Abstract
Gene editing by homology-directed recombination (HDR) can be used to couple delivery of a therapeutic gene cassette with targeted genomic modifications to generate engineered human T cells with clinically useful profiles. Here, we explore the functionality of therapeutic cassettes delivered by these means and test the flexibility of this approach to clinically relevant alleles. Because CCR5-negative T cells are resistant to HIV-1 infection, CCR5-negative anti-CD19 chimeric antigen receptor (CAR) T cells could be used to treat patients with HIV-associated B cell malignancies. We show that targeted delivery of an anti-CD19 CAR cassette to the CCR5 locus using a recombinant AAV homology template and an engineered megaTAL nuclease results in T cells that are functionally equivalent, in both in vitro and in vivo tumor models, to CAR T cells generated by random integration using lentiviral delivery. With the goal of developing off-the-shelf CAR T cell therapies, we next targeted CARs to the T cell receptor alpha constant (TRAC) locus by HDR, producing TCR-negative anti-CD19 CAR and anti-B cell maturation antigen (BCMA) CAR T cells. These novel cell products exhibited in vitro cytolytic activity against both tumor cell lines and primary cell targets. Our combined results indicate that high-efficiency HDR delivery of therapeutic genes may provide a flexible and robust method that can extend the clinical utility of cell therapeutics.
Keywords: chimeric antigen receptor, gene editing, T cell, TCR, CCR5, CD19, BCMA, cell therapy
Introduction
Among the most promising cell therapies currently in clinical trials are chimeric antigen receptor (CAR) T cells, which express an artificial receptor that redirects the effector functions of a T cell to a desired antigen through an HLA-independent targeting moiety, typically a single-chain antibody fragment (scFv), coupled to one or more intracellular signaling and co-stimulatory domains.1, 2 There are currently dozens of ongoing phase I and II clinical trials using anti-CD19, anti-BCMA, and other CARs to treat hematologic and solid tumor malignancies.3 CAR gene delivery to T cells in these trials is accomplished using randomly integrating retroviruses including γ-retroviral4, 5, 6, 7 or lentiviral vectors,8, 9 transposons,10, 11 or by transient mRNA transfection.12, 13
Recent advances in gene editing techniques allow highly efficient modifications of specific genetic loci in primary human cells, expanding the options for engineering cellular therapeutics.14, 15 We and others have previously shown a method for high-efficiency integration of functional gene cassettes by homology-directed recombination (HDR) in primary human hematopoietic cells at target loci including CCR5, AAVS1, and CD40L.16, 17, 18, 19 After DNA cleavage of the target sequence using an engineered nuclease, a gene expression cassette, flanked by regions of target gene homology, is delivered at a high copy number using adeno-associated virus (AAV). The endogenous DNA damage response machinery, activated by the double-stranded DNA break, then seamlessly integrates the gene expression cassette at the break site by HDR. If desired, the target site and template can be designed such that introduction of the engineered gene cassette simultaneously disrupts expression of the endogenous gene.
HDR gene editing to deliver a CAR could be used strategically to enhance the treatment potential for CAR T cells by coupling CAR delivery to other modifications of the T cell genome. For example, HIV-positive (HIV+) patients are at increased risk for B cell lymphomas,20 and the extended use of anti-retroviral therapies (ART) is also associated with an increased risk of plasma cell disorders.21, 22 These malignancies are theoretically targetable by anti-CD19 and anti-BCMA CAR T cells, respectively. However, HIV+ patients are excluded from ongoing clinical trials,9 in part due to the inherent vulnerability of a T cell therapy to HIV infection. Because null mutations within the gene encoding an HIV-1 co-receptor, CCR5, render T cells resistant to infection by the most prevalent strains of HIV-1,23, 24, 25 disruption of CCR5 by HDR delivery of anti-CD19- or anti-BCMA-CAR expression cassettes into the CCR5 locus could be an advantageous strategy for treating B cell and plasma cell neoplasms in these patients. Importantly, the adoptive transfer of cells with nuclease-induced CCR5 disruption has been used in clinical trials for HIV therapy with an acceptable safety profile.26
Another candidate locus for this simultaneous gene delivery/knockout approach is the T cell receptor (TCR) of αβ T cells. The alpha and beta chains of the TCR are expressed as heterodimers on the cell surface, and mutation of only one of these chains is necessary for disruption of surface TCR expression.27 HDR-mediated introduction of a CAR expression cassette in the T cell receptor alpha constant (TRAC) locus could thus be used to create TCR-negative CAR T cells that have modifications at a single genetic locus. A TCR-negative cell therapy could enable production of an off-the-shelf allogeneic immunotherapy product, improving cost effectiveness, and enabling use in patients with few or low-functioning T cells.28 Here, we demonstrate the efficiency and effectiveness of this approach to generate gene-targeted CAR T cells with concurrent disruption in these clinically relevant candidate genes.
