Abstract
Lactococcus lactis is well documented as a promising candidate for development of novel oral live vaccines. It has been broadly engineered for heterologous expression, as well as for plasmid expression vector delivery, directly inside eukaryotic cells, for DNA vaccine, or as therapeutic vehicle. This work describes the characteristics of a new plasmid, pExu (extra chromosomal unit), for DNA delivery using L. lactis and evaluates its functionality both by in vitro and in vivo assays. This plasmid exhibits the following features: (1) a theta origin of replication and (2) an expression cassette containing a multiple cloning site and a eukaryotic promoter, the cytomegalovirus (pCMV). The functionality of pExu:egfp was evaluated by fluorescence microscopy. The L. lactis MG1363 (pExu:egfp) strains were administered by gavage to Balb/C mice and the eGFP expression was monitored by fluorescence microscopy. The pExu vector has demonstrated an excellent stability either in L. lactis or in Escherichia coli. The eGFP expression at different times in in vitro assay showed that 15.8% of CHO cells were able to express the protein after transfection. The enterocytes of mice showed the expression of eGFP protein. Thus, L. lactis carrying the pExu is a good candidate to deliver genes into eukaryotic cells.
Keywords: lactic acid bacteria, Lactococcus lactis, DNA mucosal delivering, shuttle plasmid
Introduction
Lactic acid bacteria (LAB) have been reported to be useful for mucosal delivery of different molecules, like heterologous proteins, vaccine, and plasmids.1 Humans have been consuming this LAB for centuries through fermented foods, LAB have the capacity to transform sugar in lactic acid. Lactobacilli, lactococci, enterococci, streptococci, leuconostoc, and pediococci are the broad genera of these bacterial group; habitats, morphology, optimum temperature, pH and salt tolerance, and pathogenic potential are characteristics in which they differ.2 Lactobacilli and Lactococcus lactis are considered “generally recognized as safe” (GRAS) according to the U.S. Food and Drug Administration (FDA). They also fulfill criteria of the competent Qualified Presumption of Safety (QPS) according to the European Food Safety Authority (EFSA). These Gram-positive bacteria are non-pathogenic and non-invasive and inhabit different ecological niches (plant surfaces and the digestive tract of animals and human).3 L. lactis, has been intensively explored as delivery vector for antigens or therapeutics proteins.4, 5, 6, 7 Antigens delivered by recombinant LAB at the mucosal site would avoid the massive degradation of the antigens observed in the gut when purified antigens are used. The strategy of bactoinfection (live bacterial vectors for transfection of mammalian cells) opened the field to use L. lactis as a DNA delivery vehicle.8, 9 DNA delivery by bacteria into eukaryotic cells leads to host expression of post-translational modified antigens and consequently the presentation of conformational-restricted epitopes to the immune system.10 Several studies have used native11 or recombinant L. lactis expressing different invasin, describing their potential uses as DNA delivery vectors either in vivo or in vitro assays.12, 13, 14
Here, we developed a new plasmid called pExu (extra chromosomal unit) for DNA delivery. To construct this new plasmid, we used as backbone pOri253, a shuttle E. coli/L. lactis plasmid (6.09 Kb) derived from pIL253 plasmid.15, 16 This plasmid provided the theta-type replication origin and the ermAM gene conferring erythromycin resistance in both E. coli and Gram-positive bacteria. The eukaryotic region, derived from pCDNA3.1 (Invitrogen), contains the cytomegalovirus promoter (pCMV), a multiple cloning site, and the polyadenylation signal of bovine growth hormone (BGH polyA), which is essential for gene expression, with an important role in stability and translation of mRNA.17
To evaluate the functionality of pExu plasmid, we cloned the eGFP (egfp) open reading frame (ORF) into its multiple cloning site, and we confirmed its functionality after transfection into eukaryotic cells in the Chinese hamster ovarian cell line [Flp-In-CHO (Invitrogen)] (CRL 12023)-ATCC and flow cytometry in vitro test. After oral administration of L. lactis (pExu:egfp) in Balb/C mice, we were able to detect green fluorescent enterocytes, thus showing its functionality in vivo.
Results
Construction of pExu
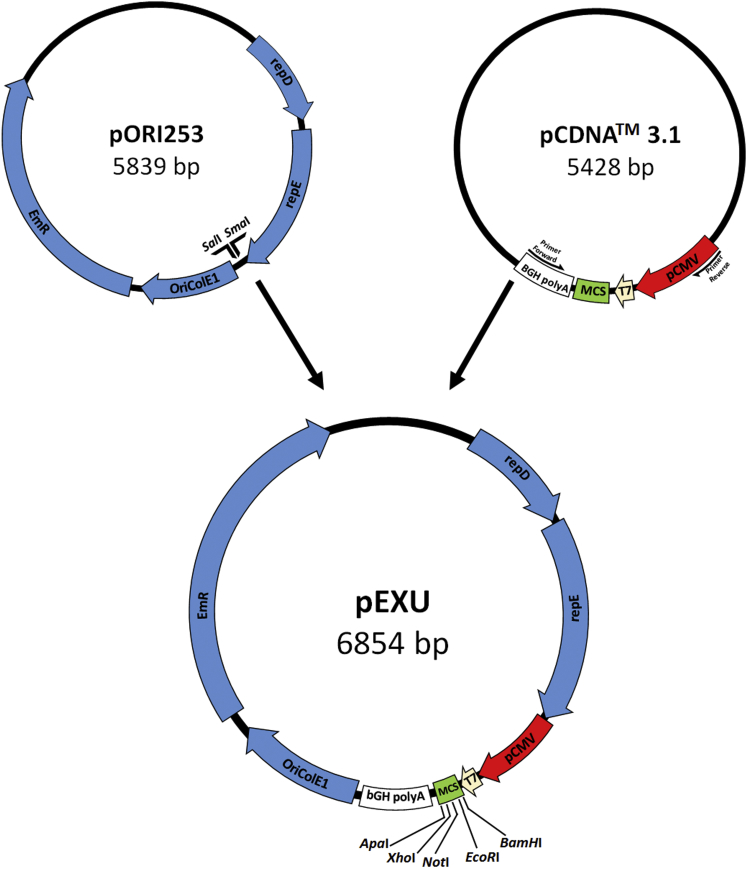
The shuttle pExu plasmid (6.854 Kb) was constructed using the PCR amplicon of PCDNA3.1, which was cloned in the pOri 253 plasmid as outlined in the Materials and Methods section. As illustrated in Figure 1, the new plasmid harbors an eukaryotic region containing the pCMV, a multiple cloning site (MCS), as well as the T7 primer binding site for sequencing and the polyadenylation signal of BGH polyA necessary for a correct mRNA maturation. The prokaryotic region contains repD/repE replication origin for L. lactis, OriColE1 replication origin for E. coli, and erythromycin resistance gene (Ery) for bacterial selection. The new plasmid was successfully stabilized in E. coli Top10 and in L. lactis subsp cremoris MG1363.
