Abstract
Immune responses elicited against cancer using existing therapies such as vaccines or immune stimulatory antibodies are often not curative. One way to potentiate antitumor immunity is to enhance the long-term persistence of anti-tumor CD8+ T cells. Studies have shown that the persistence of activated CD8+ T cells is negatively impacted by the strength of interleukin 2 (IL-2) signaling. Here, we used small interfering RNAs (siRNAs) against CD25 (IL-2Rα) to attenuate IL-2 signaling in CD8+ T cells. The siRNAs were targeted to 4-1BB-expressing CD8+ T cells by conjugation to a 4-1BB-binding oligonucleotide aptamer. Systemic administration of the 4-1BB aptamer-CD25 siRNA conjugate downregulated CD25 mRNA only in 4-1BB-expressing CD8+ T cells promoting their differentiation into memory cells. Treatment with the 4-1BB aptamer-CD25 siRNA conjugates enhanced the antitumor response of a cellular vaccine or local radiation therapy. Indicative of the generality of this approach, 4-1BB aptamer-targeted delivery of an Axin-1 siRNA, a rate-limiting component of the β-catenin destruction complex, enhanced CD8+ T cell memory development and antitumor activity. These findings show that aptamer-targeted siRNA therapeutics can be used to modulate the function of circulating CD8+ T cells, skewing their development into long-lasting memory CD8+ T cells, and thereby potentiating antitumor immunity.
Keywords: T cell memory, tumor immunotherapy, IL-2 receptor, Axin-1, aptamer, siRNA
Rajagopalan et al. show that aptamer-targeted downregulation of IL-2 receptor or Axin-1 in 4-1BB-expressing cells promotes the differentiation of antigen-activated 4-1BB-expressing CD8+ T cells into long lasting memory cells and inhibits tumor growth in mice.
Introduction
Immunotherapy of cancer using immune stimulatory antibodies has shown promise, but the therapeutic effects have been moderate, calling for the development of complementary strategies to enhance the immune response elicited by the current treatments. Recent studies have underscored the importance of CD8+ memory subsets in mediating protective immune responses, especially against chronic diseases and cancer.1 The enhanced ability of CD8+ memory T cells to confer host protection has been attributed to their greater proliferative capacity upon antigen re-encounter and their ability to mediate systemic immunity by residing in lymph nodes. Thus, promoting the memory CD8+ T cells response in the setting of cancer immunotherapy may be beneficial in controlling tumor growth.2
Antigen-activated CD8+ T cells develop into short-lived effector cells (SLECs) and memory precursor effector cells (MPECs), which give rise to long-lasting memory cells.3 The differentiation into SLECs or MPECs is controlled by a balance of intracellular mediators, many of which are transcription factors.4, 5 For example, T-bet, Blimp-1, and mTOR promote the generation of SLECs, whereas TCF7, Eomes, or BCL-6 favor the generation of MPECs. Indeed, reducing the intracellular levels of SLEC-promoting mediators like Blimp-1, T-bet, or mTOR using genetic means or pharmacological agents promoted the development of memory CD8+ T cells and long-term protective antitumor immunity.6 IL-2 signaling in activated CD8+ T cells promotes the development of SLECs via the coordinated expression of intracellular mediators, such as Blimp-1, and reduced IL-2 signaling was shown to favor the development of memory CD8+ T cells.7, 8, 9 Although blocking anti-CD25 antibodies can be used to reduce IL-2 signaling, systemic administration of CD25 antibodies could exert off-target effects, such as interfering with the function of CD25-expressing CD4+Foxp3+ regulatory cells, and therefore increase the risk of autoimmune pathology.10 Here, we describe an approach that targets, and hence limits, CD25 downregulation to activated CD8+ T cells in mice. CD25 downregulation was achieved using corresponding small interfering RNAs (siRNAs) that were targeted to activated CD8+ T cells by conjugation to a non-stimulatory 4-1BB-binding oligonucleotide aptamer.11, 12 4-1BB is a major immune stimulatory receptor transiently expressed on activated CD8+ T cells.13 The chemically synthesized oligonucleotide aptamers represent an emerging platform for generating ligands with the desired specificity that offers potential advantages over antibody- or peptide-based platforms in terms of development, manufacturing, and conjugation to its (oligonucleotide) cargo and cost.14
Results
Aptamer-Targeted siRNA Downregulation of CD25 in Activated CD8+ T Cells In Vitro
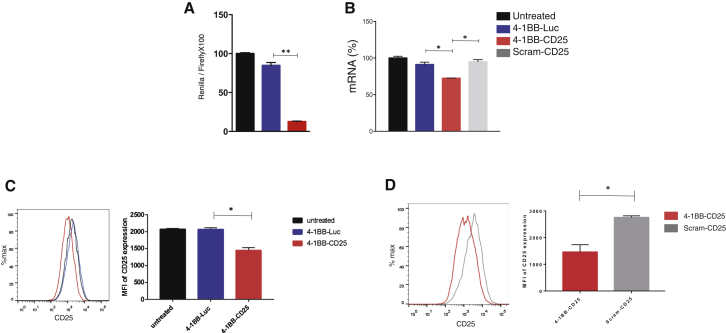
siRNAs to murine CD25 were isolated and characterized as previously described and conjugated to a monovalent 4-1BB-binding aptamer as previously described.11 Conjugation of siRNA cargos to aptamers often compromises activity of conjugated siRNA. Here, we used an algorithm whereby siRNAs with reduced melting temperature (TM) are chosen for conjugation to aptamers, which was shown to significantly increase the success of identifying siRNAs that retain bioactivity upon conjugation to aptamer.15 Aptamer-conjugated CD25, but not luciferase (luc), siRNA downregulated CD25 in chinese hamster ovary (CHO) cells as measured in the dual-luciferase ΨCHECK assay, whereby CD25 target sequences are cloned in the 3′ end of the Renilla luciferase gene (Figure 1A; Materials and Methods). siRNA activity was not negatively affected by conjugation to aptamer (Figure S1A). In the ΨCHECK assay, the siRNAs and aptamer-siRNA conjugates are introduced in the CHO cells by transfection. To determine whether the 4-1BB aptamer-CD25 siRNA conjugate can downregulate endogenous CD25 in a 4-1BB dependent manner, polyclonally activated splenocytes that transiently upregulate 4-1BB were incubated with aptamer-siRNA conjugates in the absence of transfection reagents. The specificity of 4-1BB aptamers is shown in Figure S2 by virtue of the fact that 4-1BB antibody and 4-1BB aptamer, but not isotype-matched antibody or control aptamer, bind to polyconally activated, but not naive, T cells. As shown in Figures 1B and 1C, CD25, but not luc, siRNAs conjugated to 4-1BB, but not a scrambled aptamer-downregulated CD25 RNA and protein, respectively. CD25 downregulation was 4-1BB mediated because scrambled aptamer-CD25 siRNA conjugates did not downregulate CD25 (Figures 1B and 1D).
