Abstract
Myotonic dystrophy type 1 (DM1) is caused by (CTG⋅CAG)n-repeat expansion within the DMPK gene and thought to be mediated by a toxic RNA gain of function. Current attempts to develop therapy for this disease mainly aim at destroying or blocking abnormal properties of mutant DMPK (CUG)n RNA. Here, we explored a DNA-directed strategy and demonstrate that single clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-cleavage in either its 5′ or 3′ unique flank promotes uncontrollable deletion of large segments from the expanded trinucleotide repeat, rather than formation of short indels usually seen after double-strand break repair. Complete and precise excision of the repeat tract from normal and large expanded DMPK alleles in myoblasts from unaffected individuals, DM1 patients, and a DM1 mouse model could be achieved at high frequency by dual CRISPR/Cas9-cleavage at either side of the (CTG⋅CAG)n sequence. Importantly, removal of the repeat appeared to have no detrimental effects on the expression of genes in the DM1 locus. Moreover, myogenic capacity, nucleocytoplasmic distribution, and abnormal RNP-binding behavior of transcripts from the edited DMPK gene were normalized. Dual sgRNA-guided excision of the (CTG⋅CAG)n tract by CRISPR/Cas9 technology is applicable for developing isogenic cell lines for research and may provide new therapeutic opportunities for patients with DM1.
Keywords: myotonic dystrophy, trinucleotide instability, (CTG⋅CAG)n repeat, DM1 myoblasts, CRISPR/Cas9, therapeutic genome editing, NHEJ, dsDNA break repair, somatic cell therapy
van Agtmaal et al. demonstrate that excision of the expanded (CTG⋅CAG)n repeat from the DMPK gene in myotonic dystrophy patients can be reliably achieved by dual CRISPR/Cas9 cleavage at either side of the repeat and that this offers a strategy to adjust anomalous effects of expanded DMPK transcripts in myoblasts.
Introduction
Myotonic dystrophy type 1 (DM1) is an inherited multisystemic disorder that manifests itself at different ages, with variable expression of progressive skeletal muscle wasting, myotonia, dysfunction of the heart, gastrointestinal problems, insulin resistance, cataract, and alterations in cognitive functions and behavior associated with white matter loss in the central nervous system.1, 2 DM1’s autosomal-dominant character, complex symptoms, and progression are caused by expansion of a (CTG⋅CAG)n-triplet repeat located in the 3′ UTR of the DMPK gene3, 4, 5 and in a partially overlapping antisense (DM1-AS) gene.6, 7 In DM1 families, the repeat contains more than 37 to up to several thousands of triplets and is unstable, both somatically8, 9 and intergenerationally,10, 11, 12 with a bias toward expansion, causing an increase in severity and an earlier onset of disease symptoms during aging and over successive generations.
Several mechanisms may contribute to the molecular pathogenesis of DM1, but the prevailing idea is that expanded (CUG)n-containing DMPK transcripts are dominant in disease etiology. In cells where the gene is expressed, expanded DMPK transcripts may abnormally associate with RNA-binding proteins, like members of the muscleblind-like (MBNL1–3), DEAD-box helicase (DDX), and heterogeneous ribonucleoprotein particle (hnRNP) families, causing sequestration in ribonucleoprotein (RNP) complexes that occur as distinct foci or remain in a diffuse soluble state. Other anomalies in the ribonucleoprotein network of DM1 cells are caused by altered phosphorylation of RNA-binding proteins like CELF1 or Staufen 1,13, 14 triggered by kinase activation in stress responses. In turn, these imbalances have serious in trans consequences for faithful alternative splicing,15, 16 polyadenylation,17 and expression of miRNAs,18, 19, 20 creating a network of cellular dysfunction. Additional problems may emerge from the production of toxic homopolymeric polypeptides, which are formed by decoding of the normally untranslated (CUG)n repeat tract in DMPK mRNA by repeat-associated non-ATG (RAN) translation.21, 22 Similar toxic mechanisms may be active in tissues that express DM1-AS transcripts with expanded (CAG)n repeats. Finally, (CTG⋅CAG)n expansion may also modify nearby chromatin structure,23 which is associated with epigenetic marking or altered expression of other genes in the DM1 locus like the SIX5 gene.23, 24, 25, 26, 27, 28
Due to this enormous complexity and our still unripe knowledge about the significance of these pathobiological mechanisms, it is not surprising that the development of therapy that could stop the cellular problems and thereby delay the onset or slow the progression of muscle wasting, white matter loss in brain, and other disease features seen in DM1 patients is still an unmet medical goal. From DM1 cell and mouse model studies, there is significant support for considering the RNA gain-of-function toxicity the prime therapeutic target, and proof-of-concept testing has already demonstrated that antisense oligonucleotide (AON)-mediated degradation of DMPK (CUG)n transcripts or disruption of abnormal RNP complexes by RNA binding or MBNL displacement has potential therapeutic utility.29, 30, 31, 32 Hurdles that still have to be overcome for use in vivo relate to modes of administration, cell-type specificity of action, and possible immune effects of repeated treatment with AONs or small molecule drugs. Also, more fundamental questions about repeat length effects on DMPK mRNA structure and accessibility in abnormal RNP complexes, AON, or drug effects on intracellular (re)distribution of repeat-containing RNAs and their involvement in RAN translation need attention for further progress. Moreover, therapies that degrade the (CUG)n transcript or destabilize ribonuclear foci are expected to have no impact on the modification of local chromatin structure, the dysregulation of DM1-AS transcripts,6 or SIX524, 25, 33 and possibly other in cis pathobiological effects at the DNA level.
Here, we have started to evaluate the use of somatic gene editing with endonucleases as a promising alternative for the correction of DM1 problems because this strategy offers the opportunity to drive permanent correction of the (CTG⋅CAG)n expansion mutation and cancel out DM1-associated problems at all levels, including the cis epigenetic effects and trans effects on the transcriptome and proteome.34 Specifically, we have sought to test in muscle cells whether the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system can be used to excise the expanded (CTG⋅CAG)n repeat and negate its negative effects by normalization of the expression and nucleocytoplasmic transport of long (CUG)n RNA from the mutant DMPK allele, without compromising the expression of genes like SIX5 and DM1-AS. By conducting tests in myoblasts from a transgenic DM1 mouse model, DM1 patients, and unaffected individuals, we analyzed how the abnormal topology of the repeat affects the efficacy of cleavage and repair of double-strand DNA breaks (DSBs) and gaps introduced in the DM1 locus by single and dual CRISPR/Cas9 genome editing approaches. Our findings on the gene editing outcome are applicable in the development of isogenic cell lines for research and in developing new strategies for future therapeutic intervention options in the tissues of patients with myotonic dystrophy and perhaps other types of repeat expansion disorders.
Results and Discussion
CRISPR/Cas9 Genome Editing in the DM1 Locus
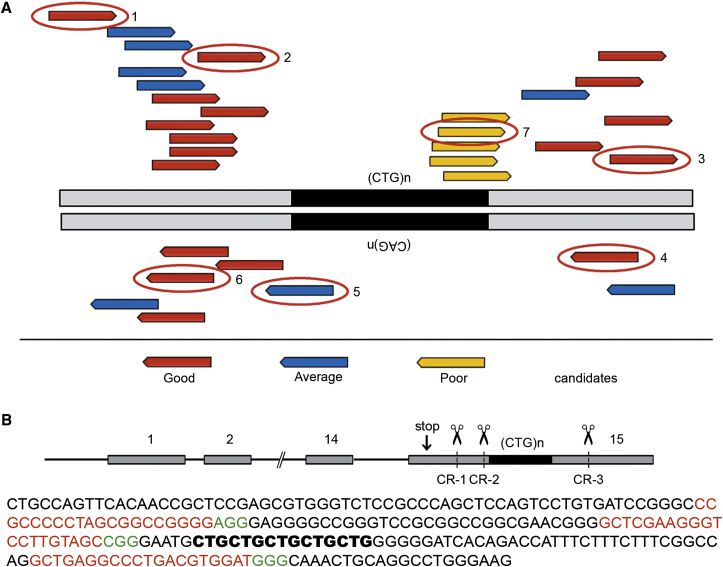
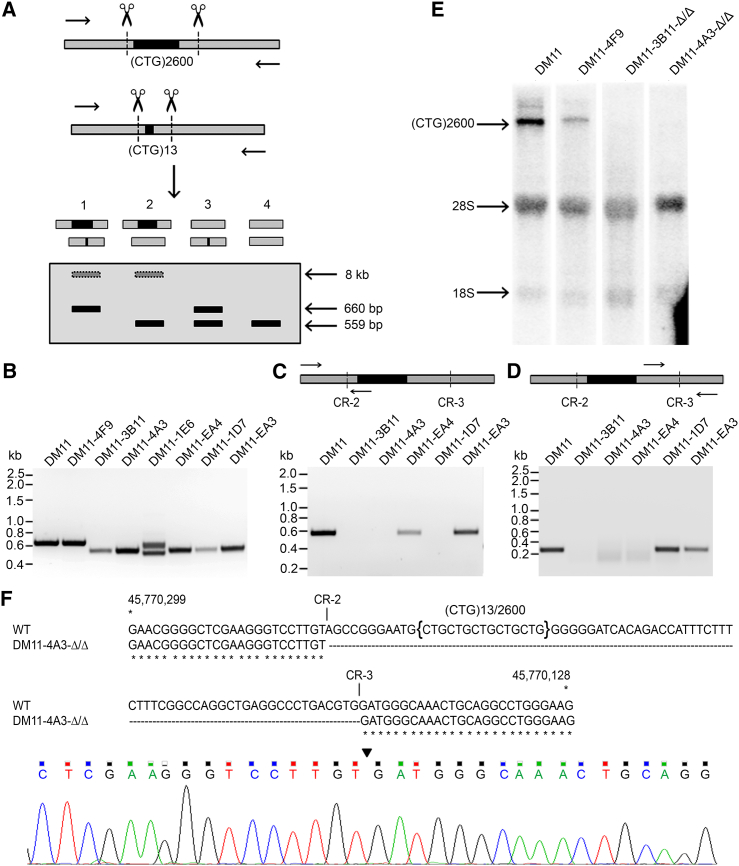
To select target sequences for CRISPR/Cas9 nucleases in the 3′ UTR in DMPK exon 15, we used different versions of guide RNA (gRNA) design software, allowing prediction of performance in the context of a human genomic background. Multiple candidate target sequences with low probability for off-target recognition and a high capacity for promoting double-stranded DNA (dsDNA) cleavage at unique sequence sites upstream and downstream of the (CTG⋅CAG)n repeat were identified (Figure 1A). The repeat tract itself cannot be targeted internally because of the absence of a protospacer adjacent motif (PAM) sequence. Sequences at its periphery were poor targets (yellow arrows in Figure 1A) because prediction indicated a high probability of targeting to other (CTG⋅CAG)n repeats elsewhere in the genome. Moreover, pilot experiments revealed a conspicuous low efficiency for CRISPR/Cas9 nucleases with repeat-directed gRNAs, so they were not considered further (Table S1; CRISPR-5 and -7 and data not shown).
Figure 1.
CRISPR Design
(A) Schematic overview of CRISPR/Cas9 target sites across part of the 3′ UTR in DMPK exon 15. The (CTG⋅CAG)n repeat is indicated in black, and flanking regions in the DMPK gene are in gray. Using different web tools, multiple candidate gRNA target sequences were identified upstream and downstream of the repeat. Positions of guide RNA target sites are displayed by arrows in different colors, representing a good (red), average (blue), or poor (yellow) utility score. Target sites that were chosen for further experiments are encircled and numbered as CRISPR-1, CRISPR-2, etc., in the text. (B) Schematic overview of the structural organization of the DMPK gene, including the (CTG)n repeat in exon 15 in black. Positions of cleavage sites for CRISPR-1, -2, and -3, i.e., the gRNA target sites that were most intensely used in this study, are indicated. The corresponding sequence in exon 15 is displayed below, with the (CTG)n repeat in bold black, CRISPR sites in red, and PAM sequences in green letters.
For comparison of the efficacy of various CRISPR/Cas9 nuclease-gRNA expression plasmid combinations, we chose to work with muscle cells, a cell type that is highly relevant for DM1 manifestation. Immortalized myoblast lines from unaffected individuals (LHCN and KM155C25),35 lineages that have overcome replicative aging because of retroviral expression of hTERT and CDK4,35 were employed. LHCN myoblasts carry two identical DMPK alleles with a (CTG⋅CAG)5 repeat, which facilitated analysis of efficiency of CRISPR/Cas9-mediated DSB formation by circumventing the need for discrimination between allelic repeat-specific effects on indel formation after break repair. Cleavage efficiency in nucleofector-transfected cells was assessed by a T7 endonuclease I (T7EI) assay36 on genomic DNA from pools of these cells (Figure 2B). As controls in the assay, untreated LHCN myoblasts showed a single, distinct T7 endonuclease-resistant fragment, whereas for KM155C25 myoblasts, carrying one (CTG⋅CAG)5 and one (CTG⋅CAG)14 allele, cleavage across CTG5⋅CAG14 or CTG14⋅CAG5 misaligned tracts in hybrid PCR products resulted in the appearance of two additional fragments (Figures 2A and 2B). Based on signal strength in the T7EI assay, two gRNA/Cas9 nucleases that showed the best ability to direct cleavage were chosen, with target sites located 11 bp upstream of the first CTG triplet (CRISPR-2, 8%–21% efficiency) and 51 bp downstream of the last CTG triplet (CRISPR-3, ∼14% efficiency). Other gRNA vectors with lower efficiencies (e.g., CRISPR-1 or candidates in Figure 1A; Table S1) were not used further in this study.
Figure 2.
