Abstract
Chagas disease, caused by the protozoan Trypanosoma cruzi, is a life-long and debilitating illness of major significance throughout Latin America, and an emergent threat to global public health. Being a neglected disease, the vast majority of Chagasic patients have limited access to proper diagnosis and treatment, and there is only a marginal investment into R&D for drug and vaccine development. In this context, identification of novel biomarkers able to transcend the current limits of diagnostic methods surfaces as a main priority in Chagas disease applied research. The expectation is that these novel biomarkers will provide reliable, reproducible and accurate results irrespective of the genetic background, infecting parasite strain, stage of disease, and clinical-associated features of Chagasic populations. In addition, they should be able to address other still unmet diagnostic needs, including early detection of congenital T. cruzi transmission, rapid assessment of treatment efficiency or failure, indication/prediction of disease progression and direct parasite typification in clinical samples. The lack of access of poor and neglected populations to essential diagnostics also stress the necessity of developing new methods operational in Point-of-Care (PoC) settings. In summary, emergent diagnostic tests integrating these novel and tailored tools should provide a significant impact on the effectiveness of current intervention schemes and on the clinical management of Chagasic patients. In this chapter, we discuss the present knowledge and possible future steps in Chagas disease diagnostic applications, as well as the opportunity provided by recent advances in high-throughput methods for biomarker discovery.
Keywords: Diagnostic applications, Chagas disease, Trypanosoma cruzi, biomarker discovery, strain polymorphisms, serotyping
INTRODUCTION
Chagas disease or American Trypanosomiasis, caused by the parasitic protozoan Trypanosoma cruzi (Kinetoplastida, Trypanosomatidae), is a life-long, neglected tropical disease, and leading cause of cardiomyopathy in endemic areas (Rassi, Rassi and Marin-Neto, 2010). With 8 to 10 million people already infected and up to 120 million individuals at risk of infection, Chagas disease constitutes the most important parasitic disease in Latin America and one of the most common globally (Stanaway and Roth, 2015). Its exact burden is however difficult to assess due to several factors including the widespread geographic distribution of T. cruzi vector-borne transmission, the decades-long lag between infection and appearance of symptoms, certain pitfalls of current diagnostic methods, biased prevalence data, and incomplete recognition of Chagas disease-attributable symptoms (Stanaway and Roth, 2015). The most recent estimates indicate that Chagas disease is responsible for ~550,000 disability adjusted life years (DALY), a measure that captures both premature mortality (~12,000 deaths per year) and non-fatal health losses (Stanaway and Roth, 2015). Despite this enormous toll, only two trypanocydal drugs, benznidazole and nifurtimox are currently available for chemotherapy. Both are nitroheterocyclic, oral compounds that require prolonged administration, may display severe adverse effects, cannot be used to treat pregnant women due to their uncertain teratogenic risks and, most importantly, show high efficacy solely if administered at the onset of infection (Carlier and Truyens, 2015; Rassi, Rassi and Marin-Neto, 2010; Viotti et al., 2006). The prospects for the development of an effective vaccine for prophylactic and/or therapeutic purposes, on the other hand are still clouded by substantial scientific and socioeconomic challenges (Beaumier et al., 2016; Bustamante and Tarleton, 2015).
T. cruzi transmission primarily occurs when humans are exposed to the contaminated feces of infected, hematophagous triatomine vectors. Large-scale intervention schemes launched in different regions of Latin America in the 1990’s have successfully shrunk the geographic limits and prevalence of vector-borne parasite transmission, and led to an overall ~40% reduction of disease prevalence (Schofield, Jannin and Salvatella, 2006). However, different ecological and demographic issues converged in the last decades to shift the epidemiological landscape for this disease. For instance, recent outbreaks of acute cases in certain regions from Brazil and Venezuela were not strictly vector-borne but rather due to accidental ingestion of T. cruzi-tainted food and fluids (Alarcon de Noya et al., 2010; Segovia et al., 2013). This 'food-borne' transmission mode likely constitutes an ancient epidemiological trait, very important to the zoonotic spreading of the parasite (Gurtler and Cardinal, 2015), and appears to be associated with increased virulence and a higher case-fatality rate in humans (Alarcon de Noya et al., 2010; Segovia et al., 2013). In addition, migratory trends of infected populations from rural areas to urban centers and/or to non-endemic regions along with changes in the eco-geographical distribution of vector populations have led to the gradual urbanization and globalization of Chagas disease, which is now recognized as an emerging worldwide threat to public health (Eisenstein, 2016). Indeed, the risk of acquiring Chagas disease through infected blood transfusion and organ transplantation is becoming a major problem even in areas of non-endemicity, such as the United States, Australia and Europe (Requena-Mendez et al., 2015; Schmunis and Yadon, 2010). Moreover, the congenital route of infection, which constitutes the main transmission mode of T. cruzi in non-endemic areas is now estimated to be responsible for 22% of new annual infections in endemic countries with active programs for home vector infestations control (Carlier and Truyens, 2015).
In this scenario, a strong and global partnership aimed to coordinate actions to control parasite transmission is urgently needed. In particular, we need to redouble our efforts to control home vector infestation, to screen blood supplies and to identify and subsequently treat T. cruzi-infected people who are still in the early stages of the disease to avoid sequelae, morbidity and economic losses. As a major step towards these goals, we ought to develop novel biomarkers able to overcome the limitations of current diagnostic applications. In this chapter, we critically appraise what has so far been achieved in this area. We also discuss possible ways to proceed in order to address major and still unmet diagnostic demands, and the opportunity provided by recent advances in high-throughput methods (i.e. peptide synthesis technology, genomics, and proteomics) in Chagas disease biomarker discovery.
TRYPANOSOMA CRUZI, AN 'ALL-WHEEL DRIVE' PARASITE
T. cruzi is a promiscuous parasite that traverses a complex life-cycle involving extracellular proliferation and differentiation inside hematophagous insect vectors from different genera, and intracellular proliferation and differentiation in a variety of vertebrate hosts (De Souza, 2002). Host switching, immune pressure as well as constant transition from intracellular to extracellular niches (and vice versa) pose significant adaptation challenges, and are concomitantly accompanied by extensive remodeling of different aspects of T. cruzi such as intracellular transport, primary metabolism, gene expression profiling and overall cellular architecture (De Souza, 2002). This striking plasticity can be also readily recognized in the diverse genetic, phenotypic and epidemiological features displayed by different strains and field isolates comprised within the T. cruzi taxon (Zingales et al., 2012). In this first section, we outline some aspects that underlie the biological flexibility of this 'all-wheel drive' parasite, and that may be relevant in terms of biomarker discovery for Chagas disease diagnostic purposes.
EPIDEMIOLOGICAL FEATURES
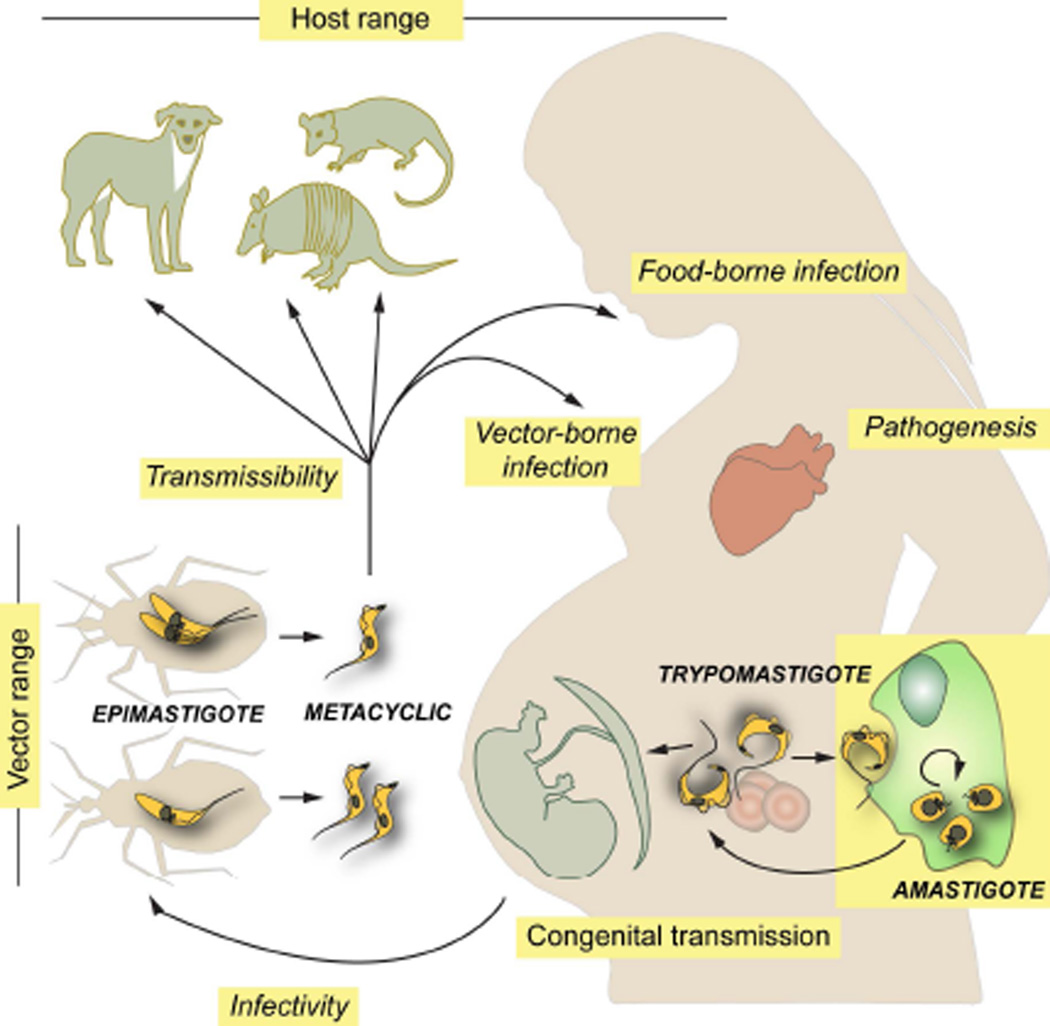
Potential T. cruzi vectors present a broad geographic distribution (from central Argentina and Chile to southern USA) and include more than 140 species of 'kissing bugs' from the subfamily Triatominae (Hemiptera, Reduviidae). Of these, only a few (i.e. Triatoma infestans, T. dimidiata, T. brasiliensis, Rhodnius prolixus and Pastrongylus megistus) have adapted to live in domiciliary setting and to blood-feed on humans and/or domestic animals and thus define the 'domestic/peridomestic' cycle of Chagas disease (Gurtler and Cardinal, 2015; Zingales et al., 2012). The 'sylvatic' cycle of T. cruzi, on the other hand, is actually an array of poorly understood cycles with different eco-epidemiological properties, each one involving multiple sylvatic and/or synanthropic triatomine species, which in turn feed on a variable range of animals. The latter include a variety of rodents, primates, c arnivores, bats, marsupials (i.e. opossums) and xenarthrans (i.e. armadillos, sloths, anteaters) (Figure 1) (Fernandes et al., 1999; Noireau, Diosque and Jansen, 2009; Zingales et al., 2012). In general terms, and although not yet fully established, all mammals are considered susceptible whereas birds and reptiles are considered refractory to T. cruzi. From an epidemiological standpoint, these non-human hosts may play key roles as parasite reservoirs and/or as determinant factors affecting T. cruzi transmission dynamics in endemic areas (Gurtler and Cardinal, 2015; Noireau, Diosque and Jansen, 2009). Importantly, they may also work as complex selective systems leading to the emergence of novel parasite traits (Noireau, Diosque and Jansen, 2009).
FIGURE 1.
Schematic diagram showing the T. cruzi life cycle and different biological features that contribute to ensure its transmission and the establishment of multiple interactions with insect vectors and infected humans. Those features for which there is direct or indirect experimental evidence suggesting inter-strain variability are denoted in italics.
Interestingly, distinct though partially overlapping sets of strains circulate in the ‘domestic/peridomestic’ and the ‘sylvatic’ cycles of T. cruzi (Gurtler and Cardinal, 2015; Noireau, Diosque and Jansen, 2009; Zingales et al., 2012). In the last decades, environmental alterations and demographic issues converged in favoring the intermingling of the two cycles. This translates into an steady increase of emergent transmission patterns involving 'exotic' T. cruzi genotypes, with the possible occurrence of atypical disease physiopathologies (Coura et al., 2002; Zingales et al., 2012).
GENETIC AND PHENOTYPIC VARIABILITY
As revealed for several pathogenic protozoa and fungi, T. cruzi displays a basically clonal reproduction mode, with occasional events of genetic exchange leading to the emergence of hybrid genotypes (Messenger and Miles, 2015). These features led to a complex population structure, made up of multiple 'clonal' strains showing remarkable genetic diversity (Tibayrenc and Ayala, 2015). Inter-strain variations may be grasped at the nucleotide level (Ackermann et al., 2012) but also structurally, in terms of dosage/diversification of antigenic gene families (Campo et al., 2004; Cerqueira et al., 2008; Llewellyn et al., 2015; Urban et al., 2011), DNA content and overall genome architecture (Lewis et al., 2009a; Minning et al., 2011; Reis-Cunha et al., 2015; Souza et al., 2011). Importantly, biochemical and genetic typing schemes developed throughout the last decades converged in the delineation of six major T. cruzi evolutionary lineages or discrete typing units (DTUs) termed TcI to TcVI, with multiple strains and even cryptic sub-lineages within each DTU (Tibayrenc and Ayala, 2015; Zingales et al., 2012). A potential seventh lineage, termed TcBat, has been recently identified in South and Central American bats (Marcili et al., 2009; Pinto et al., 2012). So far, and although all six (or seven, including TcBat) T. cruzi DTUs are capable of infecting humans, certain DTUs such as TcI, TcII, TcV and TcVI are most frequently isolated from clinical samples (Ramirez et al., 2014; Zingales et al., 2012). The reasons for this skewed distribution are unclear, although current evidence suggest that parasite strains detected in patients reflect the principal DTUs circulating among 'domestic/peridomestic' cycles in that geographical area (Messenger, Miles and Bern, 2015).
T. cruzi genotypic heterogeneity could also be grasped at the phenotypic level when different biological parameters are studied. These include, for instance, the rate of epimastigote proliferation in the vector midgut (Castro et al., 2012; de Lana et al., 1998; Vieira et al., 2016); and the extent of epimastigote differentiation into metacyclic trypomastigotes, the developmental form that bring the infection into vertebrates (Figure 1) (da Silveira Pinto et al., 2000; de Lana et al., 1998). The molecular basis for these differences are not yet understood but they might be related to the dissimilar resistance capacity of parasite strains to antimicrobial peptides or hemolytic factors and/or to their differential interaction with receptor(s) inside the crop of triatomines (Castro et al., 2012; Gonzalez et al., 2013; Vieira et al., 2016). Importantly, these biological traits define both T. cruzi infectivity toward insect vectors and its potential transmissibility to vertebrate hosts (Figure 1). These, together with differential eco-geographical distribution and certain preference of triatomids for their blood-source are in turn major determinants of Chagas disease epidemiology (Gurtler and Cardinal, 2015; Noireau, Diosque and Jansen, 2009; Zingales et al., 2012).
Parasite genotypic heterogeneity also seems to modulate key aspects of its interaction with vertebrate hosts, including humans. For instance, the capacity of metacyclic trypomastigotes to resist the harsh conditions of the gastric milieu and to invade gastric epithelium following oral infection is largely dependent on the strain-specific glycoprotein composition of their surface coat (Camandaroba, Pinheiro Lima and Andrade, 2002; Hoft et al., 1996; Maeda et al., 2016). In the same line, inter-strain genetic variations underpin a variety of biological traits involved in parasite infectivity and long-term persistence such as antigenic profile, subversion of the immune system, host cell invasion capacity, intracellular growth rate and survival of amastigotes, and sensitivity to anti-Chagasic drugs (Figure 1) (Magalhaes et al., 2015; Martin et al., 2006; Moraes et al., 2014; Mortara et al., 2008; Nagajyothi et al., 2012; Ruiz et al., 1998; Toledo et al., 2003). In contrast, no clear association between a particular T. cruzi genotype and an eventual tropism for congenital transmission could be yet established. Even though distinct strains may display subtle differences in their ability to invade trophoblasts or chorionic villi explants in vitro (Castillo et al., 2013), genetic profiling experiments have conclusively shown that i) the same set of strains circulate in the bloodstream of transmitting and non-transmitting mothers, and ii) nearly identical T. cruzi genetic signatures are recovered from infected infants born to Chagasic mothers coursing concurrent, multi-strain infections (Figure 1) (Burgos et al., 2007; del Puerto et al., 2010; Virreira et al., 2006a). Overall, the actual consensus is that maternal parasite load and human polymorphisms constitute the main risk factors for T. cruzi congenital transmission (Bua et al., 2012; Fabbro et al., 2014; Juiz et al., 2016; Kaplinski et al., 2015; Rendell et al., 2015).
Interestingly, certain (though not all) epidemiological studies have shown a partial correlation between the prevalence of particular clinical manifestations of Chagas disease and the genotype of the infecting strain (Andrade, Brodskyn and Andrade, 1983; D'Avila et al., 2009; Luquetti et al., 1986; Macedo and Pena, 1998; Virreira et al., 2006b; Zafra et al., 2011; Zingales et al., 2012). This may be attributed in part to the genetic aspects and immune competence of local human populations (Ayo et al., 2013; Deng et al., 2013; Frade et al., 2013; Luz et al., 2016; Nogueira et al., 2012) and/or to parasite genetic heterogeneities. The latter hypothesis finds support in animal studies (that do not strictly recapitulate Chagas disease-associated physiopathologies), which revealed inter-strain variations in complex phenotypes such as parasitemia, virulence, tissue tropism/distribution and pathogenicity (Figure 1) (Andrade et al., 1999; Andrade, 1990; Camandaroba, Pinheiro Lima and Andrade, 2002; de Souza et al., 1996; Laurent et al., 1997; Monteiro et al., 2013; Revollo et al., 1998; Roellig et al., 2010). However, generalized conclusions are difficult to derive, particularly because these epidemiological studies might have been skewed by a number of intrinsic shortcomings. Briefly, i) they often lacked detailed genetic/clinical information on the studied populations; ii) the infecting genotype has been in some cases inferred based on the prevailing parasite genotypes circulating in the area and not typed directly from patients; iii) patients might have been co-infected with other co-endemic pathogens that impact on the clinical presentation of Chagas disease (Salvador et al., 2016); iv) patients might have been infected with multiple parasite strains, which is usually the case in endemic areas (Perez, Lymbery and Thompson, 2014); and v) these studies might have been biased due to parasite typing pitfalls (i.e. samples were collected only from peripheral blood, which may not be representative of the situation within affected organs (Manoel-Caetano Fda et al., 2008; Vago et al., 2000)) and/or associations between local parasites and disease, making it difficult to determine whether the absence of a specific strain/DTU in patients with a given disease phenotype is due to parasite factors or to lack of patient exposure to this DTU.
Overall, and although this issue may have major implications for Chagas disease diagnosis and treatment, the existence of particular associations between T. cruzi genotype and susceptibility to different clinical presentations on Chagasic patients remains to be addressed (Messenger, Miles and Bern, 2015).