Figure 1.
Aptamer-Targeted Delivery of CD25 siRNAs to Activated CD8+ T Cells In Vitro
(A) CHO cells were co-transfected with 4-1BB-CD25 or 4-1BB-luciferase (4-1BB-luc) conjugates and with the ΨCHECK reporter plasmid containing short sequences corresponding to the murine CD25 siRNA target cloned into the 3′-untranslated region of Renilla luciferase. After 24 hr, the normalized Renilla luciferase activity was determined (Materials and Methods). (B) Purified CD8+ cells from C57BL/6 mice were polyclonally activated with a mixture of CD3 and CD28 antibodies and treated with conjugates three times at 500 nM concentration every 6 hr, starting 24 hr post activation. 24 hr after the last treatment, the cells were harvested and the levels of CD25 mRNA were assessed by qPCR. (C) 48 hr after the last treatment, the levels of CD25 protein on the cell surface were assessed by flow cytometry (n = 2). (D) CD25 expression on polyclonally activated CD8+ T cells incubated with 4-1BB or scrambled aptamer conjugated to CD25 siRNA (Scram-CD25).
4-1BB-Targeted Downregulation of CD25 in CD8+ T Cells Promotes the Acquisition of a Memory Phenotype
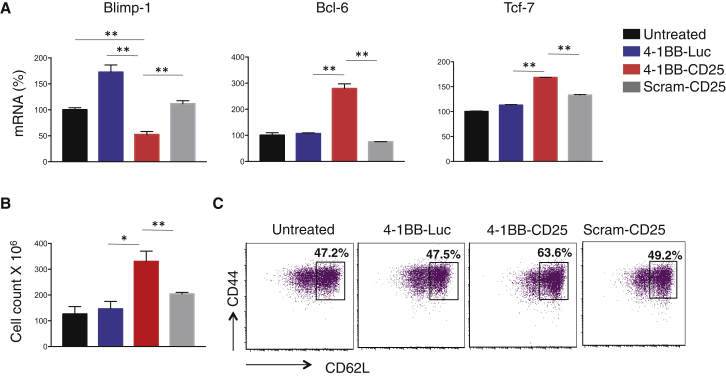
Transcription factors play an important role in effector versus memory differentiation of antigen-activated CD8+ T cells. For example, whereas Blimp-1 promotes effector differentiation, Bcl-6 and Tcf-7 favor the development of memory cells.4, 5 The level of Blimp-1, Bcl-6, and Tcf-7 mRNA were evaluated in polyclonally activated CD8+ T cells by qRT-PCR. Incubation with 4-1BB aptamer-CD25 siRNA, but neither 4-1BB aptamer-luc siRNA nor scrambled aptamer-CD25 siRNA (Figure 2A), led to the downregulation of Blimp-1 and the upregulation of Bcl-6 and Tcf-7. Thus, the transcriptional profile of 4-1BB aptamer-targeted CD25 downregulation is consistent with a memory precursor CD8+ T cell (MPEC). Interestingly we observed an increase in Blimp-1 levels with 4-1BB-luc siRNA treatment (Figure 2A), and we are currently investigating the causal factors. Consistent with the increased responsiveness of MPECs and memory cells to IL-7 due to increased expression of IL-7 receptor (IL-7R), incubation of the activated CD8+ T cells with the 4-1BB aptamer-CD25 siRNA conjugate exhibited heightened proliferation and/or reduced death in response to IL-7 (Figure 2B). Developing memory cells re-express L-selectin (CD62L) several days after activation, which enables their recirculation to the secondary lymph nodes. As shown in Figure 2C, 4-1BB aptamer-mediated reduction of CD25 expression increased the proportion of activated (CD44+) T cells expressing CD62L.
Figure 2.