Figure 1.
Schematic Representation of pExu Plasmid Construction
pOri253 - RepD and RepE replication origin for L. lactis; OriColE1 replication origin for E. coli; and Ery; pCDNA3.1 - pCMV, the T7 promoter (T7); BGH polyA signal; MSC. Not to scale.
Stability of pExu in Bacterial Strains
This shuttle-cloning vector was successfully introduced into E. coli Top10; L. lactis MG1363; and Lactobacillus delbrueckii CNRZ 327 strains. L. delbrueckii CNRZ 327 strains were used in this study to show that the pExu vector can replicate in these strains as well as in Top10 and L. lactis. As well documented by Rocha and colleagues,18 L. delbrueckii subsp. lactis CNRZ 327 has in vitro and in vivo anti-inflammatory activity. Thereby, different strains of genetically modified (GM) LAB, with inherent anti-inflammatory, can have this characteristic increased and also new ones can be incorporated.19, 20 Plasmid analysis of transformants confirmed that this plasmid was able to be entered and replicated without any structural rearrangements after 120 hr for E. coli and 240 hr for L. lactis (data not shown).
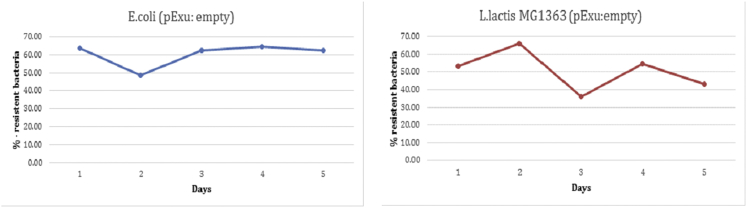
Structural and segregational analysis in E. coli and L. lactis MG1363 indicated that the vector pExu was maintained in 62% and in 42% of the cells for 135 generations in E. coli and 37.5 generations in L. lactis respectively, in absence of selective pressure (without erythromycin) as shown in Figure 2.
Figure 2.
Segregational Analysis of the Shuttle Vector pExu in E. coli and L. lactis MG1363
(Left) E. coli and L. lactis harboring pExu plasmid were cultured in LB medium or GM17, respectively, in the absence of selective pressure and plated both with and without antibiotics. The assays of selective pressure were done during 5 days (135 generations for E.coli and 37.5 generations for L. lactis). The percentages of total CFU on LB or GM17 without Ery were calculated according to the formula: %Ery-resistant colonies = (Ery-resistant CFU/total CFU) 100).
pExu:egfp Is Able to Express GFP In Vitro in Mammalian Cells
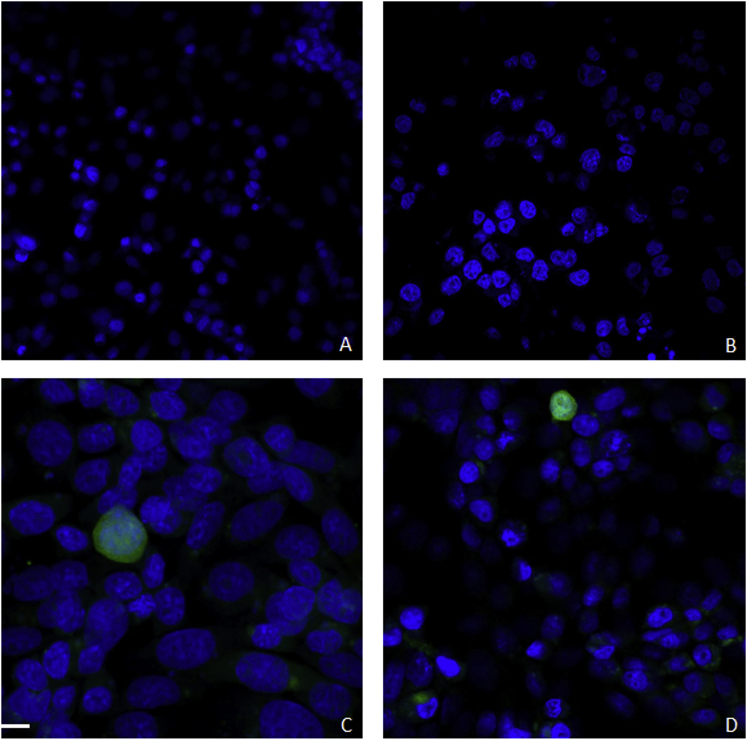
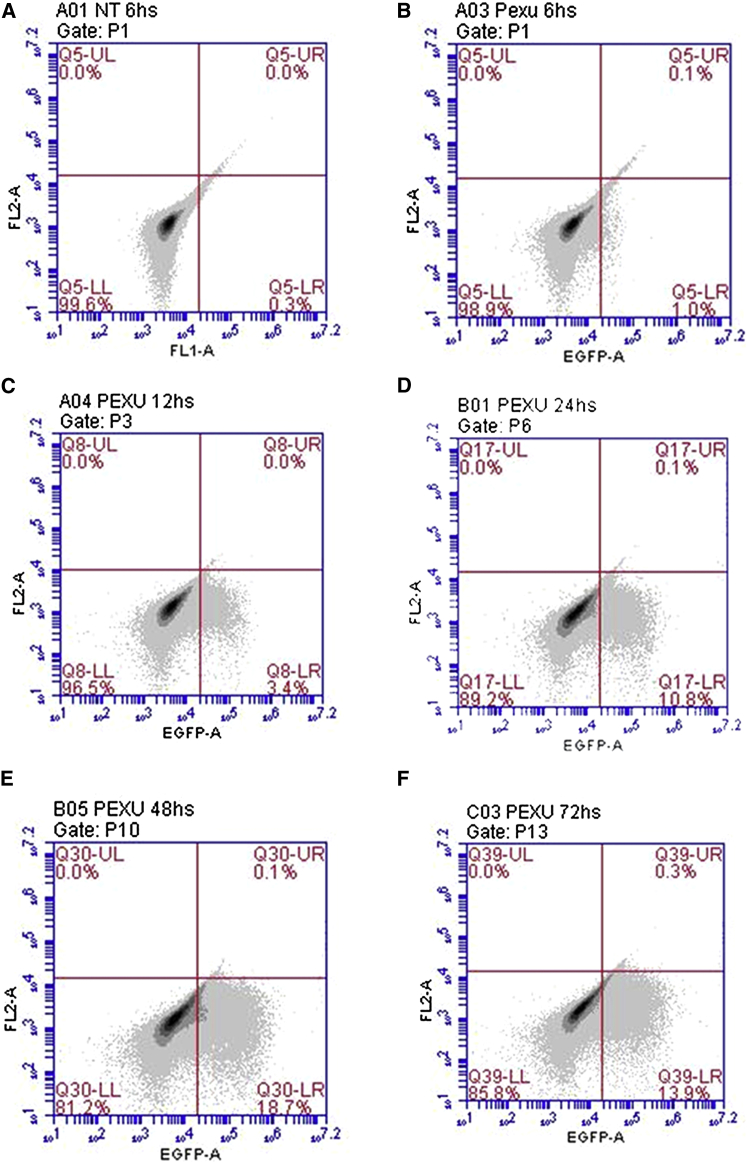