DIAGNOSTIC APPLICATIONS FOR CHAGAS D ISEASE: PRESENT KNOWLEDGE
PARASITOLOGICAL AND CLINICAL METHODS
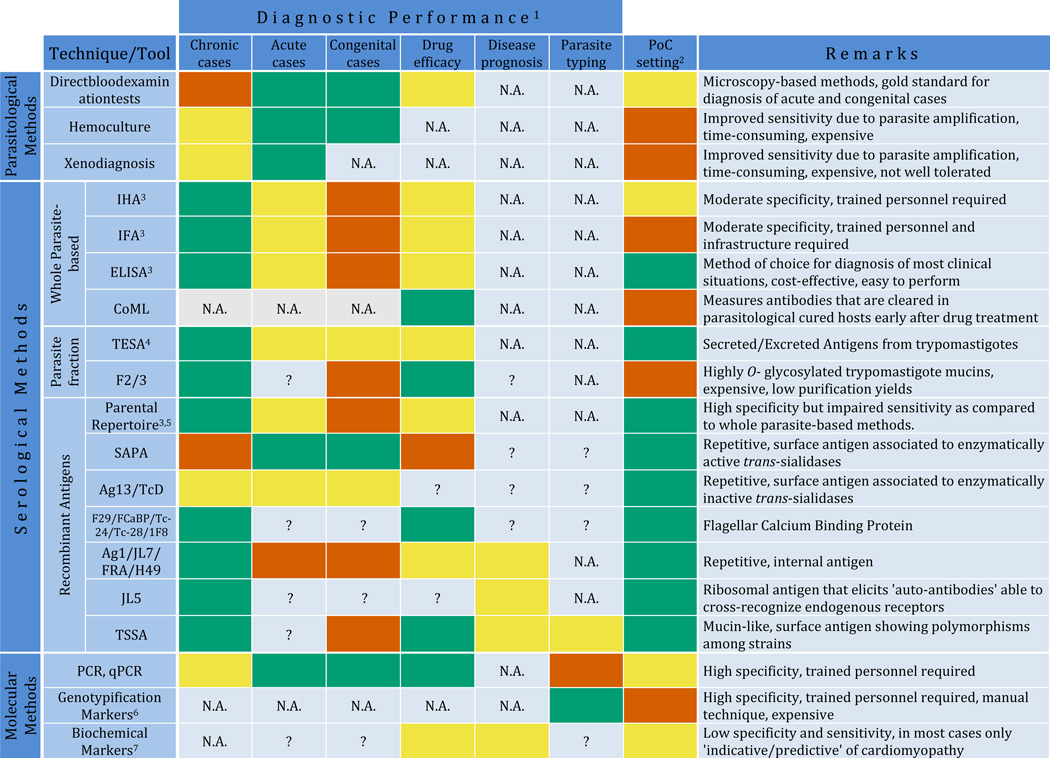
Upon T. cruzi infection, patients undergo the acute phase of Chagas disease, which extends for 40–60 days. Symptoms, if indeed occur, are usually very mild and atypical, thus often misleading its clinical recognition (Rassi, Rassi and Marin-Neto, 2010). In rare cases of vector-borne transmission, a skin nodule (called 'chagoma') or painless prolonged eyelid edema (called the 'Romanha's sign') may indicate the site of parasite inoculation. Due to the patent parasitemia verified at this initial phase, conventional microscopy (i.e. visualization of circulating trypomastigotes in peripheral blood films or buffy coat smears) remains the gold standard for diagnosis, both in acute cases and in newborns that were infected congenitally (Freilij and Altcheh, 1995; Gomes, Lorena and Luquetti, 2009). Either direct tests or concentration tests (i.e. microhematocrit or Strout test) are routinely used for this purpose. These techniques, however present certain limitations in terms of sensitivity (~80–90%), and commonly require highly trained personnel (Table 1) (Freilij and Altcheh, 1995; Gomes, Lorena and Luquetti, 2009).
Table 1.
Overview of performance and features of available diagnostic applications for Chagas disease.
Performance is arbitrarily indicated as appropriate (green boxes), non-appropriate (vermillion boxes) or intermediate/needs to be improved (yellow boxes) according to available data. N.A. and question marks (?) stand for non-applicable or not enough experimental data available to assess the performance, respectively.
Adaptability to be deployed in PoC settings, which depends on specific features of the tool/technique and which is arbitrarily indicated as above.
Commercially available from different vendors.
TESA assays include diverse techniques that measure anti-TESA antibodies in serum samples (i.e. TESA-ELISA, TESA-dot blot, TESA-blot) and techniques that capture TESA antigens directly in urine samples (i.e. Chunap assays)
The term 'parental' repertoire refers to T. cruzi antigens identified in large-scale initiatives carried out in the 80's, and includes: Ag1/JL7/FRA/H49; Ag2/B13/TCR39/PEP-2; Ag10; Ag13/TcD; Ag19; Ag26; Ag30/CRA/JL8/TCR27; Ag36/JL9/MAP; Ag54; JL1; JL5; JL9; SAPA; B12; TcE; A13; F29/FCaBP/Tc-24/Tc-28/1F8; Tc-40; HSP70; HSP78 and FL-160, according to current nomenclature. These antigens were assayed for conventional diagnosis in different combinations using a variety of technological platforms (ELISA, LFIA, Western blot, dot blot), some of which are commercially available from different vendors. The individual performance of some of these antigens showing particular features is indicated below.
Include RFLP, PCR-RFLP, RAPD, Southern blot and other DNA hybridization methods, karyotyping methods and sequence-based methods either using a single locus or MLST.
Include TNF-α, ACE2, BNP, ANP, ET-1 and other biochemical markers indicated in text.
All of abbreviations are defined in the text. Briefly: PoC, Point-of-Care; HIA, Hemagglutination Inhibition Assay; IFA, Immunofluorescence Assay; ELISA, Enzyme Linked Immunosorbent Assay; F2/3: Purified highly O-glycosylated and antigenic trypomastigote mucins; CoML, Complement-Mediated Lysis; TESA, Trypomastigote Excreted-Secreted Antigens; LFIA, Lateral-Flow Immunochromatographic Assays; SAPA, Shed Acute-Phase Antigen; TSSA, Trypomastigote Small Surface Antigen; PCR, Polymerase Chain Reaction; PCR-RFLP, PCR-based Restriction Fragment Length Polymorphism assay; MLST, Multi-Locus Sequence Typing assay; RAPD, Random Amplification of Polymorphic DNA; TNF-α, Tumor Necrosis Factor α; ACE2, Angiotensin-Converting Enzyme 2; BNP, Brain Natriuretic Peptide; ANP, Atrial Natriuretic Peptide; ET-1, Endothelin-1.
Following the initial, acute phase, if untreated, patients enter the indeterminate form of the chronic phase that may last for several years or persist indefinitely (Rassi, Rassi and Marin-Neto, 2010). This phase is characterized by the absence of relevant clinical symptoms and very low and intermittent or null parasitemia. During this phase, parasite replication is maintained in check by the elicitation of a strong and parasite-specific B- and T-cell-mediated immunity (Tarleton, 2015), being the latter the most important in terms of controlling the infection. However, elaborate pathogen immune evasion systems (Albareda et al., 2009; Giraldo et al., 2013; Padilla, Simpson and Tarleton, 2009; Paiva et al., 2012; Vasconcelos et al., 2012) and their ability to quickly invade host cells (Mortara et al., 2008; Nagajyothi et al., 2012) turn this immune response only partially effective, and most patients maintain a sub-patent infection for life. T. cruzi reactivation in immunocompromised Chagasic patients provides solid support to this hypothesis (Tarleton, 2015). Direct T. cruzi detection during the chronic phase requires biological amplification methods, such as hemoculture and xenodiagnosis (Brener, 1962), which are also difficult, expensive, time-consuming and require special laboratory bio-security conditions. In the case of xenodiagnosis, in addition, it is not applicable to certain patient populations. Most importantly, these methods yield positive results in only a proportion of serologically positive patients, thus limiting their usefulness in diagnosis and/or in monitoring drug efficacy (Table 1).
Up to 20 years after the infection, ~35% of patients develop pathological signs characteristic of Chagas disease such as cardiomyopathy, peripheral nervous system damage or dysfunction of the digestive tract often leading to megaesophagus and/or megacolon (Rassi, Rassi and Marin-Neto, 2010). These pathological changes can be revealed by electrocardiogram clinical diagnosis, X-rays and ultrasound. The nature and relative contribution of the multiple factors involved in this quite broad pathogenic range has been the subject of intense debate (Bonney and Engman, 2015; Machado et al., 2012; Teixeira et al., 2011). The current consensus states that a failure to down-regulate the inflammatory response, which is maintained by parasite persistence in tissues, appears to play a predominant role. Other contributory factors include sex, age and genotypic features of the patient, route of infection, sustained vector exposure, auto-immune responses and the existence of co-infections (Ayo et al., 2013; Bonney and Engman, 2015; Deng et al., 2013; Frade et al., 2013; Luz et al., 2016; Machado et al., 2012; Nogueira et al., 2012; Tarleton, 2015; Teixeira et al., 2011). As mentioned, a possible role of parasite genotype on Chagas disease progression/outcome has been proposed, though further research supported by novel and robust epidemiologic and diagnostic tools, and appropriate animal models are needed to address this issue.
SEROLOGICAL METHODS
Considering that patients seroconvert early upon infection, detection of anti-T. cruzi antibodies remains the most effective method for demonstrating direct exposure to the parasite (Gomes, Lorena and Luquetti, 2009). At present, the most widely used serologic methods are Indirect Hemagglutination Assay (IHA), Indirect Immunofluorescence Assay (IFA), and Enzyme-Linked ImmunoSorbent Assay (ELISA) (Table 1). First generation ELISA tests were originally developed using total parasite homogenates or, later on, using biochemically purified, parasite antigenic fractions. Among the latter, TESA (Trypomastigote Excreted/Secreted Antigens) and F2/3 (highly O-glycosylated trypomastigote mucins obtained by sequential solvent partitions) demonstrated the highest diagnostic potential (Table 1) (Almeida et al., 1997; Umezawa, Shikanai-Yasuda and Stolf, 1996). In addition to ELISA, multiple techniques including dot blot and Western blot were developed for the evaluation of these reagents.
In the late 1980's, the advent of recombinant DNA technology allowed the generation of parasite DNA or cDNA expression libraries, which were then coupled to high-throughput screenings using serum samples from Chagasic patients or experimentally infected animals. These approaches provided the first glimpses into the genomic make up of this parasite and, most importantly, led to a 'golden era' of T. cruzi antigen discovery. Indeed, by using these methods several of the immunodominant T. cruzi antigens that are still in use were identified and characterized (Burns et al., 1992; Cotrim et al., 1990; Hoft et al., 1989; Houghton et al., 1999; Ibanez, Affranchino and Frasch, 1987; Ibanez et al., 1988; Lafaille et al., 1989; Levin et al., 1989). For a complete list of these antigens (henceforth the 'parental repertoire'), see Table 1. The fact that many of them emerged from independent screenings carried out in different laboratories further supported their diagnostic potential but, unfortunately also led to a confusing nomenclature that still persist (Table 1) (reviewed in (da Silveira, Umezawa and Luquetti, 2001; Frasch et al., 1991)). Some of the antigens included in the T. cruzi 'parental repertoire' were extensively validated, in certain cases by way of multicenter trials (Caballero et al., 2007; Reithinger et al., 2010), and set the stage for the development of second-generation Chagas disease diagnostic methods. A variety of antigen expression procedures (i.e. using full-length or partially deleted recombinant, fusion proteins or synthetic peptides functionalized in different ways) and antigen display strategies (i.e. using mono- or multi-epitopic molecules) were evaluated before them being incorporated either individually or in defined mixtures into in-house and/or commercial kits (Godsel et al., 1995; Hernandez et al., 2010; Houghton et al., 2000; Krieger et al., 1992; Longhi et al., 2012; Oelemann et al., 1999; Pastini et al., 1994). Some of these tests have shown very good performances, and were thus extensively used in basic research and/or as confirmatory tests in clinical practice up to this day.
The major advantage of recombinant antigens-based tests is that they minimize the extent of specificity problems. As previously shown, sera from individuals with other co-endemic infections such as leishmaniasis and/or afflicted of certain autoimmune disorders cross-react with crude preparations of T. cruzi antigens (Araujo, 1986; Gomes, Lorena and Luquetti, 2009; Schnaidman et al., 1986). On the down side, they present lower sensitivity than whole parasite-based techniques. In addition, it should be said that the proven success of this 'parental repertoire', along with a shift in R&D priorities converged in curbing the enthusiasm for subsequent large-scale antigen-discovery efforts for Chagas disease. Indeed, most of the additional antigens/biomarkers with moderate-to-excellent diagnostic performance that emerged in the last decades were identified either as by-products of basic hypothesis-driven research (Buchovsky et al., 2001; Di Noia et al., 2002; Martinez et al., 1991; Saborio et al., 1990) or using low-to-medium throughput antigen expression/synthesis approaches (see below). Most importantly, just a few of these novel antigens have been incorporated into the already existing diagnostic application platforms.
New generation tests displaying potentially improved accuracy such as the Chemoluminescent Microparticle ImmunoAssay (CMIA) and its improved version, Architect Chagas (both from Abbott Laboratories, Wiesbaden, Germany) (Abras et al., 2016; Praast et al., 2011), or the Multi-cruzi test (InfYnity Biomarkers, Lyon, France) (Granjon et al., 2016) have been recently developed. Confirming the above mentioned trend, both use a large panel of T. cruzi antigens belonging to the 'parental repertoire' -those discovered in the 80's-, which in the case of the Multi-cruzi test is supplemented with TSSA (Trypomastigote Small Surface Antigen (Di Noia et al., 2002)). Despite the virtual lack of antigen innovation, these tests incorporate a large degree of automation and highly sensitive detection systems (Abras et al., 2016) or major technical improvements, such as a multiplexed printing method inside ELISA microplates (Granjon et al., 2016).
Overall, currently available serodiagnostic tests are simple, affordable and display quite good results in terms of large-scale population diagnosis. However, they all present certain minor concerns with regards to their reproducibility, reliability, specificity (those using whole parasites) and sensitivity (those using parasite fractions and/or recombinant antigens) which, in turn affect their overall performance. In addition, and likely due to their biased antigenic composition, they also show suboptimal performance with particular Chagasic populations (i.e. acute and/or congenital infections) (Gomes, Lorena and Luquetti, 2009), and often discordant results between assays depending on the geographic origin of the patients (Caballero et al., 2007; Guzman-Gomez et al., 2015). The latter may be attributed to variations in the local human immune responses and/or, as stated above, to differences in the antigenic constitution of T. cruzi DTUs that prevail in different endemic areas. Importantly, even after successive improvements, none of the available tests emerged as the ‘gold standard’, i.e. able to show ~100% specificity and sensitivity. In this context, current guidelines developed by the World Health Organization advice the use of at least two serological tests based on different principles for reaching a 'conclusive' diagnosis. In the case of ambiguous or discordant results, a third technique should be conducted. These guidelines thereby increase the cost of diagnosis and risk of patient ‘loss’, particularly in endemic areas. Most importantly, these diagnostic limitations delay the initiation of chemotherapy, thus limiting its efficacy.
MOLECULAR METHODS
In the late 1980's, DNA-based molecular methods emerged as an appealing alternative for the diagnosis of Chagas disease, mainly to overcome the low sensitivity of direct parasitological approaches (Moser, Kirchhoff and Donelson, 1989; Sturm et al., 1989). The introduction of the recently developed polymerase chain reaction (PCR) assay, in particular, held promises of high sensitivity, specificity, and high-throughput potential. The first targets for PCR amplification were focused on the major molecular component of the mitochondrial DNA (also known as kinetoplast DNA or kDNA). In T. cruzi and other kinetoplastid parasites, this component is present in the form of circular DNA of short length (mini-circles), which add up to several thousand copies per cell (Simpson, 1986). Later on, a large repertoire of kDNA or nuclear DNA targets including single- or multi-copy genes and satellite sequences, as well as different multi-target strategies have been explored (Schijman et al., 2011). More recently, a parasite concentration approach based on short and stable RNA aptamers was developed to facilitate PCR-based detection methods, and proposed as a potential alternative tool in monitoring parasite load in Chagasic patients (Nagarkatti et al., 2012; Nagarkatti et al., 2014).
Major challenges for the clinical implementation of PCR-based techniques derive from a number of technical factors such as source (i.e. cord blood, umbilical tissue, placental, tissue), volume, conservation and transportation of the samples, and underlying molecular biology protocols (i.e. DNA isolation, purification, pre-treatment, thermo-cycling conditions, etc.) (Schijman et al., 2011). In addition, the reproducibility and overall performance of PCR-based methods is significantly affected by the fluctuations in parasitemia that characterize the chronic phase of Chagas disease, and by inter-DTU variations in dosage and/or sequence of the targets of amplification (Lewis et al., 2009b). Despite these issues, several PCR- and, more recently, quantitative real-time PCR (qPCR)-based procedures have been developed. These allowed detection and quantification of parasite DNA from clinical samples with variable levels of reliability, complexity, selectivity and analytical sensitivity (Duffy et al., 2009; Duffy et al., 2013; Piron et al., 2007; Qvarnstrom et al., 2012; Valadares et al., 2008). In a recent work, a multicentre trial optimized and evaluated in parallel two last generation qPCR methods targeting either satellite or kDNA targets (Ramirez et al., 2015). These methods were tested using a large and heterogeneous panel of blood samples from acute and chronic patients either asymptomatic or showing different clinical manifestations, and infected through different transmission modes (Ramirez et al., 2015). Though highly specific and reproducible, both methods showed clinical sensitivities of ~80%, which is still not good enough for their application as confirmatory testing of blood donors or in clinical settings. In addition, it is worth noting that DNA-based molecular methods are expensive and difficult to be implemented in endemic areas, e.g. in PoC sites with limited infrastructure (Table 1).
DIAGNOSTIC APPLICATIONS FOR CHAGAS D ISEASE: PENDING ISSUES
As described above, current diagnostic methods and particularly those based on serology are highly accurate in detecting most of T. cruzi infections in humans. However, there are some clinical and/or epidemiological situations, discussed in this section, in which their performance is significantly hampered by methodological and/or biological issues.
EARLY DIAGNOSIS OF CONGENITAL TRANSMISSION
According to epidemiological data, maternal-fetal transmission occurs in an average of 5% of infected mothers in endemic areas, thus leading to ~15,000 congenital cases per year (Carlier and Truyens, 2015). Diagnosis in newborns is commonly based on the microscopic observation of trypomastigotes in peripheral and/or umbilical cord blood, which is more effective by the microhematocrit concentration technique (Freilij and Altcheh, 1995). Considering the usually high parasitemia at the initial stage, PCR-based analyses provide a valuable supporting tool to detect infection and to evaluate treatment outcome in these patients (Table 1) (Duffy et al., 2009; Russomando et al., 1998; Schijman et al., 2003). Standard serodiagnostic methods, though routinely used have low positive predictive value until 8–9 months after birth due to passive transfer of maternal IgG antibodies (Freilij and Altcheh, 1995). In this context, identification of novel antigens recognized by fetal IgM or IgA appeared as an appealing approach. However, and despite initial promising results (Antas et al., 2000; Betonico et al., 1999; Corral, Altcheh and Freilij, 1998; Lorca et al., 1995), these efforts have been discontinued due to the high number of false-negatives (Blanco et al., 2000; Freilij and Altcheh, 1995; Gomes, Lorena and Luquetti, 2009). It should be noted though, that a serious, high-content and unbiased screening for congenital IgM specificities have not been yet undertaken.