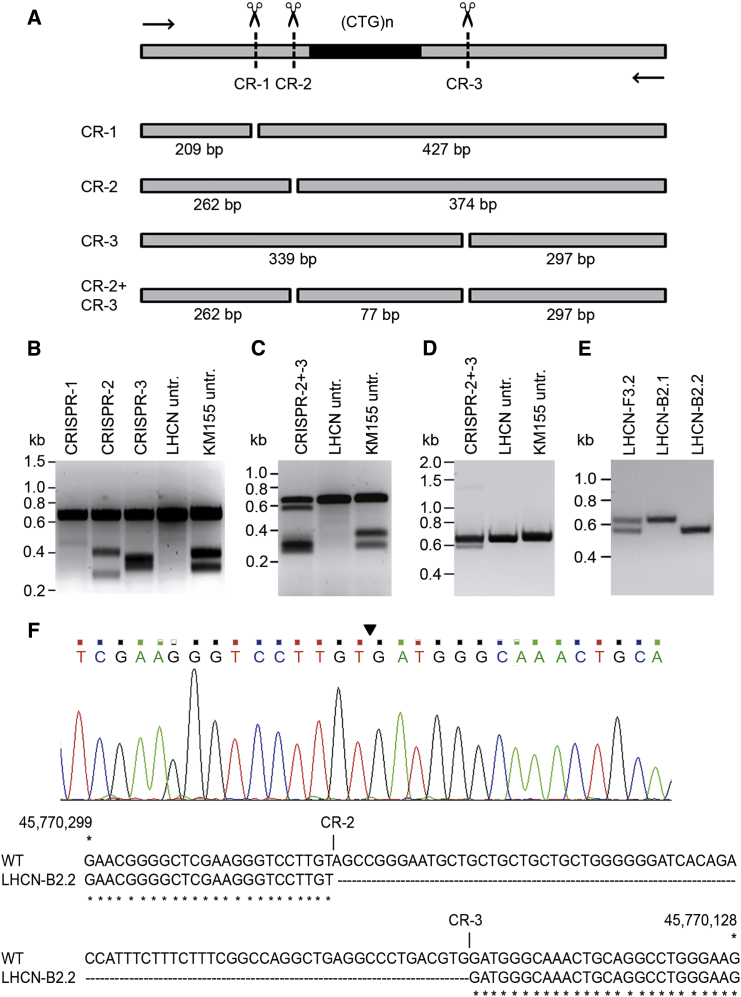
CRISPR Activity in Myoblasts with Normal-Size (CTG⋅CAG)n Repeats
(A) Schematic outline of the T7EI assay for determination of CRISPR cleavage efficiency. Part of DMPK exon 15 ([CTG⋅CAG]n repeat in black) containing CRISPR-1, -2, and -3 recognition sites and positioning of PCR primers used for amplification of the relevant segment are shown on top. Possible fragments formed in the assay are depicted, with sizes given underneath. (B) T7EI assay of DNA from small pools of transfection-positive LHCN cells. Quantification of signal strength (assessed by scanning of fluoresce signal intensity upon UV illumination for all assays shown in B–E) revealed target efficiencies of <1% for CRISPR-1, 8%–21% for CRISPR-2, and 14% for CRISPR-3. DNAs of non-transfected (untr.) LHCN (two alleles with equal DMPK repeat lengths) and KM155C25 myoblasts (one [CTG⋅CAG]5 and one [CTG⋅CAG]14 allele) were used as negative and positive control, respectively. (C) T7E1 assay of DNA from a pool of LHCN myoblasts treated with CRISPR-2 and -3 simultaneously. Untransfected LHCN and KM155C25 were included as controls. Note that we could not differentiate between deletion of the entire region between the two CRISPR sites or simultaneous formation of small indels at each of the two CRISPR sites. (D) PCR analysis of the relevant DMPK genomic segment after dual genome-editing with CRISPR-2 and -3. The upper band represents the unmodified PCR product. The lower band is indicative for deletion of the 77 bp (CTG⋅CAG)5 repeat and flanking regions in a small portion of CRISPR-2- and CRISPR-3-treated cells. (E) PCR analysis of genome changes in three CRISPR-2- and CRISPR-3-treated LHCN cell clones. Clone LHCN-B2.1 contains two unmodified repeats (single signal at 636 bp), whereas clone LHCN-B2.2 has a repeat deletion on both alleles (single signal at 559 bp). Clone LHCN-F3.2 carries one unmodified and one edited allele (signals at 559 bp and 636 bp). (F) Sequence verification of excision of the repeat-containing segment. Top: sequencing profile of the DMPK exon-15 gene region in clonally expanded LHCN cells after dual gene editing with CRISPR-2 and -3. The site at which the DSBs are fused is indicated by an arrowhead. No indels were found. Bottom: the exon-15 sequence lacking the 77-bp repeat-containing segment aligned with the normal DMPK sequence.
As a next step, we assessed activity of CRISPR-2 and -3 nucleases toward potential off-target sites elsewhere in the genome. By use of computational target prediction, potential sites were identified on the basis of similarity to the gRNA sequences. Among the sites with the highest prediction values were the CARMIL2, EBF3, DVL1, and ALK loci (Table S2). Verification with the T7E1 assay revealed no indels within these loci, indicating that no or very low cleavage activity outside the DMPK locus occurred upon use of CRISPR-2 and -3 (Figure S1). We realize, however, that indel formation at these and non-predicted off-target cleavage sites in the genome may depend on the timing and level of expression and the cell type in which these CRISPRs are being used. Precise monitoring of their fidelity and specificity would require whole genome sequencing for every variable, which is not realistic at this point. Of note, while this work was in progress, development of a new high-fidelity version of CRISPR/Cas9 nuclease was reported.37 Switching to this new tool, in combination with the two gRNAs identified here, may thus become a useful strategy for the avoidance of potential problems. Because our observations made it sufficiently clear that the occurrence of off-target DSBs constitutes only a minor problem with gene editing in the DMPK gene in myoblasts, we decided not to pursue this issue further here.
Deletion of a Short (CTG)n Repeat in Unaffected Human Myoblasts
Having observed that CRISPR-2 and -3 were effective tools for cleavage in the 3′ UTR of the DMPK gene, we next wondered whether combined use could be applied for removal of the entire repeat-containing segment. This strategy was first tested in (CTG)5/(CTG)5 LHCN myoblasts, in which this segment has the smallest possible size, spanning only 77 bp. T7E1 analysis of editing events (Figure 2C) combined with conventional PCR analysis (Figure 2D) revealed the appropriate size changes across the genomic area. Importantly, in most cases in which cleavage had occurred, both CRISPR-2 and -3 must have been simultaneously active on one allele. PCR amplification gave the original and edited products of 636 and 559 bp, respectively, in a 5:1 ratio (Figure 2D). This confirmed that the 77-bp repeat-containing segment had been deleted in a fair percentage of cells and also that the area was unmodified in other cells within the population.
Conventional PCR analysis of a series of 18 single-cell clones (representative clones shown in Figure 2E), obtained by limiting dilution after initial enrichment in small cell pools, showed that in ∼67% of cells, the (CTG⋅CAG)5-containing segment had been completely removed from at least one allele. Sequence analysis of PCR products confirmed that this excision had largely occurred cleanly, without additional deletions or insertions in 83% of the modified target sites (Figure 2F; Table S3). LHCN cell clones with double clean deletions were designated LHCN-(clone#)-Δ/Δ cells (Table S4) and used for later biological typing (see below). Only in ∼17% of alleles, small 2–27 nt indels (deletions only) were identified at CRISPR-2 and/or -3 recognition sites. Among the cells with these changes, small deletions were slightly more frequent at the CRISPR-3 site, presumably because of better guidance efficiency associated with more frequent cleavage.
Removal of a Long (CTG⋅CAG)n Repeat Segment in Myoblasts Derived from a DM1 Mouse Model
To characterize the editing potential of the CRISPR-2/-3 nuclease combination for DMPK genes with large (CTG⋅CAG)n expansions, we performed an initial series of tests in DM500 myoblasts, an immortalized cell lineage derived from the calf muscle of a compound-hemizygous DM1 model mouse that carried one cosmid-sized transgenic insert with expanded (CTG⋅CAG)330 DMPK allele38 and one H-2Kb-SV40tsA58 transgene.31 We reasoned that the best view on authentic genome and transcript changes after dual CRISPR cleavage would be obtained in cells with only one target chromosome, without the possibility of target competition or gene conversion between DMPK alleles of different lengths, as might occur in patient cells with two chromosomes 19. Unfortunately, this criterion was not fully met because the DM500 myoblast lineage had undergone tetraploidization during propagation in culture at the permissive temperature (Figure S2), a common feature of cells that express the SV40 large T antigen during cell cycle progression.39, 40 Small-pool PCR analysis of repeat length variation in the DM500 cell population revealed three types of main alleles with ∼540, ∼570, and ∼610 CTG⋅CAG repeats (Figures 3A–3D) and a subset of rarer expansion and contraction events around these two alleles. We assume that the three main alleles detected represent somatic variants that arose early after, or possibly during, the tetraploidization event. Overall, the degree of somatic instability in the DM500 cell population appeared relatively low, likely reflecting the comparatively low level of somatic instability observed in muscle in vivo in the original transgenic mice38 and the close correlation between the tissue specificity of somatic mosaicism in vivo and that observed in vitro in tissue culture.41 Although the presence of two chromosomes per cell containing large expanded alleles does not reflect the normal situation in patients, we nonetheless considered this cell line useful for determining the efficiency of CRISPR-mediated cutting in modifying the length of large expanded (CTG⋅CAG)n repeats at the DM1 locus.
Figure 3.
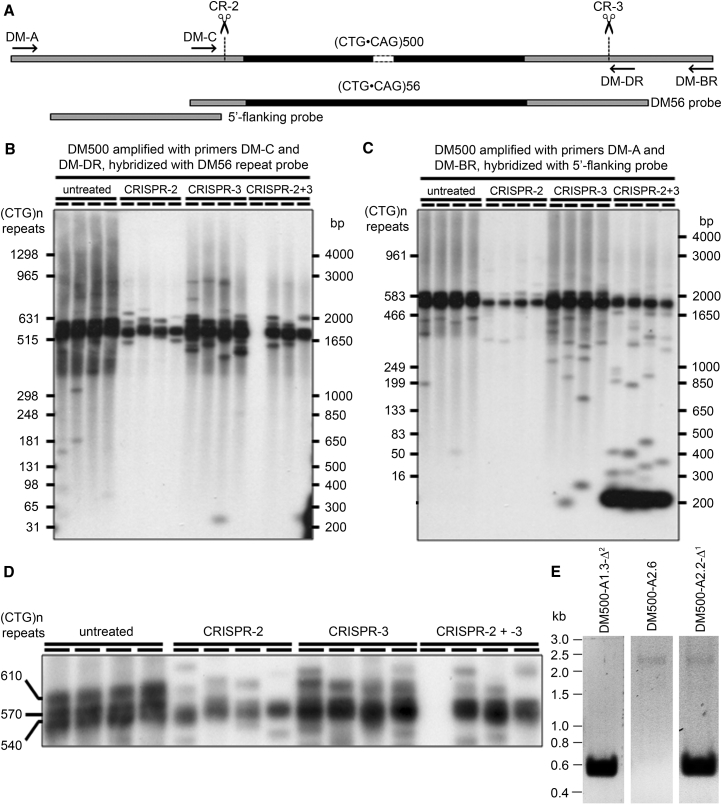
Treatment of DM500 Cells with CRISPR-2 and/or -3
(A) Schematic overview of CRISPR/Cas9 target sites relative to the (CTG⋅CAG)n repeat (black) and the flanking DNA (gray) PCR primers (arrows) and probes used for small pool PCR analysis. (B–D) Small-pool PCR analysis of genomic DNA from DM500 cells treated with CRISPR-2 and/or CRISPR-3. (B) shows amplicons from control and treated DM500 cells, generated using the inner primers (DM-C−/−DR) and hybridized using the (CTG⋅CAG)n probe DM56. (C) shows amplicons from control and treated cells, generated using the outer primers (DM-A/-BR) and hybridized using the 5′-flanking probe. For the untreated and treated cells, four replicate PCRs containing ∼50 molecules of template DNA are shown. Note that a clean full-excision DM-C/-DR product is only 85 bp long and is not expected to be detected with the DM56 probe in (B). In (B) and (C), the molecular weight markers (right-hand scale) have been converted to the number of (CTG⋅CAG)n repeats on the left-hand scale. (D) A zoomed-in shorter exposure of the autoradiograph in (B) reveals that the DM500 cell line comprises three primary alleles of ∼540, ∼570, and ∼610 (CTG⋅CAG)n repeats and that the ∼570 and ∼610 alleles are preferentially modified by both CRISPR-2 and -3. (E) PCR analysis of genome changes in DM500 clones treated with CRISPR-2 and -3 using the DMPK e15 primers described in Materials and Methods. Clone DM500-A2.6 contains unmodified (CTG⋅CAG)540/570/610 repeats (single signal at ∼2.2 kb). Clone DM500-A1.3-Δ2 has a repeat deletion on both chromosome copies (single signal at 559 bp). Clone DM500-A2.2-Δ1 carries one unmodified allele and one edited allele (signals at ∼2.2 kb and 559 bp, respectively). Note that the enormous difference in signal intensity is due to relative inefficient amplification of the expanded repeat-containing allele.
Encouragingly, small-pool PCR analysis of the entire population of quadruple-transfected cells (CRISPR-2 and -3 gRNAs, Cas9 vector, and EGFP-reporter plasmid) suggested the ∼1.7-kbp deletions (∼540/570/610 CTG⋅CAG triplets plus 62 bp of flanking sequences) were very efficient, yielding a predominant product of ∼200 bp when amplified with the distal DM-A/-BR primer pair (Figures 3A and 3C). Although the primary product appeared to be the hoped for clean excision of the repeat tract, a subset of molecules with CRISPR-2 and -3 treatment yielded variably sized fragments, presumably carrying repeat tracts intermediate between (CTG⋅CAG)0 and (CTG⋅CAG)500 (Figure 3B). The failure to detect most of these products with the inner DM-C/-DR primer pair (Figure 3B) suggests that many of these targeted loci acquired deletions that have removed the DM-C or DM-DR primer site (note the clean full-excision DM-C/DM-DR product is only 85 bp long and is not expected to be detected with the DM56 probe).