To assess the functionality of pExu plasmid, the egfp ORF was cloned into the MCS between XhoI and NotI enzyme sites. The structure of pExu:egfp was confirmed by PCR, enzymatic digestion, and sequencing. CHO cells were transfected with pExu:egfp. The confocal microscopy showed the expression of eGFP protein by eukaryotic cells (Figure 3). Also, the kinetic analysis by fluorescent microscopy after 6, 12, 24, 48, and 72 hr post-transfection showed that eGFP was expressed from 12 hr until at least 72 hr (Figure 4). The expression of eGFP protein by eukaryotic cells was quantified by flow cytometry at the same times, and the percentages of expressing cell were 1%, 3.4%, 10.8%, 18.7%, and 13.9%, respectively (Figure 5).
Figure 3.
Analysis of eGFP Expression by Eukaryotic Cells: eGFP Production by CHO Cells Were Evaluated by Confocal Microscopy
(A–D) Non-transfected CHO cells (negative control), transfected CHO cells with the pExu:empty plasmid, (negative control) (B), and transfected cells with the pExu:egfp (C and D). The cells were incubated with DAPI for nuclear staining. The images were captured using a Nikon Eclipse Ti with a C2 laser-scanning confocal with 40× (A, B, and D) and 63× (C) objective.
Figure 4.
eGFP Kinetics Expression by Transfected CHO Cells with pExu:egfp Vector
(A–F) Non-transfected cells and CHO cells transfected with pExu:egfp analyzed at 6, 12, 24, 48, and 72 hr post-transfection, respectively (B–F). The images were captured by fluorescence microscopy Carl Zeiss Axiovert 200 10×.
Figure 5.
Kinetic Expression of the eGFP Protein by CHO Cells Transfected with the pExu:egfp Plasmid
(A–F) Dot plot graphs: pExu:empty (negative control) (A); pExu:egfp transfected cell analyzed from 6 to 72 hr post-transfection (B–F). The dot plots showing the cell count on the y axis and the FL1 detector (Argon laser, 488 nm) and on the x axis are examined through flow cytometry (FACScan - BD Bioscience). The data were analyzed using the software BD CFlow.
L. lactis MG1363, Delivering the pExu:egfp Vector, Is Able to Express eGFP Protein after Oral Administration in Mice
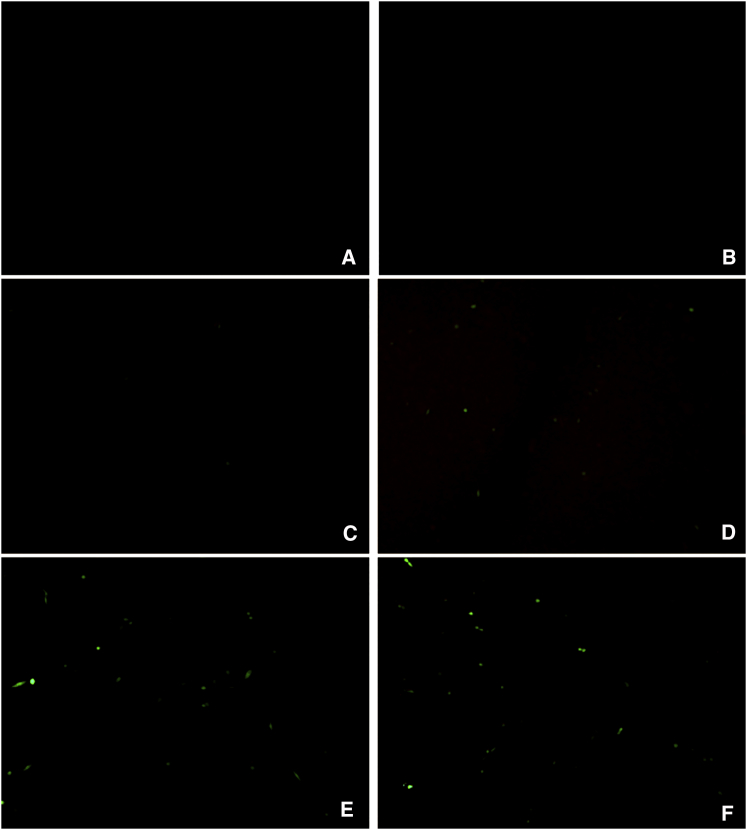
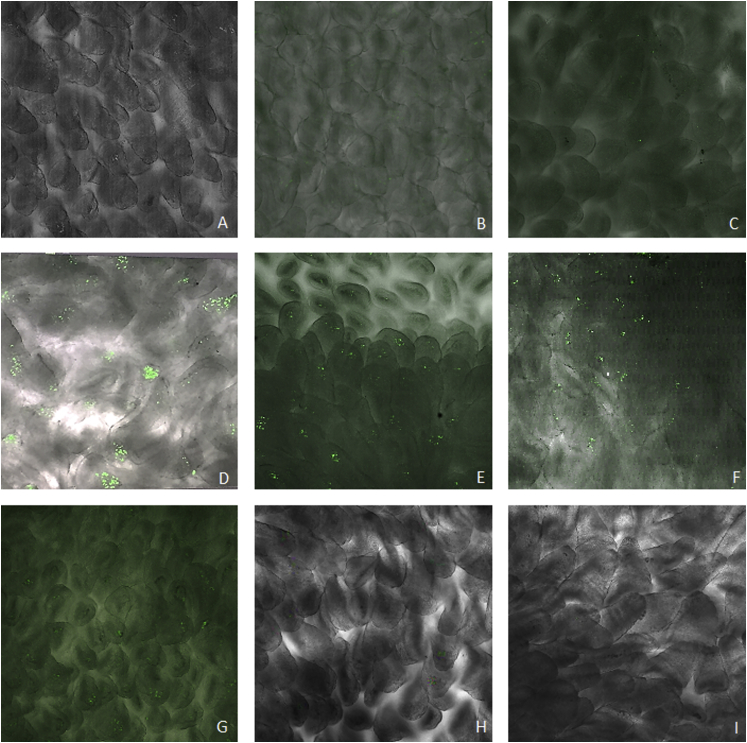
The oral administration of L. lactis MG1363 (pExu:egfp) to Balb/C mice elicited eGFP expression in the eukaryotic cells of the duodenal part of the gut 12, 24, 48, and 72 hr after gavage. It was not possible to detect eGFP expression until 6 hr and after 72 hr post gavage (Figure 6). Furthermore, we were not able to detect expression of eGFP protein by eukaryotic cells in the ileum portion at the same times points (data not shown).
