Abstract
Recently, an engineered Homeobox-nucleoporin fusion gene, NUP98-HOXA10HD or NA10HD, was reported to expand and maintain murine hematopoietic stem cells (HSCs). We postulated that NA10HD would increase the number of human γ-globin-expressing cells to therapeutic levels. We developed a double gene lentiviral vector encoding both human γ-globin and NA10HD, which was used to transduce human peripheral blood CD34+ cells and increased engraftment 2- to 2.5-fold at 15 weeks post-transplantation in immunodeficient mice. In β-thalassemic mice transplanted with β-thalassemic HSCs transduced with the γ-globin/NA10HD vector, the number of fetal hemoglobin (HbF)-expressing cells was significantly increased after 3 months, leading to resolution of the anemia. Furthermore, the increases in HbF were maintained at 6 months and persisted after secondary transplantation. In addition, NA10HD enrichment of transduced HSCs led to HbF increases without affecting homeostasis of the white blood cell lineages. Our results suggest that NA10HD increases the number of γ-globin-transduced HSCs that engraft, leading to an elevated number of fetal hemoglobin-containing red cells. These effects of NA10HD provide an improved platform for testing of the therapeutic efficacy of novel globin vectors and provide further impetus to develop safe and effective methods for selective expansion of genetically modified cells.
Keywords: gene therapy, γ-globin, transplantation, transduction, NUP98HOXA10-HD fusion protein, β-thalassemic mice, resolution of the anemia
In hemoglobinopathies, consistent expression of globin genes at therapeutic levels after lentiviral vector transfer remains challenging. Zhao et al. demonstrate that a fusion gene, NA10HD, confers enhanced self-renewal to gamma-globin-transduced beta-thalassemic mouse hematopoietic stem cells, leading to higher rates of disease resolution.
Introduction
Lentiviral targeted hematopoietic stem cell (HSC) gene therapy has recently achieved steadily accelerating progress for the treatment of hematological diseases,1, 2, 3 which includes hemoglobinopathies (sickle cell disease and β-thalassemia).4, 5, 6, 7 The first clinical trial for β-thalassemia was initiated in 2007.8 One of three patients demonstrated clinical benefit and became transfusion independent with stable hemoglobin (Hb) of greater than 8 g/dL. Over the past 10 years, even though clinical trials from several other groups are being developed or are open and enrolling participants,9, 10, 11 uniform success has not been achieved. In one of the most recent trials, four patients with sickle cell anemia have been treated. The initial patient experienced substantial clinical benefit and remains completely free of disease-related adverse events.10 However, three patients treated in the United States subsequently had minimal clinical improvement. Variables that correlated with clinical outcome again included the number of hematopoietic stem cells infused, the vector copy number (VCN) achieved during in vitro transduction, and the dose of busulfan. Generally, full myeloablation was necessary to achieve clinical benefit.10
Many different approaches were investigated to improve the efficiency of transduction and engraftment with the goal of increasing the number of genetically modified cells in peripheral blood.11, 12, 13, 14, 15, 16 One such approach is to express a HOX protein that confers a benign proliferative advantage to the modified cells over the non-transduced cells in vivo.15, 17, 18 HOXB4 was the first HOX family member found to enhance the expansion of human and mouse HSCs by promoting self-renewal divisions without losing stemness.19, 20
Certain fusion proteins of HOXB4 have also been shown to induce expansion of hematopoietic stem cells in vitro. For example, a fusion of the HIV-encoded TAT protein with HOXB4 led to significant expansion in vitro. TAT in this case was thought to facilitate transmembrane transfer of the TAT-HOXB4 protein.21 Transduction of a fusion gene encoding the N-terminal half of NUP98 and the 60 aa homeodomain of HOXA10 (NUP98-HOXA10HD or NA10HD) has proved to be extremely potent in promoting expansion of murine long-term hematopoietic stem cells both in vitro and in vivo.22, 23 It increased engraftment of human short-term and long-term repopulating cells in immunodeficient mouse models24, 25 and in non-human primate models.26 The properties of NA10HD have thus provided a powerful new tool for manipulating and investigating the self-renewing behavior of primitive murine, non-human primate, or human hematopoietic cells.
Our hypothesis was that co-expression of NA10HD and γ-globin in the lentiviral vector would expand the transduced cells without causing them to lose their primitive cell nature, thereby increasing the number of genetically corrected erythroid cells to a therapeutic level. We show here that the number of globin-expressing red blood cells and the amount of fetal hemoglobin were significantly increased, leading to the complete cure of the anemia in all mice. This increase did not affect the hematopoietic homeostasis of the white blood cell (WBC) lineages, suggesting that the NA10HD effect on transduced hematopoietic stem cells is self-controlled.
Results
A Lentiviral Vector Encoding Both Human γ-Globin and NA10HD Genes Increased Engraftment after NSG Mouse Transplantation with Transduced Human CD34+ Cells
Two different vectors were designed: a modified MLV long terminal repeat promoter (MND U3) was used to drive the NA10HD or mCherry expression cassette (Figures S1A and S1B). Both vectors contain an identical γ-globin expression cassette.15 Significant NA10HD expression in vitro from the γ-globin/NA10HD vector was demonstrated by flow cytometric detection of the intra-cellular FLAG-tagged NA10HD protein in 293T cells (Figures S2A and S2B).
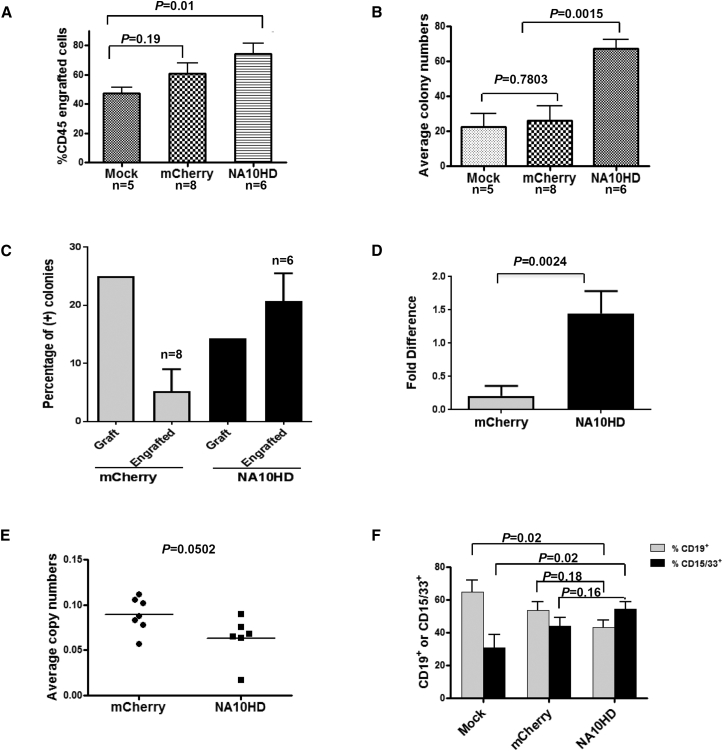
To determine whether the γ-globin/N10HD vector was biologically functional, we transduced human mobilized peripheral blood CD34+ cells with the γ-globin/NA10HD vector and γ-globin/mCherry as a control vector, respectively. Transduced cells were transplanted into NOD/SCID/IL2rγnull mice (10 NSG/group), and the percentage of CD45+ cells in the bone marrow (BM) was assessed 4 months post-transplantation. The CD45+ engrafted cell percentage in the NA10HD group but not in the mCherry group was significantly higher than the untransduced control (Figure 1A).
Figure 1.
