Abstract
Next generation sequencing based genetic association study (GAS) is a powerful tool to identify candidate disease variants and genomic regions. While low coverage sequencing offers low cost but inadequacy in calling rare variants, high coverage is able to detect essentially every variant but at a high cost. Two-stage sequencing may be an economical way to conduct GAS without losing power. In two-stage sequencing, an affordable number of samples are sequenced at high coverage as the reference panel, then to impute in a larger sample is sequenced at low coverage. As unit sequencing costs continue to decrease, investigators can now conduct GAS with more flexible sequencing depths. Here, we systematically evaluate the effect of the read depth and sample size on the variant discovery power and association power for study designs using low coverage, high coverage and two-stage sequencing. We consider 12 low coverage, 12 high coverage and 50 two-stage design scenarios with the read depth varying from 0.5x to 80x. With state-of-the-art simulation and analysis packages and in-house scripts, we simulate the complete study process from DNA sequencing to SNP calling and association testing. Our results show that with appropriate allocation of sequencing effort, two-stage sequencing is an effective approach for conducting genetic association studies. We provide practical guidelines for investigators to plan the optimum sequencing based genetic association study including two-stage sequencing design given their specific constraints of sequencing investment.
Keywords: next-generation sequencing, sequencing cost, study design, rare variant
Introduction
Genetic association study is used to identify candidate variants, genes or genomic regions which contribute to specific diseases by testing the correlations between variant frequency and disease status(Lewis and Knight, 2012). DNA sequencing has emerged as a powerful technology for disease gene discovery since 2010(Li et al., 2010; Metzker, 2010; Pasaniuc et al., 2012). With the capability to sequence the whole genome, DNA sequencing is expected to detect variants across the full minor allele frequency (MAF) spectrum and associations which may explain some of the missing heritability(Goldstein et al., 2013; Lee et al., 2014; Manolio et al., 2009; Sham and Purcell, 2014). Over the past few years, it has brought great success to genetic epidemiology studies of both rare diseases(Bamshad et al., 2011; Gilissen et al., 2014; Muona et al., 2015) and common complex diseases(Iossifov et al., 2014; O’Roak et al., 2011; Wang et al., 2015).
Next-generation sequencing (NGS) is a reliable and widely used method for DNA sequencing (Metzker, 2010). A key consideration in NGS-based study is the sequencing coverage or depth, which is commonly defined as the average number of reads representing a given nucleotide in the reconstructed sequence(Sims et al., 2014). In a genetic association study, a high-coverage (>20x) sequencing is able to find rare variants and further explain some of the missing heritability with high confidence(Lee et al., 2014; Manolio et al., 2009). However, high-coverage sequencing is much costlier. Despite the decrease in unit sequencing costs in recent years(Wetterstrand KA, 2016), the cost of high-coverage sequencing of thousands of individuals remains substantial(Sims et al., 2014). It is usually used in resequencing, exome-sequencing or targeted sequencing(Beaudoin et al., 2013; Kiezun et al., 2012; Rivas et al., 2011) and has achieved great success for the study of rare and de novo mutations(Wang et al., 2015). Low-coverage (<10x) sequencing is used more often in studies sequencing large samples, such as the 1000 Genomes Project (~7.4x)(Abecasis et al., 2012; Auton et al., 2015), the UK10K-cohorts arm (~7x)(Walter et al., 2015) and others. In addition to low and high-coverage sequencing, two-stage sequencing has been applied to achieve a tradeoff of study power and cost(Gudbjartsson et al., 2015; Pasaniuc et al., 2012; Sham and Purcell, 2014; Steinthorsdottir et al., 2014). Two-stage design usually sequences an affordable number of individuals at a high coverage, followed by a large sample of low coverage sequencing. The high coverage stage serves as a reference panel for the imputation of the low coverage stage in order to identify more rare and low-frequency variants.
Many discussions can be found in the literature exploring the cost-effective design of sequencing based genetic association studies. Shen et al. illustrated theoretical and empirical design considerations to maximize the association power under the constraint of study-wide cost in sequencing based association studies(Shen et al., 2011). Via a series of simulated and real data analyses, Li et al. evaluated the performance of low-coverage sequencing in association power, SNP discovery and genotyping accuracy for genetic association studies(Li et al., 2011). Comparisons with array and high-coverage designs were also discussed, and design tools based on these two studies are available online (Kang et al., 2013; Shen et al., 2011). Flannick et al. analyzed the efficiency and accuracy of low-coverage and SNP array and their joint analysis(Flannick et al., 2012). Another study on extremely low-coverage (0.1–0.5x) sequencing showed that it might be a viable alternative to SNP array(Pasaniuc et al., 2012). With further imputation, it can increase power for genome-wide association studies. In another type of two-stage sequencing(Yang and Thomas, 2011), the high coverage stage is used as a discovery panel to provide a subset of potential causal variants for the low coverage stage sequencing, rather than a reference panel for imputation. Some investigations have focused on the performance of these kinds of two-stage sequencing studies for association testing by varying the sample size of the two stages, testing methods, and disease models(Kang et al., 2012; Yang and Thomas, 2011; Yang and Thomas, 2014).