Figure 6.
eGFP Production in Eukaryotic Cells, Duodenal Portion, of Mice Orally Administered with Recombinant L. lactis MG1363
(A) PBS group; (B) L. lactis MG1363 (pExu:empty); (C–I) L.lactis MG1363 (pExu:egfp) analyzed at different times post-oral administration (6, 12, 24, 48, 72, 96, and 120 hr, respectively). The images were obtained using a C2 Eclipse Ti confocal microscope (Nikon) with a 40× objective.
Discussion
Genetically modified LAB can be used as vectors for local delivery of biologically active molecules (protein or DNA) either to the gastrointestinal tract or other mucosal surfaces improving the targeting of recombinant antigens.11, 13, 21, 22, 23, 24 The first report using live recombinant L. lactis was in 1993 by Wells and colleagues.25 They showed that subcutaneous injection of L. lactis producing tetanus fragment C (TTFC) protected mice against a lethal challenge with tetanus toxin.25
Since L. lactis has been described as a vehicle to deliver DNA vaccines, different approaches and vectors to increase the delivery efficiency have been developed. In 2009, our group developed a plasmid to be used in E. coli and in L. lactis, called pValac vector.26 Different genes were cloned in this vector and used in native or invasive L. lactis strains.11, 13, 14, 27, 28 Even though this plasmid has interesting features, it has some limitations to be highlighted, for example, the only one gram-negative bacterial host is the E. coli TG1 strain (V.A., unpublished data); added to the RCR (rolling-circle replication) origin of this plasmid, offering instability in cloning process and also recombination processes.29
In 2011, Tao and colleagues30 showed by in vitro assays that the treatment with glycine, which is able to fragilize the cell wall of L. lactis, increases the uptake of L. lactis by mammalian cells and thus the plasmid delivery. Nonetheless, it was reported that integrity of the whole membrane structure is essential for fruitful delivery of DNA in eukaryotic cells.31
Another intelligent approach using L. lactis as an efficient vehicle vector was done by Yagnick and colleagues,32 where they constructed a pPERDBY plasmid, which contains the egfp gene besides the multiple cloning site. This strategy allowed the possibility to clone the gene of interest in frame with a reporter gene and to evaluate the expression of the target gene by simple observation of the reporter. They did not use any chemical enhancers. In fact, this plasmid has an RCR origin; it is not considered to have such a high stability as a plasmid with theta origin.16, 33 They found good results in efficiency transferring this plasmid in human intestinal Caco-2 cells. However, they did not test its functionality in in vivo experiments. Moreover, as we showed in this report, the results can be different due to different environments and conditions.
In this report, we presented a new shuttle vector, called pExu, to be used in native L. lactis and other species. This vector is suitable for use in E. coli, in L. lactis, as well as in other LAB such as Lactococcus spp., Lactobacillus spp., Pediococcus sp., and, moreover, many other gram-positive bacteria. pExu comes from a pOri253 plasmid, a derivate of pAMβ1, which is considered as a large conjugative plasmid replicating by theta mechanisms.34, 35, 36 It offers thus higher segregational stability than RCR plasmids and, for this reason, can accommodate and maintain large DNA inserts.16, 33
Our results confirmed that the pExu plasmid carried either by E. coli or by L. lactis showed an exceptional structural stability. There were no structural changes after 120 and 240 hr for E. coli and L. lactis, respectively. Moreover, this plasmid has shown a high segregational stability. Then, we cloned the egfp ORF successfully in the pExu vector.
The confocal analysis, as well as flow cytometry performed in this study with transfected CHO cells, revealed the functionality of the pExu:egfp vector. The mammalian cells were capable of producing the reporter protein. Our images obtained by confocal or fluorescence microscopy after transfection showed a low expression of eGFP protein by mammalian cells. We awarded these results to the large size of the new plasmid (6,854 bp) added to the reagent for transfection applied to in vitro assays.
Although the eGFP expression was low in in vitro tests, we decided to transform L. lactis MG1363 with the pExu:egfp vector. The epithelial cells of the duodenal region of mice orally gavaged with L. lactis MG1363 (pExu:egfp) were able to express the eGFP protein. It was not possible to see the expression of eGFP protein in epithelial cells of the ileum region and colon (data not shown). These results were not in accordance with Almeida and colleagues.37 They describe that the administration of invasive recombinant L. lactis FnBPA+ delivering the pValac:gfp vector in mice allowed the GFP expression both on small and large intestine cells, as the authors describe in their report, the expression of FnBPA protein can help to the DNA delivery by this LAB. Our results allowed to confirm that the oral administration of L. lactis MG1363 (pExu:egfp) was able to produce recombinant protein expression in eukaryotic host cells in the duodenal region of the small intestine with good expression.