As an alternative approach to discover surrogate markers of congenital infection, the group of Dr. Frasch in Argentina undertook the identification of T. cruzi antigens exclusively and/or preferentially recognized by fetal IgGs. This approach relied on the parallel evaluation of paired mother/newborn serum samples against a multiplexed antigen panel followed by the comparison between signatures of recognition. Using this strategy, they identified SAPA (Shed Acute-Phase Antigen) and, to a lesser extent, Ag13/TcD as suitable markers to detect congenital transmission (Table 1) (Reyes et al., 1990). Both antigens belonged to the above mentioned T. cruzi 'parental repertoire' and, interestingly, both (particularly SAPA) had been previously found to display a biased, though not exclusive recognition towards acute infection sera (Affranchino et al., 1989). This is consistent with the assumption that congenital T. cruzi infection constitutes in fact an acute infection in newborns (Carlier and Truyens, 2015).
Further research disclosed that SAPA is a repetitive sequence displaying a complex antigenic structure (Alvarez et al., 2001) and involved in improving the pharmacokinetics of trans-sialidase, a major T. cruzi virulence factor (Buscaglia et al., 1999; Dc-Rubin and Schenkman, 2012; Frasch, 2000). Up to this day, SAPA remains the only available serological marker of early infection and has been widely used to diagnose recently-acquired vector-borne infections and congenitally transmitted cases (Mallimaci et al., 2010; Russomando et al., 2010; Volta et al., 2015).
More sophisticated, laborious and difficult to interpret methods were also developed. These include testing of the newborn sample by immunoblot against the TESA fraction (Table 1) (Umezawa et al., 1996) and, more recently, the Chunap (Chagas urine nanoparticle) test (Castro-Sesquen et al., 2014). In the latter method, instead of T. cruzi-specific antibodies, active infection is indirectly assessed by the capture and concentration of T. cruzi TESA antigens from patient's urine, which are then revealed using a monoclonal antibody directed to a parasite lipophosphopeptideglycan, included in the test (Castro-Sesquen et al., 2014). When evaluated on children living in endemic areas and tested at the peak of parasitemia (~1-month old) Chunap was able to accurately diagnose congenital infections (Castro-Sesquen et al., 2014), and is now being evaluated for other diagnostic applications (Castro-Sesquen et al., 2016).
On a final note, it should be stressed that given the high efficacy of trypanocidal drugs in infected newborns, their rapid diagnosis and subsequent treatment is essential. Intensive screening for distinctive immune responses (i.e. those based on IgM and/or IgA antibodies) and/or for specific molecular signatures that may translate into surrogate biomarkers for early detection of congenital T. cruzi transmission should be thus considered a top-ranking priority in Chagas disease applied research.
RAPID ASSESSMENT OF THERAPY EFFICACY
Overall, the best chemotherapy results have been achieved in acute or early chronic infections. However, even in children the cure rate is up to 62% at 2 years of follow-up, although this figure may vary according to population and geographical location (Ribeiro et al., 2009). This variability may be attributed in part to differences in the prevalence of human-associated genotypes across endemic areas. Indeed, substantial inter-strain variations in drug resistance have been ascertained both in in vitro systems and in animal infection models (Figure 1) (Moraes et al., 2014; Toledo et al., 2003).
During the chronic stage, the efficacy of drug treatment is lower and variable; and is also difficult to assess because most studies used different treatment regimens and outcome evaluation methods (variable assays, frequency and duration of follow-up) (for a recent review, see (Pinazo et al., 2014)). Even in successful treatments, the gold standard for evaluating drug efficacy, which relies on consistent negative results using conventional parasitological and whole parasite-based serological tests, may take years to decades to assess, thus stressing the necessity of novel and improved therapeutic response markers.
Several therapeutic studies support the usefulness of PCR-based strategies to evaluate treatment outcome in congenital or acute cases of Chagas disease. Moreover, PCR-based methods may also assist, though with sub-optimal performance in cases of lower parasitemias, in evaluating chemotherapy in chronic cases of Chagas disease (Britto et al., 1995; Galvao et al., 2003; Munoz et al., 2013; Russomando et al., 1998; Schijman et al., 2003). In particular, PCR-based methods seem to be a rapid indicator of the parasite’s susceptibility to drugs, allowing early therapy modification in cases of resistance or reactivation of Chagasic infection (Lages-Silva et al., 2002; Pinazo et al., 2014; Schijman et al., 2000).
In 1982, Drs. Krettli and Brener in Brazil demonstrated the existence of a special type of antibodies, which they termed 'antibodies against living blood forms', that were absent in parasitological cured hosts early after drug treatment (Krettli and Brener, 1982). These antibodies were shown to participate in resistance against T. cruzi and were solely detectable by complement-mediated lysis (CoML) assays (Krettli and Brener, 1982). Upon these findings, the authors proposed the inclusion of the CoML technique in the context of cure criteria. Indeed, the CoML technique offered an important contribution for post-therapeutic monitoring of Chagas disease in clinical trials, mostly by significantly reducing the required follow-up periods (Table 1) (Galvao et al., 1993). Numerous improvements in terms of the sensitivity and operational safety of the CoML method have been later on introduced (Alessio et al., 2014; Martins-Filho et al., 2002; Vitelli-Avelar et al., 2007). Most importantly, this concept of 'non-conventional' serological responses set the basis for multiple screenings looking for parasite molecules that could be the target of antibody responses showing different qualitative, quantitative and/or kinetic properties. Several potential biomarkers have been thereby identified and evaluated for their post-therapeutic application (Andrade et al., 2004; Fernandez-Villegas et al., 2011; Gazzinelli et al., 1993; Laucella et al., 2009; Machado-de-Assis et al., 2012; Sanchez Negrette et al., 2008; Silva et al., 2002). They included different parasite fractions (i.e. TESA, F2/3), individual recombinant antigens belonging to the 'parental repertoire' such as F29/FCaBP/Tc-24/Tc-28/1F8 (Sosa Estani et al., 1998), and other molecules such as TSSA (our unpublished results) (Table 1). Overall, the best results were obtained with the F2/3 fraction (Table 1) (Andrade et al., 2004; de Andrade et al., 1996). Interestingly, antibodies directed to α-galactosyl-containing epitopes (i.e. the main antigenic determinant in the F2/3 fraction), were later on shown to induce trypomastigote lysis independently of complement (Pereira-Chioccola et al., 2000).
More recently, the group of Dr. Tarleton explored the use of a multi-pronged approach based on the simultaneous evaluation of T. cruzi-specific B- and T-cell responses as an alternative way of measuring treatment efficacy (Alvarez et al., 2016; Laucella et al., 2009). By doing so, both a decline in the frequency of IFNγ-producing T cells and in antibody titers measured by a previously developed recombinant multiplex serological assay (Cooley et al., 2008) were observed shortly after benznidazole treatment and were thus proposed as surrogate markers for refining the post-therapeutic cure criterion (Albareda and Laucella, 2015; Alvarez et al., 2016; Laucella et al., 2009). Identification of novel biomarkers for early evaluation of anti-trypanocidal drug efficacy will fasten and improve the assessment of current chemotherapy treatments in clinical trials and, importantly, will be instrumental for the development of novel and improved treatments.
INDICATION/PREDICTION OF CHAGAS DISEASE PROGRESSION
As mentioned, Chagas disease evolves into a wide range of pathological symptoms, ranging from subclinical to potentially fatal megasyndromes (Rassi, Rassi and Marin-Neto, 2010). Identification of diagnostic and/or predictive biomarkers of disease progression would therefore represent a major achievement towards improving the clinical management of Chagasic patients. Some authors proposed serological alternatives to tackle this issue, such as quantifying the antibody titers to Ag1/JL7/FRA/H49 to distinguish between Chagas disease-associated cardiac pathology and other non-Chagasic cardiological dysfunctions (Table 1) (Abraham and Derk, 2015; Bhattacharyya et al., 2014; Kaplan et al., 1997; Levin and Hoebeke, 2008; Thomas et al., 2012). On the other hand, antibodies to another 'parental repertoire' antigen, termed JL5, were shown to cross-recognize endogenous β1-adrenergic and M2 muscarinic receptors. Such 'auto-antibodies' were shown to be associated with arrhythmogenic anomalies, which may contribute to cardiac alterations in Chagasic patients (Kaplan et al., 1997), and were thus proposed to be included in the context of disease prognosis (Table 1). More recently, a certain correlation between serological responses to TSSA and electrocardiogram abnormalities was also noted (Table 1) (Bhattacharyya et al., 2014).
Alternatively, several groups attempted to identify biochemical markers of cardiac damage and/or inflammation such as TNF-α (Talvani et al., 2004a), angiotensin-converting enzyme 2 (ACE2) (Wang et al., 2010), Brain and Atrial Natriuretic Peptides (BNP and ANP, respectively) (Garcia-Alvarez et al., 2010; Heringer-Walther et al., 2005; Ribeiro et al., 2002) as candidates for disease prognosis (Table 1). In particular, the concentration of BNP and ANP in serum were systematically studied as markers of heart damage in Chagasic patients (Garcia-Alvarez et al., 2010; Heringer-Walther et al., 2005; Ribeiro et al., 2002) since they had been previously related with cardiovascular diseases (Wang et al., 2006; Wondergem et al., 2001). Increased concentrations of BNP and ANP strongly correlated with the severity of Chagas-associated cardiac damage, being BNP more sensitive than ANP (Fernandes et al., 2007; Heringer-Walther et al., 2005; Talvani et al., 2004b). Upon these findings, the authors proposed that BNP could be measured periodically in asymptomatic patients as screening test to detect incipient ventricular dysfunction (Heringer-Walther et al., 2005). In the same line, higher levels of plasma ACE2, that catalyzes the conversion from angiotensin II to angiotensin 1–7 (Keidar, Kaplan and Gamliel-Lazarovich, 2007), were shown to correlate with clinical severity and worsening echocardiographic parameters in Chagasis patients (Wang et al., 2010). More importantly, given that the combined determination of BNP concentration and ACE2 activity had better positive predicted value than when separately analyzed, authors encouraged the use both markers to predict fatal outcomes (Wang et al., 2010).
In the same line, the role of endothelin-1 (ET-1) as a prognostic marker for T. cruzi-induced pathogenesis has also been extensively studied, though in animal models. ET-1 is a vasoactive peptide synthesized by many cell types including cardiac myocytes and cardiac fibroblasts associated with vasospasm, vascular damage, cardiovascular remodeling and inflammation (Kedzierski and Yanagisawa, 2001). Mice infected with T. cruzi exhibit increased levels of ET-1 and endothelin converting enzyme (ECE), the enzyme responsible for the conversion of the precursor to ET-1, in plasma, in the vasculature and in T. cruzi-infected myocardial cells (Huang et al., 2000; Petkova et al., 2000). Interestingly, phosphoramidon, an inhibitor of ECE, ameliorated the pathology and reduced the extent of cardiac remodeling in these animals (Jelicks et al., 2002). Moreover, ET-1 KO mice showed certain protection against chronic Chagasic cardiomyopathy (Tanowitz et al., 2005).
Overall, and despite multiple and disparate attempts, none of the serological and/or biochemical markers that have been explored so far translated into a reliable, easy to assay and interpret marker to asses cardiac damage and/or disease prognosis (recently reviewed in (Pinazo et al., 2015)). The complex and likely multifactorial nature of Chagas disease pathogenesis together with intrinsic difficulties in establishing appropriate experimental/epidemiological models converge in turning this area as the most challenging in terms of T. cruzi biomarker discovery.
TYPING OF INFECTING PARASITE STRAIN(S)
Pioneer studies aimed at fingerprinting the infecting parasite strain(s) directly in clinical samples were based on biochemical markers (i.e. MLEE, Multi-Locus Enzyme Electrophoresis) (Tibayrenc and Ayala, 2015). Recent advances in typing schemes based on RFLP (Restriction Fragment Length Polymorphism), PCR-RFLP, RAPD (Random Amplification of Polymorphic DNA), DNA hybridization, karyotyping and, particularly, sequence-based markers either using a single locus or multiple loci have greatly improved parasite genotypic resolution in vitro (Table 1) (Tibayrenc and Ayala, 2015). However, in vivo, these genotyping methods are time-consuming and costly, and require parasite isolation and amplification or a high quantity of DNA, therefore necessitating invasive sampling with medical risks. Moreover, the fact that concurrent infections with multiple T. cruzi strains appears to be the norm rather than the anomaly (Perez, Lymbery and Thompson, 2014), and the discovery that the most prevalent T. cruzi genotype present in the bloodstream can differ from the strain(s) found sequestered within organs (Manoel-Caetano Fda et al., 2008; Vago et al., 2000) further complicate this task. In this context, serotyping methods emerge as an appealing alternative to overcome these limitations, as demonstrated in other human infectious diseases (Dunbar et al., 2015; Maksimov et al., 2012). Serotyping is based on the use of specific antigens with qualitative and/or quantitative differences among parasite strains/DTUs to detect strain-specific antibody signatures.
TSSA (Di Noia et al., 2002) is a parasite adhesin displaying significant sequence homology to TcMUC, a huge family of polymorphic genes that code for the polypeptide backbones of the trypomastigote mucin molecules (Buscaglia et al., 2004; Campo et al., 2006), some of which are terminally decorated with α-galactosyl residues and constitute the F2/3 antigenic fraction. Interestingly, detailed genetic characterization of the TSSA locus disclosed minor sequence variations between TSSA variants expressed by different parasite DTUs (Bhattacharyya et al., 2010; Di Noia et al., 2002). Some of these variations were shown to have major impact on TSSA biological function and antigenicity, thereby leading to differential antibody profiles between variants (Balouz et al., 2015; Bhattacharyya et al., 2014; Canepa et al., 2012; De Marchi et al., 2011; Di Noia et al., 2002). So far, TSSA remains the only polymorphic antigen that has been successfully used for the development of DTU-specific serology methods for T. cruzi infections of humans (Table 1) (Bhattacharyya et al., 2014; Bisio et al., 2011; Burgos et al., 2010; Longhi et al., 2014; Risso et al., 2011). Despite this, the resolution and specificity of TSSA-based serotyping assays need to be improved. Discrimination between certain DTUs remains challenging as they possess identical or almost identical TSSA alleles or they are poorly immunogenic (Bhattacharyya et al., 2014; Canepa et al., 2012).
Serotyping could be a rapid, sensitive, cost-effective and relatively non-invasive alternative to stringent T. cruzi genotyping in humans, and may also be used in animal reservoirs for epidemiological studies (Bhattacharyya et al., 2015; Cimino et al., 2011). Most importantly, development of novel serotyping tools will facilitate the unraveling of possible relationships between parasite genetic variability and clinical features, a major issue in Chagas disease applied research.
POINT-OF-CARE DIAGNOSIS
People living in Chagas disease endemic areas have restricted access to laboratory facilities and/or to appropriate health centers. This, together with the associated costs and expertise needed for conventional diagnostic methods point to a number of technological and economic barriers that further stress the need to deploy PoC tests for T. cruzi infection diagnosis. PoC devices, in addition, allow the patients to see the results for themselves, which contributes to a better working relationship between local communities and people carrying out the testing (e.g., during field surveys). Moreover, the need for follow-up visits to surveyed individuals and therefore the operational costs and risks of possible attrition bias are reduced. From an epidemiological point of view, a reliable PoC test would allow intervention strategies to be implemented in situ, such as for serologic surveillance, vaccine or clinical trials; as well as for rapid initiation of treatment of infected individuals during outbreaks of acute cases, such as those recently reported (Alarcon de Noya et al., 2010; Segovia et al., 2013).
Several rapid diagnostic and PoC tests to detect infection based on immunochromatography, particle agglutination, immunofiltration, immunodot, lateral-flow immunochromatographic assays (LFIA) or DNA detection are available for a variety of infectious diseases (Nair et al., 2016; Natoli et al., 2014; Teles and Fonseca, 2015). These may be either qualitative or semi-quantitative and are characterized by the delivery of quick results, in most cases without the need for electrical equipment. In the case of Chagas disease, several LFIAs based on antigenic fractions or recombinant antigens and a single PoC test aimed at the capture and concentration of T. cruzi TESA antigens in urine samples (Chunap, see above) were developed (Table 1) (Castro-Sesquen et al., 2014; Houghton et al., 2009; Reithinger et al., 2010). Serological tests, however show substantial variations in their sensitivity in different geographical areas (Verani et al., 2009), and display sub-optimal performances (Sanchez-Camargo et al., 2014). Unfortunately, and despite being widely used in different parasitic diseases (Mondal et al., 2016; Sriworarat et al., 2015), PCR-based methods applicable in PoC settings such as those based on simple colorimetric loop-mediated isothermal amplification or Recombinase Polymerase Amplification (RPA) techniques are not yet available for T. cruzi detection.
As a first step towards the development of alternative PoC devices for diagnosis, a new diagnostic platform based on superparamagnetic microbeads coated with recombinant antigens and fluorescent (FMBIA) or electrochemical (EMBIA) detection was recently developed (Cortina et al., 2016). The EMBIA platform, including antigen-functionalized magnetic microbeads, disposable electrochemical cells-electrode cartridges and a portable potentiostat, was evaluated for serodiagnosis of human and cattle infectious diseases. In the particular case of Chagas disease, a more extensive validation was performed showing that the EMBIA platform displayed an excellent diagnostic performance almost indistinguishable from the well-established ELISA methods (Cortina et al., 2016).
DIAGNOSTIC APPLICATIONS FOR CHAGAS D ISEASE: THE ROAD AHEAD
As discussed throughout this chapter, parasite-specific immune signatures provided a prime source of biomarker candidates for development of Chagas disease diagnostic applications (see Table 1). However, they may now be re-explored using modern and powerful ‘omics’-based fingerprint approaches. Indeed, the availability of complete T. cruzi genomes from several strains together with a variety of recent genome mining exercises were performed to identify potential serodiagnostic reagents and vaccine candidates (Bhatia et al., 2004; Carmona et al., 2012; Cooley et al., 2008; Goto, Carter and Reed, 2008; Reis-Cunha et al., 2014). Importantly, most of the antigens that emerged from these in silico-guided screenings were not included in the ‘parental repertoire' (Table 1), and could thereby entail novel diagnostic capabilities, such as early assessment of drug efficacy (Alvarez et al., 2016; Laucella et al., 2009). Genome-wide approaches could be also used as a starting point to identify novel DNA-based DTU-resolution markers (Cosentino and Aguero, 2012) and/or novel serotyping reagents. As previously shown, genetic variation among T. cruzi DTUs translates into differential proteomes (Telleria et al., 2010) and, therefore also likely in distinct epitope collections ('epitomes') recognized during human infections. By carrying out genome-wide B-cell epitope prediction on proteins derived from allelic pairs of the hybrid T. cruzi CL Brener genome, the group of Dr. Bartholomeu in Brazil was able to identify three novel polymorphic epitopes potentially able to discriminate between parasite DTUs (Mendes et al., 2013). Unfortunately, this study and those mentioned above were somehow limited by the use of low-to-medium throughput antigen expression/synthesis approaches, and/or by the use of non-human sources of serum samples.
Recent advances in computerized photolithography and photochemistry, however have allowed the development of a novel peptide-chip technology where up to 1 million individual peptides are synthesized in situ on a glass slide (Buus et al., 2012). This in situ synthesis makes the production of the chip highly cost-effective and allows, for the first time, to interrogate complete proteomes. Recent work in our laboratories demonstrated both the high technical reproducibility as well as epitope mapping consistency of this platform when compared with earlier technologies (Balouz et al., 2015; Carmona et al., 2015). Most importantly, by screening the complete length of 457 parasite proteins (~7% of the T. cruzi deduced proteome) we were able to identify 2,031 Chagas disease-specific peptides and 97 novel parasite antigens (Carmona et al., 2015). Together with above mentioned studies, and besides emphasizing the huge potential of genomic/proteomic methods as major driving forces in antigen discovery, these findings indicate that a vast majority of the T. cruzi antigenic repertoire remains uncharacterized. We aim now at interrogating the entire T. cruzi deduced proteome, including inter-strain polymorphisms revealed by previously developed genetic diversity maps (Ackermann et al., 2012; Panunzi and Aguero, 2014). The use of samples collected from particular patients populations such as infected with different parasite DTUs or showing distinct Chagas disease-associated pathologies followed by a final integration of the emerging data will put us in position to discriminate between the common linear B-cell 'epitome' of T. cruzi and sets of epitopes showing differential recognition among distinct Chagas disease populations.