As unit sequencing costs continue to decrease, investigators are more flexible in their choice of sequencing depth for genetic association studies. However, how to make full and efficient use of low-coverage, high-coverage and/or two-stage sequencing designs remains unclear. In this paper, we systematically compare the effects of the read depth and sample size on the power of both variant discovery and association testing for low-coverage, high-coverage and two-stage sequencing designs. With state of art simulation and analytical packages, we simulate the entire study process from DNA sequencing to SNP calling and association testing. Our results show that, with appropriate allocation of sequencing effort, two-stage sequencing is an effective approach for genetic association studies. Based on extensive simulations, we attempt to provide some practical guidelines for the cost-effective design of sequencing based genetic association studies. In addition, we intend to publish our simulation package pipelines so that individual investigators may explore the optimum study design according to their specific technical and sample specifications, and their funding situations.
Method
Simulation framework
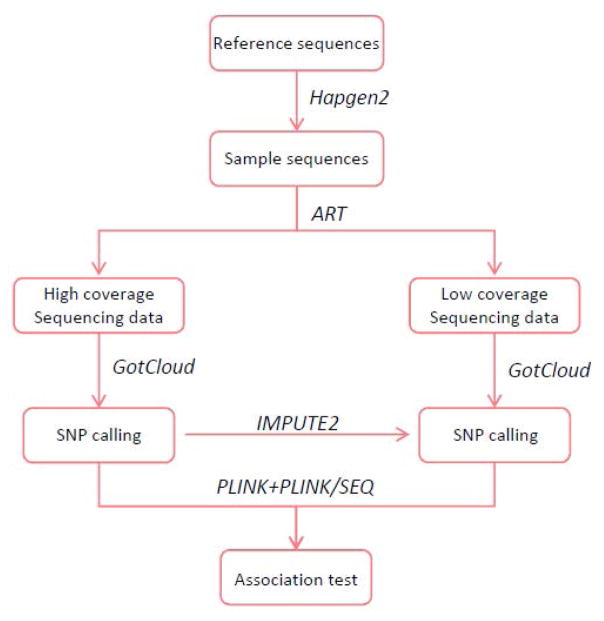
To evaluate the performance of low-coverage, high-coverage and two-stage sequencing studies with respect to SNP discovery and genetic association power, we mimicked a genetic association study in its entirety (Figure 1). First, samples of sequences were generated by Hapgen2 based on a reference genome. Hapgen2 is a program that simulates case control datasets of sequences with SNPs(Su et al., 2011) including the linkage disequilibrium (LD) between markers, and simulates multiple independent disease SNPs. For each sample, the sequencing data were produced by ART, which is a set of tools to simulate synthetic next-generation sequencing reads(Huang et al., 2012). Default empirical error models for Illumina pair-end sequencing platform and read length of 125 bp were adapted to run ART with other parameters set to default, such as the first- and second-read insertion rate of 0.00009 and 0.00015 respectively. After generating the sequencing reads, a variant calling pipeline of GotCloud (v1.13.2)(Jun et al., 2015) was employed to identify SNPs. The default and suggested parameters in GotCloud were used, such as the --minMapQuality 30, --minQual 30. The imputation in the two-stage sequencing study was conducted by IMPUTE2(Howie et al., 2012; Howie et al., 2009). Finally, the disease associations for single- and two-stage SNP genotypes were tested with the package PLINK (v1.07)(Purcell et al., 2007) and PLINK/SEQ (v0.10, https://atgu.mgh.harvard.edu/plinkseq/). The multiple testing adjusted significance level of 0.05 was used to claim significant association. The simulation was replicated 1000 times for each scenario.
Fig. 1.
Simulation pipeline
Disease model
We considered the study scenarios with equal sizes of independent case-control samples from European populations based on an additive genetic model. The disease prevalence was set to 9.3%, which is the typical prevalence of type 2 diabetes in the US(Centers for Disease Control and Prevention, 2014). Limited by the stability of Hapgen2, 15 causal SNPs were randomly picked from all the SNPs across the full MAF spectrum in each region. Only deleterious effects were considered. It was a typical assumption in previous studies(Moutsianas et al., 2015; Navon et al., 2013). In order to make the power comparable across a broad range of scenarios with different types of sequencing depth, the effect size of each causal variant was determined by controlling the variance it explained to be ~1%, which resulted in a negative correlation between variant frequency and effect size. This parameter was set to facilitate the comparison of the different designs and should not affect the results. In addition, the effect size was bounded (single allele relative risk ≤5.5) for rare variants to prevent unrealistically large effect sizes of very rare variants. Correspondingly, the variance explained by all the causal variants is ~ 10.9%. The effect size calculation was conducted with the program VarExplained (So et al., 2011).
Samples of sequences
The reference genome was taken from the 1000 Genomes haplotypes Phase 3 release (https://mathgen.stats.ox.ac.uk/impute/1000GP_Phase3.html). We randomly sampled 1000 sequences from chromosome 22 of a European population (EUR) for each of the 1000 simulation replications of each scenario. The sequence length was 100 kb: sufficient to cover the LD block, which typically extends 60 kb for EUR (Reich et al., 2001). SNPs with MAF ≥ 0.05 are defined as common SNPs. SNPs with MAF between 0.01 ≤ MAF < 0.05 are defined as low-frequency SNPs. Rare variants are those SNPs with MAF < 0.01.
Model evaluation
The sequencing investment was measured by the sequencing effort, which was the sequencing coverage multiplied by the sample size of that study(Li et al., 2011). The unit sequencing cost per mega base estimated by National Human Genome Research Institute (NHGRI) consists of direct sequencing cost and miscellaneous costs, such as sample preparations, utilities, reagents, and consumables(Wetterstrand KA, 2016). Consequently, given the same read length, the sequencing investment can be represented by the sequencing effort. We used person depth (pd) as the unit of the sequencing effort. A sequencing effort of 1,000 person depth is denoted by 1 kpd.