Increased Engraftment of Human CD34+ Cells Transduced with NA10HD in NSG Mice
(A) Human nucleated hematopoietic cell engraftment (CD45+) as percentage of total bone marrow harvested from NSG mice 4 months post-transplantation with lentiviral transduced CD34+ cells. n, number of animals. (B) CFU-C assay of human CD45+ engrafted cells were sorted from bone marrow of NSG mice 4 months post-transplantation with lentiviral transduced CD34+ cells. (C) Percentage of positive colonies which were determined to contain an integrated virus fragment by using PCR in CFU-C assay of grafts and engrafts of NSG mice. (D) Fold differences of positivity of CFU-C colonies of the transplant graft versus engrafting cells in NSG mice. (E) Comparison of copy number between mCherry and NA10HD groups. The genomic DNA was derived from sorted CD45+ engrafted cells whose were derived from bone marrow of NSG mice. (F) Distribution of CD19+ and CD15/33+ cells among three groups of CD45+ engrafted cells. Mock n = 5, mCherry n = 8, NA10HD n = 6.
The effect of the NA10HD vector was further studied using the CFU-C assay, in which equal numbers of human CD45+ cells sorted from the bone marrow of the NSG mice were plated, and the average colony number of each group was enumerated. The colony number of the NA10HD group was approximately 2- to 2.5-fold higher than the γ-globin/mCherry and Mock groups (Figure 1B), indicating that NA10HD affects the proliferative capacity of cells in a CFU-C assay. Further analysis of positive colonies in the CFU-C assay before and 15 weeks after transplantation into NSG mice showed that the percentage of positive colonies in the NA10HD group was significantly increased (p = 0.0024) (Figures 1C and 1D). The vector copy number of the NA10HD group was slightly lower than the control group, indicating that the NA10HD gene possesses expanding activity in the engrafted CD45+ cells (Figure 1E). Using antibodies to CD19 and CD15/33 to measure both lymphoid and myeloid cells, respectively, among the CD45+ cells, we found little difference between the NA10HD and Mock groups and no significant difference between the mCherry and the NA10HD groups (Figure 1F).
Addition of NA10HD to the γ-Globin Vector Improves Therapeutic Activity in β-Thalassemic Mice
To understand whether the lentiviral vector encoding both γ-globin and NA10HD can be used to enrich a minority population of β-thalassemic red blood cells (RBCs) expressing human γ-globin, we transplanted β-thalassemic mice with lineage-depleted β-thalassemic BM cells transduced with either the γ-globin/NA10HD or the γ-globin/mCherry dual-gene lentiviral vector. In order to confirm similarity of transduction efficiency between two groups, Southern blot was used to analyze the numbers of insertions in genomic DNA extracted from spleen colonies. As indicated, the transduction efficiency of both vectors was 85% with an average insertion number of 2.6 among transduced colonies in the γ-globin/NA10HD vector group and 90% with an average insertion number of 2.0 in the γ-globin/mCherry vector group (Figures S3A and S3B), indicating similar transduction efficiencies by both vectors.
F Cells
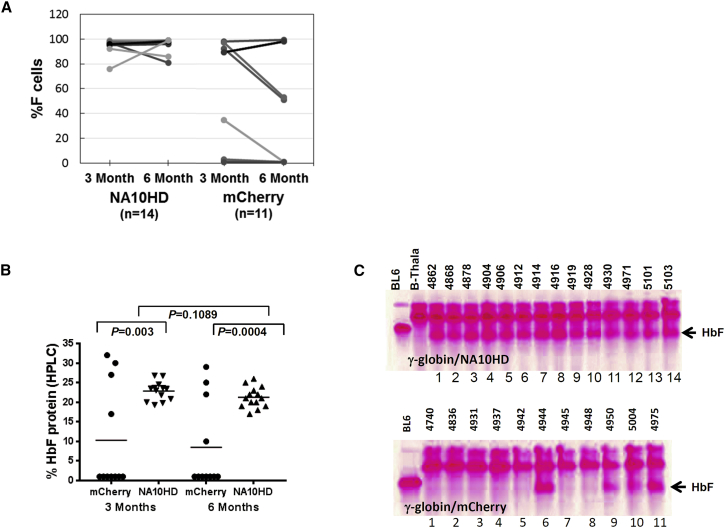
At 14 days following transplantation, the HbF-positive cells (F cells) were detected by flow cytometry in both the γ-globin/NA10HD and the γ-globin/mCherry groups after spleen colony formation (Figures S4A and S4B). At 3 and 6 months post-transplantation, the percentage of F cells in all animals in the γ-globin/NA10HD group remained high (Figure 2A). The F cell percentage in the γ-globin/NA10HD was significantly higher than in the control group at both 3 and 6 months (Figure 2A).
Figure 2.
Significant Increase of F Cell Percentage and HbF Protein in β-Thalassemic Mice Transplanted with Thalassemic Marrow Transduced with γ-Globin/NA10HD Lentiviral Vector
(A) F cell percentage in the γ-globin/NA10HD of 14 mice and the γ-globin/mCherry of 11 mice transduced groups at 3 and 6 months post-transplantation. N, number of mice. (B) Comparison of HbF protein by HPLC between γ-globin/mCherry and γ-globin/NA10HD vector groups at either 3 or 6 months post-transplantation. Statistically significant differences are indicated by p values < 0.05 as determined using Student’s t test. (C) Cellulose acetate gel electrophoresis of RBC lysates derived from individually transplanted animals. Two controls were used in the gels in Figure 2D, BL6 (C57/BL6) and β-Thal, which was from the thalassemic mouse. Top: the animal that received γ-globin/NA10HD vector; bottom: the samples derived from γ-globin/mCherry vector-transduced animals. The HbF protein band is marked by an arrow. Animal identification numbers are shown at the top of the respective gel bands.
HbF Protein
The quantitative analysis of HbF protein levels using high-performance liquid chromatography (HPLC) at 3 and 6 months showed that animals that received the γ-globin/NA10HD vector-transduced BM cells expressed HbF protein at a high level at both time points (Figures 2B, S5A, and S5B) indicating that the increased HbF level persisted (p = 0.1089). This increase in HbF occurred significantly less frequently in the control group (Figure 2B). These data clearly indicate that the NA10HD gene significantly enhanced HbF protein levels at both 3 and 6 months after transplantation (p = 0.003 and p = 0.0004, respectively). Cellulose acetate gel electrophoresis of individual transplanted RBC lysates revealed a clear HbF band (Figure 2C).
γ-Globin/NA10HD Vector Corrects the Anemia in β-Thalassemic Mice
Hemoglobin Level
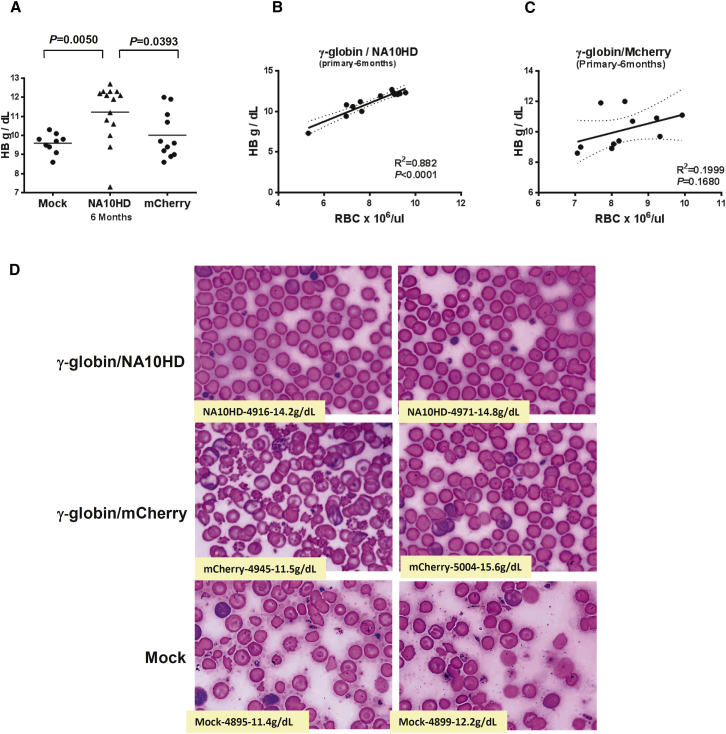
To determine whether the anemia of the β-thalassemic mice was corrected by the γ-globin/NA10HD lentiviral vector, hemoglobin levels were measured. As indicated in Figures S5C and S5D, the Hb levels in most animals of the NA10HD group averaged 11.23 ± 0.40 g/dL, while Hb levels in the mCherry group averaged 10.01 ± 0.37 g/dL (p = 0.039). The Hb level of the mCherry group was similar to that of Mock control group (Figure 3A). In order to clarify the correlation between Hb levels and RBC counts in the γ-globin/NA10HD group at 6 months following transplantation (Figure 3B), the Hb levels and RBCs were plotted, and both significantly fitted the regression line (p < 0.0001), as opposed to the γ-globin/mCherry group, for which the Hb level and RBC had no correlation (p = 0.168) (Figure 3C).
