Abstract
Electrotransfection is a widely used method for delivering genes into cells with electric pulses. Although different hypotheses have been proposed, the mechanism of electrotransfection remains controversial. Previous studies have indicated that uptake and intracellular trafficking of plasmid DNA (pDNA) are mediated by endocytic pathways, but it is still unclear which pathways are directly involved in the delivery. To this end, the present study investigated the dependence of electrotransfection on macropinocytosis. Data from the study demonstrated that electric pulses induced cell membrane ruffling and actin cytoskeleton remodeling. Using fluorescently labeled pDNA and a macropinocytosis marker (i.e., dextran), the study showed that electrotransfected pDNA co-localized with dextran in intracellular vesicles. Furthermore, electrotransfection efficiency could be decreased significantly by reducing temperature or treatment of cells with a pharmacological inhibitor of Rac1 and could be altered by changing Rac1 activity. Taken together, the findings suggested that electrotransfection of pDNA involved Rac1-dependent macropinocytosis.
Keywords: endocytosis, electroporation, electrogene transfer
Mao et al. demonstrated that macropinocytosis of plasmid DNA was important for electrotransfection of cells in vitro. They further showed that the macropinocytosis was Rac1 dependent.
Introduction
Gene therapy has shown promising results in treatment of various genetic disorders and acquired diseases.1, 2, 3, 4, 5 The treatment requires delivery of therapeutic genes into nucleus of specific cells in tissues, which must overcome several physical and biological barriers.6 One such barrier is the plasma membrane of cell, which physically limits material exchange between intra- and extracellular environments. In addition, both DNA and plasma membrane are negatively charged, which means that the repulsive electrostatic force could hinder delivery of naked DNA into cells as well.7
Various gene delivery methods have been developed to circumvent the barriers.8 In general, viral-based methods are more efficient and capable of integrating certain desired genes into the host genome for sustained gene expression. However, the viral-based systems have a number of drawbacks, including immunogenicity, cytotoxicity, and high cost.9 To overcome these problems, non-viral methods have been developed,10, 11, 12, 13, 14 and one such method is electrotransfection (or electro-gene transfer).15, 16, 17, 18, 19, 20, 21 Comparing with viral-based methods, electrotransfection does not require packing of genes into vectors and is less immunogenic. The technique relies on delivery of a sequence of electric pulses to target cells or tissues placed between electrodes, which leads to the entry of cell-impermeable molecules, including plasmid DNA (pDNA).22, 23 Although electrotransfection has been implemented as a powerful tool in both basic research and clinical applications,24, 25, 26, 27, 28, 29, 30, 31, 32 the exact mechanism of electro-gene transfer is still largely unknown.33, 34, 35, 36 One of the most popular mechanisms, known as the “pore theory,” states that, when electric-field-induced transmembrane potential exceeds a certain threshold, transient pores will form in the plasma membrane,37, 38, 39, 40, 41, 42 allowing extracellular molecules to enter cytoplasm through diffusion, electrophoresis, and/or electro-osmosis.43 Thus, the technique has also been called electroporation in the literature.17 Although the “pore theory” may explain how small molecules enter cells, it has failed to explain cellular uptake of larger biopolymers, especially pDNA. The lifetime and the size of electrically induced pores that would allow pDNA to pass through are still under debate because it is currently infeasible to measure them directly.17, 34, 44, 45
Whereas controversies remain over the biophysical nature of electrotransfection, there has been increasing evidence from various studies implicating the involvement of biological processes in the uptake and intracellular transport of electrotransfected pDNA.46, 47 Specifically, electrotransfection may involve endocytic pathways,48 which are essential for nutrient uptake and regulation of receptor-mediated cell signaling. Based on the types of vesicles involved in molecular transport, endocytosis can be divided into several categories, and one of them is macropinocytosis, which is a mode of non-selective fluid phase endocytosis.49, 50 The process requires ATP-fueled, actin-dependent membrane ruffling. Vesicles associated with macropinocytosis are called macropinosomes, which are large (0.2–5 μm in diameter) uncoated vesicles that allow cells to internalize a significant amount of extracellular solutes, especially macromolecules.51, 52 Macropinocytosis is involved in many important cellular events, including amino acid uptake,53 entry of viral particles,54 and antigen responses of immune cells.49 The present study was designed to determine the role played by macropinocytosis in electrotransfection and how the small GTPase Rac1 regulates the endocytic process.
Results
Electric Pulses Induced Actin Remodeling
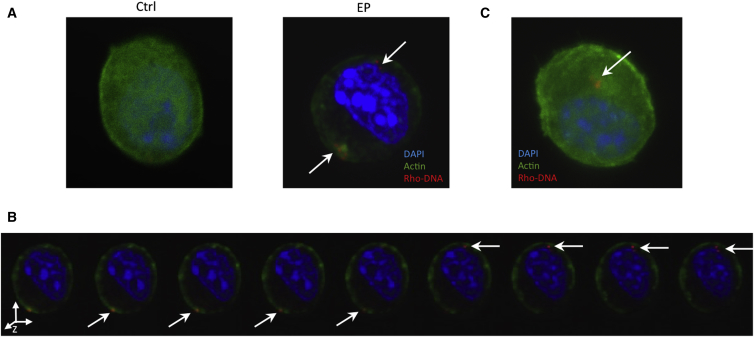
We first examined whether electric pulses could lead to actin remodeling because macropinocytosis is actin-dependent. To visualize potential changes in cytoskeletal structures, we used a B16.F10 melanoma cell line stably transfected with Lifeact-GFP, a fluorescent probe for F-actin.55 The pDNA molecules were covalently labeled with a red fluorescent dye, tetramethylrhodamine, to track their locations. Comparing with non-treated control cells, in which actin filaments were evenly distributed in the cytosol, exposure of cells to electric pulses in the presence of DNA led to formation of various structures of F-actin, including punctate-like aggregates, membrane ruffles and filopodia-like protrusions (see Figures 1A and S1A–S1C). The actin remodeling was also observed in the absence of pDNA (Figure S1D), confirming that it was caused by electric pulses. The results suggested that the electric pulse might trigger macropinocytosis because F-actin remodeling is a prerequisite for the process.
Figure 1.
Electric-Pulse-Induced Actin Remodeling and Uptake of Plasmid DNA
pDNA (pEGFP-N1) covalently labeled with rhodamine (red) was electrotransfected (450 V/cm; 5 ms; eight pulses; 1 Hz) into B16.F10 cells stably expressing the F-actin probe Lifeact-GFP (green). (A) The panel shows single optical slices from confocal images of a non-pulsed, control cell and a cell electrotransfected with pDNA at 37°C, respectively. (B) The z stack images show three-dimensional distribution of pDNA in the cytoplasm of an electrotransfected cell at 37°C. (C) The panel shows single slice from a confocal image of electrotransfected cell subjected to cold medium treatment for 10 min is shown. Arrows in all images denote pDNA internalized by cells.
Dual-fluorescence channel analysis revealed that, after electrotransfection, pDNA was enriched in multiple vesicle-like structures. Notably, co-localization of actin and pDNA was observed near the plasma membrane shortly after electrotransfection, and some pDNA aggregates were surrounded by shell-like actin structures (see Figure 1B). It was also worth to mention that the fluorescence intensity of Lifeact-GFP in cytoplasm decreased after electrotransfection, except for regions adjacent to the plasma membrane, suggesting that electric pulsing caused de-polymerization of actin because Lifeact binds mainly to filamentous, polymerized actin (F-actin). Overall, these results suggested that actin was likely to be responsible for engulfment of pDNA.
Both F-actin remodeling and endocytosis depend on energy generated from ATP hydrolysis. Therefore, we hypothesized that these processes should be halted if the cells were placed on ice for a short period (i.e., 10 min) after electric pulse application. As predicted, the results from Figure 1C showed that F-actin was ubiquitously distributed within the cell, which meant that de-polymerization was inhibited at least partially by the cold temperature. In parallel, less pDNA uptake was observed in these cells, compared to those at 37°C.
Electric Pulse-Induced Macropinocytosis of Macromolecules
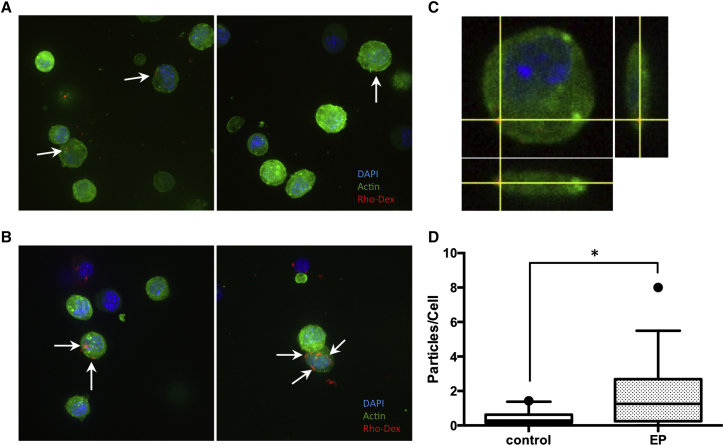
To further confirm that the electrotransfection-induced actin remodeling was responsible for macropinocytosis, we investigated cellular uptake of a fluorescently labeled dextran, a fluid phase marker for macropinocytosis.56 Because the pDNA used in our study had a molecular weight of 3,000,000 and was negatively charged in solution, we chose an anionic dextran with molecular weight of 2,000,000 as the molecular probe. Due to relatively large size and lacking of membrane receptors, dextran molecules could only be taken up by macropinocytosis. If electrotransfection caused upregulation of macropinocytosis, we would expect more uptake of dextran after cells were electrically pulsed in the presence of dextran. Data shown in Figure 2A indicated that, without application of electric pulses, a small amount of dextran molecules were internalized by cells. The punctate distribution suggested that they were internalized by macropinocytosis. When cells were treated with electric pulses in the presence of dextran, we observed remodeling of actin and an increase in the uptake of dextran over the same period as that for the non-pulsed controls (Figures 2B–2D). The pattern of dextran punctate distribution (Figure 2B) was similar to that of pDNA shown in Figure 1A, suggesting that electrotransfection could promote actin remodeling and enhance macropinocytosis.