The variant discovery power was defined as the proportion of the identified variants among the total variants. The association power was defined as the proportion of statistically significant variants identified among the total true causal variants. A logistic regression model was employed to perform an association test of the variants with Bonferroni correction for multiple testing. In addition, the rare variants were examined by the region-based association test SKAT (Wu et al., 2011) as follows: We divided each of the simulated 100 kb sequences into 9 regions with a length of 20kb by a sliding step of 10kb. Multiple testing was adjusted. Only rare variants were included. The test result for a specific region using SKAT was checked against whether the region contained any rare causal variants.
Results
Simulated data
To compare the performance of low-coverage, high-coverage and two-stage sequencing studies on variant discovery and association testing, we simulated 12 scenarios for each single-stage low and high sequencing design. The combination of low and high sequencing scenarios comprises the two-stage sequencing scenarios, which are compared to single-stage sequencing with sequencing effort. We selected 6 levels of sequencing depth to cover the typical settings: 0.5x, 2x, 8x for low coverage and 30x, 60x, 80x for high coverage (Table 1). The total sample sizes considered for low coverage were 1000, 2000, 4000, and 6000, while the sample sizes for high coverage were 100, 200, 400 and 600 (Table 1). The combination of the parameter settings results in 24 single-stage and 51 two-stage sequencing scenarios.
Table 1.
Single- and two-stage sequencing scenarios considered
| Two stage sequencing | |||
|---|---|---|---|
|
| |||
| Low coverage sequencing | High coverage sequencing | ||
|
| |||
| Depth | Sample size | Depth | Sample size |
| 0.5 | 1000 | 30 | 100 |
| 2 | 2000 | 60 | 200 |
| 8 | 4000 | 80 | 400 |
| 6000 | 600 | ||
Based on the reference data from the 1000 Genomes Project, we generated 1000 sequences with a length of 100 kb in chromosome 22. On average, there were 951 SNPs within each sequence. Nearly 60% (563/951) were rare variants (MAF<0.01). The details of the proportion of simulated SNPs are listed in Table 2. Of the 9 regions divided by the sliding window within each sequence, about 8 contain rare causal variants. The average number of rare causal variants contained in those regions was ~2.23.
Table 2.
Summary of simulated SNPs
| Data | Number of SNPs | Common | Low frequency | Rare |
|---|---|---|---|---|
| Reference | 941 | 273 (29.0%) | 107 (11.4%) | 561 (59.6%) |
| Simulated | 951 | 279 (29.4%) | 108 (11.4%) | 563 (59.2%) |
Discovery power
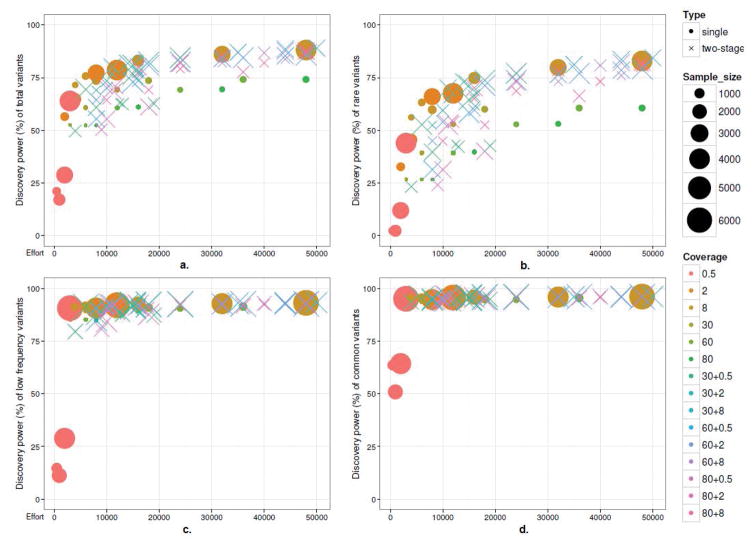
We first assessed the performance of single- and two-stage sequencing in SNP discovery using the proposed simulation methods. Figure 2a shows the discovery power of the total variants for all scenarios. There is generally a monotonically increasing quadratic relationship between discovery power and sequencing investment. The discovery power increases rapidly when the sequencing effort is below 10 kpd (kilo person depth), which includes most of the single-stage scenarios with depth ≤ 2. We call the corresponding area in Figure 2a the discovery power fast growth (DPFG) region. In order to investigate this region thoroughly, we added 2 more single stage scenarios within sequencing effort ≤ 6 kpd, which are included in Figure 2 and following results. They are 500@8 (sequencing 500 individuals at depth 8) and 750@8. Beyond the sequencing effort of 10 kpd, the gain in discovery power is limited. For example, when the sequencing effort increases from 12 kpd (6000@2x) to 48 kpd (6000@8x), the gain in discovery power is less than 10%.
Fig. 2.
Low-, high-coverage and two-stage sequencing on discovery power and sequencing effort (pd): a) total variants; b) rare variants; c) low frequency variants; d) common variants
Among those scenarios within the DPFG region, low coverage sequencing outperforms high coverage and two-stage sequencing in terms of high discovery power and low sequencing effort. The scenarios with depth 2 have a stable and steady increase in discovery power. The extremely low coverage of 0.5 has the maximum marginal gain in power. However, there is a decrease of power when the sample size increased (inverse trend) from 1000 to 2000 for coverage 0.5. The limited reads in the extremely low coverage (≤0.5x) may fail to provide sufficient information to call all the variants in the region, which could be compensated by increasing the sample size. The trend disappears when the sample size increases to 4000. It shows a minimum sample size is necessary to achieve a stable outcome in variant discovery using extremely low coverage sequencing.