Considering the huge functional and diagnostic significance of carbohydrates on T. cruzi biology (de Lederkremer and Agusti, 2009), it could be hypothesized that similar high-throughput approaches carried out on glycan and/or lectin microarrays (Fernandez-Tejada, Canada and Jimenez-Barbero, 2015) will have a major impact on Chagas disease biomarker discovery. These methodologies have been successfully explored in different parasitic infections (Anish et al., 2013; Aranzamendi et al., 2011; Martin et al., 2013).
From a wider perspective, high-throughput strategies may be also pursued to develop novel biomarkers based on the detection of T. cruzi-derived molecules (Castro-Sesquen et al., 2014; de Titto and Araujo, 1988) and/or parasite-induced modifications o n host molecules and/or cells (Li et al., 2016; Mucci et al., 2002; Muia et al., 2010; Risso et al., 2007; Trocoli Torrecilhas et al., 2009). Theoretically, these strategies would have the additional advantage to discriminate between active from past infections and/or to assist in monitoring disease progression, as validated in other human diseases (Karsdal et al., 2010; Leeansyah et al., 2013; Tritten et al., 2014). In this context, it is important to keep in mind i) the extent of variability due to host genetic background on setting these putative biochemical markers and ii) that T. cruzi-derived molecules, and particularly during the chronic phase of Chagas disease, are present in biological fluids at extremely low concentrations, and likely aggregated in membrane-coated vesicles (Bayer-Santos et al., 2013; Fernandez-Calero et al., 2015; Lantos et al., 2016; Trocoli Torrecilhas et al., 2009).
Nevertheless, modern ‘-omic’ technologies are offering alternative strategies for such a difficult system biology exploration (Cantacessi et al., 2015; Hockl et al., 2016; Preidis and Hotez, 2015). In particular, mass spectrometry (MS) methods provide a robust, versatile and sensitive analytical technology allowing high-throughput detection with mass accuracy, precise quantitation and verification of protein variants, splice isoforms, metabolites and disease-specific post-translational modifications (Crutchfield et al., 2016). Indeed, using MS-based approaches, global changes in metabolic profiles in Chagasic patients showing acute myocarditis (Girones et al., 2014) and serological biomarkers showing differences between Chagasic and healthy subjects (Santamaria et al., 2014) were recently identified. In the latter case, biomarker peaks with the best discriminatory power were further characterized by a range of proteomic and immunological techniques, indicating that specific fragments derived from proteolysis of apolipoprotein A-I and one fragment of fibronectin are specifically upregulated in Chagasic patients (Santamaria et al., 2014). Interestingly, these biomarkers returned to normal values more rapidly than whole parasite-based serological tests in patients treated with nifurtimox, thus supporting their potential use for evaluation of therapeutic efficacy (Table 1) (Santamaria et al., 2014).
In addition to the obvious impact in the diagnostic application field, it is expected that the utilization of these and other high-throughput, '-omic' techniques will provide a vast amount of putative biomarkers to be explored in other Chagas disease research areas such as molecular epidemiology and prioritization of targets for vaccine development. Most importantly, together with appropriate animal models and robust bioinformatics, molecular and cellular tools (Aguero et al., 2008; Crowther et al., 2010; Katsuno et al., 2015; Lewis et al., 2015; Magarinos et al., 2012; Moraes et al., 2014), these biomarkers will be instrumental to screen, prioritize and evaluate safety and efficacy of novel drugs/treatments.
CONCLUDING REMARKS
Over 100 years after its discovery, and despite its huge medical, economic and social burden, Chagas disease remains a major threat in several countries of Latin America and an emergent global health problem. Great efforts have been made and are still being made in Latin America and other developed countries to halt T. cruzi transmission. However, one of the key issues concerning Chagas disease control remains that of diagnosis. Without accessible and effective diagnostics tools and methods, infected individuals cannot be timely identified and hence treated. Even if treated, the success of treatment cannot be efficiently assessed.
As discussed here, current diagnostic methods are highly accurate in detecting most of T. cruzi infections in humans. However, there are still some clinical and/or epidemiological situations in which their performance is severely impaired. In addition, the complex epidemiological features of Chagas disease and the remarkable phenotypic diversity displayed by distinct T. cruzi strains, which may be associated to certain aspects of disease progression/outcome, also stress the necessity of developing new methods able to be deployed in PoC settings and capable of typing the infecting parasite(s) directly in clinical samples. The implementation of modern ‘-omic’, high-throughput and aggressive strategies constitutes an appealing alternative to move on towards filling current diagnostic gaps. Emergent diagnostic tests integrating these novel and tailored tools will provide a significant impact on the effectiveness of current intervention schemes and, most importantly, will improve the clinical management of Chagasic patients by providing the intervening physician with an accurate and integrated diagnosis.
Acknowledgments
We express our gratitude to Dr. Carlos Frasch (IIB-INTECh) for critical reading of the manuscript, and to Dr. Javier Di Noia (IRCM, Canada) for the enthusiasm and superb graphic art. We apologize to people whose work was not referenced due to limited space. Research carried out in our labs is supported by grants and contracts from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, Argentina) (to FA and CAB), the National Institutes of Health (NIAID/NIH, USA) (to FA), and Fundación Bunge y Born (Argentina) (to CAB). VB holds a PhD fellowship and FA and CAB are career investigators from the National Research Council of Argentina (CONICET).
REFERENCES
- Abraham M, Derk CT. Anti-ribosomal-P antibodies in lupus nephritis, neuropsychiatric lupus, lupus hepatitis, and Chagas' disease: promising yet limited in clinical utility. Rheumatol Int. 2015;35:27–33. doi: 10.1007/s00296-014-3058-3. [DOI] [PubMed] [Google Scholar]
- Abras A, Gallego M, Llovet T, Tebar S, Herrero M, Berenguer P, Ballart C, Marti C, Munoz C. Serological diagnosis of chronic Chagas disease: Is it time for a change? J Clin Microbiol. 2016;54:1566–1572. doi: 10.1128/JCM.00142-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann AA, Panunzi LG, Cosentino RO, Sanchez DO, Aguero F. A genomic scale map of genetic diversity in Trypanosoma cruzi. BMC Genomics. 2012;13:736. doi: 10.1186/1471-2164-13-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affranchino JL, Ibanez CF, Luquetti AO, Rassi A, Reyes MB, Macina RA, Aslund L, Pettersson U, Frasch AC. Identification of a Trypanosoma cruzi antigen that is shed during the acute phase of Chagas' disease. Mol Biochem Parasitol. 1989;34:221–228. doi: 10.1016/0166-6851(89)90050-9. [DOI] [PubMed] [Google Scholar]
- Aguero F, Al-Lazikani B, Aslett M, Berriman M, Buckner FS, Campbell RK, Carmona S, Carruthers IM, Chan AW, Chen F, Crowther GJ, Doyle MA, Hertz-Fowler C, Hopkins AL, McAllister G, Nwaka S, Overington JP, Pain A, Paolini GV, Pieper U, Ralph SA, Riechers A, Roos DS, Sali A, Shanmugam D, Suzuki T, Van Voorhis WC, Verlinde CL. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat Rev Drug Discov. 2008;7:900–907. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon de Noya B, Diaz-Bello Z, Colmenares C, Ruiz-Guevara R, Mauriello L, Zavala-Jaspe R, Suarez JA, Abate T, Naranjo L, Paiva M, Rivas L, Castro J, Marques J, Mendoza I, Acquatella H, Torres J, Noya O. Large urban outbreak of orally acquired acute Chagas disease at a school in Caracas, Venezuela. J Infect Dis. 2010;201:1308–1315. doi: 10.1086/651608. [DOI] [PubMed] [Google Scholar]
- Albareda MC, Laucella SA. Modulation of Trypanosoma cruzi-specific T-cell responses after chemotherapy for chronic Chagas disease. Mem Inst Oswaldo Cruz. 2015;110:414–421. doi: 10.1590/0074-02760140386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albareda MC, Olivera GC, Laucella SA, Alvarez MG, Fernandez ER, Lococo B, Viotti R, Tarleton RL, Postan M. Chronic human infection with Trypanosoma cruzi drives CD4+ T cells to immune senescence. J Immunol. 2009;183:4103–4108. doi: 10.4049/jimmunol.0900852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessio GD, Cortes DF, Machado de Assis GF, Junior PA, Ferro EA, Antonelli LR, Teixeira-Carvalho A, Martins-Filho OA, de Lana M. Innovations in diagnosis and post-therapeutic monitoring of Chagas disease: Simultaneous flow cytometric detection of IgG1 antibodies anti-live amastigote, anti-live trypomastigote, and anti-fixed epimastigote forms of Trypanosoma cruzi. J Immunol Methods. 2014;413:32–44. doi: 10.1016/j.jim.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Almeida IC, Covas DT, Soussumi LM, Travassos LR. A highly sensitive and specific chemiluminescent enzyme-linked immunosorbent assay for diagnosis of active Trypanosoma cruzi infection. Transfusion. 1997;37:850–857. doi: 10.1046/j.1537-2995.1997.37897424410.x. [DOI] [PubMed] [Google Scholar]
- Alvarez MG, Bertocchi GL, Cooley G, Albareda MC, Viotti R, Perez-Mazliah DE, Lococo B, Castro Eiro M, Laucella SA, Tarleton RL. Treatment Success in Trypanosoma cruzi Infection Is Predicted by Early Changes in Serially Monitored Parasite-Specific T and B Cell Responses. PLoS Negl Trop Dis. 2016;10:e0004657. doi: 10.1371/journal.pntd.0004657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez P, Leguizamon MS, Buscaglia CA, Pitcovsky TA, Campetella O. Multiple overlapping epitopes in the repetitive unit of the shed acute-phase antigen from Trypanosoma cruzi enhance its immunogenic properties. Infect Immun. 2001;69:7946–7949. doi: 10.1128/IAI.69.12.7946-7949.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade AL, Martelli CM, Oliveira RM, Silva SA, Aires AI, Soussumi LM, Covas DT, Silva LS, Andrade JG, Travassos LR, Almeida IC. Short report: benznidazole efficacy among Trypanosoma cruzi-infected adolescents after a six-year follow-up. Am J Trop Med Hyg. 2004;71:594–597. [PubMed] [Google Scholar]
- Andrade LO, Machado CR, Chiari E, Pena SD, Macedo AM. Differential tissue distribution of diverse clones of Trypanosoma cruzi in infected mice. Mol Biochem Parasitol. 1999;100:163–172. doi: 10.1016/s0166-6851(99)90035-x. [DOI] [PubMed] [Google Scholar]
- Andrade SG. Influence of Trypanosoma cruzi strain on the pathogenesis of chronic myocardiopathy in mice. Mem Inst Oswaldo Cruz. 1990;85:17–27. doi: 10.1590/s0074-02761990000100003. [DOI] [PubMed] [Google Scholar]
- Andrade V, Brodskyn C, Andrade SG. Correlation between isoenzyme patterns and biological behaviour of different strains of Trypanosoma cruzi. Trans R Soc Trop Med Hyg. 1983;77:796–799. doi: 10.1016/0035-9203(83)90292-4. [DOI] [PubMed] [Google Scholar]
- Anish C, Martin CE, Wahlbrink A, Bogdan C, Ntais P, Antoniou M, Seeberger PH. Immunogenicity and diagnostic potential of synthetic antigenic cell surface glycans of Leishmania. ACS Chem Biol. 2013;8:2412–2422. doi: 10.1021/cb400602k. [DOI] [PubMed] [Google Scholar]
- Antas PR, Azevedo EN, Luz MR, Medrano-Mercado N, Chaves AC, Vidigal PG, Volpini AC, Romanha AJ, Araujo-Jorge TC. A reliable and specific enzyme-linked immunosorbent assay for the capture of IgM from human chagasic sera using fixed epimastigotes of Trypanosoma cruzi. Parasitol Res. 2000;86:813–820. doi: 10.1007/pl00008507. [DOI] [PubMed] [Google Scholar]
- Aranzamendi C, Tefsen B, Jansen M, Chiumiento L, Bruschi F, Kortbeek T, Smith DF, Cummings RD, Pinelli E, Van Die I. Glycan microarray profiling of parasite infection sera identifies the LDNF glycan as a potential antigen for serodiagnosis of trichinellosis. Exp Parasitol. 2011;129:221–226. doi: 10.1016/j.exppara.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo FG. Analysis of Trypanosoma cruzi antigens bound by specific antibodies and by antibodies to related trypanosomatids. Infect Immun. 1986;53:179–185. doi: 10.1128/iai.53.1.179-185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayo CM, Dalalio MM, Visentainer JE, Reis PG, Sippert EA, Jarduli LR, Alves HV, Sell AM. Genetic susceptibility to Chagas disease: an overview about the infection and about the association between disease and the immune response genes. Biomed Res Int. 2013;2013:284729. doi: 10.1155/2013/284729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balouz V, Camara Mde L, Canepa GE, Carmona SJ, Volcovich R, Gonzalez N, Altcheh J, Aguero F, Buscaglia CA. Mapping Antigenic Motifs in the Trypomastigote Small Surface Antigen from Trypanosoma cruzi. Clin Vaccine Immunol. 2015;22:304–312. doi: 10.1128/CVI.00684-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer-Santos E, Aguilar-Bonavides C, Rodrigues SP, Cordero EM, Marques AF, Varela-Ramirez A, Choi H, Yoshida N, da Silveira JF, Almeida IC. Proteomic analysis of Trypanosoma cruzi secretome: characterization of two populations of extracellular vesicles and soluble proteins. J Proteome Res. 2013;12:883–897. doi: 10.1021/pr300947g. [DOI] [PubMed] [Google Scholar]
- Beaumier CM, Gillespie PM, Strych U, Hayward T, Hotez PJ, Bottazzi ME. Status of vaccine research and development of vaccines for Chagas disease. Vaccine. 2016;34:3001–3005. doi: 10.1016/j.vaccine.2016.03.074. [DOI] [PubMed] [Google Scholar]
- Betonico GN, Miranda EO, Silva DA, Houghton R, Reed SG, Campos-Neto A, Mineo JR. Evaluation of a synthetic tripeptide as antigen for detection of IgM and IgG antibodies to Trypanosoma cruzi in serum samples from patients with Chagas disease or viral diseases. Trans R Soc Trop Med Hyg. 1999;93:603–606. doi: 10.1016/s0035-9203(99)90064-0. [DOI] [PubMed] [Google Scholar]
- Bhatia V, Sinha M, Luxon B, Garg N. Utility of the Trypanosoma cruzi sequence database for identification of potential vaccine candidates by in silico and in vitro screening. Infect Immun. 2004;72:6245–6254. doi: 10.1128/IAI.72.11.6245-6254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T, Brooks J, Yeo M, Carrasco HJ, Lewis MD, Llewellyn MS, Miles MA. Analysis of molecular diversity of the Trypanosoma cruzi trypomastigote small surface antigen reveals novel epitopes, evidence of positive selection and potential implications for lineage-specific serology. Int J Parasitol. 2010;40:921–928. doi: 10.1016/j.ijpara.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya T, Falconar AK, Luquetti AO, Costales JA, Grijalva MJ, Lewis MD, Messenger LA, Tran TT, Ramirez JD, Guhl F, Carrasco HJ, Diosque P, Garcia L, Litvinov SV, Miles MA. Development of peptide-based lineage-specific serology for chronic Chagas disease: geographical and clinical distribution of epitope recognition. PLoS Negl Trop Dis. 2014;8:e2892. doi: 10.1371/journal.pntd.0002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T, Mills EA, Jansen AM, Miles MA. Prospects for T. cruzi lineage-specific serological surveillance of wild mammals. Acta Trop. 2015 doi: 10.1016/j.actatropica.2015.06.017. [DOI] [PubMed] [Google Scholar]
- Bisio M, Seidenstein ME, Burgos JM, Ballering G, Risso M, Pontoriero R, Moreau M, Altcheh J, Leguizamon MS, Freilij H, Marceillac M, Schijman AG. Urbanization of congenital transmission of Trypanosoma cruzi : prospective polymerase chain reaction study in pregnancy. Trans R Soc Trop Med Hyg. 2011;105:543–549. doi: 10.1016/j.trstmh.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Blanco SB, Segura EL, Cura EN, Chuit R, Tulian L, Flores I, Garbarino G, Villalonga JF, Gurtler RE. Congenital transmission of Trypanosoma cruzi: an operational outline for detecting and treating infected infants in north-western Argentina. Trop Med Int Health. 2000;5:293–301. doi: 10.1046/j.1365-3156.2000.00548.x. [DOI] [PubMed] [Google Scholar]
- Bonney KM, Engman DM. Autoimmune pathogenesis of Chagas heart disease: looking back, looking ahead. Am J Pathol. 2015;185:1537–1547. doi: 10.1016/j.ajpath.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brener Z. Therapeutic activity and criterion of cure on mice experimentally infected with Trypanosoma cruzi. Rev Inst Med Trop Sao Paulo. 1962;4:389–396. [PubMed] [Google Scholar]
- Britto C, Cardoso MA, Vanni CM, Hasslocher-Moreno A, Xavier SS, Oelemann W, Santoro A, Pirmez C, Morel CM, Wincker P. Polymerase chain reaction detection of Trypanosoma cruzi in human blood samples as a tool for diagnosis and treatment evaluation. Parasitology. 1995;110(Pt 3):241–247. doi: 10.1017/s0031182000080823. [DOI] [PubMed] [Google Scholar]