Figure 3.
Higher Hemoglobin Level in γ-Globin/NA10HD Lentiviral Vector Group Compared with Both γ-globin/mCherry and Mock Control Groups
(A) Comparison of the Hb levels among γ-globin/NA10HD, γ-globin/mCherry and Mock groups in blood collected 6 months post-transplantation. Statistically significant differences are indicated by p values < 0.05, as determined by Student’s t test. (B) Correlation between Hb level and RBC count in γ-globin/NA10HD-transduced group. (C) Correlation between Hb level and RBC in γ-globin/mCherry control group. (D) Blood smear slides stained by Giemsa method were derived from γ-globin/NA10HD, γ-globin/mCherry-transduced β-thalassemic cells, and mock control 3 months post-transplantation.
Erythrocyte Morphology
As shown in Figure 3D, the morphology of red cells in the γ-globin/NA10HD group was completely corrected to normal at 3 months following transplantation. In the γ-globin/mCherry group, if an animal had high HbF protein and Hb levels (Figures 2C and 3A), the RBCs also displayed normal morphology. The red blood cells observed in the animals with low or undetectable HbF protein were characterized by residual β-thalassemic red blood cell features similar to the mock group (Figure 3D).
NA10HD Enriches Transduced HSCs and Leads to a Fetal Hemoglobin Increase, Possibly through a Self-Controlled Mechanism, without Affecting White Blood Cell Numbers
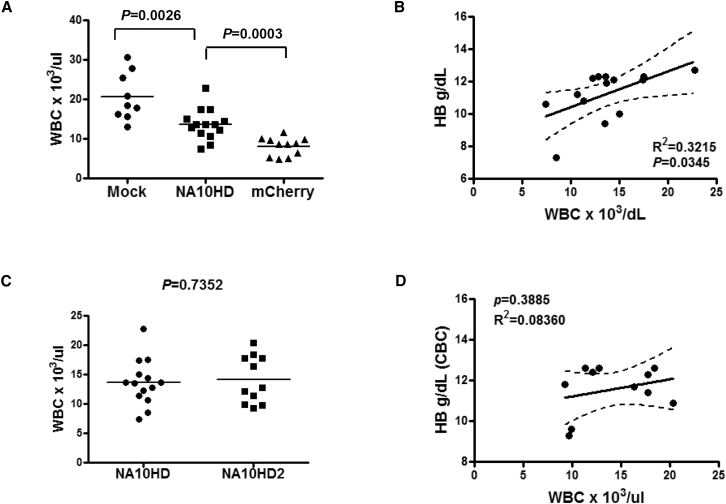
In order to determine whether both the RBC and WBC lineages were affected by the γ-globin/NA10HD vector, the blood cells were quantitated in the animals transplanted with the γ-globin/NA10HD vector, the γ-globin/mCherry vector, and the Mock group at 6 months. As indicated in Figure 4A, the number of WBCs in the γ-globin/NA10HD was intermediate and significantly lower than those in the mock control group but higher than in the mCherry group. To determine if there was a correlation between hemoglobin level and total WBCs in the γ-globin/NA10HD vector group, a regression analysis with a 95% confidence interval was performed (Figure 4B). No significant fit of Hb level or total WBCs was observed, suggesting that NA10HD enriches the erythroid progeny of transduced HSCs through increasing fetal hemoglobin. As for the increased number of WBCs in the NA10HD group compared with the mCherry group, a lower multiplicity of infection (MOI) (50% lower, designated as NA10HD2) was introduced in the next experiment. As indicated in Figures 4C and 4D, the total number of WBCs remained at the same level in both experiments, and no significant fit to the Hb level and total WBC was observed. On hemoglobin gel analysis, an obvious HbF band was identified in the lower MOI group, and their HbF protein levels reached those of the high-MOI group also (Figures S6A–S6C). The Hb levels and RBC counts showed a good correlation (Figure S6D).
Figure 4.
Correlation between White Blood Cell Count and Hemoglobin Level in Mice That Had Correction of Anemia
(A) Comparison of white blood cell count among γ-globin/NA10HD, γ-globin/mCherry, and Mock groups. The p value is indicated on each graph. (B) Correlation of hemoglobin level and total WBC count derived from β-thalassemic mice transplanted 6 months prior using γ-globin/NA10HD-transduced marrow (p = 0.0345). (C) Comparison of WBC count between 3 and 6 month post-transplantation time points of γ-globin/NA10HD and 6 months of experiment 2 using the same vector but with 50% MOI and labeled as NA10HD2. (D) Correlation of hemoglobin level and WBC count from β-thalassemic mice 6 months post-transplantation in experiment 2 using lower MOI γ-globin/NA10HD vector (p = 0.7209). The linear regression lines and 95% confidence band interval were plotted with data from the γ-globin/NA10HD and NA10HD2 groups.
Three individual experiments were performed. The γ-globin vector alone was used to transduce β-thalassemia lineage-negative bone marrow cells, which were then transplanted into β-thalassemic mice (V5m3-Thal) or BL6 mice (V5m3-BL6). The γ-globin/NA10HD vector was used to transduce the same type cells, which were then transplanted into β-thalassemic mice (NA10HD). The number of WBCs was similar in all three groups and intact β-thalassemic (Thala) mice (Figure S7A). We noticed that the WBC counts in all three Mock groups were significant higher than in the intact Thala mice (Figure S7B), suggesting that the γ-globin gene therapy possibly reduced organ damage and inflammation, as we have found previously.27
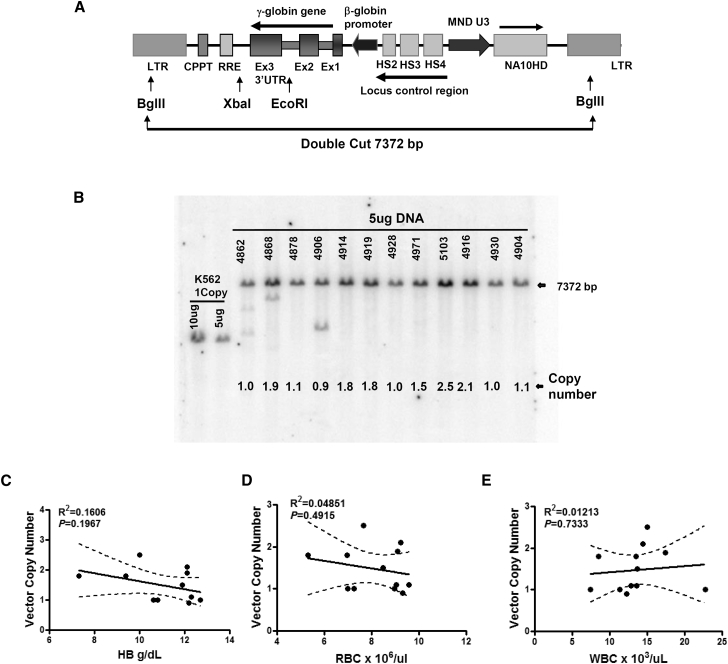
To determine whether the NA10HD enrichment of transduced HSCs was affected by the degree of gene transfer, the BM cells from primary transplanted β-thalassemic mice that received the γ-globin/NA10HD or the γ-globin/mCherry transplanted cells were collected at 6 months for vector copy number analysis. Each BM DNA sample derived from individual animals was digested using BglII (Figure 5A). The intensity of the band, relative to the standards of vector copy number, was consistent in all of the BM cells from the animals transplanted with cells transduced with the γ-globin/NA10HD-containing vector; the copy number ranged from 0.9–2.5. The average vector copy number was 1.48. As indicated, all the animals transplanted with cells transduced with the γ-globin/NA10HD vector displayed an expected virus insertion band (Figure 5B). Interestingly, additional bands were observed in three animals to indicate that the DNA rearrangement may be present in their engrafted cells.