Beyond the DPFG region, low coverage sequencing still works better than high coverage. For all of these low coverage scenarios, we can find some two-stage scenarios with a higher or at least comparable discovery power using the same or less sequencing effort (Table 3). For instance, a low coverage scenario of 6000@8x results a power of 88.01%, while a two stage scenario of 400@30+4000@8 produces a higher power of 88.54% with less effort (44 kpd vs 48 kpd). On the other hand, the power performance of some two-stage scenarios is not comparable to low coverage sequencing. For example, a two-stage scenario 400@80+2000@2 produced a detection power of only 77.71%, which is less than scenario 4000@8x (Table 3). The type I error rate for the scenarios are all well controlled under 0.05% except the single/two-stage designs involving sequencing @0.5 or 6000@2, which range from 0.06% to 1.4% (Supplementary Table 1).
Table 3.
Discovery power of total, rare, low frequency and common variants for top 20 scenarios by sequencing effort
| Scenario | Total | Rare | Low frequency | Common | Effort (kpd) | Type |
|---|---|---|---|---|---|---|
| 600@30+4000@8 | 89.04% | 84.44% | 93.34% | 96.17% | 50 | two-stage |
| 6000@8 | 88.01% | 82.82% | 93.10% | 96.06% | 48 | single |
| 200@80+4000@8 | 87.70% | 82.27% | 93.13% | 96.10% | 48 | two-stage |
| 400@80+2000@8 | 86.77% | 80.79% | 92.93% | 96.06% | 48 | two-stage |
| 600@60+6000@2 | 85.75% | 79.12% | 92.76% | 96.08% | 48 | two-stage |
| 600@80 | 74.11% | 60.49% | 91.33% | 95.41% | 48 | single |
| 400@30+4000@8 | 88.54% | 83.61% | 93.22% | 96.18% | 44 | two-stage |
| 200@60+4000@8 | 87.65% | 82.19% | 93.14% | 96.11% | 44 | two-stage |
| 600@60+4000@2 | 85.37% | 78.56% | 92.74% | 95.99% | 44 | two-stage |
| 600@60+1000@8 | 83.78% | 76.02% | 92.50% | 95.89% | 44 | two-stage |
| 400@60+2000@8 | 86.78% | 80.80% | 92.96% | 96.09% | 40 | two-stage |
| 400@80+1000@8 | 82.03% | 73.20% | 92.37% | 95.80% | 40 | two-stage |
| 400@60+6000@2 | 84.50% | 77.08% | 92.70% | 96.04% | 36 | two-stage |
| 400@80+2000@2 | 77.71% | 66.29% | 91.82% | 95.59% | 36 | two-stage |
| 600@60 | 74.12% | 60.51% | 91.32% | 95.41% | 36 | single |
| 100@30+4000@8 | 87.03% | 81.18% | 93.08% | 96.08% | 35 | two-stage |
| 4000@8 | 86.05% | 79.72% | 92.73% | 95.88% | 32 | single |
| 200@80+2000@8 | 85.43% | 78.64% | 92.82% | 95.98% | 32 | two-stage |
| 400@60+4000@2 | 84.07% | 76.44% | 92.69% | 95.91% | 32 | two-stage |
| 400@60+1000@8 | 82.05% | 73.23% | 92.36% | 95.84% | 32 | two-stage |
Full info of all scenarios available in Supplementary Table 1.
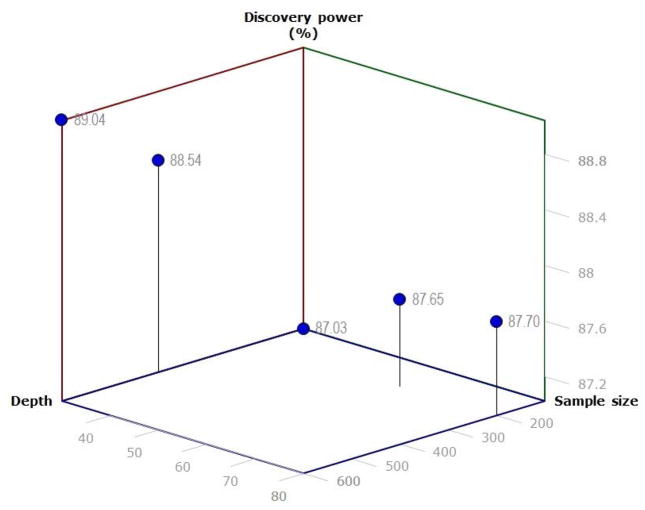
According to our results, to make an efficient two-stage design given certain sequencing investment, the stage of low coverage sequencing should utilize relatively high depth and/or sequence a large population (Table 3). Then for the high coverage stage, a large sample size is preferred rather than a high depth (Figure 3). Figure 3 shows the total variant discovery power comparison among 5 two-stage scenarios with the same design of the low-coverage stage (4000@8) but different depth and sample size for the high-coverage stage. The larger samples in high-coverage stage lead to higher discovery power of the total variants, while increasing the depth does not increase the discovery power much with fixed sample sizes.
Fig. 3.