- Bua J, Volta BJ, Velazquez EB, Ruiz AM, Rissio AM, Cardoni RL. Vertical transmission of Trypanosoma cruzi infection: quantification of parasite burden in mothers and their children by parasite DNA amplification. Trans R Soc Trop Med Hyg. 2012;106:623–628. doi: 10.1016/j.trstmh.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Buchovsky AS, Campetella O, Russomando G, Franco L, Oddone R, Candia N, Luquetti A, Gonzalez Cappa SM, Leguizamon MS. trans-sialidase inhibition assay, a highly sensitive and specific diagnostic test for Chagas' disease. Clin Diagn Lab Immunol. 2001;8:187–189. doi: 10.1128/CDLI.8.1.187-189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos JM, Altcheh J, Bisio M, Duffy T, Valadares HM, Seidenstein ME, Piccinali R, Freitas JM, Levin MJ, Macchi L, Macedo AM, Freilij H, Schijman AG. Direct molecular profiling of minicircle signatures and lineages of Trypanosoma cruzi bloodstream populations causing congenital Chagas disease. Int J Parasitol. 2007;37:1319–1327. doi: 10.1016/j.ijpara.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Burgos JM, Diez M, Vigliano C, Bisio M, Risso M, Duffy T, Cura C, Brusses B, Favaloro L, Leguizamon MS, Lucero RH, Laguens R, Levin MJ, Favaloro R, Schijman AG. Molecular identification of Trypanosoma cruzi discrete typing units in end stage chronic Chagas heart disease and reactivation after heart transplantation. Clin Infect Dis. 2010;51:485–495. doi: 10.1086/655680. [DOI] [PubMed] [Google Scholar]
- Burns JM, Jr, Shreffler WG, Rosman DE, Sleath PR, March CJ, Reed SG. Identification and synthesis of a major conserved antigenic epitope of Trypanosoma cruzi. Proc Natl Acad Sci U S A. 1992;89:1239–1243. doi: 10.1073/pnas.89.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia CA, Alfonso J, Campetella O, Frasch AC. Tandem amino acid repeats from Trypanosoma cruzi shed antigens increase the half-life of proteins in blood. Blood. 1999;93:2025–2032. [PubMed] [Google Scholar]
- Buscaglia CA, Campo VA, Di Noia JM, Torrecilhas AC, De Marchi CR, Ferguson MA, Frasch AC, Almeida IC. The surface coat of the mammal-dwelling infective trypomastigote stage of Trypanosoma cruzi is formed by highly diverse immunogenic mucins. J Biol Chem. 2004;279:15860–15869. doi: 10.1074/jbc.M314051200. [DOI] [PubMed] [Google Scholar]
- Bustamante J, Tarleton R. Reaching for the Holy Grail: insights from infection/cure models on the prospects for vaccines for Trypanosoma cruzi infection. Mem Inst Oswaldo Cruz. 2015;110:445–451. doi: 10.1590/0074-02760140440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buus S, Rockberg J, Forsstrom B, Nilsson P, Uhlen M, Schafer-Nielsen C. High-resolution mapping of linear antibody epitopes using ultrahigh-density peptide microarrays. Mol Cell Proteomics. 2012;11:1790–1800. doi: 10.1074/mcp.M112.020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero ZC, Sousa OE, Marques WP, Saez-Alquezar A, Umezawa ES. Evaluation of serological tests to identify Trypanosoma cruzi infection in humans and determine cross-reactivity with Trypanosoma rangeli and Leishmania spp. Clin Vaccine Immunol. 2007;14:1045–1049. doi: 10.1128/CVI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camandaroba EL, Pinheiro Lima CM, Andrade SG. Oral transmission of Chagas disease: importance of Trypanosoma cruzi biodeme in the intragastric experimental infection. Rev Inst Med Trop Sao Paulo. 2002;44:97–103. doi: 10.1590/s0036-46652002000200008. [DOI] [PubMed] [Google Scholar]
- Campo V, Di Noia JM, Buscaglia CA, Aguero F, Sanchez DO, Frasch AC. Differential accumulation of mutations localized in particular domains of the mucin genes expressed in the vertebrate host stage of Trypanosoma cruzi. Mol Biochem Parasitol. 2004;133:81–91. doi: 10.1016/j.molbiopara.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Campo VA, Buscaglia CA, Di Noia JM, Frasch AC. Immunocharacterization of the mucin-type proteins from the intracellular stage of Trypanosoma cruzi. Microbes Infect. 2006;8:401–409. doi: 10.1016/j.micinf.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Canepa GE, Degese MS, Budu A, Garcia CR, Buscaglia CA. Involvement of TSSA (trypomastigote small surface antigen) in Trypanosoma cruzi invasion of mammalian cells. Biochem J. 2012;444:211–218. doi: 10.1042/BJ20120074. [DOI] [PubMed] [Google Scholar]
- Cantacessi C, Hofmann A, Campbell BE, Gasser RB. Impact of next-generation technologies on exploring socioeconomically important parasites and developing new interventions. Methods Mol Biol. 2015;1247:437–474. doi: 10.1007/978-1-4939-2004-4_31. [DOI] [PubMed] [Google Scholar]
- Carlier Y, Truyens C. Congenital Chagas disease as an ecological model of interactions between Trypanosoma cruzi parasites, pregnant women, placenta and fetuses. Acta Trop. 2015;151:103–115. doi: 10.1016/j.actatropica.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Carmona SJ, Nielsen M, Schafer-Nielsen C, Mucci J, Altcheh J, Balouz V, Tekiel V, Frasch AC, Campetella O, Buscaglia CA, Aguero F. Towards High-throughput Immunomics for Infectious Diseases: Use of Next-generation Peptide Microarrays for Rapid Discovery and Mapping of Antigenic Determinants. Mol Cell Proteomics. 2015;14:1871–1884. doi: 10.1074/mcp.M114.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona SJ, Sartor PA, Leguizamon MS, Campetella OE, Aguero F. Diagnostic peptide discovery: prioritization of pathogen diagnostic markers using multiple features. PLoS ONE. 2012;7:e50748. doi: 10.1371/journal.pone.0050748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo C, Villarroel A, Duaso J, Galanti N, Cabrera G, Maya JD, Kemmerling U. Phospholipase C gamma and ERK1/2 mitogen activated kinase pathways are differentially modulated by Trypanosoma cruzi during tissue invasion in human placenta. Exp Parasitol. 2013;133:12–17. doi: 10.1016/j.exppara.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Castro-Sesquen YE, Gilman RH, Galdos-Cardenas G, Ferrufino L, Sanchez G, Valencia Ayala E, Liotta L, Bern C, Luchini A. Use of a novel chagas urine nanoparticle test (chunap) for diagnosis of congenital chagas disease. PLoS Negl Trop Dis. 2014;8:e3211. doi: 10.1371/journal.pntd.0003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Sesquen YE, Gilman RH, Mejia C, Clark DE, Choi J, Reimer-McAtee MJ, Castro R, Valencia-Ayala E, Flores J, Bowman N, Castillo-Neyra R, Torrico F, Liotta L, Bern C, Luchini A. Use of a Chagas Urine Nanoparticle Test (Chunap) to Correlate with Parasitemia Levels in T. cruzi/HIV Co-infected Patients. PLoS Negl Trop Dis. 2016;10:e0004407. doi: 10.1371/journal.pntd.0004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DP, Moraes CS, Gonzalez MS, Ratcliffe NA, Azambuja P, Garcia ES. Trypanosoma cruzi immune response modulation decreases microbiota in Rhodnius prolixus gut and is crucial for parasite survival and development. PLoS ONE. 2012;7:e36591. doi: 10.1371/journal.pone.0036591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira GC, Bartholomeu DC, DaRocha WD, Hou L, Freitas-Silva DM, Machado CR, El-Sayed NM, Teixeira SM. Sequence diversity and evolution of multigene families in Trypanosoma cruzi. Mol Biochem Parasitol. 2008;157:65–72. doi: 10.1016/j.molbiopara.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Cimino RO, Rumi MM, Ragone P, Lauthier J, D'Amato AA, Quiroga IR, Gil JF, Cajal SP, Acosta N, Juarez M, Krolewiecki A, Orellana V, Zacca R, Marcipar I, Diosque P, Nasser JR. Immuno-enzymatic evaluation of the recombinant TSSA-II protein of Trypanosoma cruzi in dogs and human sera: a tool for epidemiological studies. Parasitology. 2011;138:995–1002. doi: 10.1017/S0031182011000540. [DOI] [PubMed] [Google Scholar]
- Cooley G, Etheridge RD, Boehlke C, Bundy B, Weatherly DB, Minning T, Haney M, Postan M, Laucella S, Tarleton RL. High throughput selection of effective serodiagnostics for Trypanosoma cruzi infection. PLoS Negl Trop Dis. 2008;2:e316. doi: 10.1371/journal.pntd.0000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral RS, Altcheh JM, Freilij HL. Presence of IgM antibodies to Trypanosoma cruzi urinary antigen in sera from patients with acute Chagas' disease. Int J Parasitol. 1998;28:589–594. doi: 10.1016/s0020-7519(98)00017-4. [DOI] [PubMed] [Google Scholar]
- Cortina ME, Melli LJ, Roberti M, Mass M, Longinotti G, Tropea S, Lloret P, Serantes DA, Salomon F, Lloret M, Caillava AJ, Restuccia S, Altcheh J, Buscaglia CA, Malatto L, Ugalde JE, Fraigi L, Moina C, Ybarra G, Ciocchini AE, Comerci DJ. Electrochemical magnetic microbeads-based biosensor for point-of-care serodiagnosis of infectious diseases. Biosens Bioelectron. 2016;80:24–33. doi: 10.1016/j.bios.2016.01.021. [DOI] [PubMed] [Google Scholar]
- Cosentino RO, Aguero F. A simple strain typing assay for Trypanosoma cruzi: discrimination of major evolutionary lineages from a single amplification product. PLoS Negl Trop Dis. 2012;6:e1777. doi: 10.1371/journal.pntd.0001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotrim PC, Paranhos GS, Mortara RA, Wanderley J, Rassi A, Camargo ME, da Silveira JF. Expression in Escherichia coli of a dominant immunogen of Trypanosoma cruzi recognized by human chagasic sera. J Clin Microbiol. 1990;28:519–524. doi: 10.1128/jcm.28.3.519-524.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coura JR, Junqueira AC, Fernandes O, Valente SA, Miles MA. Emerging Chagas disease in Amazonian Brazil. Trends Parasitol. 2002;18:171–176. doi: 10.1016/s1471-4922(01)02200-0. [DOI] [PubMed] [Google Scholar]
- Crowther GJ, Shanmugam D, Carmona SJ, Doyle MA, Hertz-Fowler C, Berriman M, Nwaka S, Ralph SA, Roos DS, Van Voorhis WC, Aguero F. Identification of attractive drug targets in neglected-disease pathogens using an in silico approach. PLoS Negl Trop Dis. 2010;4:e804. doi: 10.1371/journal.pntd.0000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crutchfield CA, Thomas SN, Sokoll LJ, Chan DW. Advances in mass spectrometry-based clinical biomarker discovery. Clin Proteomics. 2016;13:1. doi: 10.1186/s12014-015-9102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Avila DA, Macedo AM, Valadares HM, Gontijo ED, de Castro AM, Machado CR, Chiari E, Galvao LM. Probing population dynamics of Trypanosoma cruzi during progression of the chronic phase in chagasic patients. J Clin Microbiol. 2009;47:1718–1725. doi: 10.1128/JCM.01658-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira JF, Umezawa ES, Luquetti AO. Chagas disease: recombinant Trypanosoma cruzi antigens for serological diagnosis. Trends Parasitol. 2001;17:286–291. doi: 10.1016/s1471-4922(01)01897-9. [DOI] [PubMed] [Google Scholar]
- da Silveira Pinto A, de Lana M, Britto C, Bastrenta B, Tibayrenc M. Experimental Trypanosoma cruzi biclonal infection in Triatoma infestans: detection of distinct clonal genotypes using kinetoplast DNA probes. Int J Parasitol. 2000;30:843–848. doi: 10.1016/s0020-7519(00)00058-8. [DOI] [PubMed] [Google Scholar]
- Dc-Rubin SS, Schenkman S. T rypanosoma cruzi trans-sialidase as a multifunctional enzyme in Chagas' disease. Cell Microbiol. 2012;14:1522–1530. doi: 10.1111/j.1462-5822.2012.01831.x. [DOI] [PubMed] [Google Scholar]
- de Andrade AL, Zicker F, de Oliveira RM, Almeida Silva S, Luquetti A, Travassos LR, Almeida IC, de Andrade SS, de Andrade JG, Martelli CM. Randomised trial of efficacy of benznidazole in treatment of early Trypanosoma cruzi infection. Lancet. 1996;348:1407–1413. doi: 10.1016/s0140-6736(96)04128-1. [DOI] [PubMed] [Google Scholar]
- de Lana M, da Silveira Pinto A, Barnabe C, Quesney V, Noel S, Tibayrenc M. Trypanosoma cruzi: compared vectorial transmissibility of three major clonal genotypes by Triatoma infestans. Exp Parasitol. 1998;90:20–25. doi: 10.1006/expr.1998.4304. [DOI] [PubMed] [Google Scholar]
- de Lederkremer RM, Agusti R. Glycobiology of Trypanosoma cruzi. Adv Carbohydr Chem Biochem. 2009;62:311–366. doi: 10.1016/S0065-2318(09)00007-9. [DOI] [PubMed] [Google Scholar]
- De Marchi CR, Di Noia JM, Frasch AC, Amato Neto V, Almeida IC, Buscaglia CA. Evaluation of a recombinant Trypanosoma cruzi mucin-like antigen for serodiagnosis of Chagas' disease. Clin Vaccine Immunol. 2011;18:1850–1855. doi: 10.1128/CVI.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza MM, Andrade SG, Barbosa AA, Jr, Macedo Santos RT, Alves VA, Andrade ZA. Trypanosoma cruzi strains and autonomic nervous system pathology in experimental Chagas disease. Mem Inst Oswaldo Cruz. 1996;91:217–224. doi: 10.1590/s0074-02761996000200018. [DOI] [PubMed] [Google Scholar]
- De Souza W. Basic cell biology of Trypanosoma cruzi. Curr Pharm Des. 2002;8:269–285. doi: 10.2174/1381612023396276. [DOI] [PubMed] [Google Scholar]
- de Titto EH, Araujo FG. Serum neuraminidase activity and hematological alterations in acute human Chagas' disease. Clin Immunol Immunopathol. 1988;46:157–161. doi: 10.1016/0090-1229(88)90016-5. [DOI] [PubMed] [Google Scholar]
- del Puerto F, Sanchez Z, Nara E, Meza G, Paredes B, Ferreira E, Russomando G. Trypanosoma cruzi lineages detected in congenitally infected infants and Triatoma infestans from the same disease-endemic region under entomologic surveillance in Paraguay. Am J Trop Med Hyg. 2010;82:386–390. doi: 10.4269/ajtmh.2010.09-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Sabino EC, Cunha-Neto E, Ribeiro AL, Ianni B, Mady C, Busch MP, Seielstad M. Genome wide association study (GWAS) of Chagas cardiomyopathy in Trypanosoma cruzi seropositive subjects. PLoS ONE. 2013;8:e79629. doi: 10.1371/journal.pone.0079629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Buscaglia CA, De Marchi CR, Almeida IC, Frasch AC. A Trypanosoma cruzi small surface molecule provides the first immunological evidence that Chagas' disease is due to a single parasite lineage. J Exp Med. 2002;195:401–413. doi: 10.1084/jem.20011433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy T, Bisio M, Altcheh J, Burgos JM, Diez M, Levin MJ, Favaloro RR, Freilij H, Schijman AG. Accurate real-time PCR strategy for monitoring bloodstream parasitic loads in chagas disease patients. PLoS Negl Trop Dis. 2009;3:e419. doi: 10.1371/journal.pntd.0000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy T, Cura CI, Ramirez JC, Abate T, Cayo NM, Parrado R, Bello ZD, Velazquez E, Munoz-Calderon A, Juiz NA, Basile J, Garcia L, Riarte A, Nasser JR, Ocampo SB, Yadon ZE, Torrico F, de Noya BA, Ribeiro I, Schijman AG. Analytical performance of a multiplex Real-Time PCR assay using TaqMan probes for quantification of Trypanosoma cruzi satellite DNA in blood samples. PLoS Negl Trop Dis. 2013;7:e2000. doi: 10.1371/journal.pntd.0002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar SA, Ritchie VB, Hoffmeyer MR, Rana GS, Zhang H. Luminex((R)) multiplex bead suspension arrays for the detection and serotyping of Salmonella spp. Methods Mol Biol. 2015;1225:1–27. doi: 10.1007/978-1-4939-1625-2_1. [DOI] [PubMed] [Google Scholar]
- Eisenstein M. Disease: Poverty and pathogens. Nature. 2016;531:S61–S63. doi: 10.1038/531S61a. [DOI] [PubMed] [Google Scholar]
- Fabbro DL, Danesi E, Olivera V, Codebo MO, Denner S, Heredia C, Streiger M, Sosa-Estani S. Trypanocide treatment of women infected with Trypanosoma cruzi and its effect on preventing congenital Chagas. PLoS Negl Trop Dis. 2014;8:e3312. doi: 10.1371/journal.pntd.0003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes F, Dantas S, Ianni BM, Ramires FJ, Buck P, Salemi VM, Lopes HF, Mady C. Leptin levels in different forms of Chagas' disease. Braz J Med Biol Res. 2007;40:1631–1636. doi: 10.1590/s0100-879x2006005000152. [DOI] [PubMed] [Google Scholar]
- Fernandes O, Mangia RH, Lisboa CV, Pinho AP, Morel CM, Zingales B, Campbell DA, Jansen AM. The complexity of the sylvatic cycle of Trypanosoma cruzi in Rio de Janeiro state (Brazil) revealed by the non-transcribed spacer of the mini-exon gene. Parasitology. 1999;118(Pt 2):161–166. doi: 10.1017/s0031182098003709. [DOI] [PubMed] [Google Scholar]
- Fernandez-Calero T, Garcia-Silva R, Pena A, Robello C, Persson H, Rovira C, Naya H, Cayota A. Profiling of small RNA cargo of extracellular vesicles shed by Trypanosoma cruzi reveals a specific extracellular signature. Mol Biochem Parasitol. 2015;199:19–28. doi: 10.1016/j.molbiopara.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Fernandez-Tejada A, Canada FJ, Jimenez-Barbero J. Recent Developments in Synthetic Carbohydrate-Based Diagnostics, Vaccines, and Therapeutics. Chemistry. 2015;21:10616–10628. doi: 10.1002/chem.201500831. [DOI] [PubMed] [Google Scholar]
- Fernandez-Villegas A, Pinazo MJ, Maranon C, Thomas MC, Posada E, Carrilero B, Segovia M, Gascon J, Lopez MC. Short-term follow-up of chagasic patients after benzonidazole treatment using multiple serological markers. BMC Infect Dis. 2011;11:206. doi: 10.1186/1471-2334-11-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade AF, Teixeira PC, Ianni BM, Pissetti CW, Saba B, Wang LH, Kuramoto A, Nogueira LG, Buck P, Dias F, Giniaux H, Llored A, Alves S, Schmidt A, Donadi E, Marin-Neto JA, Hirata M, Sampaio M, Fragata A, Bocchi EA, Stolf AN, Fiorelli AI, Santos RH, Rodrigues V, Pereira AC, Kalil J, Cunha-Neto E, Chevillard C. Polymorphism in the alpha cardiac muscle actin 1 gene is associated to susceptibility to chronic inflammatory cardiomyopathy. PLoS ONE. 2013;8:e83446. doi: 10.1371/journal.pone.0083446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch AC. Functional diversity in the trans-sialidase and mucin families in Trypanosoma cruzi. Parasitol Today. 2000;16:282–286. doi: 10.1016/s0169-4758(00)01698-7. [DOI] [PubMed] [Google Scholar]
- Frasch AC, Cazzulo JJ, Aslund L, Pettersson U. Comparison of genes encoding Trypanosoma cruzi antigens. Parasitol Today. 1991;7:148–151. doi: 10.1016/0169-4758(91)90284-u. [DOI] [PubMed] [Google Scholar]
- Freilij H, Altcheh J. Congenital Chagas' disease: diagnostic and clinical aspects. Clin Infect Dis. 1995;21:551–555. doi: 10.1093/clinids/21.3.551. [DOI] [PubMed] [Google Scholar]