Figure 5.
Vector Copy Numbers Correlate with Hemoglobin Level and the Counts of RBCs and WBCs in Thalassemic Mice Transplanted with γ-Globin/NA10HD Expressing Lentivirus
(A) Schematic diagram of the γ-globin/NA10HD lentiviral vector indicating the restriction enzyme sites used for Southern blot analysis. (B) Analysis of genomic DNA pattern of mice in γ-globin/NA10HD group as measured by Southern blot analysis. The DNA was digested using BglII, and the size of the DNA band is indicated by arrow. The copy number of each lane represents the vector copy number quantified by densitometry relative to the “standard one-copy”; a K562 clone containing a single copy of an integrated GFP lentiviral vector was used as the “standard one-copy” reference. (C–E) Correlations of copy number of individual animals of the γ-globin/NA10HD lentiviral vector with Hb level (C), RBC count (D), and WBC count (E).
A statistically significant correlation between the copy number in RBCs and WBCs and the Hb levels in individual animals was not observed (Figures 5C–5E). The copy number was plotted against RBC, WBC, and Hb levels. This suggested that the observed improvement in RBC and Hb with the NA10HD vector occurred over a range of VCNs up to a certain point and that there are likely mechanisms that regulate expression of HbF once a corrective level was achieved.
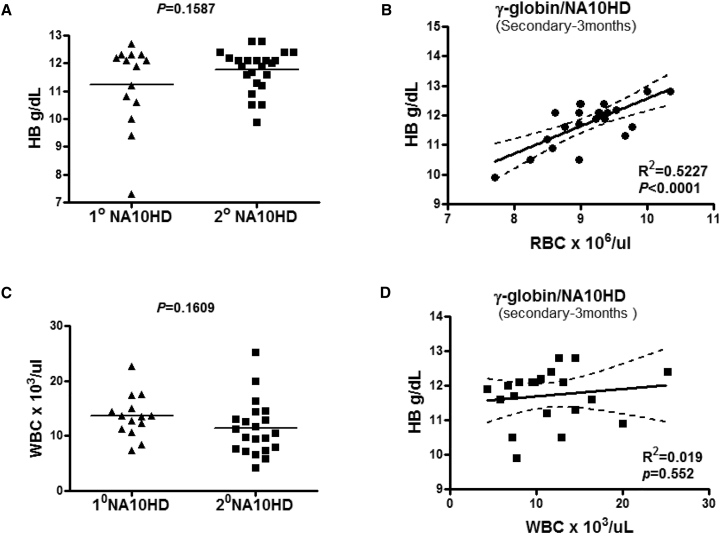
Secondary Transplantation Using Primary BM Cells Showed Sustained High-Level Expression of HbF
The BM cells from primary transplanted mice were transplanted into B6 wild-type recipients in a secondary transplantation experiment for a long-term study. The HbF protein and hemoglobin levels from secondary recipients were analyzed at 3 months after transplantation; both the HbF protein and hemoglobin levels remained stable at a high level in the γ-globin/NA10HD group (Figures 6A, 6B, and S8). To determine whether there was a correlation between Hb levels and RBCs in the γ-globin/NA10HD group (Figure 6C), Hb levels and RBC counts were analyzed and fit to the regression line at 3 months in the secondary transplant recipients (p = 0.0001). WBC counts were also analyzed in the same group and indicated that the Hb levels and WBCs did not fit a regression line, indicating that there was no correlation between Hb levels and WBCs (p = 0.552; Figure 6D).
Figure 6.
Hb Level in the γ-Globin/NA10HD Group Is Preserved in Secondary Transplanted Animals
(A) Comparison of Hb level in the γ-globin/NA10HD group from primary (n = 14) and secondary (n = 23) transplanted animals. (B) Correlation of Hb level and RBC count in the γ-globin/NA10HD group. (C) Comparison of WBC count between primary and secondary transplant of the γ-globin/NA10HD groups. (D) Correlation of Hb level and WBC count in γ-globin/NA10HD group.
Mouse ID4862, a primary transplant recipient of γ-globin-NA10HD-transduced cells, became a donor for secondary transplantation at 6 months post-transplantation. Three months after the secondary transplantation, all three recipients became ill. One animal died and the other two animals were diagnosed with small B cell leukemia. To investigate the etiology of this leukemia, we analyzed vector insertion sites from the primary animal (Figures S9A and S9B), and both oncogenes and tumor suppressor genes were not disturbed.
Vector Insertion of the γ-Globin/NA10HD Vector Occurs Mainly into Introns without Causing Clonal Dominance
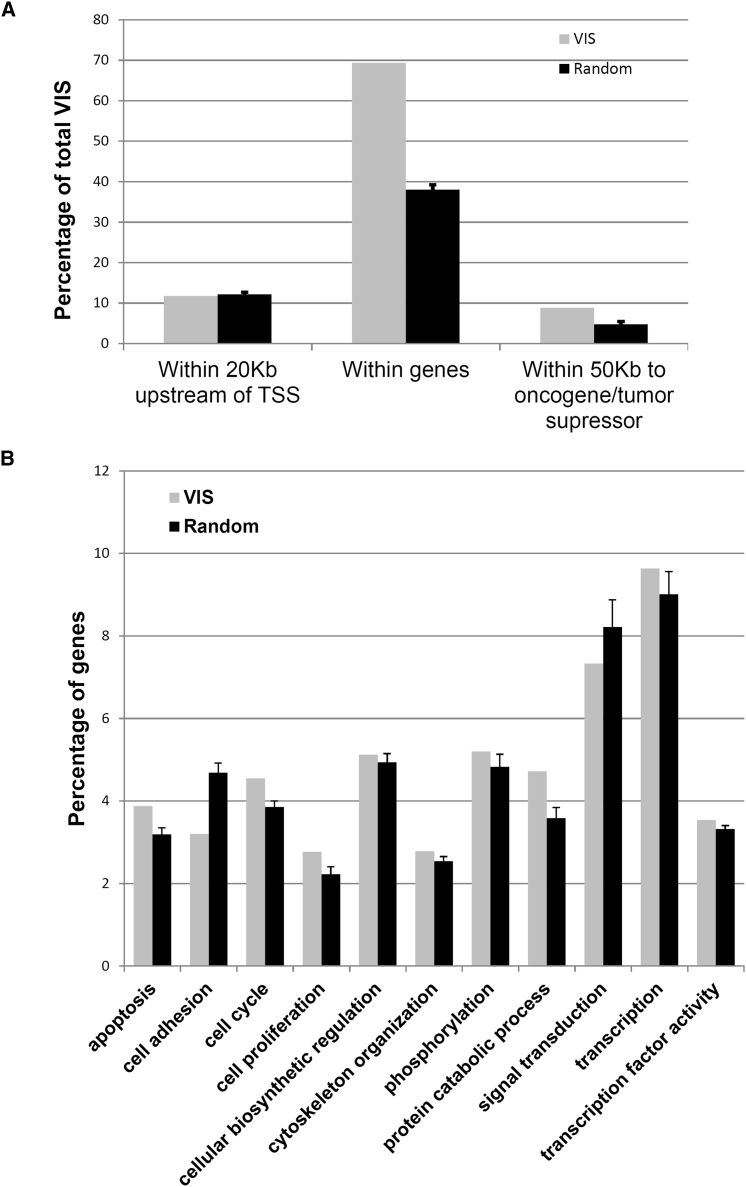
In order to profile the genomic locations of the integrated lentivirus vector, the genomic DNAs of BM cells from the primary recipients were mapped by linear amplification-mediated (LAM)-PCR, and the virus integration sites were analyzed. A total of 715 unique genomic virus integration sites were identified from 12 mice, each from three independent experiments with different restriction enzymes. As indicated in Figure 7A, about 69% of the virus integration sites in the mouse genome were within gene regions, compared with 38% of the random sets, in agreement with the observation that human immunodeficiency virus (HIV) prefers to integrate within genes.28 Further analysis indicated that 64% of the integration sites, which accounted for more than 92% of the gene-associated sites, were located within introns of the genes (Figure S10), while about 12% of integration sites were localized within 20 kb upstream of transcription start sites, and another 9% were within 50 kb of proto-oncogenes or tumor suppressor genes (Figure 7A). In addition, gene ontology analysis was performed using the DAVID functional annotation tool. The results demonstrated that no classes of genes were enriched within 50 kb of an integration site (Bonferroni p = 0.05) compared with three random sets of genes as background. Interestingly, by defining at least four integration sites within a 100 kb window (Figure 7B), we identified 15 clusters of integration sites, hotspot regions, and as indicated by Table 1, no oncogenes or tumor suppressor genes were found within 50 kb of any of these hotspot regions.