Interestingly, the small-pool PCR analysis revealed that both CRISPR-2 and -3, singly and together, preferentially targeted the (CTG⋅CAG)570 and (CTG⋅CAG)610 repeat tracts (Figures 3C and 3D). We assume that the chromatin configuration at the two transgenic DM1 loci differed between the duplicated chromosomes and that this generated differential accessibility for CRISPR/Cas9 complexes, consistent with recent data suggesting a role for chromatin structure in mediating CRISPR activity.42, 43 This may have important implications for the targeting efficiency of the normal and mutant chromosome in DM1 patient cells, given the known effect of the (CTG⋅CAG)n repeat expansion in altering the local chromatin structure.23, 27, 33
To better define the efficacy of CRISPR-mediated removal of large genomic regions, we also performed PCR (Figure 3E) and sequence analysis of the DMPK exon 15 region on a number of individually propagated genome-edited DM500 cell clones. From this analysis, we estimated that clean loss of the segment between the CRISPR-2 and -3 cleavage sites had occurred in approximately one-third (30%) of the modified transgenic chromosomes (Table S3). Imperfect deletions extending beyond the CRISPR sites were seen in 4% of the modified transgenic chromosomes. Other mutations (7%) involved large insertions and deletions and inversions. A representative subset of clones with clean repeat loss, i.e., with complete deletion in either one or both of the identical transgenic chromosomes (designated DM500-[clone#]-Δ1 or DM500-[clone#]-Δ2, respectively; Table S4), was used for further biological study (detailed below). In a number of clones that still contained at least one transgenic chromosome with an expanded (CTG⋅CAG)540/570/610 repeat, small indels of 1–24 bp were found at the CRISPR-2 or -3 recognition sites. These indels were present in 37% of all modified transgenic chromosomes, which is higher than the percentage of indels found in CRISPR-edited LHCN cells (17%; Table S3). About 90% of the indels consisted of small deletions, and 10% consisted of small insertions (data not shown). This finding may reflect species (i.e., human versus mouse), chromosome, or cell-type-specific differences in efficacy or modes of DNA repair. Also, cis effects of the long (CTG⋅CAG)540/570/610 repeat on the repair ability of CRISPR-2 or -3 breaks may have been involved. DSBs stimulate DNA repair by at least two distinct mechanisms—non-homologous end joining (NHEJ) and homology-directed repair (HDR)—of which the former is error prone.44 Differential activities of NHEJ or HDR on any of the two chromosomes 19 of different parental origin in the LHCN cells or on the two essentially identical transgenic chromosomes of maternal origin in DM500 cells or simply timing differences needed for sealing of small 77-bp or large ∼1.7-kbp gaps by these mechanisms may thus have influenced the steps used for sealing of the CRISPR cut sites by DNA-repair enzymes.
Single DSBs Near a Long (CTG⋅CAG)n Segment Frequently Induce Loss of the Entire Repeat
In order to gain further insight into the mechanism(s) that direct healing of the large gap between CRISPR-2- and CRISPR-3-induced DSBs in DM500 myoblasts, we examined the fate of the repeat segment after a single cleavage upstream or downstream by either CRISPR-2 or -3, respectively. Triple transfection (gRNA vector, Cas9 vector, and EGFP reporter), followed by cell sorting for GFP, was therefore executed. For global monitoring of sequence alterations within the target region, we PCR amplified genomic DNA isolated from small batches of 20–30 individual clones, propagated until day 28 after transfection, and combined this with detection by Southern blot analysis. To cover a broad range of possible indels, a large region was PCR amplified, bracketing the entire repeat and 273 bp and 348 bp of unique sequences upstream and downstream of the repeat, including CRISPR-2 and -3 sites. PCR products were hybridized with two independent probes (Figure 4). Even though segments with long repeats amplified with poor efficiency, these analyses were informative and demonstrated that introduction of one DSB at either side induced loss of almost the entire repeat surprisingly often, occurring once or multiple times in almost every batch of pooled cells (Figure 4). Small-pool PCR, carried out on genomic DNA from these batches of CRISPR-2- or CRISP-3-transfected cells, corroborated this observation and again demonstrated that repeat tracts were primarily lost from the transgenic chromosome that carried the larger (CTG⋅CAG)570/610 repeat tract (Figure 3). Sequence determination across the newly formed junctions within DMPK exon 15 confirmed that healing of the initial DSB induced formation of large gaps across the entire repeat (Figures 4B and S3). The application of single CRISPR cleavage adjacent to the repeat may thus have utility in generating isogenic cell lines with variable (CTG⋅CAG)n repeat sizes.
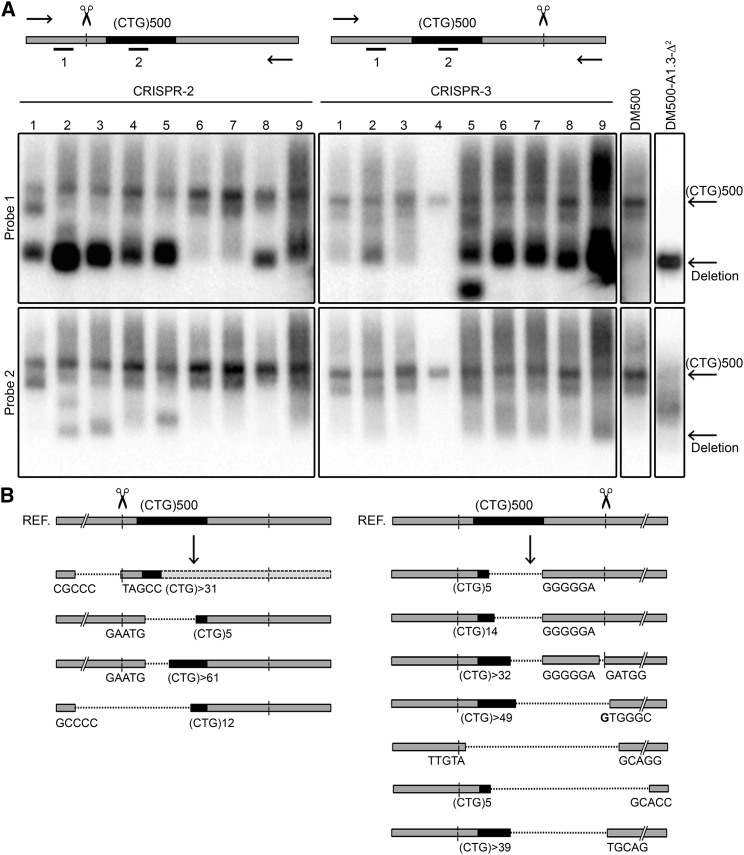
Figure 4.
Alterations in the DM500 Repeat Region after Cleavage with a Single CRISPR
DM500 cells were treated with either CRISPR-2 or -3, after which a 2.2-kb region (including the (CTG⋅CAG)530/580 repeat) was PCR amplified and subsequently analyzed on blot using 32P-labeled probes. (A) Top: outline of the relevant region in DMPK exon 15. Shown are the (CTG⋅CAG)500 repeat in black, cleavage sites for CRISPR-2 and -3, and locations of PCR primers (arrows) and probes used for hybridization detection (bars). Bottom: four large panels showing signals from PCR products from nine pools of DM500 cells (∼20–30 clones per pool) treated with CRISPR-2 (left panel) or CRISPR-3 (right panel), hybridized with probe 1, a 32P-labeled DMPK oligonucleotide located 5′ of the CRISPR cleavage sites (upper panels), or probe 2, a 32P-labeled (CAG)9 oligonucleotide (lower panels). Besides, PCR products of untreated DM500 cells and clone DM500-A1.3-Δ2 carrying a verified deletion of both repeats were used as controls (small panels on far right). Presence of small PCR products in the upper panels indicates deletion of large parts of the repeat in a fair proportion of cells. These products are invisible in the lower panels because the (CAG)9 probe will not bind if the repeat is entirely lost or considerably shortened. (B) Overview of different types of deletions induced by cleavage with single CRISPRs in isolated DM500 cell clones treated with CRISPR-2 (left; four examples) or CRISPR-3 (right, seven examples). Results shown are based on sequencing of the transgenic DMPK exon-15 gene region of these clones. Sizes of residual (CTG⋅CAG)n repeats and sequences flanking the deletions are indicated. Gray rectangles with dotted outlines indicate that no sequence data were available for that particular region.
To our knowledge, such frequent occurrence of exceptionally large deletions has never been reported before in genome-editing studies with endonucleases. Formation of small indels is commonly seen at CRISPR/Cas9-induced DSBs, but deletions in the kbp size range, preferentially formed in the unidirectional direction as we show here, seem exceptional (e.g., compare deep-sequencing assessment of indels after NHEJ and HDR events45). Instability resulting in contraction has been reported for transcription activator-like effector-based nuclease (TALEN)-cleaved short (CAG⋅CTG)30–75 repeats engineered in yeast46 and zinc finger nuclease (ZFN)-cleaved (CAG⋅CTG)23–95 repeats engineered in human cells.47 Both studies demonstrated that introduction of one DSB and the subsequent recruitment of DSB repair machinery significantly contributed to repeat instability, mostly contraction, via mechanisms in which intramolecular repair reactions prevail. Repair, in which gene conversion events between allelic segments or sister chromosomes in the S or G2 phase, could also have played a role at low frequency. Finally, as a last speculative possibility, aberrant pairing between the extremely GC-rich regions upstream and downstream of the repeat may have occurred upon single cleavage, and mechanisms that promote removal of large DNA overhangs (including the repeat!) in the single strand annealing pathway for closure of dsDNA breaks might have become active.48 Interestingly, contraction of the (CGG⋅CCG)n repeat in the FMR1 gene occurred at a low frequency in induced pluripotent stem cells (iPSCs) derived from fragile X-syndrome patients when cleaved upstream by CRISPR/Cas9,49 and deletion of a (TG⋅CA)70 dinucleotide repeat was induced by TALEN cleavage 129 bp downstream of the repeat in zebrafish.50 Extensive DNA loss may thus be a common process if simple tandem repeats are exposed to the DNA repair machinery by nearby DSBs. Precise comparison between the extent of DNA loss in the different systems remains difficult, however, because PCR primers used for analysis were differentially positioned at variable distances from the CRISPR/Cas9 or TALEN cut sites and large deletions may have been missed in some studies.
Strikingly, the gaps that were formed across the repeat in the transgenic DM1 locus in DM500 cells extended to different nucleotide positions within the repeat or its unique flanking sequences. Because remaining (CTG⋅CAG)n repeats (n > 50) were difficult to amplify and sequence, an exact repeat number could not be precisely determined for most products, but a rough estimate from agarose gel electrophoresis indicated that considerable size contractions must have occurred, with some alleles collapsing to less than 100 triplets. In a few clones, we found, by sequencing, a deletion of the larger internal part of the CTG repeat, but a fully intact flanking region between the leftover of the repeat and the actual site of CRISPR cleavage (Figure 4B). Most likely, this type of complex genome repair must have occurred after cleavage and unidirectional trimming across the repeat segment. We propose that the unique 11-bp (for CRISPR-2) or 51-bp (for CRISPR-3) flanking region was thereby lost initially. Subsequently, exchange of genetic information by gene conversion between the two (essentially) identical transgenic chromosomes in the tetraploid DM500 cells may have served to restore the unique flanking region. Apparently, this reconstruction leads to incomplete repair because the repeat tract itself did not serve as a template. Direct and indirect in vitro and in vivo studies revealed that the expanded DM1 (CTG⋅CAG)n repeat can adopt unusual non-B DNA slipped strand structures.51, 52 It is thus tempting to speculate that the large deletions are facilitated by slipped strand structures already present and/or the formation of slipped strand structures during DSB processing. Occurrence of abnormal non-B DNA topology may also partly explain why CRISPRs designed to cleave inside or very close to the repeat did not work or worked less efficiently (e.g., CRISPR-5 and -7; Table S1). Interestingly, work with ZFN nucleases that cleave within the (CAG⋅CTG)n repeat suggested higher efficiency of cleavage with increasing repeat length.47 Further study is thus required to further determine how chromatin configuration, DNA topology, and sequence context contribute to target site accessibility and efficiency of different types of endonucleases.
Excision of a Long Expanded Repeat in DM1 Patient Myoblasts
Together, our findings demonstrate that gene editing with single CRISPR cleavage near the trinucleotide repeat may trigger uncontrollable repeat loss. Dual CRISPR/Cas9-directed cleavage may therefore be the preferred approach for repeat deletion in DM1 patient cells. To assess this idea in a more direct manner, we tested the mode and efficiency of removal of a large expanded (CTG⋅CAG)n repeat in cells with a fully authentic DM1 genotype in retrovirally immortalized DM11 cl5 myoblasts derived from a DM1 patient35 with (CTG⋅CAG)13 and (CTG⋅CAG)2,600 repeats. Due to the large difference in allelic repeat size, any anomalous exchange of DNA between these chromosomes, during or after genome editing, can be easily monitored with this cell line.
Dual expression of CRISPR-2 and -3 nucleases resulted in fairly efficient cleavage and loss of both the 101-bp fragment ([CTG⋅CAG]13 repeat plus 62-bp flanking sequences) from the normal allele as well as the large ∼8-kb fragment ([CTG⋅CAG]2,600 plus flanking sequences) from the mutant allele. Through single-cell fluorescence-activated cell sorting (FACS and clonal propagation, we managed to derive 103 different myoblast clones from transfected DM11 cells, allowing precise analysis of editing events at the level of individual cells (Figure 5). Due to difficulties with PCR amplification across the large repeat, we had to adapt our analysis strategy and used a combination of PCRs to monitor the editing fate of the expanded and normal allele in the same cell lineage (Figures 5A–5D). In addition, Northern blot analysis was performed on a few representative clones to verify PCR findings, showing normal dosage or complete lack of allelic DMPK mRNA products in the clones shown in Figure 5E. Table S3 provides an overview of genome alterations that were observed for the (CTG⋅CAG)13 and (CTG⋅CAG)2,600 allele. Because of the above-mentioned difficulties with sequencing, small changes in the (CTG⋅CAG)2,600 allele could not be detected. Altogether, in 36 myoblast clones (out of 103) the CRISPR-2/-3 segment was cleanly removed from both DMPK alleles (Figure 5F). Only one myoblast line had a deleted expanded repeat segment and a (CTG⋅CAG)13 allele that was still fully intact. Altogether, 69% of the (CTG⋅CAG)13 alleles had lost the CRISPR-2/-3 fragment cleanly (62%) or with small imperfections (7%), while 51% of the (CTG⋅CAG)2,600 alleles had a deletion of the entire repeat segment (46% clean and 5% imperfect deletions). Together, these results corroborate findings with (CTG⋅CAG)540/570/610 repeats in DM500 cells and demonstrate that even repeats of the largest size class, associated with severe DM1 forms, can be efficiently excised from the DMPK gene by dual CRISPR-2/-3 cleavage. The finding that not much difference in excision efficiency is observed between wild-type (WT) and long-expanded DMPK alleles is important because it implies that CRISPR-mediated gene editing cannot be easily tuned for selective removal of the repeat segment from the mutant allele only unless combined with clonal selection of cells with the desired targeted change to their genome.