Results
We have previously published the introduction of an anti-CD19 CAR construct at the CCR5 locus by HDR-based gene editing, achieving rates of 10%–15% CAR integration via HDR in primary human T cells.18 To test in vitro function of HDR-delivered CAR constructs, we generated fluorescent protein-expressing anti-CD19CAR T cells using two methods: lentiviral delivery (LV CD19CAR-BFP) and HDR mediated by a CCR5 megaTAL nuclease and an AAV donor template with flanking regions of CCR5 homology (AAV CCR5 CD19CAR-BFP). The CD19CAR-BFP gene cassette contains an anti-CD19CAR construct driven by the γ-retroviral-derived MND promoter29 and linked by a self-cleaving T2A peptide to blue fluorescent protein (BFP) (Figure S1A). As a negative control, HDR was performed using an AAV CCR5 BFP donor template containing an MND-BFP expression cassette flanked by CCR5 homology arms. Sort-enrichment for BFP+ cells (Figure S1B) resulted in stable populations of CAR+BFP+ cells for downstream assays (Figure 1A). Pre- and post-enrichment, the MFI of CAR and BFP expression was higher in LV CD19CAR-BFP T cells (Figure S1C). Presence of the CAR construct at the CCR5 locus in HDR edited cells (CCR5[CD19CAR-BFP]) was confirmed by PCR amplification from genomic DNA (Figure S1D).
Figure 1.
Equivalent Functional Responses of LV- and HDR-Generated CD19-CAR T Cells to CD19pos Cell Line Targets In Vitro
(A) Flow cytometry showing BFP and surface CAR expression by Protein L staining in untreated (mock) T cells and BFP-enriched CCR5[BFP], CCR5[CD19CAR-BFP], and LV CD19CAR-BFP T cells. (B) Specific killing of CD19pos K562 in a mixed target assay at 48 hr post-mixing with T cells at increasing E:T ratios. The specific lysis capacity of CCR5[CD19CAR-BFP] or LV CD19CAR-BFP T cells is significant versus CCR5[BFP] T cells by ANOVA (p < 0.001), but not significantly different from each other by ANOVA, or by unpaired t test at any E:T ratio. (C–E) % CD107a+ cells of live CD3+ cells in a degranulation assay using CCR5-edited or LV T cells stimulated with CD19pos or CD19neg K562. % CD137 expression on CD3+ cells at 6–8 (D) or 24 (E) hr post-mixing with target cell lines. The significance shown is by unpaired two-tailed t test. The error bars represent SEM. *p < 0.05 and **p < 0.0001. CAR T cells were generated from three donors and each experiment was performed in triplicate.
An in vitro cytotoxicity assay showed that both CCR5[CD19CAR-BFP] and LV CD19CAR-BFP T cells demonstrated specific and dose-dependent clearing of K562 cells overexpressing CD19 (CD19pos K562) that did not differ significantly between delivery methods (Figure 1B). Specific lysis by CAR T cells generated using either method was significant compared to T cells edited using the control AAV CCR5 BFP (CCR5[BFP]) at all effector-to-target ratios (E:T) tested. In a CD107a mobilization assay, surface CD107a was detected in an equivalent percentage of CCR5[CD19CAR-BFP] and LV CD19CAR-BFP T cells (46% and 42%, respectively) after stimulation with CD19pos K562 cells (Figure 1C). The cytotoxic T cell activation marker CD137 was also upregulated equivalently at both 6–8 hr and 24 hr after co-culture with CD19pos K562 cells (Figures 1D and 1E). Importantly, these responses were not induced in CAR T cells by stimulation with CD19neg K562 cells or exhibited by CCR5[BFP] control T cells.