Figure 7.
Profile of the Virus Integration Site in Mouse Bone Marrow Genomic DNA after Transduction with the γ-Globin/NA10HD Vector
(A) The locations of 715 virus integration sites related to the gene: the percentages of total integration sites within 20 kb upstream of transcript start sites, within genes, and within 50 kb of oncogenes or tumor suppressor genes were plotted against three size-matched sets of computer-generated random sites. (B) Ontologies of genes within 50 kb of integration sites compared with size-matched set of genes within 50 kb of random generated sites, using the DAVID functional annotation tool.
Table 1.
Viral Vector γ-Globin/NA10HD Integration Hotspots
| Chr | Start | Stop | Length (bp) | VIS Count | Gene within 50 kb |
|---|---|---|---|---|---|
| chr1 | 181178372 | 181179843 | 1,471 | 5 | Cnih4//Nvl//Wdr26 |
| chr2 | 98662251 | 98667100 | 4,849 | 6 | |
| chr2 | 120513022 | 120514694 | 1,672 | 4 | 4931402G19Rik//Capn3//Zfp106 |
| chr4 | 129171134 | 129173005 | 1,871 | 8 | C77080//Fndc5//Hpca//S100pbp//Yars |
| chr4 | 134106904 | 134167961 | 61,057 | 6 | Aim1l//Catsper4//Cd52//Cep85//Gm7534//Sh3bgrl3//Ubxn11 |
| chr5 | 92157615 | 92160502 | 2,887 | 4 | U90926//Uso1 |
| chr6 | 84878244 | 84879506 | 1,262 | 4 | Exoc6b |
| chr7 | 66805807 | 66806746 | 939 | 4 | Adamts17//Cers3 |
| chr8 | 91217374 | 91218692 | 1,318 | 7 | Rpgrip1l |
| chr9 | 3000042 | 3035037 | 34,995 | 10 | Mir101c |
| chr11 | 96792129 | 96793084 | 955 | 4 | Cbx1//Gm11517//Nfe2l1//Skap1//Snx11 |
| chr13 | 55036100 | 55037292 | 1,078 | 6 | Hk3//Uimc1//Unc5a |
| chr14 | 19416508 | 19419195 | 2,687 | 5 | |
| chr15 | 89362301 | 89366163 | 3,862 | 6 | Adm2//Klhdc7b//Lmf2//Miox//Ncaph2//Odf3b//Sbf1//Sco2//Syce3//Tymp |
| chr17 | 64308442 | 64310543 | 1,985 | 7 | Pja2 |
A hotspot was defined as at least four integration sites within a 100 kb window at a certain genomic location. The size and number of integration site were shown for a total of 15 hotspots. The genes that were within 50 kb of the hotspots are listed, which include no oncogene or tumor suppressor genes.
Discussion
Gene transfer therapy for thalassemia that consistently results in sustained hemoglobin production to clinically relevant levels remains a challenge.2, 7, 8, 29 Successful transduction of HSCs faces the limitations of starting with a finite and rare cell population (i.e., HSCs after a bone marrow harvest procedure) and then maintaining the transduced cells in a primitive state resulting in the long-term hematopoiesis required for the resolution of thalassemia. Methods that focus on expanding transduced HSCs could improve the efficiency of the corrective gene transfer by increasing the number of HSCs available for transplantation as well as increasing the number of corrected HSCs after transduction. It would be important not to increase the likelihood of disturbing other cell lineages or leukemic transformation as a consequence, especially if there were an underlying predisposition to malignancy.
Ideally a transiently delivered or self-limiting method to expand transduced cells ex vivo or given as a therapeutic infusion with G-CSF to improve engraftment could be safe approaches. It is not yet clear what the minimum amount of gene corrected cells required for resolution of thalassemia is, but on the basis of limited reports of hematopoietic stem cell transplantation, stable engraftment resulting in 20% non-thalassemic cells in the bone marrow is sufficient, suggesting a similar target for the number of modified cells after gene therapy.30
Our gamma globin transfer vector without NA10HD resulted in repopulation with more than 50% of red blood cells being HbF cells in 4 of 11 animals, which was associated with improvement of the hemoglobin levels even with relatively low transduction MOI. In contrast, the addition of NA10HD to the gamma globin vector improved the efficacy to more than 80% F cells in all treated mice. There was also concurrent significant improvement in anemia, with an acceptable average vector copy number of 1.48. The initial transduction efficiencies of both vectors were similar in the 85%–90% range as measured by a splenic colony assay, but the resultant improved sustained engraftment of vector-transduced cells in the NA10HD group strongly supports the notion that the NA10HD maintained transduced cells in a relatively primitive state compared with the vector without NA10HD. This is further supported by the retransplantability of NA10HD gene-modified cells and the continued absence of thalassemia in all of the secondary transplant recipients.
Importantly, any factor that gives a survival advantage to gene modified cells should not result in an uncontrolled clonal expansion that affects other aspects of normal hematopoiesis outside of the erythropoiesis. We observed lower neutrophil and lymphocyte counts but similar monocyte counts in the NA10HD mouse groups compared with mice transplanted with untransduced thalassemic marrow. To determine whether NA10HD expression affects WBC hematopoiesis, a lower MOI was used in a parallel experiment. The lower MOI did not change the WBC count compared with the higher MOI (Figure 4C), indicating that the WBC may not be related to the expression of the NA10HD gene. In addition, when the WBC counts were analyzed in three experiments using γ-globin vectors without NA10HD (Figures S7A and S7B), the levels of WBCs were similar to the NA10HD group, supporting that the effect of lowering the WBC counts may be more related to γ-globin lentiviral gene transfer. Additionally, the total numbers of WBCs are within the normal range, as reported.16
A B cell-lineage leukemia developed in the recipients of secondary transplantation from one of the animals in the NA10HD group without a clear etiology. Previous observations of leukemic transformation with viral vector-mediated gene transfer have occurred from the vector insertion itself disturbing oncogenes. The most studied is from LMO2 associated leukemia after gammaretroviral gene transfer. In these cases, overexpression of LMO2 was shown as a major driver of leukemogenesis, but additional genetic lesions may be required to result in leukemia.31 In these cases, clonal dominance could be demonstrated with vector insertion site analysis showing that the mutations were leading to leukemia. In our case, there was no apparent dominant clone associated with oncogenesis, leading us to believe that insertional mutagenesis was not likely. A potential explanation for the leukemia is that native bone marrow cells in the primary transplant recipient may have acquired genetic alterations from the ablative radiation conditioning leading to a leukemic population that once transplanted into secondary recipients became apparent. The contribution of enforced expression of NA10HD to B lymphocytic leukemia development must be considered. HOXA10 has been associated more typically with acute myeloid leukemia in mice, in which it requires overexpression of another gene, Meis1, to develop.32 HOXA9-associated acute lymphocytic leukemia has also been shown to require Meis1.33 We did not find other genetic lesions in the leukemic cells that have been previously described with HOX-associated leukemia. We also did not find any insertion sites in the leukemic animals in any tumor suppressor genes or oncogenes and insertion sites in the secondary transplanted animals did not show amplification compared with primary cells from the same mouse, further supporting that this leukemogenesis did not occur in a manner typically associated with vector integration.