Figure 2.
Electric-Pulse-Induced Macropinocytosis
(A) B16.F10 cells expressing Lifeact-GFP in control group were incubated with tetramethylrhodamine (red)-labeled dextran at 37°C for 10 min, without exposure to electric pulses. (B) The same cells were treated with eight electric pulses at 450 V/cm, 5 ms, and 1 Hz (i.e., the EP group) in the presence of tetramethylrhodamine labeled dextran. After treatment, cells were incubated at 37°C for 10 min to allow uptake of molecules. At the end of incubation, cells were washed with PBS, fixed by paraformaldehyde, and examined with confocal microscopy. Arrows in the images denote dextran. (C) The panel shows an orthogonal view of a B16.F10 cell after treatment under the same conditions as those in (B). Two optical cross-sections in x-z and y-z planes are shown in the panel to demonstrate the internalization of dextran. (D) The panel shows quantitative measurements of cellular dextran uptake. They were performed by counting the numbers of tetramethylrhodamine-positive particles in maximum projection images of each cell from the control and EP groups. The results were presented as box-and-whisker plots showing 10th–90th percentile. n = 14; *p < 0.05 (Mann-Whitney U test).
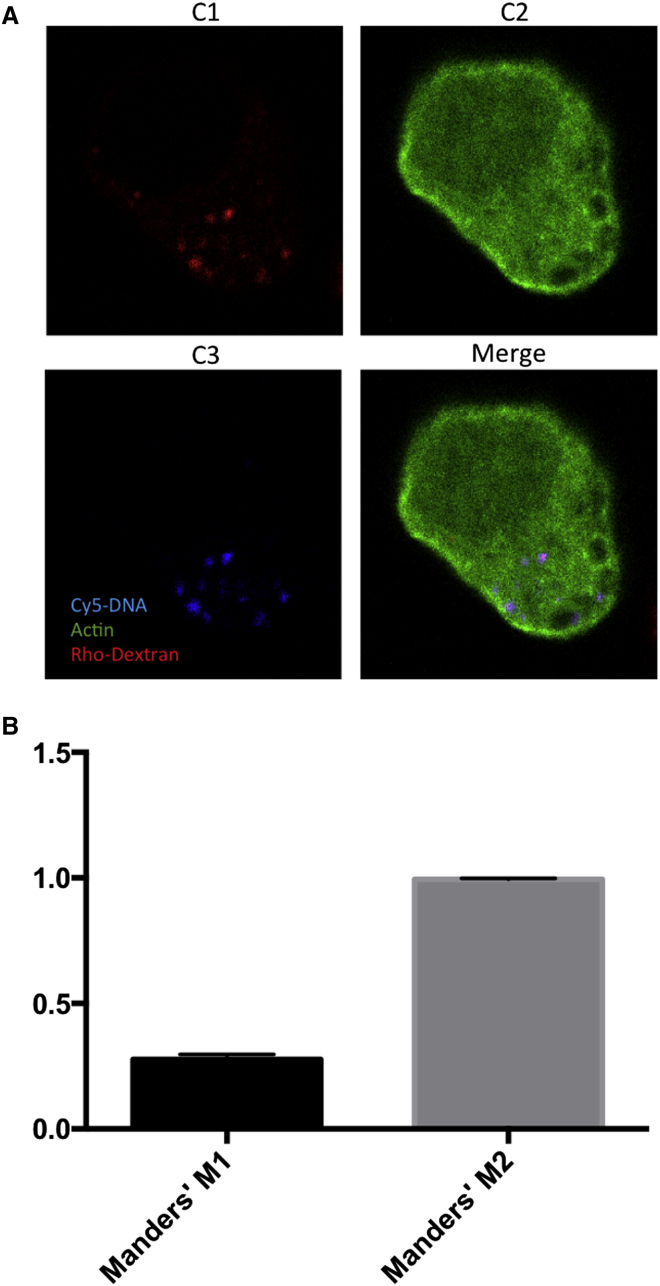
Next, we investigated whether the pDNA-containing vesicles observed in Figure 1 were identical to the vesicles observed in Figure 2. For this purpose, we electrotransfected B16.F10 Lifeact-GFP cells in the presence of a mixture of Cy5-labeled pDNA and tetramethylrhodamine-labeled dextran. The pulsed cells were examined by confocal fluorescence microscopy. If punctate structures of pDNA and dextran were of the same type, then we would expect the Cy5 fluorescence signal to overlap with the tetramethylrhodamine signal. As shown by the images in Figure 3A, the majority of the electrotransfected pDNAs (blue color) co-localized with dextran (red color). We then quantified the correlation of the two channels by Manders’ method,57, 58 which evaluates the fraction of all pixels in a z stack of confocal images from one channel that co-localizes with those from another channel by calculating the Manders’ co-localization coefficient (MCC), which varies between 0 (no co-localization) and 1 (perfect co-localization). The Manders’ M1 coefficient was 0.277 ± 0.062, indicating that approximately 28% of red pixels co-localized with blue pixels (see also Figure 3B). On the other hand, the Manders’ M2 (0.994 ± 0.011) indicated that almost 100% of pDNA co-localized with dextran. The higher value of M2 relative to M1 could be explained by the fact that we put more dextran in the pulsing buffer than pDNA. Together, the data confirmed our hypothesis that pDNA shared the same endocytic pathway as that for dextran to enter cells during electrotransfection.
Figure 3.
Co-localization of Electrotransfected Plasmid DNA with Dextran
(A) The panel shows a representative confocal image of B16.F10 cells expressing Lifeact-GFP. The cells were treated with electric pulses in the presence of both tetramethylrhodamine-labeled dextran and Cy5-labeled pEGFP. Live images were taken at 10 min after electrotransfection. Pseudo colors were assigned to each channel for visualization: red for tetramethylrhodamine; green for EGFP; and blue for Cy5, which are shown in images C1, C2, and C3, respectively. The colors are merged in the fourth image. (B) The panel shows the characterization of co-localization by Manders’ co-localization coefficient (MCC). M1 represents the fraction of all pixels in a z stack of confocal images from channel 1 (red) that co-localized with those from channel 2 (blue). M2 represents the fraction of all pixels from channel 2 (blue) that co-localize with those from channel 1 (red); n = 9. The bar and error bar represent mean and SEM, respectively.
Inhibition of Macropinocytosis Decreased the Efficiency of Electrotransfection
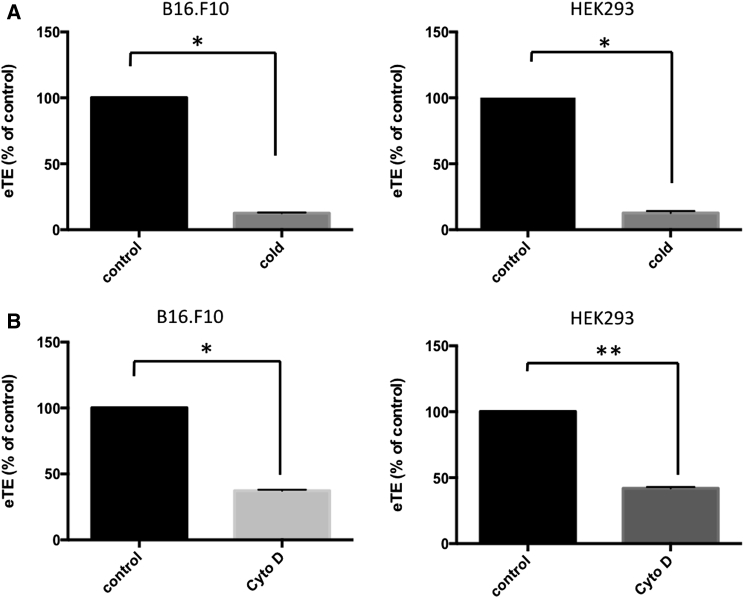
Although our results indicated that macropinocytosis was a possible route for uptake of electrotransfected pDNA, it was still unclear whether pDNA molecules carried in macropinosomes could eventually reach nuclei for expression. In other words, the results described above did not show that macropinocytosis was responsible for successful gene delivery. To address this issue, we evaluated changes in the overall transfection efficiency after inhibition of macropinocytosis, using the luciferase assay. As shown in Figures 4A and S2A, inhibition of endocytosis by treatment of cells with cold medium for 10 min resulted in decreases in electrotransfection efficiency (eTE) in different cell lines: 80% in B16.F10 cells; 90% in HEK293 cells; and 70% in COS7 cells.
Figure 4.