4-1BB Aptamer-Targeted Downregulation of CD25 in CD8+ T Cells Promotes the Acquisition of a Memory Phenotype
Purified CD8+ cells from wild-type mice were polyclonally activated with a mixture of CD3 and CD28 antibodies and treated with conjugates three times at 500 nM concentration every 6 hr, starting at 18 hr post activation. (A) 6 days after the last treatment, the levels of Blimp-1, Bcl-6, and Tcf-7 mRNA were assessed by qRT-PCR. (B) Aptamer-siRNA-treated CD8+ T cells were incubated in IL-7-containing media, and cell numbers were counted 6 days later. (C) CD62L expression of activated (CD44+) CD8+ T cells was measured by flow cytometry 6 days after the last treatment (n = 2).
4-1BB Aptamer-CD25 siRNA Conjugates Downregulate IL-2 Signaling in CD8+ T Cells In Vivo
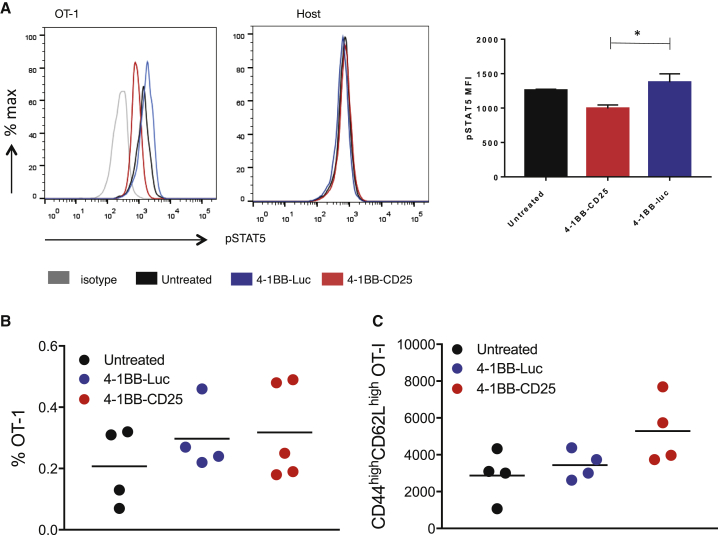
To determine if the systemic administration of 4-1BB aptamer-CD25 siRNA conjugate downregulates IL-2 signaling in circulating CD8+ T cells in mice, C57BL/6 mice were adoptively transferred with OT-I cells, transgenic CD8+ T cells specific to the dominant epitope of chicken ovalbumin (OVA), immunized with OVA peptide in the presence of lipopolysaccharide (LPS), and treated with aptamer-siRNA conjugates administered by tail-vein injection. IL-2 signaling in OT-I cells was determined by measuring the levels of phosphorylated STAT (pSTAT5) using flow cytometry. As shown in Figure 3A, 4-1BB aptamer-CD25 siRNA, but not 4-1BB aptamer-luc siRNA, treatment led to a detectable downregulation of pSTAT5 levels in the activated OT-1 that expressed 4-1BB, but not in the non-activated host CD8+ T cells that did not express 4-1BB, underscoring both the efficiency and specificity of 4-1BB aptamer-targeted delivery of siRNA in vivo. 14 days after priming, there was no overall increase in the number of OT-1 cells in the 4-1BB aptamer-CD25 siRNA-treated mice (Figure 3B), but there was a small increase that did not reach statistical significance, in the proportion of central memory CD44highCD62Lhigh OT-1 cells was seen (Figure 3C).
Figure 3.
4-1BB Aptamer-Targeted CD25 siRNA Inhibition of IL-2 Signaling in Activated CD8+ T Cells In Vivo
CD45.2 C57BL/6 mice were transferred with congenic CD45.1 OT-I cells, activated in situ with OVA peptide in the presence of LPS, and treated with the aptamer conjugates two times. 48 hr post treatment, splenocytes were analyzed by flow cytometry for pSTAT5 and CD45 expression gated on CD8+ cells. (A) Number of CD8+ (B) and CD44highCD62Lhigh (C) OT-I cells in the spleen of mice 14 days after OVA peptide priming (4–5 mice/group; n = 2).
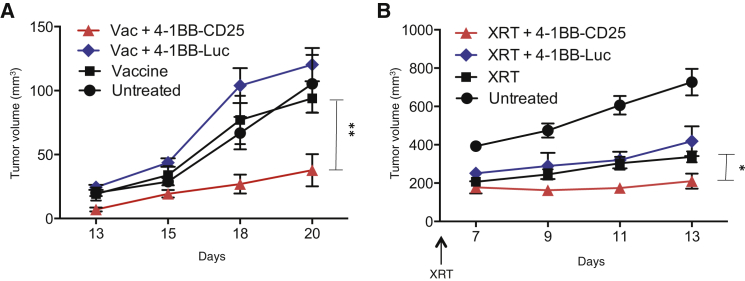
4-1BB Aptamer-CD25 siRNAs Potentiate Vaccine-Induced Antitumor Immunity in Mice
Here we tested the central hypothesis of this study: that attenuation of IL-2 signaling-mediated enhancement of the persistence of vaccine-induced antitumor immune response will engender superior protective immunity against established tumors. We used the poorly immunogenic metastatic 4T1 breast carcinoma model that was derived from a tumor induced in BALB/c mice by thioguanine treatment.16 4T1 tumors were established in mice by subcutaneous injection of tumor cells, and, 7 days post tumor inoculation, mice were either vaccinated with a cell-based vaccine consisting of a mixture of irradiated 4T1 cells expressing MHC class II and B7-1 (Figure 4A) or irradiated stereotactically at the tumor implantation site (Figure 4B). The vaccinated and irradiated mice were then treated with either 4-1BB aptamer-CD25 siRNA or 4-1BB aptamer-luc siRNA conjugates. As shown in Figures 4A and 4B, 4-1BB aptamer-CD25 siRNA, but not 4-1BB aptamer-luc siRNA, potentiated vaccine, and irradiation induced tumor inhibition. This could not have been attributed to a nonspecific proinflammatory response because administration of neither 4-1BB aptamer-CD25 siRNA nor 4-1BB aptamer-luc siRNA conjugates, in contrast to LPS, led to increased levels of tumor necrosis factor (TNF) or IL-6 in the circulation (Figure S3).