L. lactis has a passive transit through the digestive tract, less than 24 hr for mice and about 3 days for human, and it is amply reported that this bacterium does not colonize the intestine being only for delivery.38, 39 In 2008, Chatel and colleagues11 showed the capture of bacterial DNA by the epithelial membrane. To show that the bacterial capture and the protein expression is a transient process, we evaluated the eGFP expression by the host cells from 6 to 168 hr after mice gavage. We were able to see expression only between 12 and 72 hr after gavage, confirming the previous observation by Chatel and colleagues in 2008.11
Related to the 6 hr time point, it is likely a too short time for an eukaryotic cell to do the transcription, translation, and post-translational processing to express the eGFP protein in in vivo systems. Nonetheless, the no expression in the 96 to 168 hr is due to the cell extrusion process. The particularly short lifetime of IECs (intestinal epithelial cells) had shown a rebirth of the functional villus epithelium by the stem cells of the crypts every 2 to 6 days in the greatest adults mammals.40 For that reason, we are not able to see eGFP protein in the IECs of a duodenal portion after 96 hr after gavage in mice. Even more, the best turnover rates of a fixed-cell population in the body are the enterocytes.41 In this report, we can show that the egfp ORF under the control of an eukaryotic promoter carried by L. lactis MG1363 could be delivered into and expressed by host epithelial cells in the duodenal portion.
The use of food grade LAB for oral DNA vaccines is a safer alternative than the use of attenuated pathogenic bacteria, such as Shigella, Salmonella, Yersinia, and Listeria for DNA-delivering into mammalian cells.42, 43 To this end, we have reported the construction of a new shuttle vector called pExu for DNA delivery to be used in food grade bacteria, like L. lactis. Although the vector has the erythromycin resistance marker, which is not approved by the FDA, this resistance marker could be exchanged by kanamycin as well as eliminated using auxotrophic systems. The results here presented are a “proof of concept” about a design of a new vector for LAB. The in vivo results here obtained encouraged us to test it in disease models as well as in gene therapy.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacteria and plasmids used in this study are listed in Table 1. E. coli Top10 strains were aerobically grown in Luria-Bertani (LB) medium (Acumedia), incubated at 37°C with vigorous shaking. L. lactis subsp. cremoris MG1363 was grown in M17 medium (Sigma-Aldrich) containing 0.5% glucose (Synth) (GM17) at 30°C without agitation. Antibiotics were added at the indicated concentrations as necessary: kanamycin (Sigma-Aldrich) 50 μg/mL for E. coli and erythromycin (Sigma-Aldrich) 500 μg/mL for both E. coli and L. lactis.
Table 1.
Bacterial Strains and Plasmids Used in this Work
| Bacterial Strain and Plasmids | Characteristics | References |
|---|---|---|
| Escherichia coli TOP10 | E. coli K-12-derived strain; F- mcrA Δ (mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ (ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ− | Invitrogen |
| Lactococcus lactis MG1363 | L. lactis MG1363 lactis subsp. cremoris | Gasson44 |
| Lactococcus lactis MG1363 (pExu:egfp) | L. lactis MG1363 strain carrying the: pExu:egfp plasmid | this work |
| Lactobacillus delbrueckii CNRZ327 | L. delbrueckii CNRZ327 | INRA collection |
| pExu:empty | eukaryotic expression vector (pCMV/Eryr/RepA/RepC) | this work |
All pure cultures of bacteria were kept as stock cultures in 40% glycerol (Sigma-Aldrich) for E. coli and 25% glycerol for L. lactis at −80°C.
DNA Manipulations
DNA manipulation protocols were performed as described previously in Sambrook and colleagues45 with slight modifications. For plasmid DNA extraction from L. lactis, TES buffer (25% sucrose, 1 mM EDTA, and 50 mM Tris-HCl pH 8) containing lysozyme (Sigma-Aldrich) (10 mg/mL) was added for 1 hr at 37°C to prepare protoplasts. Enzymes were used as recommended by suppliers. Transformation of L. lactis was performed by means of electroporation as described in Langella and colleagues.46 L. lactis transformants were plated on GM17 agar plates containing the required antibiotic and were counted after 48 hr incubation at 30°C.
Construction of a New Shuttle Vector, pExu
The pExu vector is the fusion of two regions. The eukaryotic gene expression region comes from pCDNA3.1 vector (Invitrogen) (Table 1) containing a pCMV, a multiple cloning site (BamHI, EcoRI, NotI, XhoI, and ApaI), and a polyA tail. For this proposal, a 1,020 bp fragment was generated by PCR with a proofreading DNA polymerase (Kapa Biosystems) and the oligonucleotides PCMVFwd (5′ CCCGGGTTGACATTGATTATTGAC 3′) and PCMVRev (5′ GTCGACCCATAGAGCCCACCGCAT 3′), introducing, respectively, a SmaI and SalI (Invitrogen) (underlined) site in the fragment. The reaction mixture (final volume of 25 μL) contained: 100 ng template DNA; 1 unit of DNA polymerase; 5.0 μL Buffer Kapa CG 5X; deoxynucleoside triphosphates 25 mM, and primer forward and reverse 10 mM each. The thermal cycling program used was as follows: initial denaturation at 95°C for 3 min, 20 cycles of 98°C for 20 s, 60°C for 15 s, and 72°C for 30 s. Finally, there was an extension step at 72°C for 2 min. After analyzing the correct size and purity on 1.0% agarose gel, the PCR product was cloned into Zero Blunt TOPO vector (Invitrogen) (Table 1) getting the Topo:CMV, this construction was established by transformation in E.coli Top10 (Table 1). The nucleotide sequence of the insert was confirmed by sequencing using BigDye Terminator v3.1 no ABI 3500 (Thermo Scientific).47 Then, Topo:CMV was digested with SmaI and SalI restriction enzymes and gel purified (Kit illustra GFX PCR DNA and Gel Band Purification-GE Healthcare). The prokaryotic region of pExu plasmid comes from the pOri253 vector.15 For this proposal, pOri253 plasmid was digested with a SmaI and SalI restriction enzyme. This prokaryotic vector provides repD and repE genes responsible for replication in either enterococci or lactobacilli, as also Ery. Then the SmaI/SalI digested and purified Topo:CMV and pORI253 fragments were ligated by T4 DNA ligase (Invitrogen) to obtain the pExu vector (6,854 bp) (Table1; Figure 1). It was established by transformation in E. coli Top10 as an intermediate host and then in L. lactis MG1363 strains.
Determination of Segregational and Structural Stability of pExu Plasmid
The segregational stability of the pExu plasmid was examined by overnight growth in LB medium for E. coli and in GM17 medium for L. lactis in the presence or absence of antibiotic during 5 days. When the culture, DO600nm, reached the first point to the stationary phase, 0.1 mL of each culture was plated on LB agar containing Ery for E. coli and GM17 agar with Ery for L. lactis to determine the number of Ery resistant colonies. The Ery resistant colonies were verified as a percentage of total colony forming units (CFU) on LB or GM17 without Ery (i.e., %Ery-resistant colonies = (Ery-resistant CFU/CFU total) 100).