Reduction of eTE after Inhibition of DNA Uptake through Macropinocytosis
To determine the eTE, luciferase assay was performed on cell lysates at 24 hr after electrotransfection. Luminescence readings (LU) of each sample were normalized by total protein concentration to obtain relative luminescence units (RLU). eTE was defined as the percentage of RLU relative to control. (A) The panel shows effects of cold medium treatment on eTE in B16.F10 and HEK293 cells. After electrotransfection of pcDNA3.1(+) Luc2 = tdT, the cells were divided into two groups. The control groups were incubated at 37°C for 10 min to allow uptake of pDNA. The cold treatment group was incubated on ice for 10 min to minimize macropinocytosis. After incubation, cells were plated in 6-well plates and cultured for 24 hr to express luciferase. (B) The panel shows effects of actin inhibitor, cytochalasin D (Cyto D), on eTE in B10.F10 and HEK293 cells. Cells were pretreated with DMSO (i.e., control) or 10 μM Cyto D for 1 hr prior to electrotransfection with pcDNA3.1(+) Luc2 = tdT. n = 4–6. *p < 0.05 and **p < 0.005 (Mann-Whitney U test).
In addition, we investigated effects of ablation of actin cytoskeleton on eTE. It is known that actin remodeling requires polymerization of G-actin to form F-actin. Therefore, we hypothesized that, if we inhibited the process by a pharmacological inhibitor, cytochalasin D (Cyto D),59 we could reduce the pDNA uptake through macropinocytosis. Indeed, our experimental data revealed that pretreatment of cells with Cyto D decreased eTE in B16.F10 and HEK293 cells by 60% and 50%, respectively (Figure 4B). These results were consistent with previous reports about the inhibitory effect of Cyto D on electric-field-induced endocytosis.60
DNA Uptake by Electrotransfection-Induced Macropinocytosis Is Dependent on Rac1 Activity
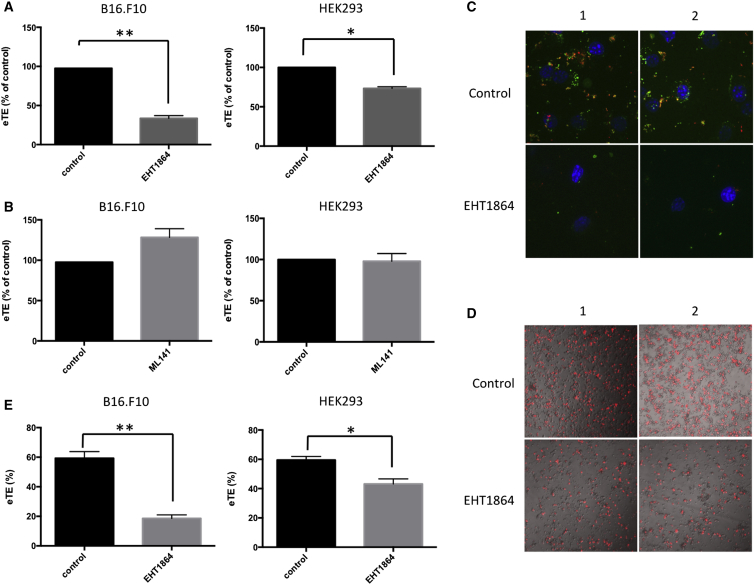
Next, we sought to elucidate molecular mechanisms of electrotransfection-induced macropinocytosis. Several signaling proteins, including PAK-1, Arf6, and Rho family GTPases, are known to regulate the initiation of macropinocytosis.61 Additionally, Rho family GTPases are well recognized for their complex interaction networks and multiple roles played in regulation of actin cytoskeleton assembly.62 It has also been reported that two of the Rho family GTPases, cell division control protein 42 (Cdc42) and Rac1, can regulate cell motility,63, 64, 65 phagocytosis,66 and macropinocytosis.67 Therefore, we hypothesized that electrotransfection could lead to actin remodeling and enhance macropinocytosis through activation of Cdc42 and Rac1. To test this hypothesis, we performed experiments with two pharmacological inhibitors, ML14168 and EHT1864,69 that targeted Cdc42 and Rac1, respectively. We chose these inhibitors because they are potent, selective, and most importantly reversible, which allowed us to observe the inhibitory effects without permanently interfering with cell signaling. Results from the luciferase assay shown in Figures 5A and S2B revealed that only the Rac1 inhibitor EHT1864 had pronounced effects on eTE. It decreased transfection efficiency by as much as 70% in B16.F10 cells, 30% in HEK293 cells, and 50% in COS7 cells. However, ML141 minimally affected the eTE in both B16.F10 and HEK293 cells (Figure 5B). These results suggested that Rac1 was more important than Cdc42 in controlling electrotransfection efficiency in these cell lines.
Figure 5.
Reduction of eTE after Pharmacological Inhibition of Rac1-Dependent Macropinocytosis
Cells were pretreated with an inhibitor or equivalent volume of DMSO (i.e., control) for 1 hr prior to electrotransfection with the pcDNA3.1 (+) Luc2 = tdT. After electrotransfection, cells were plated in 6-well plates and cultured for 24 hr. Then, eTE was quantified with the luciferase assay (see A and B). (A) The panel shows effects of a Rac1 inhibitor, EHT1864, at 10 μM on eTE in B16.F10 and HEK293 cells. (B) The panel shows effects of a Cdc42 inhibitor, ML141, at 10 μM on eTE in B16.F10 and HEK293 cells. (C) The panel shows images of cells after EHT1864 treatment. B16.F10 cells pretreated with EHT1864 (10 μM; 1 hr) were electrically pulsed in the presence of both FITC-labeled pDNA (green) and tetramethylrhodamine-labeled dextran (red). After pulsing, cells were washed and fixed for confocal microscopy examination. Yellow color indicates co-localization of green and red pixels. Cell nuclei were labeled with DAPI (blue). (D) The panel shows representative images of inhibitory effects of EHT1864 on electrotransfection in HEK293 cells. Expression of tdTomato was shown in red. The red fluorescence images were overlaid onto images of the same cells under trans-illumination. (E) The panel shows effects of EHT1864 on eTE in B16.F10 and HEK293 cells measured by flow cytometry. The data were consistent with those quantified with the luciferase assay (see A). n = 4–6. *p < 0.05 and **p < 0.005 (Mann-Whitney U test).
We then looked for direct evidence of EHT1864 inhibiting macropinocytosis in electrotransfected cells. To test whether EHT1864 treatment could prevent pDNA or dextran from entering the cells, we first inhibited Rac1 signaling in B16.F10 cells by EHT1864 and then electrically pulsed the cells in the presence of fluorescently labeled pDNA and dextran. Our hypothesis was that, if both pDNA and dextran were in the solution, they could enter the same vesicles in the macropinocytic pathway after application of electric field. To avoid the formation of pDNA-dextran complex through electrostatic interactions, we used a negatively charged dextran. Data shown in Figure 5C demonstrated a large number of pDNA and dextran molecules co-localized in cells after application of electric field. The amounts of pDNA and dextran were significantly reduced in cells pretreated with EHT1864 (Figure 5C), and few of them were co-localized. To confirm the results shown in Figure 5C, we electrotransfected HEK293 cells with pDNA encoding tdTomato and compared the transgene expression in cells pretreated with EHT1864 to that in untreated control group. Images shown in Figure 5D demonstrated that EHT1864 treatment reduced the number of tdTomato-positive cells. Meanwhile, it reduced the total number of cells in these samples. To determine whether the percent of tdTomato-positive cells or transfection efficiency was changed, we performed flow cytometry measurement with both B16.F10 and HEK293 cells. The data revealed that EHT1864 treatment indeed reduced the efficiency in B16.F10 and HEK293 cells by 60% and 15%, respectively (Figure 5E), confirming the results obtained with the luciferase assay (see Figure 5A). Taken together, these results indicated that electrotransfection could specifically trigger Rac1-dependent macropinocytosis that could be inhibited by EHT1864 treatment.
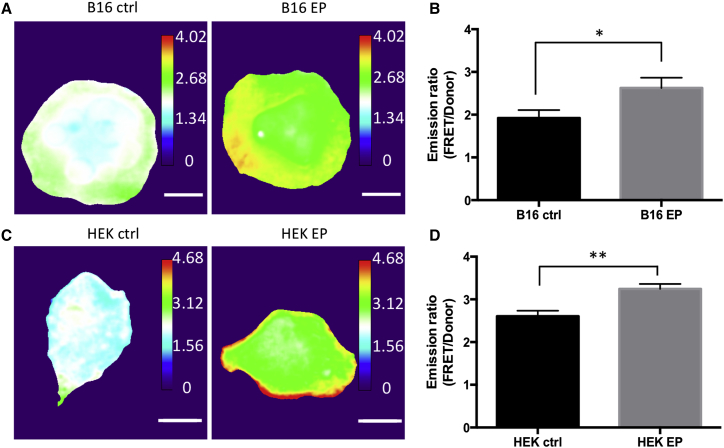
Although the pharmacological inhibitory studies had helped us identifying Rac1 as a molecular regulator, the findings from those studies might also be resulted from non-specific, off-target effects of the inhibitors. Therefore, to further confirm that inhibition of electrotransfection-induced macropinocytosis by EHT1864 could be due to the loss of Rac1 activity, we used more specific assays to test our hypothesis. First, we adopted a fluorescence resonance energy transfer (FRET)-based biosensor of Rac1 to directly examine effects of electrotransfection on cellular Rac1 activity.70 Comparing to the conventional semiquantitative pull-down activity assays,71 the FRET-based method can provide much detailed temporal and spatial information of Rac1 activity at the single-cell level. Indeed, our results in Figure 6 showed that Rac1 activities in B16.F10 and HEK293 cells were increased at 10 min after electrotransfection. The fold changes at cell edge were greater than that in the center area (Figures 6A and 6C). The average FRET/donor emission ratios were increased by 40% in B16.F10 (Figure 6B) and 20% in HEK293 cells (Figure 6D) after application of electric field. These results indicated that Rac1 was activated by electrotransfection and that the spatial pattern of activation was consistent with those of actin remodeling and macropinocytosis, which occurred mainly at cell periphery.