Figure 4.
Treatment with 4-1BB Aptamer-CD25 siRNA Conjugates Potentiates Vaccine-Induced Antitumor Immunity in the 4T1 Breast Carcinoma Model
BALB/c mice (5 mice/group) were implanted with 4T1 tumor cells. 7 days later, they were vaccinated with irradiated B7-1 and MHC class II-expressing 4T1 cells (A) or irradiated stereo tactically with 12 Gy at the site of tumor implantation (B). Vaccination was on days 10 and 13. On days 8, 11, and 14 after tumor implantation in vaccination experiments or days 7, 9, and 11 after radiation, mice were injected intravenously with 4-1BB aptamer-CD25 siRNA or 4-1BB aptamer-luc siRNA conjugates as indicated (5 mice/group; vaccination experiment, n = 3; irradiation experiment n = 2).
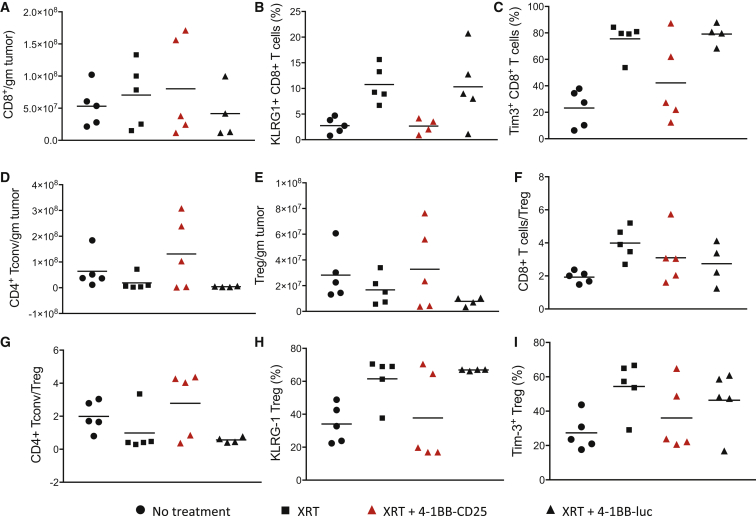
While treatment with 4-1BB-CD25 was not associated with increases in tumor-infiltrating CD8+ T cells (Figure 5A) or CD8+ T cell/regulatory T cell (Treg) ratios (Figure 5F), it prevented the radiation-induced upregulation of Tim-3 (Figure 5C) and KLRG-1 (Figure 5B), markers of advanced exhaustion in CD8+ T cells.17 Radiation led to a small reduction of CD4+ T cells in the tumor and both Foxp3 negative conventional T cells (Tconvs) (Figure 5D) and Foxp3-expressing regulatory T cells (Tregs) (Figure 5E), which was prevented by treatment with 4-1BB-CD25 conjugate. The tumor-infiltrating Tregs from mice co-treated with radiation and 4-1BB-CD25, but not 4-1BB-luc, exhibited a reduced activation status, as measured by Tim-3 (Figure 5I) and KLRG-1 expression (Figure 5H). Tim-3 expression on Tregs marks highly suppressive effector Tregs.17 The mean differences among the groups, as shown in Figure 5, have not reached statisical significance because of the large intragroup variability, commonly seen when measuring tumor infiltration of lymphocytic subsets. Nonetheless, consistent with the tumor inhibition studies (Figure 4B), these observations suggest that 4-1BB-targeted downregulation of CD25 enhances the immunogenicity of irradiated tumors in mice.
Figure 5.
Treatment with 4-1BB Aptamer-CD25 siRNA Conjugates Enhances the Immunogenicity of Irradiated Tumors
(A–I) 4T1 tumor-bearing BALB/c mice were irradiated with 10 Gy. 5 days later, they were treated with 4-1BB-CD25 or 4-1BB-luc conjugates twice 2 days apart (4–5 mice/group). 10 days after irradiation, tumors were excised and analyzed by flow cytometry (see Materials and Methods). CD4+ Tconv cells, Foxp3 negative CD4+ T cells; Treg, Foxp3-expressing CD4+ T cells (4–5 mice/group; n = 2).