The plasmid structural stability was tested by restriction analysis of plasmid isolated from colonies grown in LB medium for E. coli and in GM17 medium for L. lactis during 5 days.
Cloning Reporter Gene into pExu
In order to evaluate the functionality of pExu vector, the egfp ORF was cloned into it. The egfp ORF was amplified by PCR from pRock-eGFP plasmid with a proofreading DNA polymerase (Kapa Biosystem) using PeGFPFwd (5′ GGCGCGGCCGCAATGGTGAGCAAGGGCGAGGAG3′) and PeGFPRev (5′ GGCCTCGAGCTAGCTACTTGTACAGCTCGTC3′) oligonucleotides introducing the NotI sites in forward primers and XhoI sites in reverse primers (underlined).48 The reaction mixture (final volume of 25 μL) contained: 100 ng template DNA; 1 unit of DNA polymerase; 5.0 μL Buffer Kapa CG 5X; deoxynucleoside triphosphates 25 mM, and primer forward and reverse 10 mM each. The thermal cycling program used was as follows: initial denaturation at 95°C for 3 min, 25 cycles of 98°C for 15 s, 67°C for 20 s, and 72°C for 30 s. Finally, there was an extension step at 72°C for 2 min. After analyzing the correct size and purity on 1.0% agarose gel, the PCR product was cloned into Zero Blunt TOPO vector (Invitrogen) (Table 1) getting the Topo:egfp, this construction was established by transformation in E. coli Top10 (Table 1). The nucleotide sequence of the insert was confirmed by sequencing using BigDye Terminator v3.1 no ABI 3500 (Thermo Scientific).47 Then, Topo:egfp was digested with NotI and XhoI (Invitrogen) restriction enzymes and gel purified (Kit illustra GFX PCR DNA and Gel Band Purification-GE Healthcare). The 744 bp egfp ORF fragment was inserted into pExu MCS using the same restriction enzymes mentioned before, resulting in pExu:egfp (7,598 bp). It was established by transformation in E. coli Top10 as an intermediate host and then in L. lactis MG1363 strains.
Transfection Assays of Mammalian CHO Cells with pExu:egfp by Confocal Analyses
The pExu:egfp plasmid was examined for eGFP expression by transfection into a Chinese hamster ovary cell line (Flp-In-CHO [Invitrogen]). CHO cells were cultured in F12 Ham media (Sigma-Aldrich) supplemented with 10% fetal calf serum (Gibco), 1% L-glutamine (Sigma-Aldrich), 100 ng/mL Zeocin (Invitrogen), and 2.5% HEPES (Sigma-Aldrich). The 90% to 95% confluent CHO cells were then transfected with 4 μg of pExu:egfp, (Table 1), pExu:empty, and no plasmid (negative controls) with Lipofectamine 2000 (Invitrogen), according to supplier’s recommendation. After 48 hr, cells were washed with PBS solution, fixed for 15 min with paraformaldehyde (Sigma-Aldrich; 4% in PBS), and permeabilized with Triton X-100 (Sigma-Aldrich; 0.1% in PBS) for 10 min at room temperature. The cells were then incubated with a 1/500 dilution of DAPI (Invitrogen; 2 μg/mL) for 60 min at room temperature in the dark. After washing, the samples were mounted and the images were captured using a Nikon Eclipse Ti with a C2 laser-scanning confocal head (http://www.nikoninstruments.com/Products/Microscope-Systems/Inverted-Microscopes/Eclipse-Ti-E) equipped with three different lasers (excitation at 405, 488, and 543 nm) and emission band-pass filters at 450/50 (channel 1), 515/30 (channel 2), and 584/50 nm (channel 3). Images of each sample were collected and analyzed by NSI-Elements version 4.20 (Nikon) to drive the microscope and image acquisition.
Kinetic Analysis of eGFP Expression by Immunofluorescence
For kinetic analysis of eGFP expression, eukaryotic cells were transfected with pExu:egfp and Lipofectamine 2000, as mentioned above. The pExu:empty and non-transfected cells were used as negative controls. The expression of eGFP protein was analyzed at 6, 12, 24, 48, and 72 hr after transfection by epifluorescent microscope (Zeiss Axiovert 200) and by flow cytometry (BD Accuri C6 Flow Cytometer). Transfection assays were performed in duplicate.
Mice Handling: Administration of Recombinant L. lactis MG1363 Strain into Mice Balb/C
Conventional female Balb/C commonly inbred mice, 5–6 weeks of age, were obtained from Centro de Bioterismo (CEBIO) of Universidade Federal de Minas Gerais (UFMG-Belo Horizonte, Brazil). Procedures and manipulation of animals followed the rules of the Ethical Principles in Animal Experimentation, approved by the Ethics Committee on Animal Experimentation (CEUA/UFMG/Brazil). Animals were housed under normal husbandry conditions. Mice were kept in collective cages (four animals/cage) in an environmentally controlled room with a 12 hr light/dark cycle and given free access to water and food. We used different experimental groups: PBS group, L. lactis MG1363 (pExu:empty) as negative controls, and L. lactis MG1363 (pExu:egfp). There were two independent experiments that were performed with 36 mice in each group, altogether, we used 108 animals. The animals were separated (four animals/cage) according to each evaluated time. The strains were orally administrated to the mice by gavage with 109 CFU bacterial suspensions in a final volume of 100 μL of PBS. The administration was at one time (zero time) and after 6, 12, 24, 48, 72, 96, 120, 144, and 168 hr the animals were euthanized, and the duodenum (proximal and distal portion) and the ileum parts of animals were analyzed by fluorescence microscopy using a C2 Eclipse Ti confocal microscope (Nikon). The Images were analyzed with Volocity 3D Image Analysis Software (PerkinElmer).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 6. Statistical significance between the groups was calculated using a one-way ANOVA test, followed by the Bonferroni post test. A 95% confidence limit was considered to be significant at a value of p < 0.05.
Author Contributions
Conceptualization, P.M.-A. and F.L.R.C.; Methodology, P.M.-A. and F.L.R.C.; Investigation, P.M.-A., F.L.R.C., M.M.D., J.S.C.S., and M.M.S.; Writing - Original Draft, P.M.-A. and M.M.D.; Writing - Review & Editing, P.M.-A., M.M.D., F.V., J.-M.C., S.Y.L., and V.A.; Funding Acquisition, F.V. and V.A.; and Supervision, V.A.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This work was financially supported by the grants funding the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq/405233/2016-7), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES/88887.094350/201500), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG/02096-15).