In order to define any possible reasons that may have affected the animals which died of leukemia, we performed an additional experiment 2 (NA10HD2), in which 11 animals were further analyzed. After 7 months, histological sections of kidney, liver, brain, bone marrow, lung, heart, thyroid, and peripheral blood were carefully studied. No pathological findings were observed. Rather, the anemia was corrected in all 11 animals. Three months later, we further analyzed the secondary transplanted animals. All mice transplanted with bone marrow cells derived from the first 11 animals transplanted in experiment 2 (NA10HD2) had normal CBCs.
However, one animal containing additional bands on Southern blot analysis of its engrafted cells later developed leukemia, which might be related to the DNA rearrangement in this animal.
Because doubt may still remain about the safety of long-term expression of NA10HD in HSCs, an attractive approach may be to use an optimized NA10HD protein as an additive during the transduction process to expand the number of HSCs available for gene transfer and expansion. This would avoid the long latency period of HOX overexpression (typically months) usually needed for HOX-associated leukemia to develop. Given the relatively low average vector copy numbers we observed in our experiments, we anticipate that the amount of NA10HD protein needed in a culture system should be achievable. Additionally, short-term administration of the NA10HD to recipients of gene-modified HSCs to improve engraftment may also be worthy of exploration. Such an approach could also be tested in human HSC transduction experiments, especially because maintenance of human HSCs as opposed to murine HSCs in an undifferentiated state throughout the culturing and gene transfer process in vitro is challenging and likely is a major factor in the observed low levels of sustained gene-modified HSC engraftment. Use of NA10HD in this way may ultimately improve the number of gene-transduced HSCs produced from a finite number of cells, reducing the risk of leukemic transformation and improving the likelihood of thalassemia resolution.
Materials and Methods
Mice
Heterozygous β-thalassemic male mice (a gift from Dr. T. Townes, University of Alabama at Birmingham) were bred with HW80 (B6.C-TyrcH1b Hbbd/By; Jackson Laboratory) female mice for more than 10 years. All experimentation with mice was performed under protocols approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.15
Plasmid Construction
To generate the γ-globin/NA10HD and γ-globin/mCherry vectors, NA10HD and mCherry cDNA expression cassettes driven by the MND U3 promoter were positioned in opposite orientation from a human γ-globin expression cassette, respectively.34 The NA10HD fusion gene and the MND U3/NA10HD expression cassette have previously been described.23
Preparation and Titration of Lentiviral Vector
VSV-G pseudotyped lentiviral vector particles were prepared with a four-plasmid system by transient transfection of 293T cells, as previously described.15, 34
Transduction of Human CD34+ Cells
Human mobilized peripheral blood CD34+ cells (30 × 106) were thawed from liquid nitrogen and plated in 20 mL of Stemspan SFEMII (Catalog No. 09655; STEMCELL) supplemented with 100 ng/mL of human Flt3L (Catalog No. 130-096-476; MACS Miltenyi Biotec), human SCF (Catalog No. 130-096-693; MACS Miltenyi Biotec), and human TPO (Catalog No. 130-094-011; MACS Miltenyi Biotec). Gentamicin sulfate (Catalog No. 17-518Z; Lonza) was added at a concentration of 50 μg/mL, and the cells were incubated at 37°C for 24 hr. After that, the cells were transferred to a RetroNectin-coated plate, and cell medium was replaced by fresh medium containing both cytokines and protamine sulfate (4 μg/mL; Fresenius Kabi USA) with a final viral concentrations at 1 × 107 infectious particles/mL and incubated at 37°C overnight. The cells were then removed from the RetroNectin-coated plate, and the cell medium was refreshed under the same conditions and incubated for an additional 6 hr. The CD34+ cells were prepared at a concentration of 1 × 106 cells/300 μL in PBS containing 2% FBS, and 1 million cells per mouse were injected into adult female NOD/SCID/IL2rγnull strain mice (NSG) via the tail vein, with ten animals per group. All the recipient animals had been injected intraperitoneally (i.p.) with 35 mg/kg busulfan (Busulfex; PDL BioPharma) 24 hr prior to the transplantation.
Three hundred CD34+ cells were mixed with 3 mL of Methocult (Catalog No. H4434 GF Methocult; StemCell Technologies), and 1 mL of the mixed media was plated onto each 35-mm plate. The cells were incubated at 37°C for 10–14 days, and the proportion of transduced colonies was determined by PCR.
Fifteen weeks post-transplantation, the mice were sacrificed, and bone marrow cells were harvested from tibias and femurs into PBS supplemented with 2% FBS. Samples from individual mice were prepared as single-cell suspensions and then stained with the following monoclonal antibodies on the basis of the manufacturer’s instructions: allophycocyanin-conjugated antihuman CD45 (Clone HI30, Catalog No. 555485; BD), phycoerythrin-Cy7-conjugated antihuman CD19 (Clone SJ25C1, Catalog No. 557835; BD), phycoerythrin-conjugated antihuman CD15 (Clone HI98, Catalog No. 555402; BD), and phycoerythrin-conjugated antihuman CD33 (Clone WM-54, Catalog No. R0745; DAKO). The hCD45+ population was further analyzed for either myeloid (CD33/15) or B lymphoid (CD19) subpopulations. Dead cells stained with 4′-6-diamidino-2-phenylindole were excluded from the analysis. This analysis (four-color flow cytometric analysis) was conducted using a FACS Aria (BD Biosciences).
For CFU-C analysis, 4 × 104 sorted human CD45+ cells were mixed with 1 mL of H4434 GF Methocult and plated onto 35 mm plates and incubated at 37°C for 12–14 days. The average vector copy number was determined by real-time qPCR, using a probe set specific for the lentivirus ψ region and normalized to values obtained using a probe set specific for human RNase P, as previously described.24, 35
Transduction and Transplantation of β-Thalassemic BM Cells
Bone marrow cells obtained from 3- to 5 month-old β-thalassemic mice were harvested in PBS containing 2% FBS. Lineage-negative cells were isolated by a magnetic separation method based on the manufacturer’s protocol for the Lineage Cell Depletion Kit (Miltenyi Biotec). After magnetic separation, all lineage negative cells were pooled together and re-suspended in Stem Span medium (StemCell Technologies) supplemented with 10 μg/mL heparin (Sigma-Aldrich), 50 U/mL penicillin and streptomycin, 2 mmol/L glutamine and 10 ng/mL mouse stem cell factor, 50 ng/mL mouse thrombopoietin, 20 ng/mL mouse insulin-like growth factor 2 (all from Peprotech), and 10 ng/mL human fibroblast growth factor 1 (R&D Systems) supplemented with cytokines, incubated at 37°C and 5% CO2 for 30 hr.
Cells were harvested, re-suspended in medium containing viral vector particles, Stem Span medium supplemented with the cytokines as mentioned above, plus an additional 6 μg/mL polybrene (Sigma- Aldrich). The cells were transferred into six-well dishes coated with RetroNectin (Takara) at a multiplicity of infection (MOI) of 6–7. After 12 hr, the transduced cells were lifted and re-suspended in medium containing fresh viral vector supernatant. This procedure was performed a total of three times in 36 hr on cells for the NA10HD2 group.