To test rates of CAR delivery by HDR of a potential therapeutic vector, the self-cleaving peptide and BFP were removed from viral plasmids to generate AAV CCR5-CD19CAR and LV CD19CAR (Figure S2A). HDR rates in primary human T cells with the truncated, clinically relevant construct were 20%–35% by Protein L surface staining for the CAR kappa chain at 14 days post editing (Figure S2B).
To assess differences in function in a long-term assay, CCR5[CD19CAR] and LV CD19CAR T cells were tested in a stress test at low CAR T cell dose in an in vivo xenograft model. Luciferase-expressing Raji cells (that constitutively express CD19) were injected into NSG mice for 6 days prior to CAR T cell delivery. Intravenous injection of CCR5[CD19CAR] or LV CD19CAR T cells resulted in significant decreases in tumor burden relative to mice receiving non-transduced/edited T cells (mock T), measured by bioluminescence per a defined area (shown in Figure 2A). At the earliest time point measured (day 12 post-T cell delivery), there was a small, but significantly higher tumor burden in CCR5[CD19CAR] versus LV CD19CAR T cell cohorts (Figure 2B). However, no other significant differences in tumor burden were detected between these groups at other time points and no difference overall by repeated-measures ANOVA. The anatomical distribution of tumor bioluminescence did not appear to differ between the two groups.
Figure 2.
CCR5-CD19CAR Extends Survival in an In Vivo Tumor Model
(A) IVIS imaging of Raji-ffluc luminescence from baseline (day 5) to day 45 in mice treated at day 6 with non-transduced (mock), CCR5[CD19CAR], or LV CD19CAR T cells. The red X boxes represent deceased mice. Sensitivity settings were adjusted at each time point to maintain 600–60,000 counts per pixel. At each time point, signal intensities were assigned the same color scale for all treatment groups. (B) Tumor burden over time. Each time point is the mean bioluminescence of n (number of surviving mice in each cohort; shown in A). The significance shown is versus LV CD19CAR at each time point, using the unpaired two-tailed t test with the Holms-Sidak correction for multiple comparisons. (C) Kaplan-Meier curve showing survival of n = 8 mice in each group. (D) Engraftment of human cells (hCD45+) as a % of total live splenic cells at time of euthanasia, and expression of CAR at input and in hCD45+/hCD3+ cells from the spleen at time of euthanasia. (E) MFI of CAR expression in hCD45+hCD3+ cells from the spleen. The error bars represent SEM. The significance shown is derived from unpaired two-tailed t tests. *p < 0.05 and **p < 0.001
Survival of tumor-bearing mice was extended significantly by both CAR T cell delivery methods over mock T cells (Figure 2C). By 21 days post-tumor injection, all mice receiving mock T cells had developed hindlimb paralysis and were euthanized after imaging (median survival, 20.5 days). The median survival time for LV CD19CAR-treated mice was 38.5 days and for CCR5[CD19CAR] was 42 days. The difference in survival curves between CAR-treated groups was not significant, while each was significantly different from mock T cell treatment. At the time of euthanasia, human cell engraftment as measured by flow cytometry staining for human CD45 was slightly higher in mice receiving CCR5[CD19CAR] T cells than LV CD19CAR T cells (Figure 2D). At the time of T cell injection, CAR expression was 20% CAR+ for CCR5[CD19CAR] T cells and 16% CAR+ for LV CD19CAR T cells. At euthanasia, average CAR expression in hCD45+ hCD3+ cells in the spleen was 66% for CCR5[CD19CAR] T cells and 56% for LV CD19CAR T cells, suggesting preferential expansion of CAR+ cells in both treatment groups. Interestingly, while input CAR MFI was lower for CCR5[CD19CAR] T cells (Figure S2B), there was no statistically significant MFI difference in CAR expression between CCR5[CD19CAR] versus LV CD19CAR in tumor-bearing mouse spleens at euthanasia (Figures 2E and S2C).
As long-term HIV infection is also associated with an increased risk of plasma cell disorders, we also tested function of an anti-B cell maturation antigen (BCMA) CAR delivered by CCR5 HDR.30 In in vitro cytotoxicity assays, CCR5[BCMACAR-BFP] T cells specifically cleared BCMApos K562 in a dose-dependent manner and responded specifically to the BCMApos K562 target cell line versus CCR5[BFP] T cell controls (Figure S3).