4-1BB Aptamer-Targeted Inhibition of Axin-1 Improves Survival of CD8+ T Cells and Potentiates Antitumor Immunity
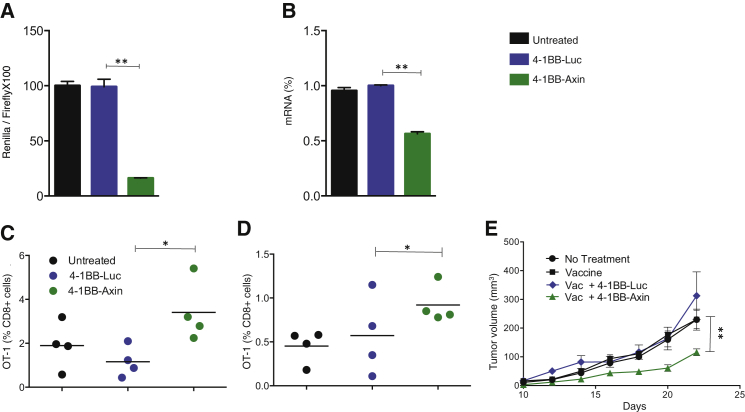
To assess the generality of 4-1BB aptamer-targeted delivery of immune modulatory siRNAs to CD8+ T cells in mice, we tested whether inhibiting Axin-1 in activated CD8+ T cells would enhance immune memory and protective antitumor immunity. Axin-1 is the rate-limiting component of the β-catenin destruction complex that negatively regulates Wnt signaling.18, 19 Wnt signaling was shown to promote the development of memory CD8+ T cells, especially memory stem cells, and improve the control of tumor growth.20 We therefore hypothesized that reduced Axin-1 expression will increase wnt signaling, leading to enhanced memory development and reduced tumor growth. We generated a 4-1BB aptamer-Axin-1 siRNA conjugate that downregulated Axin-1 targets in the ΨCHECK assay and, in a 4-1BB-dependent manner, in polyclonally activated CD8+ T cells (Figures 6A and 6B, respectively). Systemic delivery of 4-1BB aptamer-Axin-1, but not 4-1BB-luc, siRNA conjugates to mice enhanced the persistence of transferred peptide-activated OT-I cells as measured 14 and 30 days following OT-I transfer (Figures 6C and 6D, respectively) and inhibited the growth of 4T-1 tumors (Figure 6E).
Figure 6.
4-1BB Aptamer-Targeted Inhibition of Axin-1 Improves Survival of CD8+ T Cells and Enhances Antitumor Immunity
(A) 4-1BB aptamer-Axin-1 siRNA conjugate downregulation of Axin-1 targets in the ΨCHECK assay and (B) in polyclonally activated CD8+ T cells (see Materials and Methods and Figures 1A and 1B). (C and D) CD45.2 C57BL/6 mice were adoptively transferred with CD45.1 OT-I CD8+ T cells, activated with OVA peptide and LPS, and treated with 4-1BB aptamer-Axin-1 siRNA or 4-1BB aptamer-luc siRNA. OT-I cells in the blood and spleen were measured by flow cytometry 14 days (C) and 30 days (D) post transfer, respectively. (E) 4T-1 tumors were established subcutaneously in BALB/c mice, vaccinated with a mixture of irradiated 4T1 cells expressing MHC class II or B7-1, and treated with 4-1BB aptamer-Axin-1 siRNA or 4-1BB aptamer-luc siRNA as described in Materials and Methods and Figure 4A (C and D, 4 mice/group, n = 2; E, 5 mice/group, n = 3).
We have previously shown that 4-1BB aptamer-targeted downregulation of raptor, a key component of the mTORC1, in tumor-bearing mice inhibited tumor growth in the 4T1 model.11 As shown in Figure S4A, targeted inhibition of Axin-1 was more effective than that of raptor, and inhibition of Axin-1 or CD25 exhibited a comparable antitumor effect (Figure S4B).
Discussion
IL-2 signaling is a key upstream mediator of effector/memory differentiation of CD8+ T cells.7, 8, 9 Engagement of the IL-2 receptor by its ligand IL-2 leads to the upregulation of Blimp-1, a transcription factor that promotes the differentiation of antigen-activated CD8+ T cells to become short-lived effectors.9 Reduced IL-2 signaling leads to the upregulation of a number of transcription factors known to promote memory differentiation, such as Bcl-6 or Tcf-7, resulting in the development of long-lived memory cells.4, 5 Whereas IL-2 signaling can be attenuated using blocking anti-CD25 antibodies, the systemically administered antibody can also interfere with the function of the CD25-expressing CD4+Foxp3+ regulatory T cells and thereby increase the risk of autoimmune pathology. In this study, we limited CD25 downregulation to CD8+ T cells using an siRNA that was targeted to antigen-activated CD8+ T cells by conjugation to a 4-1BB-binding aptamer. Of note, in this study, we used a monovalent 4-1BB aptamer that did not costimulate 4-1BB-expressing T cells (Berezhnoy et al.11 and data not shown). In vitro, the 4-1BB aptamer-CD25 siRNA conjugate downregulated CD25 expression in polyclonally activated CD8+ T cells (Figure 1), which led to the acquisition of memory characteristics consistent with reduced IL-2 signaling, specifically downregulation of Blimp-1, upregulation of Bcl-6 and Tcf-7, enhanced response to IL-7, and upregulation of CD62L (Figure 2). In vivo, systemic administration of 4-1BB aptamer-CD25 siRNA conjugates resulted in reduced IL-2 signaling in adoptively transferred 4-1BB-expressing OT-I cells (Figure 3), but not 4-1BB negative host cells, and potentiated vaccine- and irradiation-induced tumor immunity in the 4T1 breast carcinoma tumor model (Figure 4). Our study, therefore, shows that 4-1BB aptamer delivery of CD25 siRNA to activated circulating CD8+ T cells in vivo is efficient and specific, leading to enhanced memory differentiation and improved control of tumor growth.