Figure 6.
Activation of Rac1 through Exposure of B16.F10 and HEK293 Cells to Electric Pulses
(A) The panel shows FRET/donor emission ratio images of the ECFP/YPet-based Rac1 biosensor in B16.F10 cells. (B) The panel shows the average Rac1 activity represented by FRET/donor emission ratio in control (B16 ctrl) and electrically pulsed (B16 EP) cells. (C) The panel shows FRET/donor emission ratio images of the ECFP/YPet-based Rac1 biosensor in HEK293 cells. (D) The panel shows the average Rac1 activity represented by FRET/donor emission ratio in control (HEK ctrl) and electrically pulsed (HEK EP) cells. Cells were pre-loaded with Rac1 biosensor, plated onto fibronectin-coated glass-bottom dishes, and incubated with serum-free media 1 hr prior to treatments. The bar and error bar represent mean and SEM, respectively. n = 14–50. *p < 0.05 and **p < 0.005 (Mann-Whitney U test). The scale bars represent 10 μm.
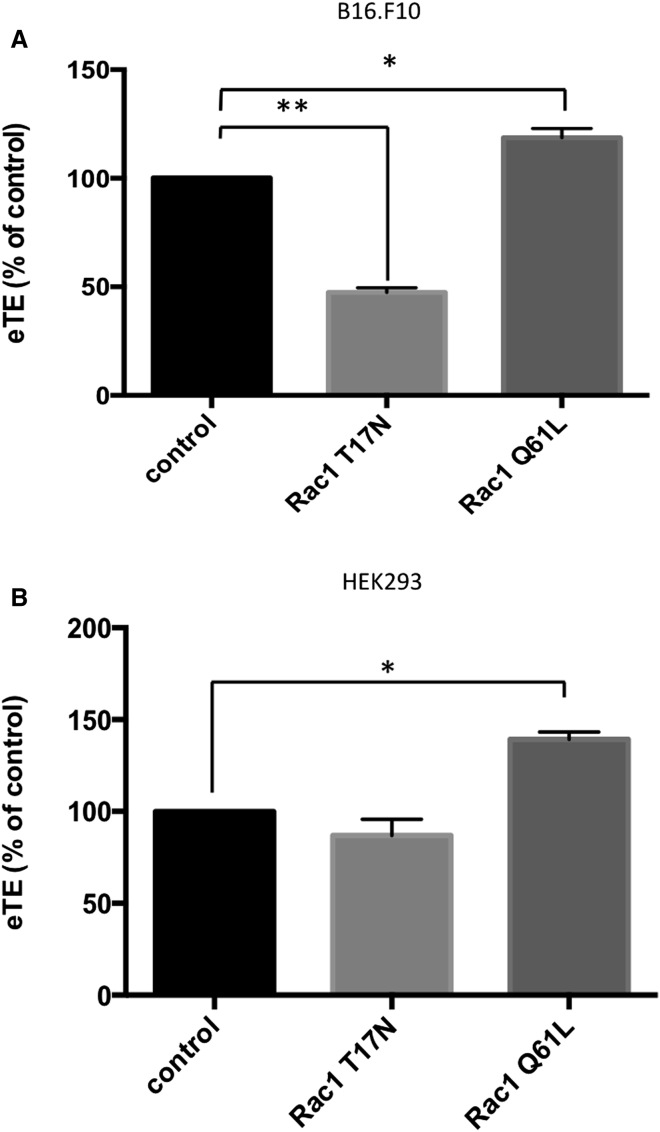
Rac1 is commonly known to be activated by receptors, but it can be also activated by nanoparticles,72 cell-penetrating peptides,73 and microRNA.74 Because mechanisms of its activation in our study are unknown, we decided not to perform the experiments using antagonists so that we could avoid non-specific activation of other targets. Instead, we introduced two mutant variants of Rac1 into cells to study the direct effects of Rac1 on electrotransfection. The first mutant was Rac1 Q61L, which carried a glutamine-to-leucine mutation at the 61st amino acid. This mutation was reported to render the enzyme constitutively active. Meanwhile, we chose a dominant negative mutant, Rac1 T17N, as the negative control, which was mutated to abandon the GTP binding affinity.75 Both mutants were first tested in B16.F10 cells, and the results are shown in Figure 7A. The transfection efficiency was increased by 20% in cells expressing Rac1 Q61L when examined by luciferase assay. In contrast, expression of Rac1 T17N resulted in 50% decrease in eTE. Similar results were observed in HEK293 cells (Figure 7B), where Rac1 Q61L expression increased eTE by 30%. Interestingly, the expression of the dominant mutant Rac1 T17N in HEK293 had no effect on eTE, which indicated that pDNA uptake in HEK293 cells may not be determined solely by Rac1 activity. Taken together, our data suggested that eTE could be altered through manipulation of Rac1 activity in vitro with either treatment of cells with pharmacological inhibitors and overexpression of Rac1 mutants.
Figure 7.
Expression of Rac1 Mutants Induced Changes in Electrotransfection Efficiency
(A) The panel shows the eTE in B16.F10 cells. The cells were pre-transfected with pDNA encoding one of the two Rac1 mutants: Rac1 Q61L and Rac1 T17N. (B) The panel shows the effects of Rac1 T17N and Q61L expression on eTE in HEK293 cells. Cells were plated in 6-well plates and transfected with pcDNA3-EGFP-Rac1-Q61L or pcDNA3-EGFP-Rac1-T17N plasmids. After 24 hr, cells were collected and electrotransfected with pcDNA3.1 (+) Luc2 = tdT. The eTE was quantified with the luciferase assay at 24 hr after electrotransfection. n = 4−10. *p < 0.05 and **p < 0.005 (Mann-Whitney U test).
Discussion
Data from the study demonstrated that macropinocytosis of DNA molecules contributed to electrotransfection of various cell lines and that the process of macropinocytosis was partly modulated by Rac1 activity. Specifically, we observed that electric pulse could induce cell membrane ruffling and actin cytoskeleton remodeling. When cells were pulsed in the presence of fluorescently labeled pDNA and a macropinocytosis marker, dextran, electrotransfected pDNA co-localized with dextran in intracellular vesicles. Furthermore, electrotransfection efficiency evaluated by luciferase assay was decreased significantly by reducing temperature for a short period (10 min) after electric pulse application, treating cells with a pharmacological inhibitor of Rac1, or expressing a dominant negative mutant of Rac1. Meanwhile, expression of a constitutively active mutant of Rac1 increased the electrotransfection efficiency. These findings suggested that electrotransfection of pDNA could be mediated by Rac1-dependent macropinocytosis.
It has been shown that inhibition of endocytosis would impede electrotransfection in vivo.76 Recent study from our lab showed that pDNA could bind to membrane during electrotransfection and continuously be internalized by cells over a period of time that was much longer than the theoretical lifetime of transient pores induced by pulsed electric fields.48 In addition, we demonstrated that a clathrin-mediated endocytosis mechanism could actively transport plasmids across the membrane and facilitate electrotransfection.77 The accumulating evidence supports the notion that electrotransfection of pDNA is mediated by endocytosis. In the present work, we further confirmed that electrotransfection could indeed induce actin remodeling, which is an essential requirement for macropinocytosis. Cytoskeleton components, such as actin filaments and microtubules, are known to be responsible for vesicle transport inside the cell.78, 79 It has been reported that electrotransfection can cause alteration of cell adhesion80 and changes in cell morphology,81 which are likely to be related to cytoskeleton as well. However, the role of actin in assisting transmembrane DNA transport has not been fully investigated. Our data shown in Figures 1 and 2 demonstrated that actin cytoskeleton was changed after electric pulse application. Interestingly, several different structures of actin were observed in electrically pulsed cells. We believed those structures to be associated with different stages of macropinocytosis: the membrane protrusions indicated local initiation of macropinocytosis, whereas the circular vesicles were likely to be macropinosomes. In addition, considering the fact that those structures were present only after pulse application, the vesicles observed inside the cytosol could also be transitioning in endocytic pathways. It was also interesting to discover that many of the DNA aggregates seemed to be transported toward the nucleus. Even within a short period (i.e., 10 min), plasmid DNA could be observed at sites that were close to the nucleus, suggesting that pDNA could quickly accumulate at certain peri-nuclear regions, possibly Golgi apparatus or endoplasmic reticulum. It is worth to mention that the rate of transport is faster than passive diffusion of large DNA molecules, e.g., pDNA, in cytoplasm.