Discovery power (total variants) comparison for 5 two-stage scenarios with same depth and sample size for the low-coverage stage
For the discovery power of rare variants, a similar pattern is observed. Figure 2b shows the discovery rate of the rare variants. The same DPFG region can be found for rare variant discovery. Although power increases rapidly in the DPFG region, extremely low coverage sequencing (0.5x) has very low absolute power (<50%) to discover rare variants, which may not be ignorable in practice. Beyond the DPFG region, two-stage sequencing expands the advantages in discovering power for rare variants compared with total variants (Table 3). Appropriate two-stage design is able to reach a higher discovery power using a lower sequencing effort compared with single-stage sequencing. The rules of how to make an appropriate two-stage design is the same as that summarized in previous analyses of the discovery of total variants.
Single-stage and two-stage sequencing show similar power in discovering common and low frequency variants. Figure 2c and 2d show that discovery power reaches a plateau for almost all scenarios with a sequencing effort exceeding 3 kpd. The few exceptions are the extremely low sequencing of 1000, 2000, and 4000 individuals at depth of 0.5. The inverse trend for extremely low coverage sequencing is present again in Figure 2c and 2d. The results suggest that extremely low sequencing may require a minimum sample size to get a reliable outcome for common and low frequency variants, for example, a sample size of 4000 for 0.5x (2000 pd).
Association power
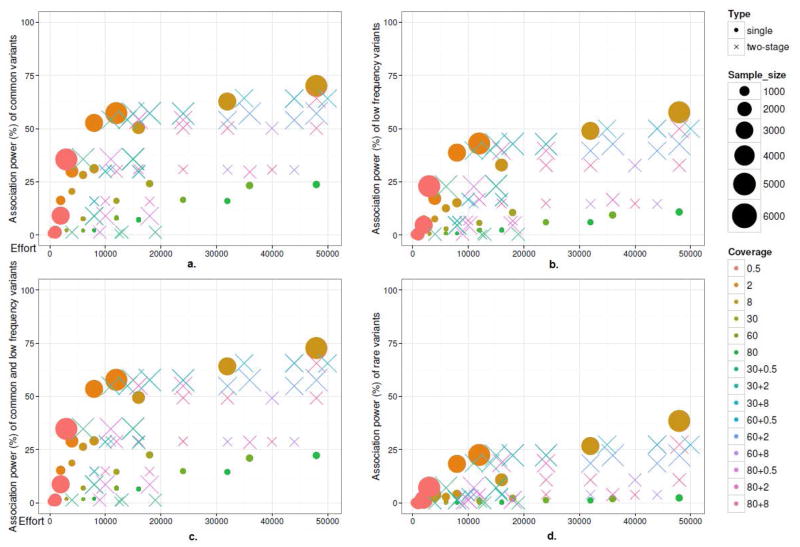
We further evaluated the association power of low, high and two stage sequencing design conditional on the variants discovered (Supplementary Figure 1, Supplementary Table 1). Figure 4 shows the power of testing common, low frequency and rare variants individually for all scenarios. There is an association power fast growth (APFG) region to the left of the sequencing effort of ~10 kpd in the plot. Within the APFG region, low coverage sequencing outperforms high coverage and two-stage scenarios. For most common variants, the power in the APFG is below 50%. Beyond the APFG region, the power gradually increases to ~75%. Most two-stage and high coverage scenarios are inferior to low coverage scenarios given a comparable sequencing effort. Some two-stage scenarios have equal or higher power than low-coverage scenarios; however, the sequencing efforts are also higher (Table 4). For example, compared with the scenario 4000@8x, a two-stage scenario of 100@30x+4000@8x reaches a higher power (64.3% vs 62.7%) with more effort (35 kpd vs 32 kpd). For low frequency and rare variants, the general conclusion is the same, but the overall power is less than common variants. Particularly for rare variants, most of the scenarios have a power under 25%. The overall type I error rate for single high-coverage scenarios are well controlled below 5%. However, the type I error rate are inflated (<19%) for single/two-stage design involving low-coverage sequencing (Supplementary Table 1). The accumulated sequencing and/or genotyping error may be the underlying reason. The extremely low-coverage (0.5x) scenarios can also control the type I error for association, but is due to the low power in discovering the candidate variants. Most of the null variants are not identified in the extremely low-coverage scenarios.
Fig. 4.
Low-, high-coverage and two-stage sequencing on association power and sequencing effort (pd): a) common variants; b) low frequency variants; c) common and low frequency variants; d) rare variants
Table 4.