- Galvao LM, Chiari E, Macedo AM, Luquetti AO, Silva SA, Andrade AL. PCR assay for monitoring Trypanosoma cruzi parasitemia in childhood after specific chemotherapy. J Clin Microbiol. 2003;41:5066–5070. doi: 10.1128/JCM.41.11.5066-5070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvao LM, Nunes RM, Cancado JR, Brener Z, Krettli AU. Lytic antibody titre as a means of assessing cure after treatment of Chagas disease: a 10 years follow-up study. Trans R Soc Trop Med Hyg. 1993;87:220–223. doi: 10.1016/0035-9203(93)90501-g. [DOI] [PubMed] [Google Scholar]
- Garcia-Alvarez A, Sitges M, Pinazo MJ, Regueiro-Cueva A, Posada E, Poyatos S, Ortiz-Perez JT, Heras M, Azqueta M, Gascon J, Sanz G. Chagas cardiomyopathy: the potential of diastolic dysfunction and brain natriuretic peptide in the early identification of cardiac damage. PLoS Negl Trop Dis. 2010;4 doi: 10.1371/journal.pntd.0000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzinelli RT, Galvao LM, Krautz G, Lima PC, Cancado JR, Scharfstein J, Krettli AU. Use of Trypanosoma cruzi purified glycoprotein (GP57/51) or trypomastigote-shed antigens to assess cure for human Chagas' disease. Am J Trop Med Hyg. 1993;49:625–635. doi: 10.4269/ajtmh.1993.49.625. [DOI] [PubMed] [Google Scholar]
- Giraldo NA, Bolanos NI, Cuellar A, Roa N, Cucunuba Z, Rosas F, Velasco V, Puerta CJ, Gonzalez JM. T lymphocytes from chagasic patients are activated but lack proliferative capacity and down-regulate CD28 and CD3zeta. PLoS Negl Trop Dis. 2013;7:e2038. doi: 10.1371/journal.pntd.0002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girones N, Carbajosa S, Guerrero NA, Poveda C, Chillon-Marinas C, Fresno M. Global metabolomic profiling of acute myocarditis caused by Trypanosoma cruzi infection. PLoS Negl Trop Dis. 2014;8:e3337. doi: 10.1371/journal.pntd.0003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsel LM, Tibbetts RS, Olson CL, Chaudoir BM, Engman DM. Utility of recombinant flagellar calcium-binding protein for serodiagnosis of Trypanosoma cruzi infection. J Clin Microbiol. 1995;33:2082–2085. doi: 10.1128/jcm.33.8.2082-2085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes YM, Lorena VM, Luquetti AO. Diagnosis of Chagas disease: what has been achieved? What remains to be done with regard to diagnosis and follow up studies? Mem Inst Oswaldo Cruz. 2009;104(Suppl 1):115–121. doi: 10.1590/s0074-02762009000900017. [DOI] [PubMed] [Google Scholar]
- Gonzalez MS, Souza MS, Garcia ES, Nogueira NF, Mello CB, Canepa GE, Bertotti S, Durante IM, Azambuja P, Buscaglia CA. Trypanosoma cruzi TcSMUG L-surface Mucins Promote Development and Infectivity in the Triatomine Vector Rhodnius prolixus. PLoS Negl Trop Dis. 2013;7:e2552. doi: 10.1371/journal.pntd.0002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Carter D, Reed SG. Immunological dominance of Trypanosoma cruzi tandem repeat proteins. Infect Immun. 2008;76:3967–3974. doi: 10.1128/IAI.00604-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granjon E, Dichtel-Danjoy ML, Saba E, Sabino E, Campos de Oliveira L, Zrein M. Development of a Novel Multiplex Immunoassay Multi-cruzi for the Serological Confirmation of Chagas Disease. PLoS Negl Trop Dis. 2016;10:e0004596. doi: 10.1371/journal.pntd.0004596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtler RE, Cardinal MV. Reservoir host competence and the role of domestic and commensal hosts in the transmission of Trypanosoma cruzi. Acta Trop. 2015;151:32–50. doi: 10.1016/j.actatropica.2015.05.029. [DOI] [PubMed] [Google Scholar]
- Guzman-Gomez D, Lopez-Monteon A, de la Soledad Lagunes-Castro M, Alvarez-Martinez C, Hernandez-Lutzon MJ, Dumonteil E, Ramos-Ligonio A. Highly discordant serology against Trypanosoma cruzi in central Veracruz, Mexico: role of the antigen used for diagnostic. Parasit Vectors. 2015;8:466. doi: 10.1186/s13071-015-1072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heringer-Walther S, Moreira MC, Wessel N, Saliba JL, Silvia-Barra J, Pena JL, Becker S, Siems WE, Schultheiss HP, Walther T. Brain natriuretic peptide predicts survival in Chagas' disease more effectively than atrial natriuretic peptide. Heart. 2005;91:385–387. doi: 10.1136/hrt.2003.026856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez P, Heimann M, Riera C, Solano M, Santalla J, Luquetti AO, Beck E. Highly effective serodiagnosis for Chagas' disease. Clin Vaccine Immunol. 2010;17:1598–1604. doi: 10.1128/CVI.00489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockl PF, Wolosiuk A, Perez-Saez JM, Bordoni AV, Croci DO, Toum-Terrones Y, Soler-Illia GJ, Rabinovich GA. Glyco-nano-oncology: Novel therapeutic opportunities by combining small and sweet. Pharmacol Res. 2016;109:45–54. doi: 10.1016/j.phrs.2016.02.005. [DOI] [PubMed] [Google Scholar]
- Hoft DF, Farrar PL, Kratz-Owens K, Shaffer D. Gastric invasion by Trypanosoma cruzi and induction of protective mucosal immune responses. Infect Immun. 1996;64:3800–3810. doi: 10.1128/iai.64.9.3800-3810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft DF, Kim KS, Otsu K, Moser DR, Yost WJ, Blumin JH, Donelson JE, Kirchhoff LV. Trypanosoma cruzi expresses diverse repetitive protein antigens. Infect Immun. 1989;57:1959–1967. doi: 10.1128/iai.57.7.1959-1967.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton RL, Benson DR, Reynolds L, McNeill P, Sleath P, Lodes M, Skeiky YA, Badaro R, Krettli AU, Reed SG. Multiepitope synthetic peptide and recombinant protein for the detection of antibodies to Trypanosoma cruzi in patients with treated or untreated Chagas' disease. J Infect Dis. 2000;181:325–330. doi: 10.1086/315165. [DOI] [PubMed] [Google Scholar]
- Houghton RL, Benson DR, Reynolds LD, McNeill PD, Sleath PR, Lodes MJ, Skeiky YA, Leiby DA, Badaro R, Reed SG. A multi-epitope synthetic peptide and recombinant protein for the detection of antibodies to Trypanosoma cruzi in radioimmunoprecipitation-confirmed and consensus-positive sera. J Infect Dis. 1999;179:1226–1234. doi: 10.1086/314723. [DOI] [PubMed] [Google Scholar]
- Houghton RL, Stevens YY, Hjerrild K, Guderian J, Okamoto M, Kabir M, Reed SG, Leiby DA, Morrow WJ, Lorca M, Raychaudhuri S. Lateral flow immunoassay for diagnosis of Trypanosoma cruzi infection with high correlation to the radioimmunoprecipitation assay. Clin Vaccine Immunol. 2009;16:515–520. doi: 10.1128/CVI.00383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Petkova SB, Pestell RG, Bouzahzah B, Chan J, Magazine H, Weiss LM, Christ GJ, Lisanti MP, Douglas SA, Shtutin V, Halonen SK, Wittner M, Tanowitz HB. Trypanosoma cruzi infection (Chagas' disease) of mice causes activation of the mitogen-activated protein kinase cascade and expression of endothelin-1 in the myocardium. J Cardiovasc Pharmacol. 2000;36:S148–S150. doi: 10.1097/00005344-200036051-00046. [DOI] [PubMed] [Google Scholar]
- Ibanez CF, Affranchino JL, Frasch AC. Antigenic determinants of Trypanosoma cruzi defined by cloning of parasite DNA. Mol Biochem Parasitol. 1987;25:175–184. doi: 10.1016/0166-6851(87)90006-5. [DOI] [PubMed] [Google Scholar]
- Ibanez CF, Affranchino JL, Macina RA, Reyes MB, Leguizamon S, Camargo ME, Aslund L, Pettersson U, Frasch AC. Multiple Trypanosoma cruzi antigens containing tandemly repeated amino acid sequence motifs. Mol Biochem Parasitol. 1988;30:27–33. doi: 10.1016/0166-6851(88)90129-6. [DOI] [PubMed] [Google Scholar]
- Jelicks LA, Chandra M, Shtutin V, Petkova SB, Tang B, Christ GJ, Factor SM, Wittner M, Huang H, Douglas SA, Weiss LM, Orleans-Juste PD, Shirani J, Tanowitz HB. Phosphoramidon treatment improves the consequences of chagasic heart disease in mice. Clin Sci (Lond) 2002;103(Suppl 48):267S–271S. doi: 10.1042/CS103S267S. [DOI] [PubMed] [Google Scholar]
- Juiz NA, Cayo NM, Burgos M, Salvo ME, Nasser JR, Bua J, Longhi SA, Schijman AG. Human Polymorphisms in Placentally Expressed Genes and Their Association With Susceptibility to Congenital Trypanosoma cruzi Infection. J Infect Dis. 2016;213:1299–1306. doi: 10.1093/infdis/jiv561. [DOI] [PubMed] [Google Scholar]
- Kaplan D, Ferrari I, Bergami PL, Mahler E, Levitus G, Chiale P, Hoebeke J, Van Regenmortel MH, Levin MJ. Antibodies to ribosomal P proteins of Trypanosoma cruzi in Chagas disease possess functional autoreactivity with heart tissue and differ from anti-P autoantibodies in lupus. Proc Natl Acad Sci U S A. 1997;94:10301–10306. doi: 10.1073/pnas.94.19.10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplinski M, Jois M, Galdos-Cardenas G, Rendell VR, Shah V, Do RQ, Marcus R, Pena MS, Abastoflor Mdel C, LaFuente C, Bozo R, Valencia E, Verastegui M, Colanzi R, Gilman RH, Bern C. Sustained Domestic Vector Exposure Is Associated With Increased Chagas Cardiomyopathy Risk but Decreased Parasitemia and Congenital Transmission Risk Among Young Women in Bolivia. Clin Infect Dis. 2015;61:918–926. doi: 10.1093/cid/civ446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsdal MA, Henriksen K, Leeming DJ, Woodworth T, Vassiliadis E, Bay-Jensen AC. Novel combinations of Post-Translational Modification (PTM) neo-epitopes provide tissue-specific biochemical markers--are they the cause or the consequence of the disease? Clin Biochem. 2010;43:793–804. doi: 10.1016/j.clinbiochem.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Katsuno K, Burrows JN, Duncan K, Hooft van Huijsduijnen R, Kaneko T, Kita K, Mowbray CE, Schmatz D, Warner P, Slingsby BT. Hit and lead criteria in drug discovery for infectious diseases of the developing world. Nat Rev Drug Discov. 2015;14:751–758. doi: 10.1038/nrd4683. [DOI] [PubMed] [Google Scholar]
- Kedzierski RM, Yanagisawa M. Endothelin system: the double-edged sword in health and disease. Annu Rev Pharmacol Toxicol. 2001;41:851–876. doi: 10.1146/annurev.pharmtox.41.1.851. [DOI] [PubMed] [Google Scholar]
- Keidar S, Kaplan M, Gamliel-Lazarovich A. ACE2 of the heart: From angiotensin I to angiotensin (1–7) Cardiovasc Res. 2007;73:463–469. doi: 10.1016/j.cardiores.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Krettli AU, Brener Z. Resistance against Trypanosoma cruzi associated to anti-living trypomastigote antibodies. J Immunol. 1982;128:2009–2012. [PubMed] [Google Scholar]
- Krieger MA, Almeida E, Oelemann W, Lafaille JJ, Pereira JB, Krieger H, Carvalho MR, Goldenberg S. Use of recombinant antigens for the accurate immunodiagnosis of Chagas' disease. Am J Trop Med Hyg. 1992;46:427–434. doi: 10.4269/ajtmh.1992.46.427. [DOI] [PubMed] [Google Scholar]
- Lafaille JJ, Linss J, Krieger MA, Souto-Padron T, de Souza W, Goldenberg S. Structure and expression of two Trypanosoma cruzi genes encoding antigenic proteins bearing repetitive epitopes. Mol Biochem Parasitol. 1989;35:127–136. doi: 10.1016/0166-6851(89)90115-1. [DOI] [PubMed] [Google Scholar]
- Lages-Silva E, Ramirez LE, Silva-Vergara ML, Chiari E. Chagasic meningoencephalitis in a patient with acquired immunodeficiency syndrome: diagnosis, follow-up, and genetic characterization of Trypanosoma cruzi. Clin Infect Dis. 2002;34:118–123. doi: 10.1086/324355. [DOI] [PubMed] [Google Scholar]
- Lantos AB, Carlevaro G, Araoz B, Ruiz Diaz P, Camara Mde L, Buscaglia CA, Bossi M, Yu H, Chen X, Bertozzi CR, Mucci J, Campetella O. Sialic Acid Glycobiology Unveils Trypanosoma cruzi Trypomastigote Membrane Physiology. PLoS Pathog. 2016;12:e1005559. doi: 10.1371/journal.ppat.1005559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laucella SA, Mazliah DP, Bertocchi G, Alvarez MG, Cooley G, Viotti R, Albareda MC, Lococo B, Postan M, Armenti A, Tarleton RL. Changes in Trypanosoma cruzi-specific immune responses after treatment: surrogate markers of treatment efficacy. Clin Infect Dis. 2009;49:1675–1684. doi: 10.1086/648072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent JP, Barnabe C, Quesney V, Noel S, Tibayrenc M. Impact of clonal evolution on the biological diversity of Trypanosoma cruzi. Parasitology. 1997;114(Pt 3):213–218. doi: 10.1017/s0031182096008414. [DOI] [PubMed] [Google Scholar]
- Leeansyah E, Malone DF, Anthony DD, Sandberg JK. Soluble biomarkers of HIV transmission, disease progression and comorbidities. Curr Opin HIV AIDS. 2013;8:117–124. doi: 10.1097/COH.0b013e32835c7134. [DOI] [PubMed] [Google Scholar]
- Levin MJ, Hoebeke J. Cross-talk between anti-beta1-adrenoceptor antibodies in dilated cardiomyopathy and Chagas' heart disease. Autoimmunity. 2008;41:429–433. doi: 10.1080/08916930802031702. [DOI] [PubMed] [Google Scholar]
- Levin MJ, Mesri E, Benarous R, Levitus G, Schijman A, Levy-Yeyati P, Chiale PA, Ruiz AM, Kahn A, Rosenbaum MB, et al. Identification of major Trypanosoma cruzi antigenic determinants in chronic Chagas' heart disease. Am J Trop Med Hyg. 1989;41:530–538. doi: 10.4269/ajtmh.1989.41.530. [DOI] [PubMed] [Google Scholar]
- Lewis MD, Francisco AF, Taylor MC, Kelly JM. A new experimental model for assessing drug efficacy against Trypanosoma cruzi infection based on highly sensitive in vivo imaging. J Biomol Screen. 2015;20:36–43. doi: 10.1177/1087057114552623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MD, Llewellyn MS, Gaunt MW, Yeo M, Carrasco HJ, Miles MA. Flow cytometric analysis and microsatellite genotyping reveal extensive DNA content variation in Trypanosoma cruzi populations and expose contrasts between natural and experimental hybrids. Int J Parasitol. 2009a;39:1305–1317. doi: 10.1016/j.ijpara.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MD, Ma J, Yeo M, Carrasco HJ, Llewellyn MS, Miles MA. Genotyping of Trypanosoma cruzi: systematic selection of assays allowing rapid and accurate discrimination of all known lineages. Am J Trop Med Hyg. 2009b;81:1041–1049. doi: 10.4269/ajtmh.2009.09-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Shah-Simpson S, Okrah K, Belew AT, Choi J, Caradonna KL, Padmanabhan P, Ndegwa DM, Temanni MR, Corrada Bravo H, El-Sayed NM, Burleigh BA. Transcriptome Remodeling in Trypanosoma cruzi and Human Cells during Intracellular Infection. PLoS Pathog. 2016;12:e1005511. doi: 10.1371/journal.ppat.1005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi SA, Atienza A, Perez Prados G, Buying A, Balouz V, Buscaglia CA, Santos R, Tasso LM, Bonato R, Chiale P, Pinilla C, Judkowski VA, Gomez KA. Cytokine Production but Lack of Proliferation in Peripheral Blood Mononuclear Cells from Chronic Chagas' Disease Cardiomyopathy Patients in Response to T. cruzi Ribosomal P Proteins. PLoS Negl Trop Dis. 2014;8:e2906. doi: 10.1371/journal.pntd.0002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi SA, Brandariz SB, Lafon SO, Niborski LL, Luquetti AO, Schijman AG, Levin MJ, Gomez KA. Evaluation of in-house ELISA using Trypanosoma cruzi lysate and recombinant antigens for diagnosis of Chagas disease and discrimination of its clinical forms. Am J Trop Med Hyg. 2012;87:267–271. doi: 10.4269/ajtmh.2012.11-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca M, Veloso C, Munoz P, Bahamonde MI, Garcia A. Diagnostic value of detecting specific IgA and IgM with recombinant Trypanosoma cruzi antigens in congenital Chagas' disease. Am J Trop Med Hyg. 1995;52:512–515. doi: 10.4269/ajtmh.1995.52.512. [DOI] [PubMed] [Google Scholar]
- Luquetti AO, Miles MA, Rassi A, de Rezende JM, de Souza AA, Povoa MM, Rodrigues I. Trypanosoma cruzi: zymodemes associated with acute and chronic Chagas' disease in central Brazil. Trans R Soc Trop Med Hyg. 1986;80:462–470. doi: 10.1016/0035-9203(86)90347-0. [DOI] [PubMed] [Google Scholar]
- Luz PR, Miyazaki MI, Chiminacio Neto N, Padeski MC, Barros AC, Boldt AB, Messias-Reason IJ. Genetically Determined MBL Deficiency Is Associated with Protection against Chronic Cardiomyopathy in Chagas Disease. PLoS Negl Trop Dis. 2016;10:e0004257. doi: 10.1371/journal.pntd.0004257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn MS, Messenger LA, Luquetti AO, Garcia L, Torrico F, Tavares SB, Cheaib B, Derome N, Delepine M, Baulard C, Deleuze JF, Sauer S, Miles MA. Deep sequencing of the Trypanosoma cruzi GP63 surface proteases reveals diversity and diversifying selection among chronic and congenital Chagas disease patients. PLoS Negl Trop Dis. 2015;9:e0003458. doi: 10.1371/journal.pntd.0003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo AM, Pena SD. Genetic Variability of Trypanosoma cruzi :Implications for the Pathogenesis of Chagas Disease. Parasitol Today. 1998;14:119–124. doi: 10.1016/s0169-4758(97)01179-4. [DOI] [PubMed] [Google Scholar]