It is known that macropinocytosis can be distinguished from other endocytic pathways, such as clathrin-mediated endocytosis (CME) and phagocytosis, because it is independent of a GTP-binding protein, dynamin 2.82 In our previous study,48 we have addressed the role of dynamin-2-dependent endocytosis played in electrotransfection. By using a specific inhibitor (i.e., dynasore), we demonstrated dynamin-2-dependent endocytosis could be responsible for electrotransfection in certain cells. As an extension of the previous work, the present study demonstrated that electric-pulse-induced macropinocytosis was also responsible for pDNA uptake during electrotransfection. These observations were consistent with reports from other research groups. For instance, Rols et al. have reported similar observation of macropinocytosis in electrically pulsed cells.83 In a more recent study by Rosazza et al.,84 the contributions of different endocytic pathways to cellular uptake of electrotransfected pDNA were quantified by using specific inhibitors of endosomal proteins and fluorescence microscopy technique. According to these results, clathrin-dependent endocytosis and macropinocytosis were each responsible for approximately 25% of pDNA uptake in CHO cells. In the current study, the inhibition of macropinocytosis indeed caused approximately 30% decrease in eTE in HEK cells. However, the decreases in eTE were much greater in B16.F10 and COS7 cells treated with EHT1864, suggesting that specific contribution of each pathway to eTE was cell line dependent. Taken together, these findings present a challenge to the conventional electro-permeabilization theory by elucidating that multiple endocytic pathways could be utilized for gene delivery during electrotransfection.
Our finding also sheds light on the actual form of pDNA being transferred into cells during electrotransfection. Several studies have indicated that particles of different sizes could enter cells via different mechanisms.8, 83, 85, 86 Vesicles in clathrin- and caveolin-mediated endocytosis are small in size (<100 nm), which can significantly limit their transport capacities for pDNA with a similar size. In contrast, macropinocytosis, which is known to be responsible for cellular uptake of viruses,87, 88 nanoparticles,89 and naked pDNA,90 can generate vesicles with sizes of up to 5 μm that could transport a significantly larger amount of pDNA per vesicle. In previous and current studies, it was observed that pDNA could form aggregates in solution with certain sizes or complexes with the plasma membrane.91, 92 Using dynamic light-scattering technology, we observed that the size of DNA aggregates grew with increasing the number of electric pulses.93 Under the electrotransfection conditions used in the current study, the average size of aggregates in solution was on the order of 500 nm, which was the preferred particle size for macropinocytosis. Single pDNA molecules or smaller aggregates could enter cells through this and other pathways. For example, pDNA molecules might be directly pushed into cytoplasm by electric field because they are negatively charged or, as discussed above, they could enter cells through clathrin- or caveolin-mediated endocytosis. As a result, inhibition of macropinocytosis should not completely block electrotransfection.
Another remarkable finding in the study was the involvement of the small GTPase Rac1 in electrotransfection. Results obtained from the pharmacological inhibitor study and the genetic gain-of-function assay revealed that Rac1 was a key regulatory GTPase for electric-pulse-induced macropinocytosis. Our discovery was consistent with previous reports showing that Rac1-mediated macropinocytosis is responsible for the entry of DNA-conjugated, single-wall carbon nanotubes into endothelial cells,94 as well as the entry of naked DNA in an in vivo study,95 which are not triggered by electric pulses. Although we did not know the exact mechanisms on how electrotransfection activated Rac1, we hypothesized that Rac1 activation and actin remodeling were required for repairing membrane damages caused by electric pulses. The hypothesis was partly supported by an observation that Rac1 inhibition by EHT1864 treatment reduced viability of pulsed cells, but not control cells (data not shown). It was also possible that Rac1 was activated in response to osmotic pressure change or ion fluxes (Na+/H+/Ca2+) during electroporation of cells. These potential mechanisms might explain why different cell lines responded differently to treatments with Rac1 inhibitors. For example, results shown in Figure 4 indicated that the electrotransfection efficiency of B16.F10 cells was more dependent on Rac1-mediated macropinocytosis than that of HEK293 cells. The data in Figure 7 showed that transient expression of a dominant negative mutant of Rac1 decreased eTE in B16.F10 cells but had little effect on eTE in HEK293 cells. Similar results have also been observed in previous studies.67, 96 Therefore, data in our study could only lead to a conclusion that Rac1 was required for electroporation-induced macropinocytosis. Future studies should be directed to address the issue on how Rac1 activation mediates macropinocytosis.
In conclusion, the significance of our study was 2-fold. First, the study provided new perspectives on electrotransfection mechanisms. Second, it demonstrated the first evidence showing the involvement of Rac1-regulated actin remodeling and macropinocytosis in electrotransfection. The finding that cytoskeletal changes could contribute to the uptake of pDNA was of importance in controlling the efficiency of electrotransfection. A better understanding in the relationship between cytoskeleton dynamics and intracellular trafficking of genes will also benefit other viral and non-viral gene delivery methods.
Experimental Procedures
Cell Lines and Cell Culture
Three cell lines were used in this study: B16.F10, a murine melanoma cell line; HEK293, a human embryonic kidney cell line; and COS7, a monkey kidney fibroblast-like cell line. Cells were obtained from Duke University Cell Culture Facility (CCF) and cultured as previously described.48, 77 In brief, cells were grown as monolayers in high-glucose DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone) and Antibiotic-Antimycotic (Invitrogen) at 37°C in a humidified incubator with 5% CO2 and passaged every 36–48 hr.
Plasmids
pEGFP-N1 was purchased from Clontech. pcDNA3.1(+) Luc2 = tdT was a gift from Christopher Contag (Addgene; plasmid no. 32904). pcDNA3-EGFP-Rac1-Q61L and pcDNA3-EGFP-Rac1-T17N plasmids were gifts from Gary Bokoch (Addgene plasmid nos. 12981 and 12982). Plasmids were amplified using DH5α E. coli and prepared from single colonies using Miniprep DNA purification kits (QIAGEN) according to manufacturer’s instructions. For fluorescence microscopy studies, pEGFP plasmids were covalently labeled with fluorescent dyes (tetramethylrhodamine for red and fluorescein isothiocyanate [FITC] for green) using the Label IT nucleic acid labeling kit (Mirus).
Pharmacological Inhibitors
Actin polymerization inhibitor Cytochalasin D was purchased from Sigma Aldrich. Cdc42 GTPase inhibitor ML141 and Rac1 inhibitor EH1864 were purchased from Santa Cruz Biotechnology. Stock solutions of the inhibitors were prepared in DMSO and stored at −20°C. For inhibitory studies, cells were seeded in 6-well plates at densities of 0.5 to 0.7 million per well and allowed to grow overnight to achieve 75%–90% confluency. Before treatment, culture medium was aspirated and cells were washed twice by PBS free of Ca2+ and Mg2+. After washing, 1 mL of serum-free DMEM was added to each well and appropriate volumes of the drugs were added to achieve final drug concentrations. In the corresponding control groups, equivalent volumes of the solvent DMSO were added. After incubation at 37°C with 5% CO2 for 1 hr, cells were collected by trypsinization and subsequently electrotransfected with plasmid DNAs to investigate effects of the drug treatment on electrotransfection efficiency.
Electrotransfection Procedures
For electrotransfection experiments, cells were plated 1 day prior to experiment and grown overnight to 75%–90% confluency. Before electrotransfection, cells were detached by 0.25% trypsin-EDTA (Invitrogen) treatment, neutralized with medium containing 10% serum, and then harvested by centrifugation. Cell pellets were then re-suspended in OptiMEM I Reduced Serum Media (Invitrogen) at a concentration of 107 cells/mL. Plasmid DNAs were then added into the suspension to achieve a final concentration of 10 μg/mL. For electrotransfection, samples were loaded into disposable 4-mm gap aluminum cuvettes (Bio-Rad) and incubated shortly before receiving an electric pulse sequence with eight pulses at 450 V/cm, 5 ms duration, and 1 Hz frequency. The pulses were generated by using BTX ECM 830 Square Wave Electroporation System (Harvard Apparatus). After electrotransfection, samples were incubated at 37°C for 10 min to promote endocytosis. In cold treatment groups, cells were incubated on ice for 10 min. Then, the cells were retrieved, seeded in fresh culture medium in 6-well plates, and cultured at 37°C with 5% CO2. Transfection efficiency was evaluated at 24 hr after electrotransfection.
Uptake of Fluorescently Labeled Plasmid DNA and Dextran
To study pDNA uptake by cells via macropinocytosis, 1 μg of tetramethylrhodamine-labeled pEGFP-N1 was mixed with 1 million cells suspended in 100 μL Opti-MEM. To study macropinocytosis induced by the same electric pulses as those for electrotransfection, 10 μg tetramethylrhodamine-labeled, anionic, lysine fixable dextran (2,000,000 molecular weight [MW]; Thermo Fisher Scientific) was mixed with 1 million cells suspended in 100 μL Opti-MEM. All samples were immediately treated with electric pulses and incubated for 10 min at different temperature per experiment requirements. Samples were then re-suspended and washed with PBS and fixed with 4% paraformaldehyde for 20 min.
Transfection of pDNA for Rac1 Mutants
B16.F10 and HEK293 cells were transfected with two Rac1 constructs (T17N and Q61L) using Genejet In Vitro DNA Transfection Reagent (Signagen Laboratories) and Lipofectamine 2000 (Invitrogen), respectively. In experiments, cells were plated in 6-well plates at 0.5 × 106 cells per well and grown overnight. The next day, cells were transfected with 1 μg plasmid in 5 μL transfection reagent according to manufacturer’s instruction. The transfected cells were further cultured for 24 hr to achieve expression of transfected proteins and then harvested and used in the study of electrotransfection.
Fluorescence Microscopy and Image Analysis
Confocal fluorescence images were acquired using either a Leica SP5 inverted confocal microscope (Leica Microsystems) with 40×/numerical aperture (NA) 1.25 objective or a XD revolution spinning disk microscope (Andor Technology) with a 60×/NA 1.2W corr UPlanApo objective. Images shown in the paper represent either optical slices near the middle plane of cells or z stack projection of maximum intensity. Regular fluorescence images were acquired with an Axio Vert A1 inverted microscope (Carl Zeiss). Image segmentation, particle counting, and co-localization analysis were performed in ImageJ using build-in functions. For counting macropinosomes formed by dextran uptake, particle size was set between 4 and 100 pixels and circularity was set to default (0.00–1.00).