The generality of aptamer-targeted siRNA delivery to promote the differentiation of CD8+ T cells was previously demonstrated using 4-1BB aptamers conjugated to raptor siRNA to reduce raptor expression, and hence mTORC1 activity, in activated CD8+ T cells as an alternative approach to promote their long-term persistence.11 In this study, we extend the generality of this approach, showing that Axin-1 expression can be downregulated in CD8+ T cells using 4-1BB aptamer-Axin siRNA conjugates, which extended the persistence of the CD8+ T cells and potentiated vaccine-induced antitumor immunity (Figure 6). siRNA delivery to hematopoietic cells has been notoriously difficult and inefficient.21 Our previous and current study suggest that aptamer-targeted siRNA delivery to hematopoietic cells could represent a highly efficient approach to manipulate the fate of normal and malignant hematopoietic cells in vivo.
Materials and Methods
6–8-week-old female C57BL/6 (H-2b), BALB/c (H-2d), and transgenic OT-I (H-2b) mice were purchased from The Jackson Laboratory. The facilities at the Division of Veterinary Resources, University of Miami, are fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care and the USDA. An Office of Laboratory Welfare (OLAW) assurance is on file, ensuring that humane animal care and use practices, as outlined in the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85-23. Revised 1985) were followed. All mice were maintained according to the guidelines established by the US Department of Agriculture and the American Association for Accreditation of Laboratory Animal Care (AAALAC). This project was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Miami School of Medicine.
Design and Characterization of Aptamer-siRNA Conjugates
The DNA template for the murine 4-1BB aptamer gggggaattctaatacgactcactataGGGAGAGAGGAAGAGGGATGGGCGACCGAACGTGCCCTTCAAAGCCGTTCACTAACCAGTGGCATAACCCAGAGGTCGATAGTACTGGATCCCCCCGTACATTCTAGATAGCC encoding a T7 promoter (lower case) and a 17 nucleotide 3′ extension (underlined) for hybridization to a complementary sequence engineered at the 5′ end of the siRNA sense strand was transcribed in vitro using the Durascribe T7 transcription kit (Epicenter). For the scrambled aptamer, gggggaattctaatacgactcactataGGGAGAGAGGAAGAGGGATGGGCAGTGCGTACGTATGGCATGCATGATCGTTACCGTATCGTCATAACCCAGAGGTCGATAGTACTGGATCCCCCCGTACATTCTAGATAGCC, as generated by Sequence Scramble algorithm (GenScript) was prepared in the same way as 4-1BB aptamer. Candidate siRNA sequences against CD25 or Axin mRNA were selected using online prediction algorithms, siRNA scales (Department of Human Genetics, University of Utah), and siDESIGN (Dharmacon, Thermo Fisher Scientific). Sequences with low melting temperature15 predicted to show a good inhibition by both algorithms were chosen and purchased as synthetic RNA oligos (Integrated DNA Technologies) with a 5′ complementary sequence to the 3′ engineered sequence in the aptamer. The siRNA was annealed to the 4-1BB aptamer and screened for target inhibition in the ΨCHECK assay (Promega). For control, GL3 duplex siRNA against luciferase mRNA was used. For psiCHECK assays, EGFP siRNA was used as a control siRNA.
ΨCHECK Assay
Synthetic DNA sequences (Integrated DNA Technologies) corresponding to the tested siRNA sequences were ligated into the 3′ end of the gene for Renilla luciferase in the ΨCHECK-2 vector. Chinese hamster ovary (CHO) cells were co-transfected with 100 ng of the ΨCHECK vector and 0.5 pmol of siRNA or aptamer-siRNA conjugates in a 24-well plate using Lipofectamine 2000 (Thermo Fisher Scientific). Renilla luciferase luminescence was measured 24 hr later (Perkin Elmer WALLAC 1420 Victor2). Firefly gene activity from the same vector was used as transfection control.
Treatment of Polyclonally Activated Cells with Aptamer-siRNA Conjugates In Vitro
Splenocytes were isolated from an 8-week-old C57BL/6 mouse. For qPCR, CD8+ T cells were purified by positive selection using CD8+ microbeads (Miltenyi Biotech). The purified CD8+ T cells were activated at 1 × 106 cells/mL in a 96-well plate using 1 μg/mL of plate-bound anti-CD3 (145-2C11) and 0.5 μg/mL of soluble anti-CD28 (37.51) antibodies (BD Pharmigen). The cells were treated with aptamer-siRNA conjugates at 500 nM three times, 6 hr apart. 24 hr after the last treatment, the cells were lysed, and the total RNA was extracted using QIAGEN RNeasy mini kit (QIAGEN). Subsequently, the extracted RNA was converted to cDNA using a high capacity cDNA reverse transcriptase kit (Applied Biosystems). cDNA obtained from 25 ng RNA was used per reaction in a TaqMan qPCR assay using the Step One qPCR machine (Applied Biosystems), with primer sets corresponding to the gene of interest or housekeeping products.
