optical Coherence Tomography. IEEE Trans Med Imag. 2015 Jun;34(6):1306–1320. doi: 10.1109/TMI.2014.2387336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baghaie A, D’Souza RM, Yu Z. Sparse and low rank decomposition based batch image alignment for speckle reduction of retinal OCT images. Proc IEEE Int Symp Biomed Imag. 2015:1141–1147. [Google Scholar]
- 34.Kopriva I, Shi F, Chen X. Enhanced low-rank+ sparsity decomposition for speckle reduction in optical coherence tomography. J biomed optics. 2016 Jul;21(7):076008–076008. doi: 10.1117/1.JBO.21.7.076008. [DOI] [PubMed] [Google Scholar]
- 35.Chiu SJ, Li XT, Nicholas P, Toth CA, Izatt JA, Farsiu S. Automatic segmentation of seven retinal layers in SDOCT images congruent with expert manual segmentation. Opt Express. 2010 Aug;18(18):19413–19428. doi: 10.1364/OE.18.019413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Hou P, Jin C, Zhu W, Luo X, Shi F, Sonka M, Chen H. Quantitative analysis of retinal layer optical intensities on threedimensional optical coherence tomography. Invest Ophthalmol Vis Set. 2013 Oct;54(10):6846–6851. doi: 10.1167/iovs.13-12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keller B, Cunefare D, Grewal DS, Mahmoud TH, Izatt JA, Farsiu S. Length-adaptive graph search for automatic segmentation of pathological features in optical coherence tomography images. J Biomed Optics. 2016 Jul;21(7):076015–076015. doi: 10.1117/1.JBO.21.7.076015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, Wong A, Bizheva K, Fieguth P, Bie H. Homotopic, non-local sparse reconstruction of optical coherence tomography imagery. Opt Exp. 2012 May;20(9):10200–10211. doi: 10.1364/OE.20.010200. [DOI] [PubMed] [Google Scholar]
- 39.Dabov K, Foi A, Katkovnik V, Egiazarian K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans Image Process. 2007 Aug;16(8):2080–2095. doi: 10.1109/tip.2007.901238. [DOI] [PubMed] [Google Scholar]
- 40.Mairal J, Bach F, Ponce J, Sapiro G, Zisserman A. Non-local sparse models for image restoration. Proc IEEE Int Conf Comput Vis. 2009:2272–2279. [Google Scholar]
- 41.Kafieh R, Rabbani H, Hajizadeh F, Ommani M. An accurate multimodal 3-D vessel segmentation method based on brightness variations on OCT layers and curvelet domain fundus image analysis. IEEE Trans Biomed Eng. 2013 Oct;60(10):2815–2823. doi: 10.1109/TBME.2013.2263844. [DOI] [PubMed] [Google Scholar]
- 42.Bowes Rickman C, Farsiu S, Toth CA, Klingeborn M. Dry age-related macular degeneration: mechanisms, therapeutic targets, and imaging. Invest Ophthalmol Vis Set. 2013 Dec;54(14):ORSF68–ORSF80. doi: 10.1167/iovs.13-12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christenbury JG, Folgar FA, O’Connell RV, Chiu SJ, Farsiu S, Toth CA. Progression of intermediate age-related macular degeneration with proliferation and inner retinal migration of hyperreflective Foci. Ophthalmol. 2013 May;120(5):1038–1045. doi: 10.1016/j.ophtha.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hedjam R, Cheriet M. Segmentation-based document image denoising. Proc Euro Workshop Vis Inf Process. 2010:61–65. [Google Scholar]
- 45.Xu Z, Baqci U, Seidel J, Thomasson D, Solomon J, Mollura DJ. Segmentation based denoising of PET images: an iterative approach via regional means and affinity propagation. Conf Med Image Comput Comput Assist Interv. 2014:698–705. doi: 10.1007/978-3-319-10404-1_87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Tanaka M, Okutomi M. Signal dependent noise removal from a single image. Proc IEEE Int Conf Image Process. 2014:2679–2683. doi: 10.1109/TIP.2014.2347204. [DOI] [PubMed] [Google Scholar]
- 47.Elad M, Aharon M. Image denoising via sparse and redundant representations over learned dictionaries. IEEE Trans Image Process. 2006 Dec;15(12):3736–3745. doi: 10.1109/tip.2006.881969. [DOI] [PubMed] [Google Scholar]
- 48.Mairal J, Bach F, Ponce J, Sapiro G. Online dictionary learning for sparse coding. Proc Int Conf Comput Vis. 2009:689–696. [Google Scholar]
- 49.Davis G, Mallat S, Avellaneda M. Adaptive greedy approximations. J Construct Approx. 1997 Mar;13(1):57–98. [Google Scholar]
- 50.Mallat SG, Zhang Z. Matching pursuits with time-frequency dictionaries. IEEE Trans Signal Process. 1993 Dec;41(12):3397–3415. [Google Scholar]
- 51.Yang J, Wright J, Huang TS, Ma Y. Image super-resolution via sparse representation. IEEE Trans Image Process. 2010 Nov;19(11):2861–2873. doi: 10.1109/TIP.2010.2050625. [DOI] [PubMed] [Google Scholar]