Rac1 Activity Assay by a FRET-Based Biosensor
The protocol is similar to that described previously.70 Cells transfected with a Rac1 FRET biosensor were plated onto fibronectin-coated glass-bottom dishes overnight in high-glucose DMEM. One hour before imaging, cells were washed and incubated in serum-free DMEM media. The electrically pulsed (EP) groups received eight pulses at 450 V/cm, 5 ms duration, 1 Hz frequency. The control group received no treatments. After incubation at 37°C for 10 min, samples were imaged at 60× magnification (Olympus UPlanSApo 60×/NA 1.35 objective) using fluorescence microscopy on an Olympus inverted fluorescence microscope (Olympus IX83) illuminated by a LambdaLS equipped with a 300 W ozone-free xenon bulb (Sutter Instruments). The images were captured using a sCMOS ORCA-Flash4.0 V2 camera (Hamamatsu). The FRET images were acquired using a custom filter set comprised of a donor excitation filter (Chroma; ET450/30×), donor emission filter (Chroma; ET485/20 m), acceptor excitation filter (Chroma; ET514/10×), acceptor emission filter (Semrock; FF01-571/72), and dichroic mirror (Chroma; T450/514rpc). The motorized filter wheels (Sutter Lambda 10-3) and automated stage (Prior H117EIX3) were controlled through MetaMorph Advanced software (Olympus). The background subtraction and calculation of pixel-by-pixel emission ratio of FRET/ECFP were performed in ImageJ using the open source plugin PixFRET.97
Luciferase Assay
Luciferase activity was quantified at 24 hr after electrotransfection. In brief, luciferin stock solution (30 mg/mL or 0.107 mol/L) was prepared by dissolving 100 mg D-luciferin in 3.3 mL ddH2O and stored as 0.25 mL aliquots at −20°C. ATP stock solution (1 mol/L) was prepared by dissolving 1 g ATP in 1.8 mL ddH2O. Cells in 6-well plates were washed twice with PBS and then lysed by a lysis buffer (25 mM Tris-HCl [pH 7.8], 4 mM EDTA, and 1% Triton X-100) and collected by scraping. After centrifugation at 10,300 g for 10 min, 50 μL of the supernatant was pipetted into 96-well plates. To each well, 150 μL of reaction solution (25 mM Tris-HCl [pH 7.8], 15 mM MgSO4, 2 mM ATP, and 5 mM D-luciferin) was added. The plate was immediately mixed and read in a Victor X4 plate reader (PerkinElmer). The luminescence was normalized by protein concentration of each sample to account for differences in cell numbers due to cell washing and variation in cell proliferation rate. The protein concentration was measured using BCA protein assay kit (Thermo Scientific). eTE in this assay was defined as the percentage of luminescence units (RLU) in experimental group relative to that in the control.
Flow Cytometry
Flow cytometry analysis was performed as previously described.77 In brief, cells were collected and re-suspended in 200–300 μL PBS and then stained with propidium iodide (PI) (5 μg/ mL). Flow cytometry analysis was performed with a BD FACSCanto II flow cytometer (Becton Dickinson). 488-nm laser was chosen for simultaneous excitation of GFP and PI. Single-cell populations were separated using front and side light scattering as independent variables. Compensation was set between 20% and 25% to resolve emission spectra overlap between the two detection channels. To access the eTE, 10,000 events were collected for each sample. Raw data acquisition was performed with the BD FACSDiva software. Data analysis was performed using FlowJo. eTE in this assay was defined as the percentage of PI−/GFP+ population over total viable (PI−) cells.
Statistical Test
Error bars in all figures represent the SEM. Differences between unpaired groups were evaluated with the Mann-Whitney U test. They were considered to be statistically significant if p < 0.05. The statistical analysis was performed using Prism (GraphPad Software).
Author Contributions
M.M. performed experiments. L.W. and J.H. performed experiments with confocal microscopy. C.-C.C. performed experiments with flow cytometry. Y.W. developed the assay for FRET analysis of Rac1 activity. K.E.R. assisted in FRET data analysis. B.D.H. and P.B.L provided expertise in data analysis and interpretation. M.M. and F.Y. conceived ideas, designed experiments, and wrote the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
The work was supported partly by grants from the NIH (GM098520) and the National Science Foundation (BES-0828630).
Footnotes
Supplemental Information includes two figures and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2016.12.009.
Supplemental Information
References
- 1.Li S.-D., Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006;13:1313–1319. doi: 10.1038/sj.gt.3302838. [DOI] [PubMed] [Google Scholar]
- 2.Kumar S.R., Markusic D.M., Biswas M., High K.A., Herzog R.W. Clinical development of gene therapy: results and lessons from recent successes. Mol. Ther. Methods Clin. Dev. 2016;3:16034. doi: 10.1038/mtm.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amer M.H. Gene therapy for cancer: present status and future perspective. Mol. Cell. Ther. 2014;2:27. doi: 10.1186/2052-8426-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bongianino R., Priori S.G. Gene therapy to treat cardiac arrhythmias. Nat. Rev. Cardiol. 2015;12:531–546. doi: 10.1038/nrcardio.2015.61. [DOI] [PubMed] [Google Scholar]
- 5.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 6.Jones C.H., Chen C.-K., Ravikrishnan A., Rane S., Pfeifer B.A. Overcoming nonviral gene delivery barriers: perspective and future. Mol. Pharm. 2013;10:4082–4098. doi: 10.1021/mp400467x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belting M., Sandgren S., Wittrup A. Nuclear delivery of macromolecules: barriers and carriers. Adv. Drug Deliv. Rev. 2005;57:505–527. doi: 10.1016/j.addr.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Pavlin M., Kandušer M. New insights into the mechanisms of gene electrotransfer--experimental and theoretical analysis. Sci. Rep. 2015;5:9132. doi: 10.1038/srep09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas C.E., Ehrhardt A., Kay M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 10.Cahill K. Cell-penetrating peptides, electroporation and drug delivery. IET Syst. Biol. 2010;4:367–378. doi: 10.1049/iet-syb.2010.0007. [DOI] [PubMed] [Google Scholar]
- 11.Veldhoen S., Laufer S.D., Trampe A., Restle T. Cellular delivery of small interfering RNA by a non-covalently attached cell-penetrating peptide: quantitative analysis of uptake and biological effect. Nucleic Acids Res. 2006;34:6561–6573. doi: 10.1093/nar/gkl941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y.-W., Lee H.-J., Tolliver L.M., Aronstam R.S. Delivery of nucleic acids and nanomaterials by cell-penetrating peptides: opportunities and challenges. BioMed Res. Int. 2015;2015:834079. doi: 10.1155/2015/834079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khalil I.A., Kogure K., Akita H., Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol. Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 14.Escoffre J.M., Kaddur K., Rols M.P., Bouakaz A. In vitro gene transfer by electrosonoporation. Ultrasound Med. Biol. 2010;36:1746–1755. doi: 10.1016/j.ultrasmedbio.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 15.Chu G., Hayakawa H., Berg P. Electroporation for the efficient transfection of mammalian cells with DNA. Nucleic Acids Res. 1987;15:1311–1326. doi: 10.1093/nar/15.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu M., Zhao D., Zhong W., Yan H., Wang X., Liang Z., Li Z. High-density distributed electrode network, a multi-functional electroporation method for delivery of molecules of different sizes. Sci. Rep. 2013;3:3370. doi: 10.1038/srep03370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weaver J.C. Electroporation theory. Concepts and mechanisms. Methods Mol. Biol. 1995;47:1–26. doi: 10.1385/0-89603-310-4:1. [DOI] [PubMed] [Google Scholar]
- 18.Gehl J. Electroporation: theory and methods, perspectives for drug delivery, gene therapy and research. Acta Physiol. Scand. 2003;177:437–447. doi: 10.1046/j.1365-201X.2003.01093.x. [DOI] [PubMed] [Google Scholar]
- 19.Prasanna G.L., Panda T. Electroporation: basic principles, practical considerations and applications in molecular biology. Bioprocess Eng. 1997;16:261. [Google Scholar]
- 20.Matsuda T., Cepko C.L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dean D.A., Machado-Aranda D., Blair-Parks K., Yeldandi A.V., Young J.L. Electroporation as a method for high-level nonviral gene transfer to the lung. Gene Ther. 2003;10:1608–1615. doi: 10.1038/sj.gt.3302053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsong T.Y. Electroporation of cell membranes. Biophys. J. 1991;60:297–306. doi: 10.1016/S0006-3495(91)82054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melkonyan H., Sorg C., Klempt M. Electroporation efficiency in mammalian cells is increased by dimethyl sulfoxide (DMSO) Nucleic Acids Res. 1996;24:4356–4357. doi: 10.1093/nar/24.21.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chicaybam L., Sodre A.L., Curzio B.A., Bonamino M.H. An efficient low cost method for gene transfer to T lymphocytes. PLoS One. 2013;8:e60298. doi: 10.1371/journal.pone.0060298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marrero B., Shirley S., Heller R. Delivery of interleukin-15 to B16 melanoma by electroporation leads to tumor regression and long-term survival. Technol. Cancer Res. Treat. 2014;13:551–560. doi: 10.7785/tcrtexpress.2013.600252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arena C.B., Sano M.B., Rylander M.N., Davalos R.V. Theoretical considerations of tissue electroporation with high-frequency bipolar pulses. IEEE Trans. Biomed. Eng. 2011;58:1474–1482. doi: 10.1109/TBME.2010.2102021. [DOI] [PubMed] [Google Scholar]