For flow cytometry, total splenocytes after RBC lysis were plated at 1 × 106 cells/mL in a 24-well plate and activated with 1 μg/ml of soluble anti-CD3 antibody (145-2C11, BD Pharmigen). Starting 24 hr after activation, the cells were treated with conjugates at 500 nM three times, 6 hr apart. Live CD8+ T cells were analyzed 48 hr after the last treatment using live/dead marker (fixable viability dye eflour780), CD8b-PerCP/Cy5.5, and CD25-PE antibodies. To assess for the levels of CD62L, cells were stained 6 days after the last treatment with live/dead marker, CD8b-PerCP/Cy5.5, CD44-eflour450, and CD62L-antigen-presenting cell (APC) antibodies.
Expression of 4-1BB on T Cells
Total splenocytes from an 8-week-old C57BL/6 mouse after RBC lysis were plated at 1 × 106 cells/mL in a 24-well plate and activated with 1 ug/mL of soluble anti-CD3 antibody (145-2C11, BD Pharmigen). 24 hr following activation, the activated cells were mixed in 1:1 ratio with fresh naive splenocytes isolated from mouse. Live CD8+ T cells were analyzed using live/dead marker (fixable viability dye eflour780), CD8b-APC, CD44-ef450, and CD25-PEcy7 antibodies. For staining with antibody, flourochrome-conjugated 4-1BB-PE antibody and its corresponding isotype-PE control was used. For aptamer staining, biotin oligo complementary to the 3′ end of the aptamer was annealed to the aptamer and incubated at room temperature with streptavidin-PE for 15 min to label the aptamer.
T Cell Proliferation in the Presence of IL-7
Total splenocytes after RBC lysis were plated at 1 × 106 cells/mL in a 24-well plate and activated with 1 μg/mL of soluble anti-CD3 antibody (145-2C11). Starting 24 hr after activation, the cells were treated with conjugates at 500 nM three times, 6 hr apart. 48 hr after the last treatment, 106 cells were incubated in 5 mL of media containing 5 ng/mL recombinant IL-7 (R&D Systems). Every alternate day, live cells were counted and 106 cells were passaged into 5 mL of fresh media containing IL-7.
Adoptive Transfer of OT-1 Cells into Wild-Type Mice and Treatment with Aptamer-siRNA Conjugates
Splenocytes from 8-week-old CD45.1 OT-I mice were isolated and enriched for CD8+ T cells using Ly-2 microbeads (Miltenyi Biotech) to greater than 90% purity, as verified by flow cytometry. CD8+ cells (0.5 × 106) were transferred into age-matched CD45.2 C57BL/6 mice by tail-vein injection. The transferred cells were activated the next day by tail-vein injection of 10 μg SIINFEKL peptide (Anaspec), the chicken OVA dominant class I epitope, and 10 μg of Escherichia coli LPS (Sigma-Aldrich) in 200 μL PBS. The animals received intravenous (i.v.) injections of 1.5 nmol of aptamer-siRNA conjugate in 200 μL PBS 6 and 24 hr after peptide administration. To measure the levels of pSTAT5, splenocytes were isolated 48 hr after the second aptamer-siRNA conjugate injection and fixed in 4% PFA for 10 min in a 37°C water bath. The cells were then centrifuged and permeabilized using 500 μL of 100% methanol for 30 min on ice. After two washes, the cells were stained using CD8-PE, CD45.1-eflour450, and pSTAT5-Alexaflour647 antibodies (BD Pharmingen).
Tumor Immunotherapy Studies
BALB/c mice were injected subcutaneously (s.c.) with 1.0 × 104 4T1 tumor cells and immunized with a mixture of irradiated B7-1 and MHC class II-expressing 4T1 cells22 on days 7, 10, and 13 after tumor implantation. They were treated with 1.5 nmol of aptamer-siRNA conjugates on days 8, 11, and 14, injected i.v. Tumor measurements were taken every other day. For experiments involving radiation, tumors of an approximate volume of 100 mm3 were radiated stereotactically with 12 Gy and treated with 1.5 nmol of aptamer-siRNA conjugates on days 7, 9, and 11 after radiation. Tumor measurements were made every other day.
Analysis of Tumor-Infiltrating T Cells
BALB/c mice were implanted subcutaneously with 4T1 tumor cells and irradiated stereotactically with 10 Gy at the site of tumor implantation when the tumors reached 75 mm3. On days 5 and 7 after radiation, mice were injected intravenously with 4-1BB aptamer-CD25 siRNA or 4-1BB aptamer-luc siRNA conjugates as indicated. 10 days after irradiation, tumors were excised. Tumor-infiltrating T cells (TILs) were isolated by dissecting tumor tissue into small fragments, followed by digestion with 1 mg/mL of collagenase in complete RPMI media (10%FBS, 1% penicillin, streptomycin) prior to using Gentle MACS Dissociator. Cell suspension was passed through a 70 μm nylon strainer to obtain a single cell population. TILs were then washed twice with FACS buffer and stained as described above and analyzed by flow cytometry.
Flow Cytometry
Cells were incubated with eFluor780 for 30 min at 4°C and washed twice in FACS buffer prior to staining in order to distinguish live cells from dead cells. Cells were then incubated with Fc block for 10 min at 4°C and incubated for 30 min at 4°C with CD3 FITC, CD4 PerCP Cy5.5, CD25 PE-Cy7, CD62L eFluor 450, Foxp3 PE, Tim3 APC, CD127 AF700, CD8 BV605, and CD44 BV510. Antibodies were obtained from eBiosciences and Bio Legend. For intracellular staining, cells were fixed and permeabilized with Foxp3 staining buffer kit (eBioscience) according to the manufacturer’s instructions. Cells were incubated for 1 hr at room temperature in the dark with anti-Foxp3 PE-labeled antibody. Cells were collected and analyzed using CytoFLex (BD Biosciences) and Kaluza software, respectively.