- Machado-de-Assis GF, Silva AR, Do Bem VA, Bahia MT, Martins-Filho OA, Dias JC, Albajar-Vinas P, Torres RM, Lana M. Posttherapeutic cure criteria in Chagas' disease: conventional serology followed by supplementary serological, parasitological, and molecular tests. Clin Vaccine Immunol. 2012;19:1283–1291. doi: 10.1128/CVI.00274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado FS, Dutra WO, Esper L, Gollob KJ, Teixeira MM, Factor SM, Weiss LM, Nagajyothi F, Tanowitz HB, Garg NJ. Current understanding of immunity to Trypanosoma cruzi infection and pathogenesis of Chagas disease. Semin Immunopathol. 2012;34:753–770. doi: 10.1007/s00281-012-0351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda FY, Clemente TM, Macedo S, Cortez C, Yoshida N. Host cell invasion and oral infection by Trypanosoma cruzi strains of genetic groups TcI and TcIV from chagasic patients. Parasit Vectors. 2016;9:189. doi: 10.1186/s13071-016-1455-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes LM, Viana A, Chiari E, Galvao LM, Gollob KJ, Dutra WO. Differential Activation of Human Monocytes and Lymphocytes by Distinct Strains of Trypanosoma cruzi. PLoS Negl Trop Dis. 2015;9:e0003816. doi: 10.1371/journal.pntd.0003816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magarinos MP, Carmona SJ, Crowther GJ, Ralph SA, Roos DS, Shanmugam D, Van Voorhis WC, Aguero F. TDR Targets: a chemogenomics resource for neglected diseases. Nucleic Acids Res. 2012;40:D1118–D1127. doi: 10.1093/nar/gkr1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimov P, Zerweck J, Maksimov A, Hotop A, Gross U, Spekker K, Daubener W, Werdermann S, Niederstrasser O, Petri E, Mertens M, Ulrich RG, Conraths FJ, Schares G. Analysis of clonal type-specific antibody reactions in Toxoplasma gondii seropositive humans from Germany by peptide-microarray. PLoS ONE. 2012;7:e34212. doi: 10.1371/journal.pone.0034212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallimaci MC, Sosa-Estani S, Russomando G, Sanchez Z, Sijvarger C, Alvarez IM, Barrionuevo L, Lopez C, Segura EL. Early diagnosis of congenital Trypanosoma cruzi infection, using shed acute phase antigen, in Ushuaia, Tierra del Fuego, Argentina. Am J Trop Med Hyg. 2010;82:55–59. doi: 10.4269/ajtmh.2010.09-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoel-Caetano Fda S, Carareto CM, Borim AA, Miyazaki K, Silva AE. kDNA gene signatures of Trypanosoma cruzi in blood and oesophageal mucosa from chronic chagasic patients. Trans R Soc Trop Med Hyg. 2008;102:1102–1107. doi: 10.1016/j.trstmh.2008.05.027. [DOI] [PubMed] [Google Scholar]
- Marcili A, Lima L, Cavazzana M, Junqueira AC, Veludo HH, Maia Da Silva F, Campaner M, Paiva F, Nunes VL, Teixeira MM. A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology. 2009;136:641–655. doi: 10.1017/S0031182009005861. [DOI] [PubMed] [Google Scholar]
- Martin CE, Broecker F, Eller S, Oberli MA, Anish C, Pereira CL, Seeberger PH. Glycan arrays containing synthetic Clostridium difficile lipoteichoic acid oligomers as tools toward a carbohydrate vaccine. Chem Commun (Camb) 2013;49:7159–7161. doi: 10.1039/c3cc43545h. [DOI] [PubMed] [Google Scholar]
- Martin DL, Weatherly DB, Laucella SA, Cabinian MA, Crim MT, Sullivan S, Heiges M, Craven SH, Rosenberg CS, Collins MH, Sette A, Postan M, Tarleton RL. CD8+ T-Cell responses to Trypanosoma cruzi are highly focused on strain-variant trans-sialidase epitopes. PLoS Pathog. 2006;2:e77. doi: 10.1371/journal.ppat.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Campetella O, Frasch AC, Cazzulo JJ. The major cysteine proteinase (cruzipain) from Trypanosoma cruzi is antigenic in human infections. Infect Immun. 1991;59:4275–4277. doi: 10.1128/iai.59.11.4275-4277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Filho OA, Eloi-Santos SM, Teixeira Carvalho A, Oliveira RC, Rassi A, Luquetti AO, Rassi GG, Brener Z. Double-blind study to evaluate flow cytometry analysis of anti-live trypomastigote antibodies for monitoring treatment efficacy in cases of human Chagas' disease. Clin Diagn Lab Immunol. 2002;9:1107–1113. doi: 10.1128/CDLI.9.5.1107-1113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes TA, Reis Cunha JL, de Almeida Lourdes R, Rodrigues Luiz GF, Lemos LD, Dos Santos AR, da Camara AC, Galvao LM, Bern C, Gilman RH, Fujiwara RT, Gazzinelli RT, Bartholomeu DC. Identification of Strain-Specific B-cell Epitopes in Trypanosoma cruzi Using Genome-Scale Epitope Prediction and High-Throughput Immunoscreening with Peptide Arrays. PLoS Negl Trop Dis. 2013;7:e2524. doi: 10.1371/journal.pntd.0002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenger LA, Miles MA. Evidence and importance of genetic exchange among field populations of Trypanosoma cruzi. Acta Trop. 2015;151:150–155. doi: 10.1016/j.actatropica.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenger LA, Miles MA, Bern C. Between a bug and a hard place: Trypanosoma cruzi genetic diversity and the clinical outcomes of Chagas disease. Expert Rev Anti Infect Ther. 2015;13:995–1029. doi: 10.1586/14787210.2015.1056158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minning TA, Weatherly DB, Flibotte S, Tarleton RL. Widespread, focal copy number variations (CNV) and whole chromosome aneuploidies in Trypanosoma cruzi strains revealed by array comparative genomic hybridization. BMC Genomics. 2011;12:139. doi: 10.1186/1471-2164-12-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal D, Ghosh P, Khan MA, Hossain F, Bohlken-Fascher S, Matlashewski G, Kroeger A, Olliaro P, Abd El Wahed A. Mobile suitcase laboratory for rapid detection of Leishmania donovani using recombinase polymerase amplification assay. Parasit Vectors. 2016;9:281. doi: 10.1186/s13071-016-1572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro WM, Margioto Teston AP, Gruendling AP, dos Reis D, Gomes ML, de Araujo SM, Bahia MT, Magalhaes LK, de Oliveira Guerra JA, Silveira H, Toledo MJ, Vale Barbosa M. Trypanosoma cruzi I and IV stocks from Brazilian Amazon are divergent in terms of biological and medical properties in mice. PLoS Negl Trop Dis. 2013;7:e2069. doi: 10.1371/journal.pntd.0002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes CB, Giardini MA, Kim H, Franco CH, Araujo-Junior AM, Schenkman S, Chatelain E, Freitas-Junior LH. Nitroheterocyclic compounds are more efficacious than CYP51 inhibitors against Trypanosoma cruzi: implications for Chagas disease drug discovery and development. Sci Rep. 2014;4:4703. doi: 10.1038/srep04703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortara RA, Andreoli WK, Fernandes MC, da Silva CV, Fernandes AB, L'Abbate C, da Silva S. Host cell actin remodeling in response to Trypanosoma cruzi: trypomastigote versus amastigote entry. Subcell Biochem. 2008;47:101–109. doi: 10.1007/978-0-387-78267-6_8. [DOI] [PubMed] [Google Scholar]
- Moser DR, Kirchhoff LV, Donelson JE. Detection of Trypanosoma cruzi by DNA amplification using the polymerase chain reaction. J Clin Microbiol. 1989;27:1477–1482. doi: 10.1128/jcm.27.7.1477-1482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucci J, Hidalgo A, Mocetti E, Argibay PF, Leguizamon MS, Campetella O. Thymocyte depletion in Trypanosoma cruzi infection is mediated by trans-sialidase-induced apoptosis on nurse cells complex. Proc Natl Acad Sci U S A. 2002;99:3896–3901. doi: 10.1073/pnas.052496399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muia RP, Yu H, Prescher JA, Hellman U, Chen X, Bertozzi CR, Campetella O. Identification of glycoproteins targeted by Trypanosoma cruzi trans-sialidase, a virulence factor that disturbs lymphocyte glycosylation. Glycobiology. 2010;20:833–842. doi: 10.1093/glycob/cwq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz C, Zulantay I, Apt W, Ortiz S, Schijman AG, Bisio M, Ferrada V, Herrera C, Martinez G, Solari A. Evaluation of nifurtimox treatment of chronic Chagas disease by means of several parasitological methods. Antimicrob Agents Chemother. 2013;57:4518–4523. doi: 10.1128/AAC.00227-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagajyothi F, Machado FS, Burleigh BA, Jelicks LA, Scherer PE, Mukherjee S, Lisanti MP, Weiss LM, Garg NJ, Tanowitz HB. Mechanisms of Trypanosoma cruzi persistence in Chagas disease. Cell Microbiol. 2012;14:634–643. doi: 10.1111/j.1462-5822.2012.01764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkatti R, Bist V, Sun S, Fortes de Araujo F, Nakhasi HL, Debrabant A. Development of an aptamer-based concentration method for the detection of Trypanosoma cruzi in blood. PLoS ONE. 2012;7:e43533. doi: 10.1371/journal.pone.0043533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkatti R, de Araujo FF, Gupta C, Debrabant A. Aptamer based, non-PCR, non-serological detection of Chagas disease biomarkers in Trypanosoma cruzi infected mice. PLoS Negl Trop Dis. 2014;8:e2650. doi: 10.1371/journal.pntd.0002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair CB, Manjula J, Subramani PA, Nagendrappa PB, Manoj MN, Malpani S, Pullela PK, Subbarao PV, Ramamoorthy S, Ghosh SK. Differential Diagnosis of Malaria on Truelab Uno(R), a Portable, Real-Time, MicroPCR Device for Point-Of-Care Applications. PLoS ONE. 2016;11:e0146961. doi: 10.1371/journal.pone.0146961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli L, Maher L, Shephard M, Hengel B, Tangey A, Badman SG, Ward J, Guy RJ. Point-of-care testing for chlamydia and gonorrhoea: implications for clinical practice. PLoS ONE. 2014;9:e100518. doi: 10.1371/journal.pone.0100518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira LG, Santos RH, Ianni BM, Fiorelli AI, Mairena EC, Benvenuti LA, Frade A, Donadi E, Dias F, Saba B, Wang HT, Fragata A, Sampaio M, Hirata MH, Buck P, Mady C, Bocchi EA, Stolf NA, Kalil J, Cunha-Neto E. Myocardial chemokine expression and intensity of myocarditis in Chagas cardiomyopathy are controlled by polymorphisms in CXCL9 and CXCL10. PLoS Negl Trop Dis. 2012;6:e1867. doi: 10.1371/journal.pntd.0001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireau F, Diosque P, Jansen AM. Trypanosoma cruzi: adaptation to its vectors and its hosts. Vet Res. 2009;40:26. doi: 10.1051/vetres/2009009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelemann WM, Vanderborght BO, Verissimo Da Costa GC, Teixeira MG, Borges-Pereira J, De Castro JA, Coura JR, Stoops E, Hulstaert F, Zrein M, Peralta JM. A recombinant peptide antigen line immunoassay optimized for the confirmation of Chagas' disease. Transfusion. 1999;39:711–717. doi: 10.1046/j.1537-2995.1999.39070711.x. [DOI] [PubMed] [Google Scholar]
- Padilla AM, Simpson LJ, Tarleton RL. Insufficient TLR activation contributes to the slow development of CD8+ T cell responses in Trypanosoma cruzi infection. J Immunol. 2009;83:1245–1252. doi: 10.4049/jimmunol.0901178. [DOI] [PubMed] [Google Scholar]
- Paiva CN, Feijo DF, Dutra FF, Carneiro VC, Freitas GB, Alves LS, Mesquita J, Fortes GB, Figueiredo RT, Souza HS, Fantappie MR, Lannes-Vieira J, Bozza MT. Oxidative stress fuels Trypanosoma cruzi infection in mice. J Clin Invest. 2012;122:2531–2542. doi: 10.1172/JCI58525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panunzi LG, Aguero F. A genome-wide analysis of genetic diversity in Trypanosoma cruzi intergenic regions. PLoS Negl Trop Dis. 2014;8:e2839. doi: 10.1371/journal.pntd.0002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastini AC, Iglesias SR, Carricarte VC, Guerin ME, Sanchez DO, Frasch AC. Immunoassay with recombinant Trypanosoma cruzi antigens potentially useful for screening donated blood and diagnosing Chagas disease. Clin Chem. 1994;40:1893–1897. [PubMed] [Google Scholar]
- Pereira-Chioccola VL, Acosta-Serrano A, Correia de Almeida I, Ferguson MA, Souto-Padron T, Rodrigues MM, Travassos LR, Schenkman S. Mucin-like molecules form a negatively charged coat that protects Trypanosoma cruzi trypomastigotes from killing by human anti-alpha-galactosyl antibodies. J Cell Sci. 2000;113(Pt 7):1299–1307. doi: 10.1242/jcs.113.7.1299. [DOI] [PubMed] [Google Scholar]
- Perez CJ, Lymbery AJ, Thompson RC. Chagas disease: the challenge of polyparasitism? Trends Parasitol. 2014;30:176–182. doi: 10.1016/j.pt.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Petkova SB, Tanowitz HB, Magazine HI, Factor SM, Chan J, Pestell RG, Bouzahzah B, Douglas SA, Shtutin V, Morris SA, Tsang E, Weiss LM, Christ GJ, Wittner M, Huang H. Myocardial expression of endothelin-1 in murine Trypanosoma cruzi infection. Cardiovasc Pathol. 2000;9:257–265. doi: 10.1016/s1054-8807(00)00045-4. [DOI] [PubMed] [Google Scholar]
- Pinazo MJ, Thomas MC, Bua J, Perrone A, Schijman AG, Viotti RJ, Ramsey JM, Ribeiro I, Sosa-Estani S, Lopez MC, Gascon J. Biological markers for evaluating therapeutic efficacy in Chagas disease, a systematic review. Expert Rev Anti Infect Ther. 2014;12:479–496. doi: 10.1586/14787210.2014.899150. [DOI] [PubMed] [Google Scholar]
- Pinazo MJ, Thomas MC, Bustamante J, Almeida IC, Lopez MC, Gascon J. Biomarkers of therapeutic responses in chronic Chagas disease: state of the art and future perspectives. Mem Inst Oswaldo Cruz. 2015;110:422–432. doi: 10.1590/0074-02760140435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto CM, Kalko EK, Cottontail I, Wellinghausen N, Cottontail VM. TcBat a bat-exclusive lineage of Trypanosoma cruzi in the Panama Canal Zone, with comments on its classification and the use of the 18S rRNA gene for lineage identification. Infect Genet Evol. 2012;12:1328–1332. doi: 10.1016/j.meegid.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Piron M, Fisa R, Casamitjana N, Lopez-Chejade P, Puig L, Verges M, Gascon J, Gomez i Prat J, Portus M, Sauleda S. Development of a real-time PCR assay for Trypanosoma cruzi detection in blood samples. Acta Trop. 2007;103:195–200. doi: 10.1016/j.actatropica.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Praast G, Herzogenrath J, Bernhardt S, Christ H, Sickinger E. Evaluation of the Abbott ARCHITECT Chagas prototype assay. Diagn Microbiol Infect Dis. 2011;69:74–81. doi: 10.1016/j.diagmicrobio.2010.08.019. [DOI] [PubMed] [Google Scholar]
- Preidis GA, Hotez PJ. The newest "omics"--metagenomics and metabolomics--enter the battle against the neglected tropical diseases. PLoS Negl Trop Dis. 2015;9:e0003382. doi: 10.1371/journal.pntd.0003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvarnstrom Y, Schijman AG, Veron V, Aznar C, Steurer F, da Silva AJ. Sensitive and specific detection of Trypanosoma cruzi DNA in clinical specimens using a multi-target real-time PCR approach. PLoS Negl Trop Dis. 2012;6:e1689. doi: 10.1371/journal.pntd.0001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez JC, Cura CI, da Cruz Moreira O, Lages-Silva E, Juiz N, Velazquez E, Ramirez JD, Alberti A, Pavia P, Flores-Chavez MD, Munoz-Calderon A, Perez-Morales D, Santalla J, Marcos da Matta Guedes P, Peneau J, Marcet P, Padilla C, Cruz-Robles D, Valencia E, Crisante GE, Greif G, Zulantay I, Costales JA, Alvarez-Martinez M, Martinez NE, Villarroel R, Villarroel S, Sanchez Z, Bisio M, Parrado R, Maria da Cunha Galvao L, Jacome da Camara AC, Espinoza B, Alarcon de Noya B, Puerta C, Riarte A, Diosque P, Sosa-Estani S, Guhl F, Ribeiro I, Aznar C, Britto C, Yadon ZE, Schijman AG. Analytical Validation of Quantitative Real-Time PCR Methods for Quantification of Trypanosoma cruzi DNA in Blood Samples from Chagas Disease Patients. J Mol Diagn. 2015;17:605–615. doi: 10.1016/j.jmoldx.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez JD, Hernandez C, Montilla M, Zambrano P, Florez AC, Parra E, Cucunuba ZM. First report of human Trypanosoma cruzi infection attributed to TcBat genotype. Zoonoses Public Health. 2014;61:477–479. doi: 10.1111/zph.12094. [DOI] [PubMed] [Google Scholar]
- Rassi A, Jr, Rassi A, Marin-Neto JA. Chagas disease. Lancet. 2010;375:1388–1402. doi: 10.1016/S0140-6736(10)60061-X. [DOI] [PubMed] [Google Scholar]
- Reis-Cunha JL, Mendes TA, de Almeida Lourdes R, Ribeiro DR, Machado-de-Avila RA, de Oliveira Tavares M, Lemos DS, Camara AC, Olortegui CC, de Lana M, da Cunha Galvao LM, Fujiwara RT, Bartholomeu DC. Genome-Wide Screening and Identification of New Trypanosoma cruzi Antigens with Potential Application for Chronic Chagas Disease Diagnosis. PLoS ONE. 2014;9:e106304. doi: 10.1371/journal.pone.0106304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Cunha JL, Rodrigues-Luiz GF, Valdivia HO, Baptista RP, Mendes TA, de Morais GL, Guedes R, Macedo AM, Bern C, Gilman RH, Lopez CT, Andersson B, Vasconcelos AT, Bartholomeu DC. Chromosomal copy number variation reveals differential levels of genomic plasticity in distinct Trypanosoma cruzi strains. BMC Genomics. 2015;16:499. doi: 10.1186/s12864-015-1680-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reithinger R, Grijalva MJ, Chiriboga RF, de Noya BA, Torres JR, Pavia-Ruz N, Manrique-Saide P, Cardinal MV, Gurtler RE. Rapid detection of Trypanosoma cruzi in human serum by use of an immunochromatographic dipstick test. J Clin Microbiol. 2010;48:3003–3007. doi: 10.1128/JCM.02474-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendell VR, Gilman RH, Valencia E, Galdos-Cardenas G, Verastegui M, Sanchez L, Acosta J, Sanchez G, Ferrufino L, LaFuente C, Abastoflor Mdel C, Colanzi R, Bern C. Trypanosoma cruzi-infected pregnant women without vector exposure have higher parasitemia levels: implications for congenital transmission risk. PLoS ONE. 2015;10:e0119527. doi: 10.1371/journal.pone.0119527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena-Mendez A, Aldasoro E, de Lazzari E, Sicuri E, Brown M, Moore DA, Gascon J, Munoz J. Prevalence of Chagas disease in Latin-American migrants living in Europe: a systematic review and meta-analysis. PLoS Negl Trop Dis. 2015;9:e0003540. doi: 10.1371/journal.pntd.0003540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo S, Oury B, Laurent JP, Barnabe C, Quesney V, Carriere V, Noel S, Tibayrenc M. Trypanosoma cruzi: impact of clonal evolution of the parasite on its biological and medical properties. Exp Parasitol. 1998;89:30–39. doi: 10.1006/expr.1998.4216. [DOI] [PubMed] [Google Scholar]
- Reyes MB, Lorca M, Munoz P, Frasch AC. Fetal IgG specificities against Trypanosoma cruzi antigens in infected newborns. Proc Natl Acad Sci U S A. 1990;87:2846–2850. doi: 10.1073/pnas.87.7.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro AL, dos Reis AM, Barros MV, de Sousa MR, Rocha AL, Perez AA, Pereira JB, Machado FS, Rocha MO. Brain natriuretic peptide and left ventricular dysfunction in Chagas' disease. Lancet. 2002;360:461–462. doi: 10.1016/S0140-6736(02)09638-1. [DOI] [PubMed] [Google Scholar]