References
- 1.LeBlanc J.G., Aubry C., Cortes-Perez N.G., de Moreno de LeBlanc A., Vergnolle N., Langella P., Azevedo V., Chatel J.M., Miyoshi A., Bermúdez-Humarán L.G. Mucosal targeting of therapeutic molecules using genetically modified lactic acid bacteria: an update. FEMS Microbiol. Lett. 2013;344:1–9. doi: 10.1111/1574-6968.12159. [DOI] [PubMed] [Google Scholar]
- 2.Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front. Cell. Infect. Microbiol. 2012;2:86. doi: 10.3389/fcimb.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin A., Quinquis B., Renault P., Sorokin A., Ehrlich S.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 2004;22:1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pontes D.S., de Azevedo M.S.P., Chatel J.M., Langella P., Azevedo V., Miyoshi A. Lactococcus lactis as a live vector: heterologous protein production and DNA delivery systems. Protein Expr. Purif. 2011;79:165–175. doi: 10.1016/j.pep.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Wells J. Mucosal vaccination and therapy with genetically modified lactic acid bacteria. Annu. Rev. Food Sci. Technol. 2011;2:423–445. doi: 10.1146/annurev-food-022510-133640. [DOI] [PubMed] [Google Scholar]
- 6.Bermúdez-Humarán L.G., Aubry C., Motta J.P., Deraison C., Steidler L., Vergnolle N., Chatel J.M., Langella P. Engineering lactococci and lactobacilli for human health. Curr. Opin. Microbiol. 2013;16:278–283. doi: 10.1016/j.mib.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Cano-Garrido O., Rueda F.L., Sànchez-García L., Ruiz-Ávila L., Bosser R., Villaverde A., García-Fruitós E. Expanding the recombinant protein quality in Lactococcus lactis. Microb. Cell Fact. 2014;13:167. doi: 10.1186/s12934-014-0167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loessner H., Weiss S. Bacteria-mediated DNA transfer in gene therapy and vaccination. Expert Opin. Biol. Ther. 2004;4:157–168. doi: 10.1517/14712598.4.2.157. [DOI] [PubMed] [Google Scholar]
- 9.Schoen C., Stritzker J., Goebel W., Pilgrim S. Bacteria as DNA vaccine carriers for genetic immunization. Int. J. Med. Microbiol. 2004;294:319–335. doi: 10.1016/j.ijmm.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Beláková J., Horynová M., Krupka M., Weigl E., Raska M. DNA vaccines: are they still just a powerful tool for the future? Arch. Immunol. Ther. Exp. (Warsz.) 2007;55:387–398. doi: 10.1007/s00005-007-0044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatel J.M., Pothelune L., Ah-Leung S., Corthier G., Wal J.-M., Langella P. In vivo transfer of plasmid from food-grade transiting lactococci to murine epithelial cells. Gene Ther. 2008;15:1184–1190. doi: 10.1038/gt.2008.59. [DOI] [PubMed] [Google Scholar]
- 12.Guimarães V.D., Gabriel J.E., Lefèvre F., Cabanes D., Gruss A., Cossart P., Azevedo V., Langella P. Internalin-expressing Lactococcus lactis is able to invade small intestine of guinea pigs and deliver DNA into mammalian epithelial cells. Microbes Infect. 2005;7:836–844. doi: 10.1016/j.micinf.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Innocentin S., Guimarães V., Miyoshi A., Azevedo V., Langella P., Chatel J.M., Lefèvre F. Lactococcus lactis expressing either Staphylococcus aureus fibronectin-binding protein A or Listeria monocytogenes internalin A can efficiently internalize and deliver DNA in human epithelial cells. Appl. Environ. Microbiol. 2009;75:4870–4878. doi: 10.1128/AEM.00825-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Azevedo M., Karczewski J., Lefévre F., Azevedo V., Miyoshi A., Wells J.M., Langella P., Chatel J.M. In vitro and in vivo characterization of DNA delivery using recombinant Lactococcus lactis expressing a mutated form of L. monocytogenes Internalin A. BMC Microbiol. 2012;12:299. doi: 10.1186/1471-2180-12-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lertcanawanichakul M. Construction of plasmid vector for expression of bacteriocin N15-encoding gene and effect of engineered bacteria on Enterococcus faecalis. Curr. Microbiol. 2007;54:108–112. doi: 10.1007/s00284-006-0186-3. [DOI] [PubMed] [Google Scholar]
- 16.Simon D., Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 17.Lutz C.S. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem. Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- 18.Santos Rocha C., Gomes-Santos A.C., Garcias Moreira T., de Azevedo M., Diniz Luerce T., Mariadassou M., Longaray Delamare A.P., Langella P., Maguin E., Azevedo V. Local and systemic immune mechanisms underlying the anti-colitis effects of the dairy bacterium Lactobacillus delbrueckii. PLoS ONE. 2014;9:e85923. doi: 10.1371/journal.pone.0085923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Carmen S., de Moreno de LeBlanc A., Martin R., Chain F., Langella P., Bermúdez-Humarán L.G., LeBlanc J.G. Genetically engineered immunomodulatory Streptococcus thermophilus strains producing antioxidant enzymes exhibit enhanced anti-inflammatory activities. Appl. Environ. Microbiol. 2014;80:869–877. doi: 10.1128/AEM.03296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.del Carmen S., Miyoshi A., Azevedo V., de Moreno de LeBlanc A., LeBlanc J.G. Evaluation of a Streptococcus thermophilus strain with innate anti-inflammatory properties as a vehicle for IL-10 cDNA delivery in an acute colitis model. Cytokine. 2015;73:177–183. doi: 10.1016/j.cyto.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 21.Steidler L., Hans W., Schotte L., Neirynck S., Obermeier F., Falk W., Fiers W., Remaut E. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–1355. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 22.Ravnikar M., Štrukelj B., Obermajer N., Lunder M., Berlec A. Engineered lactic acid bacterium Lactococcus lactis capable of binding antibodies and tumor necrosis factor alpha. Appl. Environ. Microbiol. 2010;76:6928–6932. doi: 10.1128/AEM.00190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daniel C., Roussel Y., Kleerebezem M., Pot B. Recombinant lactic acid bacteria as mucosal biotherapeutic agents. Trends Biotechnol. 2011;29:499–508. doi: 10.1016/j.tibtech.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Berlec A., Završnik J., Butinar M., Turk B., Štrukelj B. In vivo imaging of Lactococcus lactis, Lactobacillus plantarum and Escherichia coli expressing infrared fluorescent protein in mice. Microb. Cell Fact. 2015;14:181. doi: 10.1186/s12934-015-0376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells J.M., Wilson P.W., Norton P.M., Gasson M.J., Le Page R.W. Lactococcus lactis: high-level expression of tetanus toxin fragment C and protection against lethal challenge. Mol. Microbiol. 1993;8:1155–1162. doi: 10.1111/j.1365-2958.1993.tb01660.x. [DOI] [PubMed] [Google Scholar]