Bone marrow cells were harvested after the third transduction, washed in PBS containing 2% FBS, and re-suspended in the same buffer. Then 0.7 × 106 transduced bone marrow cells were injected via the tail vein into β-thalassemic mice irradiated with 1,125 or 1,100 cGy for the NA10HD2 group.
Hematological Analysis
F cells were quantified by using flow cytometry, and hemoglobin cellulose acetate gel electrophoresis and fluorescence-activated cell sorting (FACS) analysis of red cells to verify expression of human γ-globin were performed as previously described.15
Secondary Transplantation
To demonstrate the persistence of the integrated provirus, secondary transplantation was performed at 6 or 7 months after primary transplantation. A total of 10 million BM cells from primary transplanted mice were injected into BL6 irradiated with 1,125 cGy, and the blood analysis was performed after 3 months.
Spleen Colony-Forming Unit Assay
Five thousand transduced cells containing NA10HD or mCherry vector were injected into BL6 mice irradiated with 950 cGy. Mice were sacrificed 14 days later, and the individual colonies were obtained for genomic DNA preparation using EcoRI restriction enzyme for analysis of transduction efficiency.
Vector Copy Number Analysis
The presence of vector and copy number were determined by Southern blot analysis using standard laboratory procedure as described previously.15
Identification of Vector Insertion Sites Using LAM-PCR
Genomic provirus junction sequences were detected by using LAM-PCR as described previously.14, 30 Briefly, 100 ng of genomic DNA was derived from 6-month-old transplanted mice. LAM PCR was performed in the following manner: biotin labeled oligo (5′-AGGGTCTGAGGGATCTCTAGTTAC) was used for the initial 50 cycles of linear PCR. Another biotin labeled oligo (5′-CAGTGGGTTCCCTAGTTAGCCAGAGA) that contained a MseI site was used to run the same linear PCR. Biotin-labeled PCR products were purified using streptavidin-coated beads. The beads and single-strand DNA complex were washed and incubated with Klenow DNA polymerase, dNTP, and random hexanucleotides (Roche Diagnostic) to generate a double-stranded DNA fragment, which was digested with MseI, MspI, or NlaIII. The digested DNAs were ligated to a restriction site-complementary linker cassette. Following that, the DNAs were used as templates for nested PCR amplification. Viral vector-specific sequence (5′-CAGTGGGTTCCCTAGTTAGC) and linker cassette sequence (5′-ACTGACAGCGGAGATAATCG) were designed as the primers for the first PCR (PCR1) (30 cycles). In the second PCR, the PCR1 product, which had been amplified from the genomic provirus junction sequence, was used as a template. An LTR primer (5′-GGCAAAAAGCAGATCTTGTCTTCTT) and linker cassette primer (5′-GTGCGAGTAGCATACTAGAG) were applied and ligated to the 3′ end of the Illumina overhang adaptor sequences (forward overhang-specific sequence and reverse overhang-specific sequence were provided with the Illumina Miseq system) as well as for next index primer targeted. The method was provided with the Illumina Miseq system.
Mapping of Viral Integration Sites
Illumina high-throughput sequencing was used to identify the vector integration sites in the mouse genome. After the adaptor sequences were trimmed, the paired-end reads containing the LTR sequence and at least 30 nucleotides of mouse genomic DNA sequence were uniquely aligned to the mouse reference genome (UCSC assembly mm10, GRCm38) using CLC Genomic Workbench version 7.5 (CLC Bio). The viral integration sites were established on those mapped reads by confirmation of the specific restriction enzyme recognition sequences of MseI, NlaIII, or MspI, on the opposite end from the integration site of the paired-end read. A total of 715 unique integration sites were identified after combining individual experiments with three restriction enzymes from 12 mice and removing those identified in three mock experiments in the negative control. The genomic location of these integration sites was determined according to the annotation of mouse RefSeq database (GRCm38), and the ontologies of genes flanking to the integration sites were analyzed using DAVID (http://david.ncifcrf.gov). As a control data set, three size-matched sets of random sites were generated by following the criteria as described previously.36
Author Contributions
H.F.Z. designed research, performed research, collected and interpreted data, and wrote the manuscript. A.A. performed part of the research and helped in writing the manuscript. Y.-S.K. and T.P. performed research. J.Z. helped in writing the introduction. K.H. was a collaborator. Y.-D.W. performed bioinformatics. D.A.P. was the supervisor (deceased). A.W.N. is a supervisor and completed the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
We thank Dr. Andrew Wilber and Dr. Janice Riberdy for valuable discussion, the laboratory of Dr. Dennis Jay for performing the HPLC analysis of red cell lysates, the Flow Cytometry Core Facility of St. Jude Children’s Research Hospital (SJCRH), and the staff of the Animal Resource Center of SJCRH for animal care and experimental analysis. We also want to acknowledge the excellent administrative assistance provided by Pat Streich. This work was supported by National Heart, Lung, and Blood Institute program project (P01 HL053749) and the American Lebanese Syrian Associated Charities.
Footnotes
Supplemental Information includes ten figures and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.01.019.
Supplemental Information
References
- 1.Siler U., Paruzynski A., Holtgreve-Grez H., Kuzmenko E., Koehl U., Renner E.D., Alhan C., de Loosdrecht A.A., Schwäble J., Pfluger T. Successful combination of sequential gene therapy and rescue Allo-HSCT in two children with X-CGD—importance of timing. Curr. Gene Ther. 2015;15:416–427. doi: 10.2174/1566523215666150515145255. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh S., Thrasher A.J., Gaspar H.B. Gene therapy for monogenic disorders of the bone marrow. Br. J. Haematol. 2015 doi: 10.1111/bjh.13520. Published online June 5, 2015. [DOI] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S., Pai S.Y., Gaspar H.B., Armant M., Berry C.C., Blanche S., Bleesing J., Blondeau J., de Boer H., Buckland K.F. A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2014;371:1407–1417. doi: 10.1056/NEJMoa1404588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawliuk R., Westerman K.A., Fabry M.E., Payen E., Tighe R., Bouhassira E.E., Acharya S.A., Ellis J., London I.M., Eaves C.J. Correction of sickle cell disease in transgenic mouse models by gene therapy. Science. 2001;294:2368–2371. doi: 10.1126/science.1065806. [DOI] [PubMed] [Google Scholar]
- 5.Persons D.A., Hargrove P.W., Allay E.R., Hanawa H., Nienhuis A.W. The degree of phenotypic correction of murine beta-thalassemia intermedia following lentiviral-mediated transfer of a human gamma-globin gene is influenced by chromosomal position effects and vector copy number. Blood. 2003;101:2175–2183. doi: 10.1182/blood-2002-07-2211. [DOI] [PubMed] [Google Scholar]