Single variant association testing power of total, rare, low frequency and common variants for top 20 scenarios sorted by power of the common variant
| Scenario | Total | Rare | Low frequency | Common | Effort (kpd) | Type |
|---|---|---|---|---|---|---|
| 6000@8 | 51.95% | 38.45% | 57.61% | 70.04% | 48 | single |
| 4000@8+100@30 | 42.70% | 27.34% | 50.00% | 64.29% | 35 | two-stage |
| 4000@8+200@60 | 42.68% | 27.32% | 50.00% | 64.17% | 44 | two-stage |
| 4000@8+200@80 | 42.67% | 27.32% | 49.95% | 64.17% | 48 | two-stage |
| 4000@8+600@30 | 42.66% | 27.30% | 49.95% | 64.17% | 50 | two-stage |
| 4000@8+400@30 | 42.67% | 27.32% | 50.00% | 64.15% | 44 | two-stage |
| 4000@8 | 41.63% | 26.65% | 48.88% | 62.66% | 32 | single |
| 6000@2+100@60 | 36.66% | 22.42% | 42.84% | 57.28% | 18 | two-stage |
| 6000@2 | 36.69% | 22.42% | 42.98% | 57.20% | 12 | single |
| 6000@2+200@30 | 36.59% | 22.31% | 42.86% | 57.17% | 18 | two-stage |
| 6000@2+600@60 | 36.57% | 22.29% | 42.78% | 57.16% | 48 | two-stage |
| 6000@2+200@60 | 36.61% | 22.38% | 42.76% | 57.16% | 24 | two-stage |
| 6000@2+100@30 | 36.61% | 22.38% | 42.82% | 57.14% | 15 | two-stage |
| 6000@2+400@60 | 36.58% | 22.31% | 42.74% | 57.12% | 36 | two-stage |
| 6000@2+400@30 | 36.58% | 22.35% | 42.73% | 57.06% | 24 | two-stage |
| 4000@2+100@80 | 33.36% | 18.75% | 39.66% | 54.20% | 16 | two-stage |
| 4000@2+100@30 | 33.34% | 18.73% | 39.66% | 54.18% | 11 | two-stage |
| 4000@2+200@30 | 33.29% | 18.65% | 39.66% | 54.12% | 14 | two-stage |
| 4000@2+200@80 | 33.27% | 18.65% | 39.62% | 54.12% | 24 | two-stage |
| 4000@2+400@60 | 33.20% | 18.58% | 39.57% | 54.05% | 32 | two-stage |
Full info of all scenarios available in Supplementary Table 1.
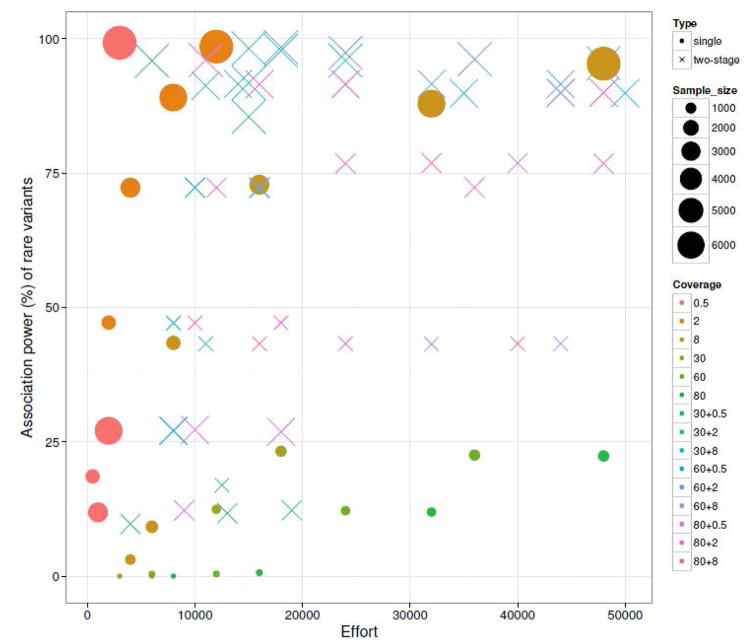
We then used a region-based test to compare association power for rare variants (Figure 5). The power of the pooled test increased dramatically compared with the single rare variant test. Low and two-stage sequencing are able to reach a power near 100%, while high coverage sequencing could only reach 25% even for very large sequencing efforts. For the extremely low designs, the power is only ~25% even for a sample size as large as 4000. The deficiency of association power is due to the limited discovery power for rare variants, which is only ~10%. However, when the discovery power increases to ~45% (6000@0.5), the association power rapidly reaches the saturation point near 100%. Further, the association power for the pooled test is likely determined by the sample size used for the low coverage stage. For example, scenarios 14–24 in Table 5 are two-stage scenarios having 4000 individuals for the low coverage stage. Scenarios 25 and 26 are single stage low coverage sequencing with 4000 samples. Although the association power for the two-stage is higher than for the single stage designs, they all result in power around 80%. Varying the sample size and depth of the high coverage stage (scenarios14–24) does not produce great differences in power. With respect to the optimum design of high power and little effort, the advanced two-stage designs all rely on a larger sample size in the low coverage stage than in single stage sequencing; for example, 100@30+6000@2 with 15 kpd effort and 98.20% power compared with 4000@8 with 32 kpd effort and 87.88% power.
Fig. 5.
Region based association power and sequencing effort for rare variants
Table 5.
Selected scenarios with top region based rare variants association power
| No. | Scenario | Power | Effort (kpd) | Type |
|---|---|---|---|---|
| 1 | 6000@0.5 | 99.22% | 3 | single |
| 2 | 6000@2 | 98.53% | 12 | single |
| 3 | 6000@2+100@60 | 98.43% | 18 | two-stage |
| 4 | 6000@2+100@30 | 98.20% | 15 | two-stage |
| 5 | 6000@2+200@30 | 97.79% | 18 | two-stage |
| 6 | 6000@2+200@60 | 97.40% | 24 | two-stage |
| 7 | 6000@2+400@60 | 96.19% | 36 | two-stage |
| 8 | 6000@0.5+100@80 | 96.02% | 11 | two-stage |
| 9 | 6000@2+400@30 | 95.94% | 24 | two-stage |
| 10 | 6000@0.5+100@30 | 95.93% | 6 | two-stage |
| 11 | 6000@2+600@60 | 95.45% | 48 | two-stage |
| 12 | 6000@8 | 95.36% | 48 | single |
| 13 | 6000@0.5+200@60 | 91.77% | 15 | two-stage |
| 14 | 4000@2+400@60 | 91.61% | 32 | two-stage |
| 15 | 4000@2+200@30 | 91.57% | 14 | two-stage |
| 16 | 4000@2+600@60 | 91.56% | 44 | two-stage |
| 17 | 4000@2+200@80 | 91.52% | 24 | two-stage |
| 18 | 4000@2+100@80 | 91.50% | 16 | two-stage |
| 19 | 4000@2+100@30 | 91.27% | 11 | two-stage |
| 20 | 4000@8+200@80 | 89.97% | 48 | two-stage |
| 21 | 4000@8+400@30 | 89.95% | 44 | two-stage |
| 22 | 4000@8+200@60 | 89.91% | 44 | two-stage |
| 23 | 4000@8+600@30 | 89.87% | 50 | two-stage |
| 24 | 4000@8+100@30 | 89.85% | 35 | two-stage |
| 25 | 4000@2 | 89.02% | 8 | single |
| 26 | 4000@8 | 87.88% | 32 | single |
Full info of all scenarios available in Supplementary Table 2.