Figure 5.
Dual Genome Editing of DM11 cells Using CRISPR-2 and -3
(A) PCR strategy used for characterization of DM11 clones treated with CRISPR-2 and -3. Four possible outcomes for PCR analysis are displayed (primer positions indicated on top). Because it is not possible to amplify the (CTG⋅CAG)2,600 repeat efficiently, the putative ∼8 kbp signal (gray band with dotted outlines) will not be visible on gel. (B) Results from the PCR for untreated DM11 cells and seven independent DM11 clones treated with CRISPR-2 and -3. Sequencing showed that the deletion of the (CTG·CAG)2,600 repeat in clone DM11-1E6 started 15 bp upstream of the CR-2 site and extended until 16 bp downstream of the CR-3 site; hence, the lower signal is somewhat smaller than the expected 559 bp (outcome 3 in A). A small deletion of 11 nt was found in the (CTG⋅CAG)13 allele. (C and D) Analysis of the fate of the (CTG⋅CAG)2,600 repeat in five different DM11 clones that yielded a single PCR fragment of 559 bp with the assay described in (A). A clean deletion of the (CTG⋅CAG)13 repeat was confirmed by sequencing for all five clones. To verify the fate of the (CTG⋅CAG)2,600 repeat, PCRs were done across the CRISPR-2 site and across the CRISPR-3 site. Absence of products in both reactions indicate deletions of the (CTG⋅CAG)2,600 repeat in both alleles (outcome 4 in A; clones DM11-3B11 and DM11-4A3). (E) Northern blot analysis of RNA isolated from DM11 clones using a 32P-labeled (CAG)9 probe to verify DMPK (CUG)2600 expression. RNA from non-treated DM11 cells was included as positive control. Absence of signal in clones DM11-3B11-Δ/Δ and DM11-4A3-Δ/Δ corroborates successful repeat removal. Clone DM11-4F9 shows a slightly larger DMPK transcript than that of DM11 cells, presumably due to expansion of the repeat during cell culture. (F) Sequence verification of excision of the repeat-containing segment in clone DM11-4A3-Δ/Δ. Top: DMPK exon 15 sequence that has lost the repeat-containing segment aligned with the normal DMPK sequence. Bottom: DNA sequencing profile of the DMPK exon 15 region. The site at which the two DSBs are fused is indicated by an arrowhead. Absence of double peaks indicates that no differences exist between the two modified alleles.
Unexpectedly, we detected three cell clones in which inversion of the 101 bp (CTG⋅CAG)13 segment had occurred (Figure S4). Inversion can only be explained if the fragment that was liberated by dual CRISPR activity was re-incorporated during repair of the DSB. One idea that may explain the relatively high frequency with which we found chromosomal fragment inversion with the (CTG⋅CAG)13 repeat is that the rearrangements are effectively controlled mechanistically by genomic proximity and nuclear organization by a process that keeps the ends of fragments formed by DSBs together in nuclear locales for DNA repair.53, 54, 55, 56 Also, others noticed a high incidence of chromosome segment inversion upon use of dual CRISPR genome cleavage.57 Although no evidence was found for inversion of the (CTG⋅CAG)2,600 repeat, our combined observations and the results of others make clear that careful analysis is essential and caution is warranted in the use of the CRISPR/Cas9 system in myoblast genome engineering.
We expect that further adaptation of the editing strategy, for example, by combining dual CRISPR/Cas9 cleavage with HDR to achieve repair of genome-editing scars in the mutant DMPK gene will not be of much help to avoid unwanted genome changes. In theory, repair of a CRISPR-induced gap or a small inversion would be possible by performing a second round of gene editing with CRISPR cleavage at the junction site(s) and co-transfection of a donor fragment derived from DMPK exon 15, for example, a single-stranded oligonucleotide45 with a normal (CTG⋅CAG)5 repeat. Insertional recombination of this donor fragment by HDR would leave no or only minor sequence alterations around the CRISPR-2 and -3 recognition sites.58 Genome editing with this two-step approach is, however, only realistic for use in cells in vitro and too elaborate for use in vivo. Any direct use of a donor fragment for HDR in muscle cells would make the procedure too highly dependent on correct timing of CRISPR/Cas9 treatment because NHEJ is a much faster process and HDR only occurs in the S and G2-M phases.59, 60, 61 Further sophistication may thus only be possible by genetic or pharmacological suppression of NHEJ62, 63 or by increasing HDR via use of special engineered Cas9 variants,64 cell-cycle manipulation of the host cell, or precise timing of CRISPR cleavage events. We thus anticipate that clean-cut repair of repeat anomalies may not be easily achievable because too many variables must be tailored for use in DM1 muscle cells in vitro and certainly in vivo.
Biological Effects of (CTG⋅CAG)n Repeat Excision: Are Biologically Relevant Sequences Lost?
Excision of the (CTG⋅CAG)n repeat and flanking regions in DMPK exon 15 can only have meaningful medical applications or provide therapeutic benefit if the loss of this gene segment has no adverse biological effects. To gain insight into possible side effects of our excision strategy, we performed a bioinformatics search using RegRNA 2.0, an integrated web server for the identification of regulatory motifs and elements in RNA.65 Analysis of a somewhat larger region than the lost segment of the DMPK RNA (nucleotides 45,770,337-45,770,127 in DMPK) revealed a binding motif for IKAROS family zinc finger 3 (IKZF3), which is involved in chromatin remodeling.66 However, IKZF3 is expressed most strongly in leukocytes, the spleen, and the thymus, and is virtually absent in other tissues (http://www.proteinatlas.org). In addition, two overlapping DNA motifs for poly ADP ribose polymerase (PARP) binding and an activating transcription factor (ATF)/cAMP response element-binding protein (CREB) activator motif were identified. Because none of these proteins has a known function in the DM1 locus, this leaves the role of the deleted tract still enigmatic. A search across the novel junction sequence generated by repair of the DSB in the edited DMPK gene yielded no evidence for newly formed regulatory motifs.
Effects of (CTG⋅CAG)n Repeat Excision on Gene Expression, DMPK mRNA Localization, and Ribonuclear Foci
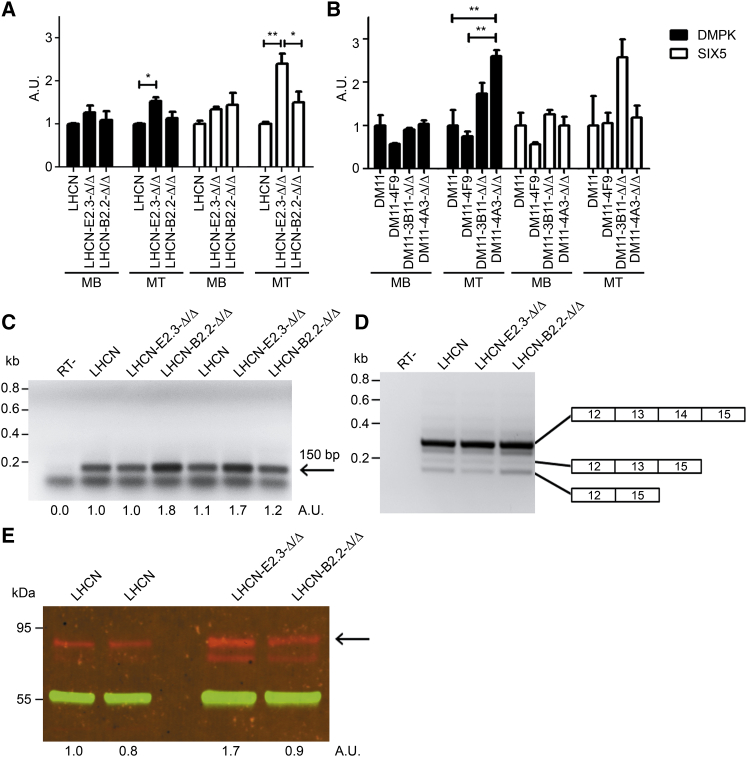
We next endeavored to test the consequences of repeat deletion for DNA coding capacity, RNA fate, integrity of cellular functions, and myogenic capacity. Deletion of the (CTG⋅CAG)n repeat and 62 bp of flanking regions appeared to have no impact on DMPK’s coding capacity or on that from its neighbor SIX5. RT-qPCR determination of steady-state levels of DMPK mRNA in healthy control LHCN myoblasts and gene-edited LHCN-E2.3-Δ/Δ and LHCN-B2.2-Δ/Δ derivatives or myotubes derived thereof (Figure 6A; Table S4 for clone designations) showed that expression did vary, but with a marginal statistical significance between pairs of cells in the series. Differences could also not be attributed to the absence or presence of the repeat tract. A similar picture was observed for DMPK mRNA expression in gene-edited myoblasts and myotubes from the DM11 series (Figure 6B). Note that variation in expression in Figures 6A and 6B is within the 1–2.5 arbitrary unit range when normalized to expression levels of DMPK in the parental LHCN and DM11 myoblast or myotube populations. Differences in the extent of myogenic differentiation, which can be attributed to clonal (epigenetic?) inequality and difficulty in precisely controlling the onset of myogenesis in culture, may explain this variation. An overall similar picture was obtained for expression from SIX5, the neighboring gene in the DM1 locus, although RT-qPCR quantification revealed that expression of this gene in myoblasts-myotubes in culture is very low, corroborating literature data.24, 25, 33 Muscle cells may thus not be the proper cell type to assess cis-acting repeat effects on transcriptional activity of SIX5 because its promoter may already be in a heterochromatic repressed state in these cells. From our findings in LHCN and DM11 clonal derivatives, we can conclude, however, that SIX5 expression by promoter leakage is not altered upon deletion of DMPK’s repeat tract. Further study of epigenetic effects of repeat loss on SIX5 expression must await gene-editing efforts in cells of other tissue origin because this is not reliably possible in our isogenic LHCN or DM11 cell series.
Figure 6.
Effects of (CTG⋅CAG)n Repeat Excision on Expression of RNA and Protein Products from Genes in the DM1 Locus
(A and B) RNA was isolated from (A) LHCN myoblasts (MBs) or LHCN- derivatives (see Table S4 for genotype specification) or myotubes (MTs) formed thereof after 5 days in differentiation medium or (B) from DM11 and CRISPR-edited derivative (see Table S4) MBs and MTs, and used for RT-qPCR analysis of expression of DMPK (black bars) and SIX5 (white bars). Bar heights in the diagram correspond to steady-state expression levels given in arbitrary units (n = 3; mean + SEM). (C) RT-PCR analysis of DM1-AS expression in myoblasts (signal strength of the specific 150-bp product is given in arbitrary units underneath; the lower band seen in all lanes represents a primer-dimer signal). (D) RT-PCR analysis of major splice isoforms of DMPK mRNA formed by alternative skipping of exon 13 to 14 or 14 regions in DMPK heterogeneous nuclear RNA (hnRNA) from myoblasts. (E) Visualization of DMPK protein production in parental and gene-edited LHCN myotubes (5 days of differentiation) by western blot analysis. The most abundant DMPK isoform, i.e., the protein produced from the longest RNA splice isoform with exons 13 to 14 included, has an apparent molecular weight of 80–85 kDa (arrow) and is present in all cells. The smaller DMPK isoform, lacking exon 13 to 14 sequences, comigrates with cross-reacting proteins.80 Variation in signal strength of immunostaining with polyvalent rabbit anti-DMPK antiserum (red) is given in arbitrary units below. Staining with monoclonal mouse-anti-β tubulin antibody (green) was used as control for loading and normalization.
Because the DNA segment comprising exon 15 of the DMPK gene is also transcribed as part of the antisense gene in the DM1 locus (DM1-AS)6 (A.E.E.G. Gudde, S.J. van Heeringen, A. de Oude, I.D.G.v.K., J. Estabrook, E.T. Wang, B.W., and D.G.W., unpublished data), we wondered whether DM1-AS expression might be affected by loss of the (CTG⋅CAG)n repeat segment. We therefore monitored untreated and edited LHCN myoblasts for expression of DM1-AS RNA using semiquantitative RT-PCR (due to the high GC content of the DM1-AS gene, reliable primer pairs for use in qPCR could not be designed [A.E.E.G. Gudde, S.J. van Heeringen, A. de Oude, I.D.G.v.K., J. Estabrook, E.T. Wang, B.W., and D.G.W., unpublished data]). DM1-AS RNA was detected in all cells (Figure 6C), with only minor variation in expression level. Collectively, our results demonstrate that gene expression in the DM1 locus is not substantially altered by deletion of the fragment containing the (CTG⋅CAG)5 repeat.
To verify that alternative splicing involving exon 12–15 of the DMPK transcript was not affected in cis, RT-PCR analysis across this region was performed (Figure 6D). Comparison of PCR products from isoforms present in untreated and edited LHCN myoblasts revealed three major splice forms that have been previously described.31, 67 No significant differences in signal ratio for these splice forms were found, confirming our expectation that alternative splicing across the exon 12–15 region would not be overtly affected by the deletion. Finally, western blot analysis via immunodetection with a DMPK-specific antibody also revealed no qualitative differences in translatability between WT and gene-edited DMPK mRNAs. As anticipated, the longest DMPK isoform product, translated from the most abundant DMPK mRNA variant with exons 12–15 included (see Figure 6D), also yielded the strongest staining signal in lysates from LHCN myotubes with and without the 77-bp repeat tract, with some clonal variation in intensity (Figure 6E). Because our genetic experiments in LHCN, DM500, and DM11 myoblasts showed that successful dual CRISPR excision of normal and long-expanded repeat tracts yields exactly identical alleles, we may thus conclude from these combined biological results that repeat editing leaves the normal coding capacity of genes in the DM1 locus in myoblasts largely intact.