- 6.Pestina T.I., Hargrove P.W., Jay D., Gray J.T., Boyd K.M., Persons D.A. Correction of murine sickle cell disease using gamma-globin lentiviral vectors to mediate high-level expression of fetal hemoglobin. Mol. Ther. 2009;17:245–252. doi: 10.1038/mt.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nienhuis A.W. Development of gene therapy for blood disorders: an update. Blood. 2013;122:1556–1564. doi: 10.1182/blood-2013-04-453209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulad F., Wang X., Qu J., Taylor C., Ferro L., Karponi G., Bartido S., Giardina P., Heller G., Prockop S.E. Safe mobilization of CD34+ cells in adults with β-thalassemia and validation of effective globin gene transfer for clinical investigation. Blood. 2014;123:1483–1486. doi: 10.1182/blood-2013-06-507178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik P. Gene therapy for hemoglobinopathies: tremendous successes and remaining caveats. Mol. Ther. 2016;24:668–670. doi: 10.1038/mt.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Negre O., Eggimann A.V., Beuzard Y., Ribeil J.A., Bourget P., Borwornpinyo S., Hongeng S., Hacein-Bey S., Cavazzana M., Leboulch P., Payen E. Gene therapy of the β-hemoglobinopathies by lentiviral transfer of the β(A(T87Q))-globin gene. Hum. Gene Ther. 2016;27:148–165. doi: 10.1089/hum.2016.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baldwin K., Urbinati F., Romero Z., Campo-Fernandez B., Kaufman M.L., Cooper A.R., Masiuk K., Hollis R.P., Kohn D.B. Enrichment of human hematopoietic stem/progenitor cells facilitates transduction for stem cell gene therapy. Stem Cells. 2015;33:1532–1542. doi: 10.1002/stem.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karponi G., Psatha N., Lederer C.W., Adair J.E., Zervou F., Zogas N., Kleanthous M., Tsatalas C., Anagnostopoulos A., Sadelain M. Plerixafor+G-CSF-mobilized CD34+ cells represent an optimal graft source for thalassemia gene therapy. Blood. 2015;126:616–619. doi: 10.1182/blood-2015-03-629618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Negre O., Bartholomae C., Beuzard Y., Cavazzana M., Christiansen L., Courne C., Deichmann A., Denaro M., de Dreuzy E., Finer M. Preclinical evaluation of efficacy and safety of an improved lentiviral vector for the treatment of β-thalassemia and sickle cell disease. Curr. Gene Ther. 2015;15:64–81. doi: 10.2174/1566523214666141127095336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H., Pestina T.I., Nasimuzzaman M., Mehta P., Hargrove P.W., Persons D.A. Amelioration of murine beta-thalassemia through drug selection of hematopoietic stem cells transduced with a lentiviral vector encoding both gamma-globin and the MGMT drug-resistance gene. Blood. 2009;113:5747–5756. doi: 10.1182/blood-2008-10-186684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negre O., Fusil F., Colomb C., Roth S., Gillet-Legrand B., Henri A., Beuzard Y., Bushman F., Leboulch P., Payen E. Correction of murine β-thalassemia after minimal lentiviral gene transfer and homeostatic in vivo erythroid expansion. Blood. 2011;117:5321–5331. doi: 10.1182/blood-2010-01-263582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seita J., Weissman I.L. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alharbi R.A., Pettengell R., Pandha H.S., Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013;27:1000–1008. doi: 10.1038/leu.2012.356. [DOI] [PubMed] [Google Scholar]
- 19.Sauvageau G., Thorsteinsdottir U., Eaves C.J., Lawrence H.J., Largman C., Lansdorp P.M., Humphries R.K. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 20.Schiedlmeier B., Klump H., Will E., Arman-Kalcek G., Li Z., Wang Z., Rimek A., Friel J., Baum C., Ostertag W. High-level ectopic HOXB4 expression confers a profound in vivo competitive growth advantage on human cord blood CD34+ cells, but impairs lymphomyeloid differentiation. Blood. 2003;101:1759–1768. doi: 10.1182/blood-2002-03-0767. [DOI] [PubMed] [Google Scholar]
- 21.Krosl J., Austin P., Beslu N., Kroon E., Humphries R.K., Sauvageau G. In vitro expansion of hematopoietic stem cells by recombinant TAT-HOXB4 protein. Nat. Med. 2003;9:1428–1432. doi: 10.1038/nm951. [DOI] [PubMed] [Google Scholar]
- 22.Ohta H., Sekulovic S., Bakovic S., Eaves C.J., Pineault N., Gasparetto M., Smith C., Sauvageau G., Humphries R.K. Near-maximal expansions of hematopoietic stem cells in culture using NUP98-HOX fusions. Exp. Hematol. 2007;35:817–830. doi: 10.1016/j.exphem.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekulovic S., Gasparetto M., Lecault V., Hoesli C.A., Kent D.G., Rosten P., Wan A., Brookes C., Hansen C.L., Piret J.M. Ontogeny stage-independent and high-level clonal expansion in vitro of mouse hematopoietic stem cells stimulated by an engineered NUP98-HOX fusion transcription factor. Blood. 2011;118:4366–4376. doi: 10.1182/blood-2011-04-350066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abraham A., Kim Y.S., Zhao H., Humphries K., Persons D.A. Increased engraftment of human short term repopulating hematopoietic cells in NOD/SCID/IL2rγnull mice by lentiviral expression of NUP98-HOXA10HD. PLoS ONE. 2016;11:e0147059. doi: 10.1371/journal.pone.0147059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sloma I., Imren S., Beer P.A., Zhao Y., Lecault V., Leung D., Raghuram K., Brimacombe C., Lambie K., Piret J. Ex vivo expansion of normal and chronic myeloid leukemic stem cells without functional alteration using a NUP98HOXA10homeodomain fusion gene. Leukemia. 2013;27:159–169. doi: 10.1038/leu.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watts K.L., Zhang X., Beard B.C., Chiu S.Y., Trobridge G.D., Humphries R.K., Kiem H.P. Differential effects of HOXB4 and NUP98-HOXA10hd on hematopoietic repopulating cells in a nonhuman primate model. Hum. Gene Ther. 2011;22:1475–1482. doi: 10.1089/hum.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pestina T.I., Hargrove P.W., Zhao H., Mead P.E., Smeltzer M.P., Weiss M.J., Wilber A., Persons D.A. Amelioration of murine sickle cell disease by nonablative conditioning and γ-globin gene-corrected bone marrow cells. Mol. Ther. Methods Clin. Dev. 2015;2:15045. doi: 10.1038/mtm.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derse D., Crise B., Li Y., Princler G., Lum N., Stewart C., McGrath C.F., Hughes S.H., Munroe D.J., Wu X. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J. Virol. 2007;81:6731–6741. doi: 10.1128/JVI.02752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finotti A., Breda L., Lederer C.W., Bianchi N., Zuccato C., Kleanthous M., Rivella S., Gambari R. Recent trends in the gene therapy of β-thalassemia. J. Blood Med. 2015;6:69–85. doi: 10.2147/JBM.S46256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreani M., Manna M., Lucarelli G., Tonucci P., Agostinelli F., Ripalti M., Rapa S., Talevi N., Galimberti M., Nesci S. Persistence of mixed chimerism in patients transplanted for the treatment of thalassemia. Blood. 1996;87:3494–3499. [PubMed] [Google Scholar]
- 31.Howe S.J., Mansour M.R., Schwarzwaelder K., Bartholomae C., Hubank M., Kempski H., Brugman M.H., Pike-Overzet K., Chatters S.J., de Ridder D. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorsteinsdottir U., Sauvageau G., Hough M.R., Dragowska W., Lansdorp P.M., Lawrence H.J., Largman C., Humphries R.K. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol. Cell. Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozovskaia T., Feinstein E., Mor O., Foa R., Blechman J., Nakamura T., Croce C.M., Cimino G., Canaani E. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4 : 11) abnormality. Oncogene. 2001;20:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- 34.Hanawa H., Hargrove P.W., Kepes S., Srivastava D.K., Nienhuis A.W., Persons D.A. Extended beta-globin locus control region elements promote consistent therapeutic expression of a gamma-globin lentiviral vector in murine beta-thalassemia. Blood. 2004;104:2281–2290. doi: 10.1182/blood-2004-03-0863. [DOI] [PubMed] [Google Scholar]
- 35.Wielgosz M.M., Kim Y.S., Carney G.G., Zhan J., Reddivari M., Coop T., Heath R.J., Brown S.A., Nienhuis A.W. Generation of a lentiviral vector producer cell clone for human Wiskott-Aldrich syndrome gene therapy. Mol. Ther. Methods Clin. Dev. 2015;2:14063. doi: 10.1038/mtm.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasimuzzaman M., Kim Y.S., Wang Y.D., Persons D.A. High-titer foamy virus vector transduction and integration sites of human CD34(+) cell-derived SCID-repopulating cells. Mol. Ther. Methods Clin. Dev. 2014;1:14020. doi: 10.1038/mtm.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.