- 27.De Vry J., Martínez-Martínez P., Losen M., Bode G.H., Temel Y., Steckler T., Steinbusch H.W., De Baets M., Prickaerts J. Low current-driven micro-electroporation allows efficient in vivo delivery of nonviral DNA into the adult mouse brain. Mol. Ther. 2010;18:1183–1191. doi: 10.1038/mt.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yadollahpour A., Rezaee Z. Electroporation as a new cancer treatment technique: a review on the mechanisms of action. Biomed. Pharmacol. J. 2014;7:53–62. [Google Scholar]
- 29.Bodles-Brakhop A.M., Heller R., Draghia-Akli R. Electroporation for the delivery of DNA-based vaccines and immunotherapeutics: current clinical developments. Mol. Ther. 2009;17:585–592. doi: 10.1038/mt.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daud A.I., Mirza N., Lenox B., Andrews S., Urbas P., Gao G.X., Lee J.H., Sondak V.K., Riker A.I., Deconti R.C., Gabrilovich D. Phenotypic and functional analysis of dendritic cells and clinical outcome in patients with high-risk melanoma treated with adjuvant granulocyte macrophage colony-stimulating factor. J. Clin. Oncol. 2008;26:3235–3241. doi: 10.1200/JCO.2007.13.9048. [DOI] [PubMed] [Google Scholar]
- 31.Cemazar M., Ambrozic Avgustin J., Pavlin D., Sersa G., Poli A., Krhac Levacic A., Tesic N., Lampreht Tratar U., Rak M., Tozon N. Efficacy and safety of electrochemotherapy combined with peritumoral IL-12 gene electrotransfer of canine mast cell tumours. Vet. Comp. Oncol. 2016 doi: 10.1111/vco.12208. Published online February 3, 2016. [DOI] [PubMed] [Google Scholar]
- 32.Wilgenhof S., Corthals J., Van Nuffel A.M.T., Benteyn D., Heirman C., Bonehill A., Thielemans K., Neyns B. Long-term clinical outcome of melanoma patients treated with messenger RNA-electroporated dendritic cell therapy following complete resection of metastases. Cancer Immunol. Immunother. 2015;64:381–388. doi: 10.1007/s00262-014-1642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C., Smye S.W., Robinson M.P., Evans J.A. Membrane electroporation theories: a review. Med. Biol. Eng. Comput. 2006;44:5–14. doi: 10.1007/s11517-005-0020-2. [DOI] [PubMed] [Google Scholar]
- 34.Escoffre J.-M., Portet T., Wasungu L., Teissié J., Dean D., Rols M.-P. What is (still not) known of the mechanism by which electroporation mediates gene transfer and expression in cells and tissues. Mol. Biotechnol. 2009;41:286–295. doi: 10.1007/s12033-008-9121-0. [DOI] [PubMed] [Google Scholar]
- 35.Henshaw J.W., Zaharoff D.A., Mossop B.J., Yuan F. Electric field-mediated transport of plasmid DNA in tumor interstitium in vivo. Bioelectrochemistry. 2007;71:233–242. doi: 10.1016/j.bioelechem.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canatella P.J., Karr J.F., Petros J.A., Prausnitz M.R. Quantitative study of electroporation-mediated molecular uptake and cell viability. Biophys. J. 2001;80:755–764. doi: 10.1016/S0006-3495(01)76055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shil P., Bidaye S., Vidyasagar P.B. Analysing the effects of surface distribution of pores in cell electroporation for a cell membrane containing cholesterol. J. Phys. D Appl. Phys. 2008;41:055502. [Google Scholar]
- 38.Mahnič-Kalamiza S., Miklavčič D., Vorobiev E. Dual-porosity model of solute diffusion in biological tissue modified by electroporation. Biochim. Biophys. Acta. 2014;1838:1950–1966. doi: 10.1016/j.bbamem.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Lee E.W., Wong D., Prikhodko S.V., Perez A., Tran C., Loh C.T., Kee S.T. Electron microscopic demonstration and evaluation of irreversible electroporation-induced nanopores on hepatocyte membranes. J. Vasc. Interv. Radiol. 2012;23:107–113. doi: 10.1016/j.jvir.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 40.Spugnini E.P., Arancia G., Porrello A., Colone M., Formisano G., Stringaro A., Citro G., Molinari A. Ultrastructural modifications of cell membranes induced by “electroporation” on melanoma xenografts. Microsc. Res. Tech. 2007;70:1041–1050. doi: 10.1002/jemt.20504. [DOI] [PubMed] [Google Scholar]
- 41.Tekle E., Oubrahim H., Dzekunov S.M., Kolb J.F., Schoenbach K.H., Chock P.B. Selective field effects on intracellular vacuoles and vesicle membranes with nanosecond electric pulses. Biophys. J. 2005;89:274–284. doi: 10.1529/biophysj.104.054494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang D.C., Reese T.S. Changes in membrane structure induced by electroporation as revealed by rapid-freezing electron microscopy. Biophys. J. 1990;58:1–12. doi: 10.1016/S0006-3495(90)82348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rols M.P., Teissié J. Electropermeabilization of mammalian cells. Quantitative analysis of the phenomenon. Biophys. J. 1990;58:1089–1098. doi: 10.1016/S0006-3495(90)82451-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie T.D., Tsong T.Y. Study of mechanisms of electric field-induced DNA transfection. II. Transfection by low-amplitude, low-frequency alternating electric fields. Biophys. J. 1990;58:897–903. doi: 10.1016/S0006-3495(90)82434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaharoff D.A., Henshaw J.W., Mossop B., Yuan F. Mechanistic analysis of electroporation-induced cellular uptake of macromolecules. Exp. Biol. Med. (Maywood) 2008;233:94–105. doi: 10.3181/0704-RM-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stapulionis R. Electric pulse-induced precipitation of biological macromolecules in electroporation. Bioelectrochem. Bioenerg. 1999;48:249–254. doi: 10.1016/s0302-4598(98)00206-2. [DOI] [PubMed] [Google Scholar]
- 47.Delgado-Cañedo A., Santos D.G.D., Chies J.A.B., Kvitko K., Nardi N.B. Optimization of an electroporation protocol using the K562 cell line as a model: role of cell cycle phase and cytoplasmic DNAses. Cytotechnology. 2006;51:141–148. doi: 10.1007/s10616-006-9028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu M., Yuan F. Membrane binding of plasmid DNA and endocytic pathways are involved in electrotransfection of mammalian cells. PLoS One. 2011;6:e20923. doi: 10.1371/journal.pone.0020923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kerr M.C., Teasdale R.D. Defining macropinocytosis. Traffic. 2009;10:364–371. doi: 10.1111/j.1600-0854.2009.00878.x. [DOI] [PubMed] [Google Scholar]
- 50.Falcone S., Cocucci E., Podini P., Kirchhausen T., Clementi E., Meldolesi J. Macropinocytosis: regulated coordination of endocytic and exocytic membrane traffic events. J. Cell Sci. 2006;119:4758–4769. doi: 10.1242/jcs.03238. [DOI] [PubMed] [Google Scholar]
- 51.Swanson J.A. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 2008;9:639–649. doi: 10.1038/nrm2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim J.P., Gleeson P.A. Macropinocytosis: an endocytic pathway for internalising large gulps. Immunol. Cell Biol. 2011;89:836–843. doi: 10.1038/icb.2011.20. [DOI] [PubMed] [Google Scholar]
- 53.Commisso C., Davidson S.M., Soydaner-Azeloglu R.G., Parker S.J., Kamphorst J.J., Hackett S., Grabocka E., Nofal M., Drebin J.A., Thompson C.B. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meier O., Boucke K., Hammer S.V., Keller S., Stidwill R.P., Hemmi S., Greber U.F. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Biol. 2002;158:1119–1131. doi: 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J.T.H., Teasdale R.D., Liebl D. Macropinosome quantitation assay. MethodsX. 2014;1:36–41. doi: 10.1016/j.mex.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costes S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manders E.M.M., Verbeek F.J., Aten J.A. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 59.Schliwa M. Action of cytochalasin D on cytoskeletal networks. J. Cell Biol. 1982;92:79–91. doi: 10.1083/jcb.92.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Glogauer M., Lee W., McCulloch C.A. Induced endocytosis in human fibroblasts by electrical fields. Exp. Cell Res. 1993;208:232–240. doi: 10.1006/excr.1993.1242. [DOI] [PubMed] [Google Scholar]
- 61.El-Sayed A., Harashima H. Endocytosis of gene delivery vectors: from clathrin-dependent to lipid raft-mediated endocytosis. Mol. Ther. 2013;21:1118–1130. doi: 10.1038/mt.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burridge K., Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 63.Jung I.D., Lee J., Yun S.Y., Park C.G., Choi W.S., Lee H.W., Choi O.H., Han J.W., Lee H.Y. Cdc42 and Rac1 are necessary for autotaxin-induced tumor cell motility in A2058 melanoma cells. FEBS Lett. 2002;532:351–356. doi: 10.1016/s0014-5793(02)03698-0. [DOI] [PubMed] [Google Scholar]