- 26.Guimarães V., Innocentin S., Chatel J.M., Lefèvre F., Langella P., Azevedo V., Miyoshi A. A new plasmid vector for DNA delivery using lactococci. Genet. Vaccines Ther. 2009;7:4. doi: 10.1186/1479-0556-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.del Carmen S., Martín Rosique R., Saraiva T., Zurita-Turk M., Miyoshi A., Azevedo V., de Moreno de LeBlanc A., Langella P., Bermúdez-Humarán L.G., LeBlanc J.G. Protective effects of lactococci strains delivering either IL-10 protein or cDNA in a TNBS-induced chronic colitis model. J. Clin. Gastroenterol. 2014;48(Suppl 1):S12–S17. doi: 10.1097/MCG.0000000000000235. [DOI] [PubMed] [Google Scholar]
- 28.Pereira V.B., Zurita-Turk M., Saraiva T.D.L., Castro C.P.D., Souza B.M., Agresti P.M., Lima F.A., Pfeiffer V.N., Azevedo M.S.V., Rocha C.S., Pontes D.S. DNA vaccines approach: From concepts to applications. World J. Vaccines. 2014;4:50–71. [Google Scholar]
- 29.del Solar G., Giraldo R., Ruiz-Echevarría M.J., Espinosa M., Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao L., Pavlova S.I., Ji X., Jin L., Spear G. A novel plasmid for delivering genes into mammalian cells with noninvasive food and commensal lactic acid bacteria. Plasmid. 2011;65:8–14. doi: 10.1016/j.plasmid.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker T., Pfannschmidt S., Guiard B., Stojanovski D., Milenkovic D., Kutik S., Pfanner N., Meisinger C., Wiedemann N. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J. Biol. Chem. 2008;283:120–127. doi: 10.1074/jbc.M706997200. [DOI] [PubMed] [Google Scholar]
- 32.Yagnik B., Padh H., Desai P. Construction of a new shuttle vector for DNA delivery into mammalian cells using non-invasive Lactococcus lactis. Microbes Infect. 2016;18:237–244. doi: 10.1016/j.micinf.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 33.Kiewiet R., Kok J., Seegers J.F.M.L., Venema G., Bron S. The mode of replication is a major factor in segregational plasmid instability in Lactococcus lactis. Appl. Environ. Microbiol. 1993;59:358–364. doi: 10.1128/aem.59.2.358-364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swinfield T.J., Oultram J.D., Thompson D.E., Brehm J.K., Minton N.P. Physical characterisation of the replication region of the Streptococcus faecalis plasmid pAM beta 1. Gene. 1990;87:79–90. [PubMed] [Google Scholar]
- 35.von Wright A., Sibakov M. Genetic modification of lactic acid bacteria. In: von Wright A., editor. Lactic Acid Bacteria. CRC Press; 1998. pp. 161–198. [Google Scholar]
- 36.Shareck J., Choi Y., Lee B., Miguez C.B. Cloning vectors based on cryptic plasmids isolated from lactic acid bacteria: their characteristics and potential applications in biotechnology. Crit. Rev. Biotechnol. 2004;24:155–208. doi: 10.1080/07388550490904288. [DOI] [PubMed] [Google Scholar]
- 37.Almeida J.F., Mariat D., Azevedo V., Miyoshi A., de Moreno de LeBlanc A., Del Carmen S., Martin R., Langella P., LeBlanc J.G., Chatel J.M. Correlation between fibronectin binding protein A expression level at the surface of recombinant lactococcus lactis and plasmid transfer in vitro and in vivo. BMC Microbiol. 2014;14:248. doi: 10.1186/s12866-014-0248-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gruzza M., Fons M., Ouriet M.F., Duval-Iflah Y., Ducluzeau R. Study of gene transfer in vitro and in the digestive tract of gnotobiotic mice from Lactococcus lactis strains to various strains belonging to human intestinal flora. Microb. Releases. 1994;2:183–189. [PubMed] [Google Scholar]
- 39.Klijn N., Weerkamp A.H., de Vos W.M. Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl. Environ. Microbiol. 1995;61:2771–2774. doi: 10.1128/aem.61.7.2771-2774.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayhew T.M., Myklebust R., Whybrow A., Jenkins R. Epithelial integrity, cell death and cell loss in mammalian small intestine. Histol. Histopathol. 1999;14:257–267. doi: 10.14670/HH-14.257. [DOI] [PubMed] [Google Scholar]
- 41.Gelberg H.B. Fourth Edition. Mosby; 2007. Pathologic Basis of Veterinary Disease. [Google Scholar]
- 42.Wells J.M., Mercenier A. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat. Rev. Microbiol. 2008;6:349–362. doi: 10.1038/nrmicro1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin I.Y.C., Van T.T.H., Smooker P.M. Live-attenuated bacterial vectors: Tools for vaccine and therapeutic agent delivery. Vaccines (Basel) 2015;3:940–972. doi: 10.3390/vaccines3040940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gasson M.J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J., Russell D.W. Third Edition. Cold Spring Harbor Press; 2001. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- 46.Langella P., Le Loir Y., Ehrlich S.D., Gruss A. Efficient plasmid mobilization by pIP501 in Lactococcus lactis subsp. lactis. J. Bacteriol. 1993;175:5806–5813. doi: 10.1128/jb.175.18.5806-5813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DaRocha W.D., Silva R.A., Bartholomeu D.C., Pires S.F., Freitas J.M., Macedo A.M., Vazquez M.P., Levin M.J., Teixeira S.M. Expression of exogenous genes in Trypanosoma cruzi: improving vectors and electroporation protocols. Parasitol. Res. 2004;92:113–120. doi: 10.1007/s00436-003-1004-5. [DOI] [PubMed] [Google Scholar]