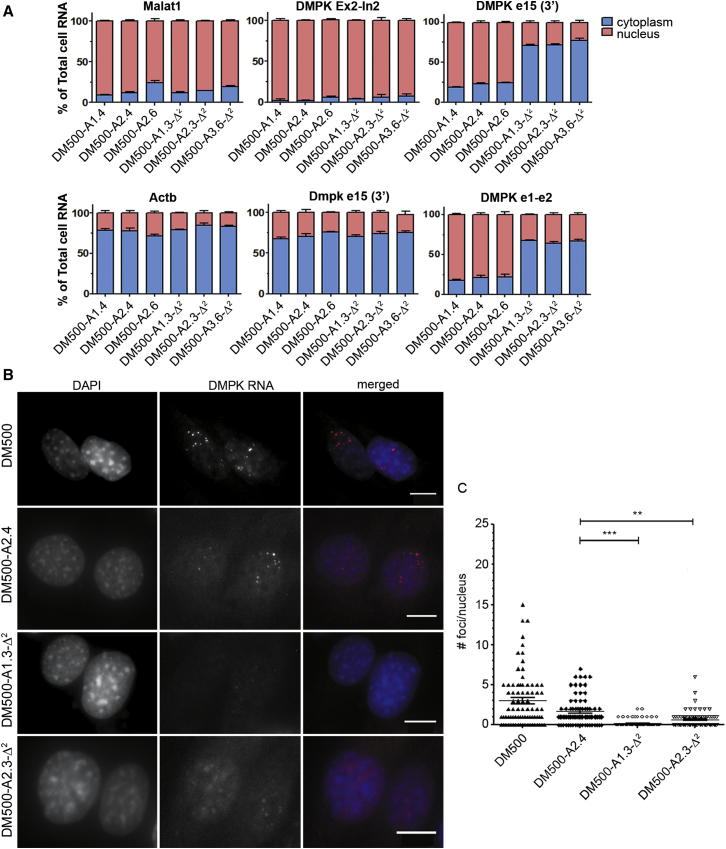
Concomitantly, we expected to see reversal of the abnormalities in the location of expanded DMPK mRNA in modified DM500 and DM11 myoblasts. Transcripts from the expanded allele in DM1 patients form stoichiometrically abnormal complexes with RNP proteins. These complexes prevent proper nucleocytoplasmic transport or could in fact be formed as a result of the impairment in nuclear export.68, 69 We therefore examined whether transcripts from the edited DMPK allele had regained a normal intracellular distribution, despite partial deletion of the 3′ UTR. We used cell fractionation to follow nuclear retention and transport of mature RNA transcripts to the cytoplasm. Genome-edited DM500 myoblasts with two original or two edited DMPK copies formed ideal substrates for comparison because no interference of normal-sized transcripts could occur in these cells. Consistent with our expectation, transgenic DMPK transcripts that lacked the entire (CUG)570–610 repeat tract of ∼1.7 kb were mainly found in the cytoplasmic fraction, whereas expanded RNAs resided predominantly in the nuclear fraction (Figure 7A). Similar tests in edited DM11 cells corroborated this picture (data not shown). To address whether removal of the trinucleotide repeat resulted in effective dissipation of abnormal RNP structures, we performed fluorescence in situ hybridization (FISH) and immunofluorescence microscopy for subcellular MBNL1 protein distribution. Comparison of nuclear RNP foci numbers between DM500 and DM11 myoblast clones with and without expanded repeats revealed a complete absence of foci in edited cells (Figures 7B, 7C, and S5). In accordance with this observation, immunodetection of MBNL1 sequestration in original DM11 cells and clone DM11-4F9 versus clones DM11-3B11-Δ/Δ and DM11 DM11-4A3-Δ/Δ provided clear supportive evidence for reversal of abnormal ribonuclear complex formation. Anti-MBNL1 stained nuclear foci were no longer seen in myoblasts without the repeat (Figure S6).
Figure 7.
Effects of (CTG·CAG)n Repeat Excision on Nuclear DMPK RNA Retention in DM500 Myoblasts
(A) Cell fractionation was used to collect nuclear and cytoplasmic RNA from three DM500 clonal cell lines containing unmodified (CTG⋅CAG)540/570/610 repeats and three independent DM500-Δ2 clonal cell lines with deletions of the repeat in both transgenic alleles. RT-qPCR analysis was used to determine expression levels for (1) nuclear markers Malat1 and pre-DMPK mRNA (exon 2-intron 2 amplicon), (2) cytoplasmic markers Actb and Dmpk from mouse, and (3) mature DMPK mRNA from the human transgene (exon 1-exon 2 and internal exon 15 amplicon). (B) RNA FISH on untreated DM500 cells, clone DM500-A2.4, containing two expanded (CTG·CAG)540/570/610 repeats, and two DM500-Δ2 clones. Foci containing DMPK (CUG)540/570/610 RNA were labeled using a (CAG)6-TYE563 LNA probe (red). Nuclei were stained with DAPI (blue). No foci were seen in the DM500-Δ2 clones. Scale bar, 10 μm. (C) Quantification of nuclear foci in cell lines shown in (B). Each symbol represents the number of foci in one nucleus. Mean + SEM. ***p < 0.005.
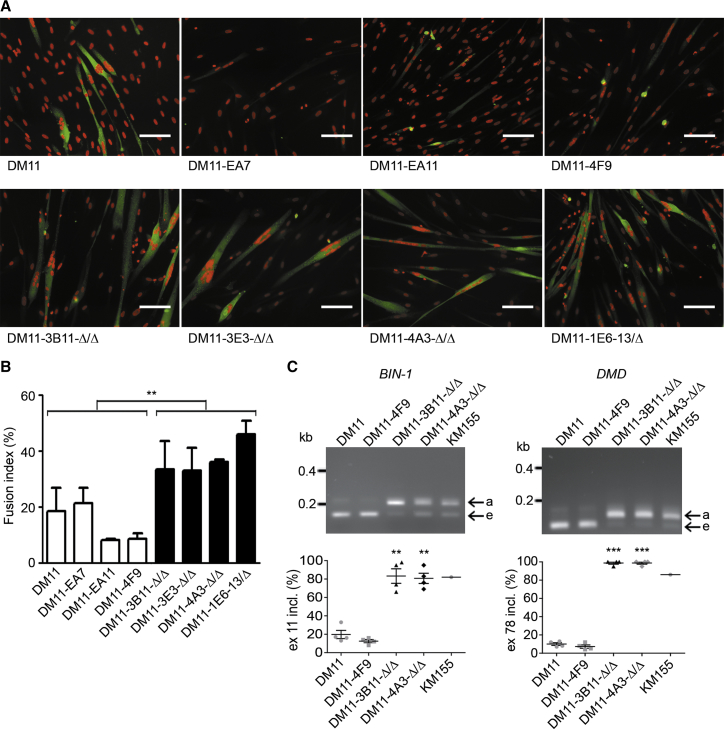
Next, we checked whether the experimental protocol of genomic engineering had not jeopardized the ability of myoblasts to form multinuclear myotubes. Panels in Figures 8A (second row) and S7 illustrate maintenance of fusion capacity for four Δ/Δ cell lineages (DM11-3B11-Δ/Δ, DM11-3E3-Δ/Δ, DM11-4A3-Δ/Δ, and DM11-1E6-Δ/Δ) and one DM500 cell clone (DM500-A3.5-Δ2). We consider the appearance of normal looking multinuclear myotubes as convincing evidence for retainment of myogenic potential after the cell manipulation and editing steps. Furthermore, more detailed quantitative analysis revealed that loss of the DMPK repeat has a true beneficial effect because a significant increase in fusion index was seen for all cell clones in the Δ/Δ group compared to the parental DM11 or CRISPR-treated myoblasts that still had a repeat. This improvement in myogenic capacity was noticeable at both day 7 (Figures 8A and 8B) and day 10 (not shown) after onset of differentiation in vitro. Thus, using our collection of isogenic human and mouse myoblasts with and without repeats (Table S4), precise comparative studies can now be done of all the different steps involved in myogenic differentiation, cell-cycle arrest, activation of the muscle-specific transcriptome, and elongation, alignment, and fusion of myoblasts into multinucleated myotubes. Finally, removal of the repeat also lead to normalization of alternative splicing of biomarker RNAs in Δ/Δ myoblasts, with reappearance of adult splice modes for both BIN-1 and DMD pre-mRNAs, as seen in control KM155 myoblasts, but different from the anomalous embryonal splicing that occurred in the parental DM11 myoblasts and in DM11-4F9 cells with the repeat (Figure 8C).
Figure 8.
Effects of (CTG⋅CAG)n Repeat Excision on Myogenic Capacity and Aberrant RNA Splicing in DM11 Myoblasts
(A) Immunostaining of MHC expression (green) in DM11 cells after 7 days of differentiation. Nuclei were stained with DAPI (red). Scale bar, 100 μm. (B) Fusion index of DM11 cells after 7 days of differentiation. The fusion index was calculated as the ratio of the number of nuclei inside MHC-positive myotubes to the number of total nuclei × 100. Note the improvement of fusion index after excision of the expanded repeat. Mean + SEM. **p < 0.01. (C) Comparative RT-PCR analysis of BIN-1 and DMD in DM11 cells after 5 days of differentiation. Typical embryonic splicing patterns (e) were reverted to the normal adult (a) modes of alternative splicing after loss of the repeat. KM155 myoblasts were used as control cells. (n = 4, mean + SEM). **p < 0.01; ***p < 0.001.
Collectively, our comparison between edited and non-edited myoblasts validates the already well-known detrimental influence that repeat expansion has on DMPK RNA location and some of the best known gain-of-function properties. More opportunities for fundamental discoveries with DM1 cells emerge now. Genome-editing approaches with selective use of single or dual CRISPR cleavage in DMPK’s 3′ UTR region, perhaps in combination with a second round of editing with HDR,58 could be applied to generate isogenic populations of myoblasts or other DM1-relevant cell types. Simple identification and cloning of cells with stepwise incremental repeat lengths or repeat sequence alterations from these populations could help to establish a series of next-generation isogenic DM1 cell models, without the modifier effects of genetic background heterogeneity.
The outcome of our genome editing efforts demonstrates that therapeutic reversal of DM1-related problems is a realistic goal and may be effectively achieved in the future. Deletion of the (CTG⋅CAG)n repeat and small flanking segments from the non-coding region of the DMPK gene had no overt consequences for in vitro cultured cells. Better tests for capacity in myogenic development and for muscle function still need to be done before editing can be evaluated for use in vivo. Our dual CRISPR editing approach may thus have value for future development of somatic gene therapy for DM1, either directly in muscles in adult tissues in vivo, in myofibers or satellite cells, or via use in stem cells prior to muscle regeneration. An alternative genome-editing strategy with therapeutic potential, based on the introduction of a premature poly(A) addition site a few kbps upstream of the repeat in the DMPK gene in iPSCs, was published by Xia et al.70 Clearly this approach was similarly aimed at loss of repeat toxicity at the RNA level. However, in addition to the requirement of a repair template and activity of HDR, this modification also invoked greater changes in the 3′ UTR of the DMPK mRNA and would not be expected to directly correct any epigenetic abnormalities across the mutant DM1 locus. A future comparison between the effectiveness and adverse biological consequences of both strategies is therefore warranted.
Testing these CRISPR/Cas9 approaches in animal models is still very much dependent on procedural progress at the cell level. Further improvements that need to be established consist of combined use of new high-fidelity Cas9 variants,37 with gene sizes that can be easily accommodated in recombinant vectors for in vivo transduction like AAV.71 Also, attention is needed to further improve the specificity of gRNA recognition by using special gRNA lengths in order to keep the off-target activity at the lowest possible level.72 Fortunately, developments that are relevant for these issues appear with revolutionary speed in the CRISPR/Cas9 field. Our analyses provide evidence that unilateral cleavage with CRISPR nucleases to only one side of an unstable repeat in the genome should probably be avoided. Deployment of intrinsic machinery for DNA end resection, NHEJ, and recombination repair may produce unpredictable genome changes across the site of cleavage and DNA closure if repeat DNA with intrinsically odd topology hangs around as a loose end. Repair functions from specialized endonuclease complexes (CtBP-CtIP-MRN proteins) for resection processing may thereby fulfill a modulatory role.73 The scheme in Figure S8 provides a simplified model and hypothetical explanation for the fate of DNA, with non-B structure exposed by unilateral DSBs. Our work therefore includes a cautionary note for all attempts to remove unstable DNA by genome editing in repeat expansion disorders.
Materials and Methods
Design and Construction of Components for the CRISPR/Cas9 Platform
A human codon-optimized version of the Streptococcus pyogenes Cas9 protein was used in combination with a custom guide RNA.74 Throughout the text, the term CRISPR-x is used to indicate the Cas9/gRNA ribonucleprotein endonuclease complex. Target sites for CRISPR/Cas9 across the trinucleotide repeat in exon 15 of DMPK (NCBI, Gene Database, GeneID: 1760, nucleotides 45,770,345–45,770,148) were selected using four CRISPR gRNA design web tools (http://omictools.com, http://crispr.mit.edu, https://chopchop.rc.fas.harvard.edu, and https://www.dna20.com). Different target sites were scored based on the number of predicted off-targets and whether off-targets were perfect hits or contained mismatches. Also, considerations like location of the target site in the gene, GC content, and presence of a guanine at position 20 in the target site, which appears to improve the cutting rate,75 were included. Only CRISPR target sites with the highest scores and with a location as close as possible (either 5′ or 3′ flanking) to the (CTG⋅CAG)n repeat were chosen. Target sites should perfectly match the PAM sequence and 8–12 nt sequence at the 3′ end of the gRNA; mismatches at the 5′ end will be tolerated.74, 76, 77 According to the rules outlined by Mali et al.,74 potential off-targets were found by identifying exact matches to the thirteen 3′-most bases of the gRNA and the PAM sequence in the human genome using BLAST searches. The best matches were reported as potential off-target sites for the CRISPR gRNAs.