Statistics
Unpaired two-tailed Student’s t tests were performed between individual treatment groups using GraphPad Prism, version 5.0 (GraphPad Software). p values ≤ 0.05 were considered statistically significant: *p < 0.05; **p < 0.005.
Author Contributions
A.R. was responsible for constructing conjugates and immunotherapy experiments; A.B. was responsible for designing conjugate and evaluating their characterization; B.S. was responsible for the flow cytometry experiments; Y.P.-D. was responsible for the qRT-PCR analysis of intracellular mediators; and E.G. was responsible for overall research design, experimental approaches, and writing the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by NIH NCI grant R01CA181598. We thank Thomas Malek (Department of Microbiology and Immunology, University of Miami), Bhavna Verma (Sylvester Comprehensive Cancer Center, University of Miami), and Shradha Patel (Sylvester Comprehensive Cancer Center, University of Miami) for their assistance and advice.
Footnotes
Supplemental Information includes four figures and can be found online at http://dx.doi.org/10.1016/j.ymthe.2016.10.021.
Supplemental Information
References
- 1.Klebanoff C.A., Gattinoni L., Restifo N.P. CD8+ T-cell memory in tumor immunology and immunotherapy. Immunol. Rev. 2006;211:214–224. doi: 10.1111/j.0105-2896.2006.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinrichs C.S., Gattinoni L., Restifo N.P. Programming CD8+ T cells for effective immunotherapy. Curr. Opin. Immunol. 2006;18:363–370. doi: 10.1016/j.coi.2006.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joshi N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., Kaech S.M. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang J.T., Wherry E.J., Goldrath A.W. Molecular regulation of effector and memory T cell differentiation. Nat. Immunol. 2014;15:1104–1115. doi: 10.1038/ni.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaech S.M., Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gattinoni L., Klebanoff C.A., Restifo N.P. Pharmacologic induction of CD8+ T cell memory: better living through chemistry. Sci. Transl. Med. 2009;1:11ps12. doi: 10.1126/scitranslmed.3000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalia V., Sarkar S., Subramaniam S., Haining W.N., Smith K.A., Ahmed R. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Pipkin M.E., Sacks J.A., Cruz-Guilloty F., Lichtenheld M.G., Bevan M.J., Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rutishauser R.L., Martins G.A., Kalachikov S., Chandele A., Parish I.A., Meffre E., Jacob J., Calame K., Kaech S.M. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi T., Wing J.B., Sakaguchi S. Two modes of immune suppression by Foxp3(+) regulatory T cells under inflammatory or non-inflammatory conditions. Semin. Immunol. 2011;23:424–430. doi: 10.1016/j.smim.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Berezhnoy A., Castro I., Levay A., Malek T.R., Gilboa E. Aptamer-targeted inhibition of mTOR in T cells enhances antitumor immunity. J. Clin. Invest. 2014;124:188–197. doi: 10.1172/JCI69856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNamara J.O., Kolonias D., Pastor F., Mittler R.S., Chen L., Giangrande P.H., Sullenger B., Gilboa E. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J. Clin. Invest. 2008;118:376–386. doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C., Lin G.H., McPherson A.J., Watts T.H. Immune regulation by 4-1BB and 4-1BBL: complexities and challenges. Immunol. Rev. 2009;229:192–215. doi: 10.1111/j.1600-065X.2009.00765.x. [DOI] [PubMed] [Google Scholar]
- 14.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berezhnoy A., Brenneman R., Bajgelman M., Seales D., Gilboa E. Thermal Stability of siRNA Modulates Aptamer- conjugated siRNA Inhibition. Mol. Ther. Nucleic Acids. 2012;1:e51–e59. doi: 10.1038/mtna.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heppner G.H., Miller F.R., Shekhar P.M. Nontransgenic models of breast cancer. Breast Cancer Res. 2000;2:331–334. doi: 10.1186/bcr77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson A.C. Tim-3: an emerging target in the cancer immunotherapy landscape. Cancer Immunol. Res. 2014;2:393–398. doi: 10.1158/2326-6066.CIR-14-0039. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi A. Roles of Axin in the Wnt signalling pathway. Cell. Signal. 1999;11:777–788. doi: 10.1016/s0898-6568(99)00054-6. [DOI] [PubMed] [Google Scholar]
- 19.Moon R.T. Wnt/beta-catenin pathway. Sci. STKE. 2005;2005:cm1. doi: 10.1126/stke.2712005cm1. [DOI] [PubMed] [Google Scholar]
- 20.Gattinoni L., Zhong X.S., Palmer D.C., Ji Y., Hinrichs C.S., Yu Z., Wrzesinski C., Boni A., Cassard L., Garvin L.M. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat. Med. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peer D. A daunting task: manipulating leukocyte function with RNAi. Immunol. Rev. 2013;253:185–197. doi: 10.1111/imr.12044. [DOI] [PubMed] [Google Scholar]
- 22.Baskar S., Glimcher L., Nabavi N., Jones R.T., Ostrand-Rosenberg S. Major histocompatibility complex class II+B7-1+ tumor cells are potent vaccines for stimulating tumor rejection in tumor-bearing mice. J. Exp. Med. 1995;181:619–629. doi: 10.1084/jem.181.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.