- Ribeiro I, Sevcsik AM, Alves F, Diap G, Don R, Harhay MO, Chang S, Pecoul B. New, improved treatments for Chagas disease: from the R&D pipeline to the patients. PLoS Negl Trop Dis. 2009;3:e484. doi: 10.1371/journal.pntd.0000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso MG, Pitcovsky TA, Caccuri RL, Campetella O, Leguizamon MS. Immune system pathogenesis is prevented by the neutralization of the systemic trans-sialidase from Trypanosoma cruzi during severe infections. Parasitology. 2007;134:503–510. doi: 10.1017/S0031182006001752. [DOI] [PubMed] [Google Scholar]
- Risso MG, Sartor PA, Burgos JM, Briceno L, Rodriguez EM, Guhl F, Chavez OT, Espinoza B, Monteon VM, Russomando G, Schijman AG, Bottasso OA, Leguizamon MS. Immunological identification of Trypanosoma cruzi lineages in human infection along the endemic area. Am J Trop Med Hyg. 2011;84:78–84. doi: 10.4269/ajtmh.2011.10-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roellig DM, McMillan K, Ellis AE, Vandeberg JL, Champagne DE, Yabsley MJ. Experimental infection of two South American reservoirs with four distinct strains of Trypanosoma cruzi. Parasitology. 2010;137:959–966. doi: 10.1017/S0031182009991995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz RC, Favoreto S, Jr, Dorta ML, Oshiro ME, Ferreira AT, Manque PM, Yoshida N. Infectivity of Trypanosoma cruzi strains is associated with differential expression of surface glycoproteins with differential Ca2+ signalling activity. Biochem J. 1998;330(Pt 1):505–511. doi: 10.1042/bj3300505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russomando G, de Tomassone MM, de Guillen I, Acosta N, Vera N, Almiron M, Candia N, Calcena MF, Figueredo A. Treatment of congenital Chagas' disease diagnosed and followed up by the polymerase chain reaction. Am J Trop Med Hyg. 1998;59:487–491. doi: 10.4269/ajtmh.1998.59.487. [DOI] [PubMed] [Google Scholar]
- Russomando G, Sanchez Z, Meza G, de Guillen Y. Shed acute-phase antigen protein in an ELISA system for unequivocal diagnosis of congenital Chagas disease. Expert Rev Mol Diagn. 2010;10:705–707. doi: 10.1586/erm.10.70. [DOI] [PubMed] [Google Scholar]
- Saborio JL, Wrightsman RA, Kazuko SG, Granger BS, Manning JE. Trypanosoma cruzi: identification of a surface antigen restricted to the flagellar region of the infective form of the parasite. Exp Parasitol. 1990;70:411–418. doi: 10.1016/0014-4894(90)90125-v. [DOI] [PubMed] [Google Scholar]
- Salvador F, Sulleiro E, Sanchez-Montalva A, Martinez-Gallo M, Carrillo E, Molina I. Impact of Helminth Infection on the Clinical and Microbiological Presentation of Chagas Diseases in Chronically Infected Patients. PLoS Negl Trop Dis. 2016;10:e0004663. doi: 10.1371/journal.pntd.0004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Camargo CL, Albajar-Vinas P, Wilkins PP, Nieto J, Leiby DA, Paris L, Scollo K, Florez C, Guzman-Bracho C, Luquetti AO, Calvo N, Tadokoro K, Saez-Alquezar A, Palma PP, Martin M, Flevaud L. Comparative evaluation of 11 commercialized rapid diagnostic tests for detecting Trypanosoma cruzi antibodies in serum banks in areas of endemicity and nonendemicity. J Clin Microbiol. 2014;52:2506–2512. doi: 10.1128/JCM.00144-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Negrette O, Sanchez Valdez FJ, Lacunza CD, Garcia Bustos MF, Mora MC, Uncos AD, Basombrio MA. Serological evaluation of specific-antibody levels in patients treated for chronic Chagas' disease. Clin Vaccine Immunol. 2008;15:297–302. doi: 10.1128/CVI.00106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria C, Chatelain E, Jackson Y, Miao Q, Ward BJ, Chappuis F, Ndao M. Serum biomarkers predictive of cure in Chagas disease patients after nifurtimox treatment. BMC Infect Dis. 2014;14:302. doi: 10.1186/1471-2334-14-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schijman AG, Altcheh J, Burgos JM, Biancardi M, Bisio M, Levin MJ, Freilij H. Aetiological treatment of congenital Chagas' disease diagnosed and monitored by the polymerase chain reaction. J Antimicrob Chemother. 2003;52:441–449. doi: 10.1093/jac/dkg338. [DOI] [PubMed] [Google Scholar]
- Schijman AG, Bisio M, Orellana L, Sued M, Duffy T, Mejia Jaramillo AM, Cura C, Auter F, Veron V, Qvarnstrom Y, Deborggraeve S, Hijar G, Zulantay I, Lucero RH, Velazquez E, Tellez T, Sanchez Leon Z, Galvao L, Nolder D, Monje Rumi M, Levi JE, Ramirez JD, Zorrilla P, Flores M, Jercic MI, Crisante G, Anez N, De Castro AM, Gonzalez CI, Acosta Viana K, Yachelini P, Torrico F, Robello C, Diosque P, Triana Chavez O, Aznar C, Russomando G, Buscher P, Assal A, Guhl F, Sosa Estani S, DaSilva A, Britto C, Luquetti A, Ladzins J. International study to evaluate PCR methods for detection of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. PLoS Negl Trop Dis. 2011;5:e931. doi: 10.1371/journal.pntd.0000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schijman AG, Vigliano C, Burgos J, Favaloro R, Perrone S, Laguens R, Levin MJ. Early diagnosis of recurrence of Trypanosoma cruzi infection by polymerase chain reaction after heart transplantation of a chronic Chagas' heart disease patient. J Heart Lung Transplant. 2000;19:1114–1117. doi: 10.1016/s1053-2498(00)00168-6. [DOI] [PubMed] [Google Scholar]
- Schmunis GA, Yadon ZE. Chagas disease: a Latin American health problem becoming a world health problem. Acta Trop. 2010;115:14–21. doi: 10.1016/j.actatropica.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Schnaidman BB, Yoshida N, Gorin PA, Travassos LR. Cross-reactive polysaccharides from Trypanosoma cruzi and fungi (especially Dactylium dendroides) J Protozool. 1986;33:186–191. doi: 10.1111/j.1550-7408.1986.tb05587.x. [DOI] [PubMed] [Google Scholar]
- Schofield CJ, Jannin J, Salvatella R. The future of Chagas disease control. Trends Parasitol. 2006;22:583–588. doi: 10.1016/j.pt.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Segovia M, Carrasco HJ, Martinez CE, Messenger LA, Nessi A, Londono JC, Espinosa R, Martinez C, Alfredo M, Bonfante-Cabarcas R, Lewis MD, de Noya BA, Miles MA, Llewellyn MS. Molecular epidemiologic source tracking of orally transmitted Chagas disease, Venezuela. Emerg Infect Dis. 2013;19:1098–1101. doi: 10.3201/eid1907.121576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva ED, Pereira VR, Gomes JA, Lorena VM, Cancado JR, Ferreira AG, Krieger MA, Goldenberg S, Correa-Oliveira R, Gomes YM. Use of the EIE-recombinant-Chagas-biomanguinhos kit to monitor cure of human Chagas' disease. J Clin Lab Anal. 2002;16:132–136. doi: 10.1002/jcla.10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L. Kinetoplast DNA in trypanosomid flagellates. Int Rev Cytol. 1986;99:119–179. doi: 10.1016/s0074-7696(08)61426-6. [DOI] [PubMed] [Google Scholar]
- Sosa Estani S, Segura EL, Ruiz AM, Velazquez E, Porcel BM, Yampotis C. Efficacy of chemotherapy with benznidazole in children in the indeterminate phase of Chagas' disease. Am J Trop Med Hyg. 1998;59:526–529. doi: 10.4269/ajtmh.1998.59.526. [DOI] [PubMed] [Google Scholar]
- Souza RT, Lima FM, Barros RM, Cortez DR, Santos MF, Cordero EM, Ruiz JC, Goldenberg S, Teixeira MM, da Silveira JF. Genome size, karyotype polymorphism and chromosomal evolution in Trypanosoma cruzi. PLoS ONE. 2011;6:e23042. doi: 10.1371/journal.pone.0023042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriworarat C, Phumee A, Mungthin M, Leelayoova S, Siriyasatien P. Development of loop-mediated isothermal amplification (LAMP) for simple detection of Leishmania infection. Parasit Vectors. 2015;8:591. doi: 10.1186/s13071-015-1202-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanaway JD, Roth G. The burden of Chagas disease: estimates and challenges. Glob Heart. 2015;10:139–144. doi: 10.1016/j.gheart.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Sturm NR, Degrave W, Morel C, Simpson L. Sensitive detection and schizodeme classification of Trypanosoma cruzi cells by amplification of kinetoplast minicircle DNA sequences: use in diagnosis of Chagas' disease. Mol Biochem Parasitol. 1989;33:205–214. doi: 10.1016/0166-6851(89)90082-0. [DOI] [PubMed] [Google Scholar]
- Talvani A, Rocha MO, Barcelos LS, Gomes YM, Ribeiro AL, Teixeira MM. Elevated concentrations of CCL2 and tumor necrosis factor-alpha in chagasic cardiomyopathy. Clin Infect Dis. 2004a;38:943–950. doi: 10.1086/381892. [DOI] [PubMed] [Google Scholar]
- Talvani A, Rocha MO, Cogan J, Maewal P, de Lemos J, Ribeiro AL, Teixeira MM. Brain natriuretic peptide and left ventricular dysfunction in chagasic cardiomyopathy. Mem Inst Oswaldo Cruz. 2004b;99:645–649. doi: 10.1590/s0074-02762004000600020. [DOI] [PubMed] [Google Scholar]
- Tanowitz HB, Huang H, Jelicks LA, Chandra M, Loredo ML, Weiss LM, Factor SM, Shtutin V, Mukherjee S, Kitsis RN, Christ GJ, Wittner M, Shirani J, Kisanuki YY, Yanagisawa M. Role of endothelin 1 in the pathogenesis of chronic chagasic heart disease. Infect Immun. 2005;73:2496–2503. doi: 10.1128/IAI.73.4.2496-2503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarleton RL. CD8+ T cells in Trypanosoma cruzi infection. Semin Immunopathol. 2015;37:233–238. doi: 10.1007/s00281-015-0481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira AR, Hecht MM, Guimaro MC, Sousa AO, Nitz N. Pathogenesis of chagas' disease: parasite persistence and autoimmunity. Clin Microbiol Rev. 2011;24:592–630. doi: 10.1128/CMR.00063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles F, Fonseca L. Nucleic-acid testing, new platforms and nanotechnology for point-of-decision diagnosis of animal pathogens. Methods Mol Biol. 2015;1247:253–283. doi: 10.1007/978-1-4939-2004-4_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telleria J, Biron DG, Brizard JP, Demettre E, Seveno M, Barnabe C, Ayala FJ, Tibayrenc M. Phylogenetic character mapping of proteomic diversity shows high correlation with subspecific phylogenetic diversity in Trypanosoma cruzi. Proc Natl Acad Sci U S A. 2010;107:20411–20416. doi: 10.1073/pnas.1015496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Fernandez-Villegas A, Carrilero B, Maranon C, Saura D, Noya O, Segovia M, Alarcon de Noya B, Alonso C, Lopez MC. Characterization of an immunodominant antigenic epitope from Trypanosoma cruzi as a biomarker of chronic Chagas' disease pathology. Clin Vaccine Immunol. 2012;19:167–173. doi: 10.1128/CVI.05566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M, Ayala FJ. The population genetics of Trypanosoma cruzi revisited in the light of the predominant clonal evolution model. Acta Trop. 2015;151:156–165. doi: 10.1016/j.actatropica.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo MJ, Bahia MT, Carneiro CM, Martins-Filho OA, Tibayrenc M, Barnabe C, Tafuri WL, de Lana M. Chemotherapy with benznidazole and itraconazole for mice infected with different Trypanosoma cruzi clonal genotypes. Antimicrob Agents Chemother. 2003;47:223–230. doi: 10.1128/AAC.47.1.223-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritten L, Burkman E, Moorhead A, Satti M, Geary J, Mackenzie C, Geary T. Detection of circulating parasite-derived microRNAs in filarial infections. PLoS Negl Trop Dis. 2014;8:e2971. doi: 10.1371/journal.pntd.0002971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trocoli Torrecilhas AC, Tonelli RR, Pavanelli WR, da Silva JS, Schumacher RI, de Souza W, NC ES, de Almeida Abrahamsohn I, Colli W, Manso Alves MJ. Trypanosoma cruzi: parasite shed vesicles increase heart parasitism and generate an intense inflammatory response. Microbes Infect. 2009;11:29–39. doi: 10.1016/j.micinf.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Umezawa ES, Nascimento MS, Kesper N, Jr, Coura JR, Borges-Pereira J, Junqueira AC, Camargo ME. Immunoblot assay using excreted-secreted antigens of Trypanosoma cruzi in serodiagnosis of congenital, acute, and chronic Chagas' disease. J Clin Microbiol. 1996;34:2143–2147. doi: 10.1128/jcm.34.9.2143-2147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa ES, Shikanai-Yasuda MA, Stolf AM. Changes in isotype composition and antigen recognition of anti-Trypanosoma cruzi antibodies from acute to chronic Chagas disease. J Clin Lab Anal. 1996;10:407–413. doi: 10.1002/(SICI)1098-2825(1996)10:6<407::AID-JCLA16>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Urban I, Santurio LB, Chidichimo A, Yu H, Chen X, Mucci J, Aguero F, Buscaglia CA. Molecular diversity of the Trypanosoma cruzi TcSMUG family of mucin genes and proteins. Biochem J. 2011;438:303–313. doi: 10.1042/BJ20110683. [DOI] [PubMed] [Google Scholar]
- Vago AR, Andrade LO, Leite AA, d'Avila Reis D, Macedo AM, Adad SJ, Tostes S, Jr, Moreira MC, Filho GB, Pena SD. Genetic characterization of Trypanosoma cruzi directly from tissues of patients with chronic Chagas disease: differential distribution of genetic types into diverse organs. Am J Pathol. 2000;156:1805–1809. doi: 10.1016/s0002-9440(10)65052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadares HM, Pimenta JR, de Freitas JM, Duffy T, Bartholomeu DC, Oliveira Rde P, Chiari E, Moreira Mda C, Filho GB, Schijman AG, Franco GR, Machado CR, Pena SD, Macedo AM. Genetic profiling of Trypanosoma cruzi directly in infected tissues using nested PCR of polymorphic microsatellites. Int J Parasitol. 2008;38:839–850. doi: 10.1016/j.ijpara.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Vasconcelos JR, Bruna-Romero O, Araujo AF, Dominguez MR, Ersching J, de Alencar BC, Machado AV, Gazzinelli RT, Bortoluci KR, Amarante-Mendes GP, Lopes MF, Rodrigues MM. Pathogen-induced proapoptotic phenotype and high CD95 (Fas) expression accompany a suboptimal CD8+ T-cell response: reversal by adenoviral vaccine. PLoS Pathog. 2012;8:e1002699. doi: 10.1371/journal.ppat.1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verani JR, Seitz A, Gilman RH, LaFuente C, Galdos-Cardenas G, Kawai V, de LaFuente E, Ferrufino L, Bowman NM, Pinedo-Cancino V, Levy MZ, Steurer F, Todd CW, Kirchhoff LV, Cabrera L, Verastegui M, Bern C. Geographic variation in the sensitivity of recombinant antigen-based rapid tests for chronic Trypanosoma cruzi infection. Am J Trop Med Hyg. 2009;80:410–415. [PubMed] [Google Scholar]
- Vieira CS, Waniek PJ, Castro DP, Mattos DP, Moreira OC, Azambuja P. Impact of Trypanosoma cruzi on antimicrobial peptide gene expression and activity in the fat body and midgut of Rhodnius prolixus. Parasit Vectors. 2016;9:119. doi: 10.1186/s13071-016-1398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti R, Vigliano C, Lococo B, Bertocchi G, Petti M, Alvarez MG, Postan M, Armenti A. Long-term cardiac outcomes of treating chronic Chagas disease with benznidazole versus no treatment: a nonrandomized trial. Ann Intern Med. 2006;144:724–734. doi: 10.7326/0003-4819-144-10-200605160-00006. [DOI] [PubMed] [Google Scholar]
- Virreira M, Alonso-Vega C, Solano M, Jijena J, Brutus L, Bustamante Z, Truyens C, Schneider D, Torrico F, Carlier Y, Svoboda M. Congenital Chagas disease in Bolivia is not associated with DNA polymorphism of Trypanosoma cruzi. Am J Trop Med Hyg. 2006a;75:871–879. [PubMed] [Google Scholar]
- Virreira M, Serrano G, Maldonado L, Svoboda M. Trypanosoma cruzi: typing of genotype (sub)lineages in megacolon samples from bolivian patients. Acta Trop. 2006b;100:252–255. doi: 10.1016/j.actatropica.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Vitelli-Avelar DM, Sathler-Avelar R, Wendling AP, Rocha RD, Teixeira-Carvalho A, Martins NE, Dias JC, Rassi A, Luquetti AO, Eloi-Santos SM, Martins-Filho OA. Non-conventional flow cytometry approaches to detect anti-Trypanosoma cruzi immunoglobulin G in the clinical laboratory. J Immunol Methods. 2007;318:102–112. doi: 10.1016/j.jim.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Volta BJ, Russomando G, Bustos PL, Scollo K, De Rissio AM, Sanchez Z, Cardoni RL, Bua J. Diagnosis of congenital Trypanosoma cruzi infection: A serologic test using Shed Acute Phase Antigen (SAPA) in mother-child binomial samples. Acta Trop. 2015;147:31–37. doi: 10.1016/j.actatropica.2015.03.026. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Gona P, Larson MG, Tofler GH, Levy D, Newton-Cheh C, Jacques PF, Rifai N, Selhub J, Robins SJ, Benjamin EJ, D'Agostino RB, Vasan RS. Multiple biomarkers for the prediction of first major cardiovascular events and death. N Engl J Med. 2006;355:2631–2639. doi: 10.1056/NEJMoa055373. [DOI] [PubMed] [Google Scholar]
- Wang Y, Moreira Mda C, Heringer-Walther S, Ebermann L, Schultheiss HP, Wessel N, Siems WE, Walther T. Plasma ACE2 activity is an independent prognostic marker in Chagas' disease and equally potent as BNP. J Card Fail. 2010;16:157–163. doi: 10.1016/j.cardfail.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Wondergem J, Strootman EG, Frolich M, Leer JW, Noordijk EM. Circulating atrial natriuretic peptide plasma levels as a marker for cardiac damage after radiotherapy. Radiother Oncol. 2001;58:295–301. doi: 10.1016/s0167-8140(00)00303-0. [DOI] [PubMed] [Google Scholar]
- Zafra G, Mantilla JC, Jacome J, Macedo AM, Gonzalez CI. Direct analysis of genetic variability in Trypanosoma cruzi populations from tissues of Colombian chagasic patients. Hum Pathol. 2011;42:1159–1168. doi: 10.1016/j.humpath.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MM, Schijman AG, Llewellyn MS, Lages-Silva E, Machado CR, Andrade SG, Sturm NR. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol. 2012;12:240–253. doi: 10.1016/j.meegid.2011.12.009. [DOI] [PubMed] [Google Scholar]