- 64.Palamidessi A., Frittoli E., Garré M., Faretta M., Mione M., Testa I., Diaspro A., Lanzetti L., Scita G., Di Fiore P.P. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134:135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 65.Yang W.-H., Lan H.-Y., Huang C.-H., Tai S.-K., Tzeng C.-H., Kao S.-Y., Wu K.J., Hung M.C., Yang M.H. RAC1 activation mediates Twist1-induced cancer cell migration. Nat. Cell Biol. 2012;14:366–374. doi: 10.1038/ncb2455. [DOI] [PubMed] [Google Scholar]
- 66.Schlam D., Bagshaw R.D., Freeman S.A., Collins R.F., Pawson T., Fairn G.D., Grinstein S. Phosphoinositide 3-kinase enables phagocytosis of large particles by terminating actin assembly through Rac/Cdc42 GTPase-activating proteins. Nat. Commun. 2015;6:8623. doi: 10.1038/ncomms9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dharmawardhane S., Schürmann A., Sells M.A., Chernoff J., Schmid S.L., Bokoch G.M. Regulation of macropinocytosis by p21-activated kinase-1. Mol. Biol. Cell. 2000;11:3341–3352. doi: 10.1091/mbc.11.10.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Surviladze, Z., Waller, A., Strouse, J.J., Bologa, C., Ursu, O., Salas, V., Parkinson, J.F., Phillips, G.K., Romero, E., Wandinger-Ness, A. et al. (2010). A potent and selective inhibitor of Cdc42 GTPase. In Probe Reports from the NIH Molecular Libraries Program (NCBI). https://www.ncbi.nlm.nih.gov/books/NBK51965/. [PubMed]
- 69.Désiré L., Bourdin J., Loiseau N., Peillon H., Picard V., De Oliveira C., Bachelot F., Leblond B., Taverne T., Beausoleil E. RAC1 inhibition targets amyloid precursor protein processing by gamma-secretase and decreases Abeta production in vitro and in vivo. J. Biol. Chem. 2005;280:37516–37525. doi: 10.1074/jbc.M507913200. [DOI] [PubMed] [Google Scholar]
- 70.Ouyang M., Sun J., Chien S., Wang Y. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc. Natl. Acad. Sci. USA. 2008;105:14353–14358. doi: 10.1073/pnas.0807537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benard V., Bokoch G.M. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 2002;345:349–359. doi: 10.1016/s0076-6879(02)45028-8. [DOI] [PubMed] [Google Scholar]
- 72.Diesel B., Hoppstädter J., Hachenthal N., Zarbock R., Cavelius C., Wahl B., Thewes N., Jacobs K., Kraegeloh A., Kiemer A.K. Activation of Rac1 GTPase by nanoparticulate structures in human macrophages. Eur. J. Pharm. Biopharm. 2013;84:315–324. doi: 10.1016/j.ejpb.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 73.Gerbal-Chaloin S., Gondeau C., Aldrian-Herrada G., Heitz F., Gauthier-Rouvière C., Divita G. First step of the cell-penetrating peptide mechanism involves Rac1 GTPase-dependent actin-network remodelling. Biol. Cell. 2007;99:223–238. doi: 10.1042/BC20060123. [DOI] [PubMed] [Google Scholar]
- 74.Sun G., Cao Y., Shi L., Sun L., Wang Y., Chen C., Wan Z., Fu L., You Y. Overexpressed miRNA-137 inhibits human glioma cells growth by targeting Rac1. Cancer Biother. Radiopharm. 2013;28:327–334. doi: 10.1089/cbr.2012.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Subauste M.C., Von Herrath M., Benard V., Chamberlain C.E., Chuang T.H., Chu K., Bokoch G.M., Hahn K.M. Rho family proteins modulate rapid apoptosis induced by cytotoxic T lymphocytes and Fas. J. Biol. Chem. 2000;275:9725–9733. doi: 10.1074/jbc.275.13.9725. [DOI] [PubMed] [Google Scholar]
- 76.Markelc B., Skvarca E., Dolinsek T., Kloboves V.P., Coer A., Sersa G., Cemazar M. Inhibitor of endocytosis impairs gene electrotransfer to mouse muscle in vivo. Bioelectrochemistry. 2015;103:111–119. doi: 10.1016/j.bioelechem.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 77.Chang C.-C., Wu M., Yuan F. Role of specific endocytic pathways in electrotransfection of cells. Mol. Ther. Methods Clin. Dev. 2014;1:14058. doi: 10.1038/mtm.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Badding M.A., Dean D.A. Highly acetylated tubulin permits enhanced interactions with and trafficking of plasmids along microtubules. Gene Ther. 2013;20:616–624. doi: 10.1038/gt.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mulcahy L.A., Pink R.C., Carter D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles. 2014;3:24641. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pehlivanova V.N., Tsoneva I.H., Tzoneva R.D. Multiple effects of electroporation on the adhesive behaviour of breast cancer cells and fibroblasts. Cancer Cell Int. 2012;12:9. doi: 10.1186/1475-2867-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meulenberg C.J., Todorovic V., Cemazar M. Differential cellular effects of electroporation and electrochemotherapy in monolayers of human microvascular endothelial cells. PLoS One. 2012;7:e52713. doi: 10.1371/journal.pone.0052713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pelkmans L., Helenius A. Insider information: what viruses tell us about endocytosis. Curr. Opin. Cell Biol. 2003;15:414–422. doi: 10.1016/s0955-0674(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 83.Rols M.P., Femenia P., Teissie J. Long-lived macropinocytosis takes place in electropermeabilized mammalian cells. Biochem. Biophys. Res. Commun. 1995;208:26–35. doi: 10.1006/bbrc.1995.1300. [DOI] [PubMed] [Google Scholar]
- 84.Rosazza C., Deschout H., Buntz A., Braeckmans K., Rols M.-P., Zumbusch A. Endocytosis and endosomal trafficking of DNA after gene electrotransfer in vitro. Mol. Ther. Nucleic Acids. 2016;5:e286. doi: 10.1038/mtna.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rudolph C., Plank C., Lausier J., Schillinger U., Müller R.H., Rosenecker J. Oligomers of the arginine-rich motif of the HIV-1 TAT protein are capable of transferring plasmid DNA into cells. J. Biol. Chem. 2003;278:11411–11418. doi: 10.1074/jbc.M211891200. [DOI] [PubMed] [Google Scholar]
- 86.Torchilin V.P., Levchenko T.S., Rammohan R., Volodina N., Papahadjopoulos-Sternberg B., D’Souza G.G.M. Cell transfection in vitro and in vivo with nontoxic TAT peptide-liposome-DNA complexes. Proc. Natl. Acad. Sci. USA. 2003;100:1972–1977. doi: 10.1073/pnas.0435906100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rasmussen I., Vilhardt F. Macropinocytosis is the entry mechanism of amphotropic murine leukemia virus. J. Virol. 2015;89:1851–1866. doi: 10.1128/JVI.02343-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sánchez E.G., Quintas A., Pérez-Núñez D., Nogal M., Barroso S., Carrascosa Á.L., Revilla Y. African swine fever virus uses macropinocytosis to enter host cells. PLoS Pathog. 2012;8:e1002754. doi: 10.1371/journal.ppat.1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun X., Yau V.K., Briggs B.J., Whittaker G.R. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology. 2005;338:53–60. doi: 10.1016/j.virol.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 90.Wittrup A., Sandgren S., Lilja J., Bratt C., Gustavsson N., Mörgelin M., Belting M. Identification of proteins released by mammalian cells that mediate DNA internalization through proteoglycan-dependent macropinocytosis. J. Biol. Chem. 2007;282:27897–27904. doi: 10.1074/jbc.M701611200. [DOI] [PubMed] [Google Scholar]
- 91.Yu M., Tan W., Lin H. A stochastic model for DNA translocation through an electropore. Biochim. Biophys. Acta. 2012;1818:2494–2501. doi: 10.1016/j.bbamem.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 92.Escoffre J.-M., Portet T., Favard C., Teissié J., Dean D.S., Rols M.-P. Electromediated formation of DNA complexes with cell membranes and its consequences for gene delivery. Biochim. Biophys. Acta. 2011;1808:1538–1543. doi: 10.1016/j.bbamem.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 93.Chang C.-C., Mao M., Liu Y., Wu M., Vo-Dinh T., Yuan F. Improvement in electrotransfection of cells using carbon-based electrodes. Cell. Mol. Bioeng. 2016;9:538–545. doi: 10.1007/s12195-016-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bhattacharya S., Roxbury D., Gong X., Mukhopadhyay D., Jagota A. DNA conjugated SWCNTs enter endothelial cells via Rac1 mediated macropinocytosis. Nano Lett. 2012;12:1826–1830. doi: 10.1021/nl204058u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fumoto S., Nishi J., Ishii H., Wang X., Miyamoto H., Yoshikawa N., Nakashima M., Nakamura J., Nishida K. Rac-mediated macropinocytosis is a critical route for naked plasmid DNA transfer in mice. Mol. Pharm. 2009;6:1170–1179. doi: 10.1021/mp900042p. [DOI] [PubMed] [Google Scholar]
- 96.West M.A., Prescott A.R., Eskelinen E.L., Ridley A.J., Watts C. Rac is required for constitutive macropinocytosis by dendritic cells but does not control its downregulation. Curr. Biol. 2000;10:839–848. doi: 10.1016/s0960-9822(00)00595-9. [DOI] [PubMed] [Google Scholar]
- 97.Feige J.N., Sage D., Wahli W., Desvergne B., Gelman L. PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microsc. Res. Tech. 2005;68:51–58. doi: 10.1002/jemt.20215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.