Discussion
Our study compared low-coverage, high-coverage and two-stage sequencing in the design of genetic association studies. In line with previous findings(Li et al., 2011; Pasaniuc et al., 2012), we found that given a certain sequencing effort, low-coverage sequencing is an efficient method to conduct genetic association studies. High-coverage sequencing is not as efficient as low-coverage with regard to sequencing investment. The advantage of high-coverage is the convincing detection of rare and de novo mutations, which may play a significant part in the heritability of complex genetic diseases(Veltman and Brunner, 2012). In our results, the false discovery rate (FDR) of rare variant detection was < 1.3% for high-coverage scenarios. Two-stage design is another way to utilize this advantage of high-sequencing. Compared with low-coverage only, two-stage sequencing is more powerful in discovering rare variants. We have summarized several basic guidelines on how to make an efficient two-stage design. Given a certain sequencing investment, the first priority under consideration is the sample size of the low-coverage stage. It determines not only the discovery power but also the final association power. The next priority is the depth of the low-coverage stage. The higher the depth, the higher the discovery power. The third priority is the sample size for the high-coverage stage, while the depth of the high-coverage stage is less impactful.
To achieve an optimum design in discovery power and association power given a certain sequencing investment, single stage using low-coverage sequencing is the best choice if only a small amount of investment (<10 kpd) is available. Use caution for extremely low-coverage sequencing (~0.5x), as a minimum sample size may be necessary for a reliable outcome, such as 4000 for 0.5x. If the available investment is sufficient (> 10 kpd), low-coverage and appropriate two-stage design are both applicable. The optimum design at specific sequencing investment can be derived through enumerating all or the major representative types of the possible designs using our simulation pipeline. It is noticeable that we focus on low (<10x) and high-coverage (20x). The mid-range coverage between 10x and 20x is not covered, but should also be considered when search for the optimal design in reality. Our conclusions are based on the unit sequencing cost estimate from NHGRI. The unit sequencing cost may not accurately predict the sample recruitment cost in past and future studies. By simply replacing the estimate of the sequencing investment, optimum design can still be found for investigators having specific cost information.
Our study only compared the discovery and association power for single variants. The efficiency for low-, high- and two-stage sequencing in detecting other structural variants remains unexplored. For instance, the power of detecting insertion, deletion, copy number variations (CNVs) are also dependent on sequencing depth(Xi et al., 2011), so our conclusions may not apply in those scenarios.
The influence of sequencing effort on discovery and association power shows different patterns. Sequencing effort >~3 kpd is sufficient to identify >90% common and low frequency variants. On the other hand, the association power of common and low frequency variants monotonically increases within the entire range (0–50 kpd) of the sequencing effort. For rare variants, the discovery power is highly correlated with the sequencing effort. An investment < 10 kpd gives a higher marginal return in discovery power, while an investment > 10 kpd still increases the power, albeit more slowly. Owing to the powerful gene/region based rare variant testing method, the power growing interval is shortened to <3 kpd (Figure 5). Beyond 3 kpd, association power reaches a plateau. In short, sequencing effort is more influential for the association testing of common and low frequency variants and discovery of rare variants, but less influential for the discovery power of common and low frequency variants and gene/region based association testing of rare variants.
Genetic architecture varies for different diseases. The disease model we considered is only one of the possible hypotheses. While the optimum design strategy under other complex disease models requires further research, our simulation pipeline can be easily extended to other models using our in-house scripts, which are available at https://github.com/xu1912/spS-Gas.git. Moreover, our conclusions here can serve as a baseline reference for other disease modeling. For instance, in our study, rare, low frequency and common variants all have deleterious effects and the magnitude of their effects is inversely correlated with their frequencies. If, for another disease, rare variants only have modest effects, two-stage sequencing may provide more advantages in association power by virtue of its ability to include more rare variants.
In the pooled rare variant association test, we noticed that given the same sample size 6000, although the power for 0.5x, 2x and 8x are similar, the lower depth results in higher power. This is because the lower depth identified more false rare variants which inflated the association power. The FDR of the rare variants for 0.5x, 2x and 8x with 6000 sample size are 73.8%, 47.9% and 3.1% respectively. While the FDR can be controlled by tuning the filters in the variant discovery package, it demonstrates that investigators should be careful in analyzing data from extremely low (~0.5x) sequencing.