Complementary DNA oligonucleotides specifying the 20-nt gRNA sequence were annealed and incorporated into gRNA cloning vector 41824 (Addgene) by use of the Gibson Assembly kit (NEB). Proper insertion of the target sequence into the vector was verified by sequencing. The vector encoding hCas9 was obtained from Addgene (plasmid 41815). Plasmid pMAX-EGFP encoding GFP, an indicator of efficiency of transient transfection, was purchased from Lonza.
Cell Culture and Nucleofection
Immortalized human myoblasts LHCN-M2 (LHCN in short)35 and KM155C2578 were derived from primary myoblasts obtained from control individuals unaffected with DM1 and with two normal-sized DMPK alleles, (CTG5/CTG5) and (CTG5/CTG14), respectively. Immortalized human DM1 myoblasts, DM11 cl5 (CTG13/CTG2600) (DM11 in short), were derived from primary myoblasts obtained from a DM1 patient. LHCN and KM155C25 myoblasts were propagated in Skeletal Muscle Cell Growth Medium (PromoCell) supplemented with 15% (v/v) FBS (Sigma-Aldrich) and glutamax (GIBCO) on dishes coated with 0.1% gelatin (Sigma-Aldrich). DM11 myoblasts were grown in a 1:1 mix of Skeletal Muscle Cell Growth Medium and F-10 Nutrient Mix (GIBCO), supplemented with 20% (v/v) FBS (Sigma). Differentiation to myotubes (fused myoblasts with ≥2 nuclei) was induced by growing myoblasts to confluency and replacing the proliferation medium by differentiation medium consisting of DMEM (GIBCO) supplemented with 4 mM L-glutamine (GIBCO), 1 mM pyruvate (Sigma), 10 μg/mL insulin (Sigma), and 100 μg/mL apo-transferrin (Sigma). All culturing of human myoblasts was performed under normal conditions by incubation under a 7.5% CO2 atmosphere at 37°C.
DM500 cells, an SV40-TAgts-immortalized DM1 model myoblast cell line expressing a transgenic human DM1 locus bearing a (CTG⋅CAG)500 repeat,31 were grown in DMEM supplemented with 4 mM L-glutamine, 1 mM pyruvate (Sigma), 20% (v/v) FBS (PAA Laboratories), 20 units of γ-IFN/mL (BD Biosciences), and 2% (v/v) chicken embryo extract (Sera Laboratories International) on gelatin-coated dishes at 7.5% CO2 and at a permissive temperature of 33°C. Differentiation to myotubes was induced by growing the myoblasts to confluency on matrigel (BD) coated dishes, replacing the proliferation medium by differentiation medium containing DMEM (GIBCO) supplemented with 4 mM L-glutamine (GIBCO), 1 mM pyruvate (Sigma), and 5% horse serum (GIBCO), and placing the cells at a temperature of 39°C (to inactivate the SV40TAgts) in an atmosphere of 7.5% CO2.
Expression vectors were introduced by nucleofection because this turned out to be the most efficient procedure for DNA delivery into these difficult to transfect cell types. Nucleofection was performed using the Amaxa P5 Primary Cell 4D-Nucleofector Kit (Lonza), according to the manufacturer’s protocol for human skeletal muscle myoblasts. For co-nucleofection of 1 × 106 cells, 10 μg of hCas9 plasmid, 10 μg total of gRNA plasmid, and 2 μg of pMAX EGFP (a reporter for transfection efficiency) were used. After nucleofection of LHCN and DM500 cells and subsequent cell sorting (BD FACS Aria Cell Sorter) for GFP-positive cells 2 days post-nucleofection, cells were diluted to about 200 cells/mL, seeded into 96-well plates (∼20 cells per well), and propagated for 11 days. DNA from these cell populations was then isolated and analyzed by PCR. Independent LHCN cell clones were obtained by limiting dilution cloning, and ring cloning was used to obtain independent DM500 lines. For further analyses, each clone was expanded to a population of ∼3.8 × 105 cells for both cell lines.
In the case of DM11 myoblasts, GFP-positive cells were collected after nucleofection by single-cell sorting 2 days post-nucleofection. Single cells were collected in 96-well plates and cultured in medium consisting of a 1:1 mix of normal and conditioned medium. Conditioned medium was harvested from untreated DM11 cells that had been grown in this medium for 3 days. Conditioned medium was filter sterilized before use. Once the cells in the 96-well plate had reached 70% confluency (several weeks, no passaging), medium was switched to normal medium and then each clonal population was expanded to about 3.8 × 105 cells.
PCR Amplification Analysis of Genuine and Off-Target Genome-Editing Events
PCRs were performed using Q5 High-Fidelity DNA Polymerase (NEB). Most loci were successfully amplified using the standard Q5 High-Fidelity DNA Polymerase protocol. PCR on the remaining loci was performed in the presence of the GC enhancer provided by the manufacturer. The following primers were used:
CARMIL2: 5′-AGTGGCTGGTCTAAGGGTGTCAGCTTCAGGA-3′ and
5′-TCCTAGACAAAGGTCAGTCAGAGCATGGTGAGGAT-3′;
EBF3: 5′-AGACAGCATAGCAGGTCAGCAGCAG-3′ and 5′-ACCCTGAGCCCATCTGGAACCCTC-3′;
ALK: 5′-CAATCT-GCTTTCTCCCAGTTTGACT-3′ and 5′-ATCTCGTGATCCGCCCACCTT-3′;
DVL1: 5′-TAACGAGCACCTACTTCATT-3′ and 5′-CACAACATAATGGGCTGG-3′;
DMPK e15: 5′-GCCAACTCACCGCAGTCTGG-3′ and 5′-TCAGCGAGTCGGAGGACGAGG-3′;
CR-2 site: 5′-GCCAATGACGAGTTCGGACGG-3′ and 5′-AGCAGCAGCAGCATTCCCG-3′;
CR-3 site: 5′-CCATTTCTTTCTTTCGGCCAGGCTG-3′ and 5′-GCGAGTCGGAGGACGAGGTCAATAA-3′.
T7 Endonuclease I Cleavage Assay
For identification of indels after CRISPR/Cas9 cleavage, the T7 endonuclease I assay, adapted from Wyvekens et al.,36 was used. Briefly, cells were collected and digested with 100 μg/mL proteinase K in lysis buffer (100 mM Tris-HCl, pH 7.5, 5 mM EDTA, 0.2% SDS, and 2 mM NaCl) for 3 hr at 55°C. Samples were centrifuged for 15 min at 16,000 × g (4°C). Supernatants were collected and equal volumes of isopropanol were added, after which the samples were centrifuged for 20 min at 16,000 × g at 4°C. Supernatants were discarded, and the pellets were washed once with 200 μL of 70% ethanol. After the washing step, the DNA pellets were dried and dissolved in TE buffer (10 mM Tris, pH 7.5, and 1 mM EDTA).
PCR primers were designed to amplify the sequence containing the CRISPR target site, such that the total length of the PCR product was about 600 bp, with the cleavage site located approximately in the middle. Pertinent segments were amplified by PCR, and the resulting products were purified using a gel extraction kit (QIAGEN). Approximately 250 ng of purified PCR product was denatured at 94°C and re-annealed in NEB buffer 2 using a thermocycler (5 min, 95°C; ramp down to 85°C at −2°C/s; ramp down to 25°C at −0.1°C/s; hold at 4°C). The re-annealed PCR products were digested with 10 U T7 endonuclease I (NEB) for 15 min at room temperature. The reaction was stopped by adding 2 uL of 0.25 M EDTA, and PCR products were resolved by electrophoresis on a 1% agarose gel. DNA fragments were stained with 0.5 μg/mL ethidium bromide (Amresco), and ImageJ software was used for quantification of band intensities. Targeting efficiencies were calculated using the following formula: % gene modification = 100 × (1−[1 − fraction cleaved]1/2).79
Small-Pool PCR
Small-pool PCR was carried out as previously described using either DM-C and DM-DR or the more distal DM-A and DM-BR.41 PCR conditions were 28 cycles of 96°C for 45 s, 68°C for 45 s, and 70°C for 3 min, followed by 68°C for 1 min and 70°C for 10 min. The PCR buffer was Custom PCR Master Mix-No Taq (Thermo Scientific #SM-0005) supplemented with 69 mM β-mercaptoethanol (Sigma-Aldrich). Taq polymerase (Sigma-Aldrich) was used at 1 unit per 10 μL of reaction. Template DNA from untreated and CRISPR/Cas9-treated DM500 cells was measured using the Qubit fluorometer and the dsDNA HS kit (Thermo Fisher Scientific). 1% agarose gels were blotted onto Hybond N membrane (GE Healthcare) and hybridized. Probes and molecular weight marker (1 kb+ ladder, Thermo Fisher Scientific) were labeled using 32P-dCTP (Perkin Elmer) and the Random Primers DNA Labeling System (Thermo Fisher Scientific). The (CTG⋅CAG)n repeat probe DM56 was a 244-bp fragment comprising (CTG⋅CAG)56 and 54-bp flanking sequences amplified using primers DM-C and DM-ER (5′-AAATGGTCTGTGATCCCCCC-3′) using template DNA from a late-onset DM1 patient. Probe H was a 99-bp fragment that hybridizes to the 5′ flanking sequence upstream of the (CTG⋅CAG)n repeat. Probe H was amplified using primers DM-H (5′-TCTCCGCCCAGCTCCAGTCC-3′) and DM-CR (5′-AGGACCCTTCGAGCCCCGTTC-3′).
Karyotyping
Karyotyping was performed to confirm tetraploidization of the DM500 cell line. DM500 cells were grown to sub-confluency in a six-well dish and incubated in 1 mL of DMEM medium containing 20 μL of 10 μg/mL colcemid (GIBCO) for 2 hr at 37°C. Cells were trypsinized and collected by centrifugation. The cell pellet was resuspended in 1 mL of 75 mM KCl, after which another 2 mL of 75 mM KCl was added to the sample. Cells were incubated for 10 min at 37°C. Three drops of fixative solution (75% methanol and 25% glacial acetic acid) were added to the sample, and cells were collected by centrifugation. The pellet was washed twice with fixative solution and resuspended in 6–20 drops of fixative solution, depending on the amount of cells. Drops of cell suspensions were dropped from a height of 30 cm onto clean wet slides and left to dry. The cells were then stained using freshly made Giemsa Stain solution (Merck). Chromosomes were analyzed using a phase contrast microscope with a 40× objective.
Southern Blot Hybridization of PCR Products
To determine the presence of the (CTG⋅CAG)500 repeat in CRISPR/Cas9 genome-edited DM500 myoblasts, we used PCR amplification of the (CTG⋅CAG)500 repeat-containing gene segment, followed by Southern blot hybridization. PCR products were resolved by electrophoresis on a 1% agarose gel, transferred by capillary transfer to a Hybond-XL nylon membrane (Amersham Pharmacia Biotech), and hybridized with 32P-labeled oligonucleotides. A DMPK oligo (5′-AGAACTGTCTTCGACTCCGGG-3′), located 5′ of the CRISPR cleavage sites, was used to visualize PCR products from both the unmodified DMPK gene and from cells with a deletion of the region between the two CRISPR sites. The oligo was 5′ end labeled using ɣ32P-ATP and T4 polynucleotide kinase and hybridized to the membrane in Church-Gilbert hybridization solution overnight at 42°C. The membrane was washed, exposed to a Bio-Rad Phosphor Imaging Screen, and imaged by Phosphor-Imager analysis (Molecular Imager FX, Bio-Rad). Analysis was performed with Quantity One (Bio-Rad) and ImageJ software. After imaging, the probe was stripped from the membrane using boiling buffer (0.1 X SSC and 0.1% SDS). Subsequently, using similar conditions for hybridization, washing, and exposure analysis, 32P-end-labeled (CAG)9 probe was used to visualize PCR products containing an expanded (CTG⋅CAG)n repeat.
Western Blot Analysis of DMPK
Analysis of DMPK protein production in myoblasts was performed by western blotting, essentially as described by Oude Ophuis et al.80 Staining with monoclonal anti-β tubulin E7 antibody (Developmental Studies, Hybridoma Bank, University of Iowa) was used as control for loading. As secondary antibodies, 1:10,000 goat-anti-mouse 800CW and goat-anti-rabbit 680RD (Thermo Fisher) were used. Detection was performed on the Odyssey CLx Infrared Western Blot Imaging System (Westburg). ImageJ software was used for quantitative protein expression analysis.
Cellular Fractionation and RNA Isolation
The protocol for RNA isolation from different cell fractions was adapted from Rio et al.81 Briefly, 0.6 × 106 cells (70% confluency) were trypsinized and collected by centrifugation. Cell pellets were washed twice with cold PBS, after which the cell pellet was resuspended in 0.5 mL of ice-cold cell disruption buffer (20 mM Tris-HCl [pH 7.5], 1.5 mM MgCl2, 10 mM KCl, and 10 mM dithiothreitol). After 10 min in cell disruption buffer on ice, the cells were transferred to a 1-mL glass Dounce Tissue Grinder (Wheaton) and homogenized using 15 strokes with a tight pestle. The crude cell lysate was transferred to a fresh tube and 5 μL of 10% Triton X-100 (Sigma-Aldrich) was added to a final concentration of 0.1% Triton X-100 and mixed by inversion five times. Cell nuclei were pelleted immediately by centrifuging the homogenate at 1,500 × g at 4°C for 5 min. The supernatant containing the cytoplasmic fractions was collected. RNA was isolated from both fractions using the Aurum Total RNA Mini Kit (Bio-Rad) according to the manufacturer’s protocol. All RNA preparations were quantified using a NanoVue spectrometer (GE Healthcare Europe). Approximately 500 ng of RNA of the cytoplasmic fractions and equal volumes of the corresponding nuclear fractions were used as a template for subsequent cDNA synthesis and RT-qPCR.