- 52.Buades A, Coll B, Morel JM. A review of image denoising algorithms, with a new one. Multiscale Model Simul. 2005 Feb;4(2):490–530. [Google Scholar]
- 53.Protter M, Elad M, Takeda H, Milanfar P. Generalizing the nonlocal-means to super-resolution reconstruction. IEEE Trans Image Process. 2009 Jan;18(1):36–51. doi: 10.1109/TIP.2008.2008067. [DOI] [PubMed] [Google Scholar]
- 54.Kumar R, Banerjee A, Vemuri BC. Volterrafaces: Discriminant analysis using volterra kernels. Proc IEEE Conf Comput Vis Pattern Recognit. 2009:150–155. [Google Scholar]
- 55.Friedman J, Hastie T, Tibshirani R. The Elements of Statistical Learning. Vol. 1. New York: Springer; 2001. [Google Scholar]
- 56.Tropp JA, Gilbert AC, Strauss MJ. Algorithms for simultaneous sparse approximation. Part I: Greedy pursuit. Signal Process. 2006 Mar;86(3):572–588. [Google Scholar]
- 57.Farsiu S, Chiu SJ, O’Connell RV, Folgar FA, Yuan E, Izatt JA, Toth CA. Quantitative Classification of Eyes with and without Intermediate Age-related Macular Degeneration Using Optical Coherence Tomography. Ophthalmology. 2014;121(l):162–172. doi: 10.1016/j.ophtha.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thévenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998 Jan;7(1):27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 59.Weiss Y, Freeman WT. What makes a good model of natural images? Proc IEEE Int Conf Comput Vis Pattern Recognit. 2007:1–8. [Google Scholar]
- 60.Xu J, Zhang L, Zuo W, Zhang D, Feng X. Patch Group Based Nonlocal Self-Similarity Prior Learning for Image Denoising. Proc IEEE Int Conf Comput Vis. 2015 [Google Scholar]
- 61.Chong GT, Farsiu S, Freedman SF, Sarin N, Koreishi AF, Izatt JA, Toth CA. Abnormal foveal morphology in ocular albinism imaged with spectral-domain optical coherence tomography. Arch Ophthalmol. 2009 Jan;127(1):37–44. doi: 10.1001/archophthalmol.2008.550. [DOI] [PubMed] [Google Scholar]
- 62.Cincotti G, Loi G, Pappalardo M. Frequency decomposition and compounding of ultrasound medical images with wavelets packets. IEEE Trans Med Imag. 2001 Aug;20(8):764–771. doi: 10.1109/42.938244. [DOI] [PubMed] [Google Scholar]
- 63.Bao P, Zhang L. Noise reduction for magnetic resonance images via adaptive multiscale products thresholding. IEEE Trans Med Imag. 2003 Sep;22(9):1089–1099. doi: 10.1109/TMI.2003.816958. [DOI] [PubMed] [Google Scholar]
- 64.Gibbons JD, Chakraborti S. Nonparametric Statistical Inference. Vol. 168. Boca Raton, FL: CRC Press; 2003. [Google Scholar]
- 65.Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Susstrunk S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell. 2012 Nov;34(11):2274–2281. doi: 10.1109/TPAMI.2012.120. [DOI] [PubMed] [Google Scholar]
- 66.Chiu SJ, Allingham MJ, Mettu PS, Cousins SW, Izatt JA, Farsiu S. Kernel regression based segmentation of optical coherence tomography images with diabetic macular edema. Biomed Opt Exp. 2015 Apr;6(4):1172–1194. doi: 10.1364/BOE.6.001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vermeer KA, van der Schoot J, Lemij HG, de Boer JF. Automated segmentation by pixel classification of retinal layers in ophthalmic OCT images. Biomed Opt Exp. 2011 Jun;2(6):1743–1756. doi: 10.1364/BOE.2.001743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rabbani H, Allingham MJ, Mettu PS, Cousins SW, Farsiu S. Fully automatic segmentation of fluorescein leakage in subjects with diabetic macular edema automatic leakage segmentation in DME. Invest Ophthalmol Vis Sci. 2015 Feb;56(3):1482–1492. doi: 10.1167/iovs.14-15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zawadzki RJ, Fuller AR, Wiley DF, Hamann B, Choi SS, Werner JS. Adaptation of a support vector machine algorithm for segmentation and visualization of retinal structures in volumetric optical coherence tomography data sets. J Biomed Opt. 2007 Jul;12(4):041206. doi: 10.1117/1.2772658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X, Niemeijer M, Zhang L, Lee K, Abràmoff MD, Sonka M. Three-dimensional segmentation of fluid-associated abnormalities in retinal OCT: probability constrained graph-search-graph-cut. IEEE Trans Med Imag. 2012 Mar;31(8):1521–1531. doi: 10.1109/TMI.2012.2191302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garvin MK, Abramoff MD, Wu X, Russell SR, Burns TL, Sonka M. Automated 3-D intraretinal layer segmentation of macular spectral-domain optical coherence tomography images. IEEE Trans Med Imag. 2009 Sep;28(9):1436–1447. doi: 10.1109/TMI.2009.2016958. [DOI] [PMC free article] [PubMed] [Google Scholar]