Another aspect to consider is missing heritability. Through simulating the causal variants by assuming the variance they explained, we find almost all scenarios with sequencing investment over 10 kpd failed to discover ~9.83% heritability, which is impossible to be identified by subsequent association testing. Advanced SNP calling methods may be helpful; the deficiency in association power leads to extra loss in heritability. The analysis of common variants shows that study design plays a role in uncovering heritability. The missing heritability in association testing may be partially recovered by improved study design and testing methods (Eichler et al., 2010; Zuk et al., 2014). A recent publication found negligible missing heritability for human height and body mass index through imputation after first stage genotyping(Yang et al., 2015), which shows a better design helps in finding missing heritability. The impact of sequencing study design on missing heritability in a genetic study merits further investigation.
Conclusion
In conclusion, through a complete simulation pipeline for sequencing based genetic association studies, we assessed the performance of low-, high- and two-stage sequencing with regard to the discovery power and association power of single variants in randomly picked 100 kb regions from human chromosome 22. Based on our results, we provide some basic guidelines for formulating a competent two-stage sequencing design and find an optimum design given a certain sequencing investment. With the high sequencing of large samples becoming more achievable, our study will facilitate the effective and efficient application of low-, high- and two-stage sequencing in genetic studies.
Supplementary Material
Acknowledgments
This work was supported by the grants from NIH (R01-AR050496-11, R01-AR057049-06, R01-AR059781-03) and Edward G. Schlieder Endowment at Tulane University. The authors would like to appreciate the assistance of Loula Burton, Office of Research in Tulane University, in editing the manuscript. We thank the editor and the two anonymous reviewers whose comments/suggestions helped improve and clarify this manuscript.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- Beaudoin M, Goyette P, Boucher G, et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013;9:e1003723. doi: 10.1371/journal.pgen.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States. US Department of Health and Human Services; Atlanta, GA: 2014. [Google Scholar]
- Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannick J, Korn JM, Fontanillas P, et al. Efficiency and power as a function of sequence coverage, SNP array density, and imputation. PLoS Comput Biol. 2012;8:e1002604. doi: 10.1371/journal.pcbi.1002604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Allen A, Keebler J, et al. Sequencing studies in human genetics: design and interpretation. Nat Rev Genet. 2013;14:460–470. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Helgason H, Gudjonsson SA, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, et al. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Li L, Myers JR, et al. ART: a next-generation sequencing read simulator. Bioinformatics. 2012;28:593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G, Wing MK, Abecasis GR, et al. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 2015;25:918–925. doi: 10.1101/gr.176552.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang G, Lin D, Hakonarson H, et al. Two-stage extreme phenotype sequencing design for discovering and testing common and rare genetic variants: efficiency and power. Hum Hered. 2012;73:139–147. doi: 10.1159/000337300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Huang KC, Xu Z, et al. AbCD: arbitrary coverage design for sequencing-based genetic studies. Bioinformatics. 2013;29:799–801. doi: 10.1093/bioinformatics/btt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, Garimella K, Do R, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Abecasis GR, Boehnke M, et al. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Knight J. Introduction to Genetic Association Studies. Cold Spring Harbor Protocols. 2012;2012 doi: 10.1101/pdb.top068163. db. [DOI] [PubMed] [Google Scholar]
- Li Y, Sidore C, Kang HM, et al. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Vinckenbosch N, Tian G, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Moutsianas L, Agarwala V, Fuchsberger C, et al. The power of gene-based rare variant methods to detect disease-associated variation and test hypotheses about complex disease. PLoS Genet. 2015;11:e1005165. doi: 10.1371/journal.pgen.1005165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muona M, Berkovic SF, Dibbens LM, et al. A recurrent de novo mutation in KCNC1 causes progressive myoclonus epilepsy. Nat Genet. 2015;47:39–46. doi: 10.1038/ng.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon O, Sul JH, Han B, et al. Rare variant association testing under low-coverage sequencing. Genetics. 2013;194:769–779. doi: 10.1534/genetics.113.150169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B, Rohland N, McLaren PJ, et al. Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nat Genet. 2012;44:631–635. doi: 10.1038/ng.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Rivas MA, Beaudoin M, Gardet A, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Purcell SM. Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet. 2014;15:335–346. doi: 10.1038/nrg3706. [DOI] [PubMed] [Google Scholar]
- Shen Y, Song R, Pe’er I. Coverage tradeoffs and power estimation in the design of whole-genome sequencing experiments for detecting association. Bioinformatics. 2011;27:1995–1997. doi: 10.1093/bioinformatics/btr305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D, Sudbery I, Ilott NE, et al. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- So HC, Gui AH, Cherny SS, et al. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol. 2011;35:310–317. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
- Steinthorsdottir V, Thorleifsson G, Sulem P, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46:294–298. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- Su Z, Marchini J, Donnelly P. HAPGEN2: simulation of multiple disease SNPs. Bioinformatics. 2011;27:2304–2305. doi: 10.1093/bioinformatics/btr341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- Walter K, Min JL, Huang J, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lu Q, Zhao H. A review of study designs and statistical methods for genomic epidemiology studies using next generation sequencing. Front Genet. 2015;6:149. doi: 10.3389/fgene.2015.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterstrand KA. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) 2016 [Google Scholar]
- Wu MC, Lee S, Cai T, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi R, Hadjipanayis AG, Luquette LJ, et al. Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc Natl Acad Sci U S A. 2011;108:E1128–E1136. doi: 10.1073/pnas.1110574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Thomas DC. Two-stage design of sequencing studies for testing association with rare variants. Hum Hered. 2011;71:209–220. doi: 10.1159/000328193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bakshi A, Zhu Z, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Thomas DC. Two-stage family-based designs for sequencing studies. BMC Proc. 2014;8:S32. doi: 10.1186/1753-6561-8-S1-S32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Schaffner SF, Samocha K, et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.