Northern Blotting
To determine the absence or presence of (CUG)n-repeat-containing RNA transcripts from the (CTG)13 and (CTG)2600 alleles in CRISPR/Cas9 genome-edited DM11 myoblasts, Northern blotting was performed according to standard procedures. RNA was isolated from the DM11 cell clones using an Aurum Total RNA Mini Kit. All RNA preparations were quantified using a NanoVue spectrometer. Depending on the RNA yield from different clonally expanded cell lineages, 5–10 μg of RNA was dissolved in loading buffer and loaded onto a 1.2% agarose-formaldehyde denaturing gel and resolved by electrophoresis. RNA was transferred to Hybond-XL nylon membrane by capillary transfer in 10X SSC and UV cross-linked using a UV Stratalinker 1800 (Invitrogen). The membrane was hybridized with a 32P-end-labeled (CAG)9 probe in Church-Gilbert hybridization solution overnight at 42°C, after which the membrane was washed and the probes were detected and analyzed as described above.
RNA FISH and Image Analysis of Ribonuclear Foci
DM500 cells were grown in eight-well IBIDI chambers coated with 0.1% gelatin (Sigma) and expanded to 90% confluency. DM11 cells were grown on coverslips coated with 0.1% gelatin (Sigma-Aldrich) to 50%–60% confluency. The cells were washed once with PBS and fixed in 4% formaldehyde, 5 mM MgCl2 in PBS for 10 min at room temperature. Cells were washed three times for 5 min with PBS, after which ice-cold 70% ethanol was added for overnight incubation. The ice-cold 70% ethanol was refreshed once, after which the fixed cells were washed two times for 5 min with PBS at room temperature. Cell-containing coverslips were prehybridized in 40% deionized formamide (Ambion) in 2X SSC (Ambion) for 20 min at room temperature, followed by overnight hybridization at 37°C with 0.1 ng/μL (CAG)6-TYE563 LNA probe (Exiqon) in hybridization buffer containing 40% deionized formamide, 2 mg/mL BSA (Sigma), 100 mg/mL dextran sulfate (Pharmacia), 0.1% Triton X-100 (Sigma-Aldrich), 1 mg/mL herring sperm DNA (Promega), 100 μg/mL yeast tRNA (Ambion), 2 mM vanadyl ribonucleoside complex (NEB), and 2 X SSC. Coverslips were washed twice for 5 min with PBS before staining cell nuclei with 1 μg/mL DAPI (Sigma) in PBS for 10 min at room temperature. Cells were then washed twice for 5 min with PBS, and preparations were mounted with Mowiol 488 (Hoechst).
Fluorescent images were acquired using a Leica DMI6000B microscope with a 63× objective in three different wavelength intervals using filter sets for DAPI, fluorescein isothiocyanate (FITC), and tetramethylrhodamine (TRITC)/Cy3. Images were subsequently analyzed using FIJI software. DAPI masks were created using auto-thresholding using Huang’s method,82 followed by a watershed. For analysis of DM11 cells, a “find maxima” option in FIJI was applied for the TRITC channel using a noise tolerance of 200, resulting in images containing single points. Positive pixels were counted in the nucleus using the previously made DAPI masks. For DM500, “find maxima” was also applied for Cy3, but using a noise tolerance of 150. Fluorescence signal intensity needed correction for aspecific maxima by subtracting the consistently high autofluorescence background signal observed in DM500 cells. Using the DAPI masks, the positive pixels in each nucleus were counted.
RT-qPCR
To quantify endogenous RNA levels in the various cell lines, RT-qPCR was performed. RNA was isolated using an Aurum Total RNA Mini Kit (Bio-Rad) according to the manufacturer’s protocol. Next, cDNA synthesis was performed using 500 ng of RNA as a template, with random hexamers and the iScript cDNA synthesis kit (Bio-Rad) in a total volume of 20 μL. For RT-qPCR, 3 μL of the 10-fold diluted cDNA sample was mixed in a final volume of 10 μL containing 5 μL of iQ SYBR Green Supermix (Bio-Rad) and 4 pmol of each primer. Samples were analyzed using the CFX96 Real-time System (Bio-Rad). As negative controls, no-RT controls (RT−) and no-template controls (NTCs) were included. Endogenous DMPK levels were normalized to GAPDH and HPRT1 levels.
Primers used: Malat1: 5′-GCTGTTGGCACGACACCTTC-3′ and 5′-ACTGTGAACCAAAGCCGCAC-3′; Actb: 5′-GCTCTGGCTCCTAGCACCAT-3′ and 5′-GCCACCGATCCACACAGAGT-3′; Dmpk e15(3′): 5′-GGATCAGCAAGACCTCTGCCAG-3′ and 5′-TGTGGCTCCGTTGTTAGAGTGC-3′; DMPK e1-e2: 5′-ACTGGCCCAGGACAAGTACG-3′ and 5′-CCTCCTTAAGCCTCACCACG-3′; DMPK e2-in2: 5′-GAGGGACGACTTCGAGATTCTGAA-3′ and 5′-CACCACGAGTCAAGTCAGGC-3′; DMPK e15(3′): 5′-TGCCTGCTTACTCGGGAAATT-3′ and 5′-GAGCAGCGCAAGTGAGGAG-3′; GAPDH: 5′-CCCGCTTCGCTCTCTGCTCC-3′ and 5′-CCTTCCCCATGGTGTCTGAGCG-3′; HPRT1: 5′-TGACACTGGCAAAACAATGCA-3′ and 5′-GGTCCTTTTCACCAGCAAGCT-3′. Primers for SIX5 have been described previously;33 however, because the efficiency of amplification for this primer pair appeared to be an artificial 113.5% in our analyses, an efficiency correction was included in the calculations.
RT-PCR Analysis of in cis and in trans Effects on Alternative Splicing and DM1-AS RNA Expression
To analyze DMPK exon 12–15 splice isoforms, 500 ng of total RNA was used as a template for cDNA synthesis with random hexamers using the Superscript III RT kit (Invitrogen) in a total volume of 20 μL, according to the manufacturer’s protocol. The cDNA was diluted 1:10, and 1 μL was used for subsequent amplification by PCR using forward and reverse DMPK e12–e15 primers: 5′-GAACCGGGACCTAGAGGCACACGT-3′ and 5′-TCGGAGCGGTTGTGAACTG-3′. As negative control, a no-RT control (RT−) was included. PCR products were separated on a 1% agarose gel. Quantification was performed using ImageJ software.
For analysis of DM1-AS expression, 1 μg of total myoblast RNA was used as a template for cDNA synthesis with 2 pmol of gene-specific primer RT AS 3CAG 5′-CGACTGGAGCACGAGGACACTGACGCCTGCCAGTTCACAACCGCTCCGAGCGT-3′ and the Superscript III reverse transcriptase kit (Invitrogen) in a total volume of 20 μL, according to the manufacturer’s protocol. Undiluted cDNA (1 μL) was used for subsequent amplification by PCR using AS DMPK: 5′-CCTTCGAGCCCCGTTCGC-3′ and RT AS 3CAG primer.83 As negative control, a no-RT control (RT−) was included. PCR products were separated on a 2% agarose gel.
To analyze BIN1 exon 11 inclusion and DMD exon 78 inclusion in alternative RNA splicing, 500 ng of total RNA from myotubes at 5 days of differentiation was used as a template for cDNA synthesis with random hexamers using the iScript cDNA synthesis kit (Bio-Rad) in a total volume of 20 μL. The cDNA was diluted 1:10, and 1 μL was used for subsequent amplification by PCR using forward and reverse primers BIN1 ex11 5′-AGAACCTCAATGATGTGCTGG-3′ and 5′-TCGTGGTTGACTCTGATCTCGG-3′ or DMD ex78 5′- TTAGAGGAGGTGATGGAGCA-3′ and 5′-GATACTAAGGACTCCATCGC-3′. As negative control, a no-RT control (RT−) was included. PCR products were separated on a 2.5% agarose gel. Quantification was performed using ImageJ software. Exon inclusion was quantified as the percentage of the total intensity of isoform signals.
Immunofluorescence Staining
For assaying myogenic capacity with myosin heavy chain (MHC) staining, DM11 clones were seeded in eight-well IBIDI chambers, propagated for 2 days in growth medium, and cultured in differentiation medium for 5 or more days. Cells were washed with PBS and fixed with 2% PFA in 0.1 M phosphate buffer for 15 min at room temperature. After fixation, cells were washed with PBS again and permeabilized with blocking buffer (0.1% Triton X-100 [Sigma-Aldrich], 0.1% glycin [Merck], and 3% BSA [Sigma] in PBS) for 30 min at room temperature. Samples were incubated overnight at 4°C with blocking buffer containing 1:10 diluted anti-MHC antibody, MF 20 (DSHB, University of Iowa). Cells were washed with PBS and incubated with 4 μg/mL goat-anti-mouse AF488 (Thermo Fisher) and 100 ng/mL DAPI (Sigma) in blocking buffer for 1 hr at room temperature in the dark. Cells were washed and kept in PBS. Fluorescent images were acquired using a Leica DMI6000B microscope with a 10× objective.
For visualization of co-accumulation of MBNL1 with (CUG)n expanded DMPK transcripts in intranuclear RNP foci, DM11 cells were grown on coverslips coated with 0.1% gelatin (Sigma-Aldrich) to 80% confluency. Cells were washed with PBS, fixed in ice-cold 1:1 acetone/methanol, incubated overnight at −20°C, washed with PBS again, and incubated for 1 hr in block buffer containing 3% BSA (Sigma), 0.1% glycin (Merck), and 0.1% Triton X-100 (Sigma) at room temperature. Samples were then incubated overnight at 4°C in the same buffer containing 1:10 diluted anti-MBNL1 antibody, MB1a (4A8) from DSHB, University of Iowa. Samples were washed and incubated with 4 μg/mL goat-anti-mouse AF488 (Thermo Fisher) and 100 ng/mL DAPI (Sigma) in blocking buffer for 1 hr at room temperature in the dark. Finally, slides with fixed cells were washed with PBS, milliQ, 70% and 100% ethanol, and mounted with Mowiol 488 (Hoechst). Images were acquired using an Olympus FV 1000 microscope with a 60×, NA 1.35 oil objective. Quantification of MBNL1 foci was performed on wide-field images taken with a Leica DMI600B microscope with a 63×, NA 0.90 dry objective.
Statistical Analysis
All statistical analyses were performed with GraphPad Prism version 5.01 software (GraphPad Software) using one-way ANOVA or Student’s t test. All values in graphs are presented as mean ± SEM. The significance level was set at 0.05. *p < 0.05; **p < 0.01; ***p < 0.001.
Author Contributions
Conceived and Designed the Experiments, E.L.v.A., D.G.W., and B.W.; Performed the Experiments, E.L.v.A., L.M.A., M.W., S.A.C., I.D.G.v.K., and W.J.A.A.v.d.B.; Analyzed the Data, E.L.v.A., G.G., D.F., V.M., D.G.M., D.G.W., and B.W.; Contributed Reagents/Materials/Analysis Tools, G.G., D.F., and V.M.; Wrote the Paper, E.L.v.A., D.G.M., D.G.W., and B.W.; Performed Critical Reading of the Manuscript, S.A.C., G.G., D.F., and V.M.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by ZonMw (TOP grant NL91212009) and by the Prinses Beatrix Spierfonds (grant number W.OR12-05; with contribution from the Stichting Spieren voor Spieren). Funding to pay Open Access charges was provided by both organizations and Radboudumc. We would like to thank Huib Croes and Jack Fransen from the DGW-BW group at Radboudumc for technical assistance and advice, the DGM group at the University of Glasgow for helpful discussions, colleagues at the Radboudumc Central Animal Laboratory, and the Imagine Institute and Center for Research in Myology, Paris, France, for advice in muscle cell derivation from transgenic mice and patients and for providing helpful information regarding experimental protocols for cell culture. We thank the Radboudumc technology platform and Glasgow Polyomics facility at the University of Glasgow for assistance with DNA sequencing, and the Myotonic Dystrophy Support Group and the Muscular Dystrophy Association for support.
Footnotes
Supplemental Information includes eight figures and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2016.10.014.
Contributor Information
Derick G. Wansink, Email: rick.wansink@radboudumc.nl.
Bé Wieringa, Email: be.wieringa@radboudumc.nl.
Supplemental Information
References
- 1.Caso F., Agosta F., Peric S., Rakočević-Stojanović V., Copetti M., Kostic V.S., Filippi M. Cognitive impairment in myotonic dystrophy type 1 is associated with white matter damage. PLoS ONE. 2014;9:e104697. doi: 10.1371/journal.pone.0104697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minnerop M., Weber B., Schoene-Bake J.C., Roeske S., Mirbach S., Anspach C., Schneider-Gold C., Betz R.C., Helmstaedter C., Tittgemeyer M. The brain in myotonic dystrophy 1 and 2: evidence for a predominant white matter disease. Brain. 2011;134:3530–3546. doi: 10.1093/brain/awr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groenen P., Wieringa B. Expanding complexity in myotonic dystrophy. BioEssays. 1998;20:901–912. doi: 10.1002/(SICI)1521-1878(199811)20:11<901::AID-BIES5>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 4.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 5.Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barceló J., O’Hoy K. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 6.Cho D.H., Thienes C.P., Mahoney S.E., Analau E., Filippova G.N., Tapscott S.J. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Michel L., Huguet-Lachon A., Gourdon G. Sense and antisense DMPK RNA foci accumulate in DM1 tissues during development. PLoS ONE. 2015;10:e0137620. doi: 10.1371/journal.pone.0137620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monckton D.G., Wong L.J., Ashizawa T., Caskey C.T. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum. Mol. Genet. 1995;4:1–8. doi: 10.1093/hmg/4.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Morales F., Couto J.M., Higham C.F., Hogg G., Cuenca P., Braida C., Wilson R.H., Adam B., del Valle G., Brian R. Somatic instability of the expanded CTG triplet repeat in myotonic dystrophy type 1 is a heritable quantitative trait and modifier of disease severity. Hum. Mol. Genet. 2012;21:3558–3567. doi: 10.1093/hmg/dds185. [DOI] [PubMed] [Google Scholar]
- 10.Harley H.G., Brook J.D., Rundle S.A., Crow S., Reardon W., Buckler A.J., Harper P.S., Housman D.E., Shaw D.J. Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature. 1992;355:545–546. doi: 10.1038/355545a0. [DOI] [PubMed] [Google Scholar]
- 11.Pratte A., Prévost C., Puymirat J., Mathieu J. Anticipation in myotonic dystrophy type 1 parents with small CTG expansions. Am. J. Med. Genet. A. 2015;167A:708–714. doi: 10.1002/ajmg.a.36950. [DOI] [PubMed] [Google Scholar]
- 12.Höweler C.J., Busch H.F.M., Geraedts J.P.M., Niermeijer M.F., Staal A. Anticipation in myotonic dystrophy: fact or fiction? Brain. 1989;112:779–797. doi: 10.1093/brain/112.3.779. [DOI] [PubMed] [Google Scholar]
- 13.Kuyumcu-Martinez N.M., Wang G.S., Cooper T.A. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei C., Jones K., Timchenko N.A., Timchenko L. GSK3β is a new therapeutic target for myotonic dystrophy type 1. Rare Dis. 2013;1:e26555. doi: 10.4161/rdis.26555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J.E., Cooper T.A. Pathogenic mechanisms of myotonic dystrophy. Biochem. Soc. Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Timchenko L. Molecular mechanisms of muscle atrophy in myotonic dystrophies. Int. J. Biochem. Cell Biol. 2013;45:2280–2287. doi: 10.1016/j.biocel.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batra R., Charizanis K., Manchanda M., Mohan A., Li M., Finn D.J., Goodwin M., Zhang C., Sobczak K., Thornton C.A. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell. 2014;56:311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalsotra A., Singh R.K., Gurha P., Ward A.J., Creighton C.J., Cooper T.A. The Mef2 transcription network is disrupted in myotonic dystrophy heart tissue, dramatically altering miRNA and mRNA expression. Cell Rep. 2014;6:336–345. doi: 10.1016/j.celrep.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rau F., Freyermuth F., Fugier C., Villemin J.P., Fischer M.C., Jost B., Dembele D., Gourdon G., Nicole A., Duboc D. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 20.Perbellini R., Greco S., Sarra-Ferraris G., Cardani R., Capogrossi M.C., Meola G., Martelli F. Dysregulation and cellular mislocalization of specific miRNAs in myotonic dystrophy type 1. Neuromuscul. Disord. 2011;21:81–88. doi: 10.1016/j.nmd.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Cleary J.D., Ranum L.P. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum. Mol. Genet. 2013;22(R1):R45–R51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otten A.D., Tapscott S.J. Triplet repeat expansion in myotonic dystrophy alters the adjacent chromatin structure. Proc. Natl. Acad. Sci. USA. 1995;92:5465–5469. doi: 10.1073/pnas.92.12.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klesert T.R., Otten A.D., Bird T.D., Tapscott S.J. Trinucleotide repeat expansion at the myotonic dystrophy locus reduces expression of DMAHP. Nat. Genet. 1997;16:402–406. doi: 10.1038/ng0897-402. [DOI] [PubMed] [Google Scholar]
- 25.Thornton C.A., Wymer J.P., Simmons Z., McClain C., Moxley R.T., 3rd Expansion of the myotonic dystrophy CTG repeat reduces expression of the flanking DMAHP gene. Nat. Genet. 1997;16:407–409. doi: 10.1038/ng0897-407. [DOI] [PubMed] [Google Scholar]
- 26.Liu G., Chen X., Gao Y., Lewis T., Barthelemy J., Leffak M. Altered replication in human cells promotes DMPK (CTG)(n) · (CAG)(n) repeat instability. Mol. Cell. Biol. 2012;32:1618–1632. doi: 10.1128/MCB.06727-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckley L., Lacey M., Ehrlich M. Epigenetics of the myotonic dystrophy-associated DMPK gene neighborhood. Epigenomics. 2016;8:13–31. doi: 10.2217/epi.15.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He F., Todd P.K. Epigenetics in nucleotide repeat expansion disorders. Semin. Neurol. 2011;31:470–483. doi: 10.1055/s-0031-1299786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.González-Barriga A., Mulders S.A., van de Giessen J., Hooijer J.D., Bijl S., van Kessel I.D., van Beers J., van Deutekom J.C., Fransen J.A., Wieringa B. Design and analysis of effects of triplet repeat oligonucleotides in cell models for myotonic dystrophy. Mol. Ther. Nucleic Acids. 2013;2:e81. doi: 10.1038/mtna.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wheeler T.M., Leger A.J., Pandey S.K., MacLeod A.R., Nakamori M., Cheng S.H., Wentworth B.M., Bennett C.F., Thornton C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulders S.A., van den Broek W.J., Wheeler T.M., Croes H.J., van Kuik-Romeijn P., de Kimpe S.J., Furling D., Platenburg G.J., Gourdon G., Thornton C.A. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rzuczek S.G., Southern M.R., Disney M.D. Studying a drug-like, RNA-focused small molecule library identifies compounds that inhibit RNA toxicity in myotonic dystrophy. ACS Chem. Biol. 2015;10:2706–2715. doi: 10.1021/acschembio.5b00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brouwer J.R., Huguet A., Nicole A., Munnich A., Gourdon G. Transcriptionally repressive chromatin remodelling and CpG methylation in the presence of expanded CTG-tepeats at the DM1 locus. J. Nucleic Acids. 2013;2013:567435. doi: 10.1155/2013/567435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richard G.F. Shortening trinucleotide repeats using highly specific endonucleases: a possible approach to gene therapy? Trends Genet. 2015;31:177–186. doi: 10.1016/j.tig.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 35.Zhu C.H., Mouly V., Cooper R.N., Mamchaoui K., Bigot A., Shay J.W., Di Santo J.P., Butler-Browne G.S., Wright W.E. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell. 2007;6:515–523. doi: 10.1111/j.1474-9726.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 36.Wyvekens N., Tsai S.Q., Joung J.K. Genome editing in human cells using CRISPR/Cas nucleases. Curr. Protoc. Mol. Biol. 2015;112:31.3.1–31.3.18. doi: 10.1002/0471142727.mb3103s112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seznec H., Lia-Baldini A.S., Duros C., Fouquet C., Lacroix C., Hofmann-Radvanyi H., Junien C., Gourdon G. Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum. Mol. Genet. 2000;9:1185–1194. doi: 10.1093/hmg/9.8.1185. [DOI] [PubMed] [Google Scholar]
- 39.Davoli T., de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–776. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinehart C.A., Mayben J.P., Butler T.D., Haskill J.S., Kaufman D.G. Alterations of DNA content in human endometrial stromal cells transfected with a temperature-sensitive SV40: tetraploidization and physiological consequences. Carcinogenesis. 1992;13:63–68. doi: 10.1093/carcin/13.1.63. [DOI] [PubMed] [Google Scholar]
- 41.Gomes-Pereira M., Fortune M.T., Monckton D.G. Mouse tissue culture models of unstable triplet repeats: in vitro selection for larger alleles, mutational expansion bias and tissue specificity, but no association with cell division rates. Hum. Mol. Genet. 2001;10:845–854. doi: 10.1093/hmg/10.8.845. [DOI] [PubMed] [Google Scholar]
- 42.Hinz J.M., Laughery M.F., Wyrick J.J. Nucleosomes inhibit Cas9 endonuclease activity in vitro. Biochemistry. 2015;54:7063–7066. doi: 10.1021/acs.biochem.5b01108. [DOI] [PubMed] [Google Scholar]
- 43.Knight S.C., Xie L., Deng W., Guglielmi B., Witkowsky L.B., Bosanac L., Zhang E.T., El Beheiry M., Masson J.B., Dahan M. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350:823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- 44.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L., Guell M., Byrne S., Yang J.L., De Los Angeles A., Mali P., Aach J., Kim-Kiselak C., Briggs A.W., Rios X. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richard G.F., Viterbo D., Khanna V., Mosbach V., Castelain L., Dujon B. Highly specific contractions of a single CAG/CTG trinucleotide repeat by TALEN in yeast. PLoS ONE. 2014;9:e95611. doi: 10.1371/journal.pone.0095611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mittelman D., Moye C., Morton J., Sykoudis K., Lin Y., Carroll D., Wilson J.H. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc. Natl. Acad. Sci. USA. 2009;106:9607–9612. doi: 10.1073/pnas.0902420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugawara N., Ira G., Haber J.E. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park C.Y., Halevy T., Lee D.R., Sung J.J., Lee J.S., Yanuka O., Benvenisty N., Kim D.W. Reversion of FMR1 methylation and silencing by editing the triplet repeats in fragile X iPSC-rerived neurons. Cell Rep. 2015;13:234–241. doi: 10.1016/j.celrep.2015.08.084. [DOI] [PubMed] [Google Scholar]
- 50.Huang W., Zheng J., He Y., Luo C. Tandem repeat modification during double-strand break repair induced by an engineered TAL effector nuclease in zebrafish genome. PLoS ONE. 2013;8:e84176. doi: 10.1371/journal.pone.0084176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Axford M.M., Wang Y.H., Nakamori M., Zannis-Hadjopoulos M., Thornton C.A., Pearson C.E. Detection of slipped-DNAs at the trinucleotide repeats of the myotonic dystrophy type I disease locus in patient tissues. PLoS Genet. 2013;9:e1003866. doi: 10.1371/journal.pgen.1003866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu G., Chen X., Bissler J.J., Sinden R.R., Leffak M. Replication-dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nat. Chem. Biol. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Renkawitz J., Lademann C.A., Jentsch S. Mechanisms and principles of homology search during recombination. Nat. Rev. Mol. Cell Biol. 2014;15:369–383. doi: 10.1038/nrm3805. [DOI] [PubMed] [Google Scholar]
- 54.Barzel A., Kupiec M. Finding a match: how do homologous sequences get together for recombination? Nat. Rev. Genet. 2008;9:27–37. doi: 10.1038/nrg2224. [DOI] [PubMed] [Google Scholar]
- 55.Jasin M., Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J., Shou J., Guo Y., Tang Y., Wu Y., Jia Z., Zhai Y., Chen Z., Xu Q., Wu Q. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J. Mol. Cell Biol. 2015;7:284–298. doi: 10.1093/jmcb/mjv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 59.Yang D., Scavuzzo M.A., Chmielowiec J., Sharp R., Bajic A., Borowiak M. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci. Rep. 2016;6:21264. doi: 10.1038/srep21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao Z., Bozzella M., Seluanov A., Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 63.Orthwein A., Noordermeer S.M., Wilson M.D., Landry S., Enchev R.I., Sherker A., Munro M., Pinder J., Salsman J., Dellaire G. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Gutschner T., Haemmerle M., Genovese G., Draetta G.F., Chin L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. 2016;14:1555–1566. doi: 10.1016/j.celrep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 65.Chang T.H., Huang H.Y., Hsu J.B., Weng S.L., Horng J.T., Huang H.D. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J., Sif S., Jones B., Jackson A., Koipally J., Heller E., Winandy S., Viel A., Sawyer A., Ikeda T. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 67.Groenen P.J., Wansink D.G., Coerwinkel M., van den Broek W., Jansen G., Wieringa B. Constitutive and regulated modes of splicing produce six major myotonic dystrophy protein kinase (DMPK) isoforms with distinct properties. Hum. Mol. Genet. 2000;9:605–616. doi: 10.1093/hmg/9.4.605. [DOI] [PubMed] [Google Scholar]
- 68.Wojciechowska M., Krzyzosiak W.J. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettersson O.J., Aagaard L., Jensen T.G., Damgaard C.K. Molecular mechanisms in DM1 - a focus on foci. Nucleic Acids Res. 2015;43:2433–2441. doi: 10.1093/nar/gkv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xia G., Gao Y., Jin S., Subramony S.H., Terada N., Ranum L.P.W., Swanson M.S., Ashizawa T. Genome modification leads to phenotype reversal in human myotonic dystrophy type 1 induced pluripotent stem cell-derived neural stem cells. Stem Cells. 2015;33:1829–1838. doi: 10.1002/stem.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Senís E., Fatouros C., Große S., Wiedtke E., Niopek D., Mueller A.-K., Börner K., Grimm D. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol. J. 2014;9:1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 72.Ren X., Yang Z., Xu J., Sun J., Mao D., Hu Y., Yang S.J., Qiao H.H., Wang X., Hu Q. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014;9:1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H., Li Y., Truong L.N., Shi L.Z., Hwang P.Y., He J., Do J., Cho M.J., Li H., Negrete A. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell. 2014;54:1012–1021. doi: 10.1016/j.molcel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mamchaoui K., Trollet C., Bigot A., Negroni E., Chaouch S., Wolff A., Kandalla P.K., Marie S., Di Santo J., St Guily J.L. Immortalized pathological human myoblasts: towards a universal tool for the study of neuromuscular disorders. Skelet. Muscle. 2011;1:34. doi: 10.1186/2044-5040-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 80.Oude Ophuis R.J., Mulders S.A., van Herpen R.E., van de Vorstenbosch R., Wieringa B., Wansink D.G. DMPK protein isoforms are differentially expressed in myogenic and neural cell lineages. Muscle Nerve. 2009;40:545–555. doi: 10.1002/mus.21352. [DOI] [PubMed] [Google Scholar]
- 81.Rio D.C., Ares M., Jr., Hannon G.J., Nilsen T.W. Preparation of cytoplasmic and nuclear RNA from tissue culture cells. Cold Spring Harb. Protoc. 2010;2010 doi: 10.1101/pdb.prot5441. pdb.prot5441. [DOI] [PubMed] [Google Scholar]
- 82.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huguet A., Medja F., Nicole A., Vignaud A., Guiraud-Dogan C., Ferry A., Decostre V., Hogrel J.Y., Metzger F., Hoeflich A. Molecular, physiological, and motor performance defects in DMSXL mice carrying >1,000 CTG repeats from the human DM1 locus. PLoS Genet. 2012;8:e1003043. doi: 10.1371/journal.pgen.1003043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.