Another gene targeting/gene disruption strategy that may be clinically useful would knock out expression of the endogenous TRAC locus in CAR T cells, allowing allogeneic use of T cell immunotherapies by removing potential endogenous TCR mediated graft-versus-host responses.28, 31, 32, 33 Combining TRAC disruption with HDR-mediated delivery of a therapeutic cassette, however, has not been previously investigated. We previously developed a TRAC megaTAL with high on-target versus off-target cutting (NHEJ) rates that resulted in efficient knock down of TCR surface expression in primary human T cells.34 We used mRNA encoding this TRAC megaTAL and AAV donor templates with TRAC homology arms (Figure 3A) to generate CD19CAR T cells by HDR at the TRAC locus (TRAC[CD19CAR]). For comparison, we again used LV to make CD19CAR T cells. We also used a more clinically translatable protocol that eliminated pre-enrichment of CD3+ cells from PBMC prior to CAR delivery by either HDR or LV and achieved CAR expression rates of ∼40% by both methods (Figures 3B and 3C). Of note, more than 90% of CAR+ TRAC[CD19CAR] T cells were CD3−. Across three donors, the phenotype of CAR+ cells did not differ significantly between CAR+ T cells generated by HDR versus LV, each being ∼50% TCM by CD62L and CD45RA surface staining (Figures 3D and 3E).
Figure 3.
Efficient Generation of CD3− CD19CAR+ T Cells by Integration of CD19CAR into the TRAC Locus
(A) Schematic of the TRAC and the homology template containing a second generation CD19CAR construct under the control of the MND promoter. The annotated TRAC-megaTAL cleavage site is located within the first exon of TRAC. The CD19CAR lentiviral construct has an MND promoter regulating the expression of a second generation CD19-targeting CAR combined with a CD8 hinge/trans-membrane (TM) domain and 41BB/CD3ζ intracellular signaling domains.42 (B) Analysis of CAR and CD3 expression on cells transduced with LV CD19CAR or gene edited to TRAC[CD19CAR]. The representative FACS plots (left) and summary data from three unique donors (right) are shown. The colored quadrants were used for T cell phenotyping in (C), as indicated by arrows. (C) Phenotypic analysis of CD45RA and CD62L T cell differentiation markers in CAR+ T cells 6 days post-HDR or LV transduction, with representative FACS plots (left) and summary data from three unique donors (right) is shown. Analysis in (B) and (C) is representative of three unique experiments and at least two different donors per experiment. The error bars represent SEM.
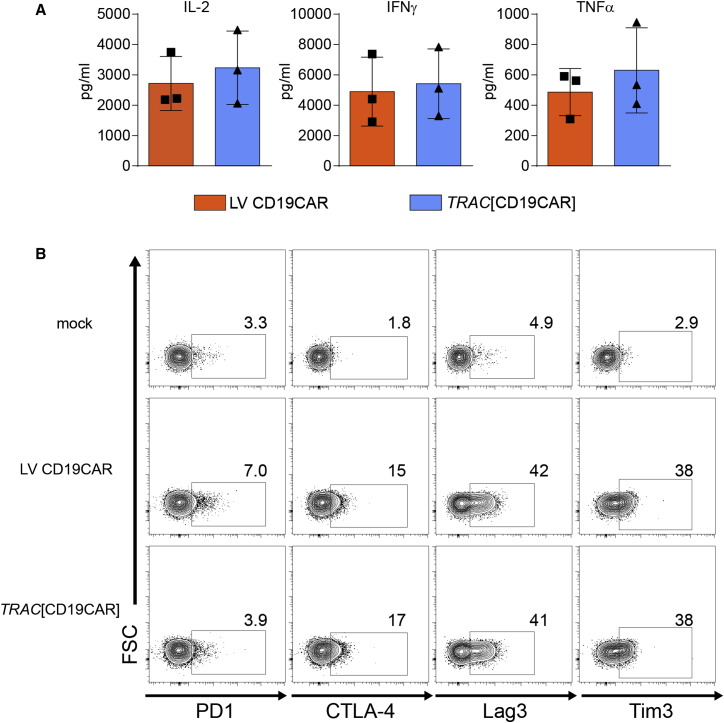
To determine whether TRAC HDR knock out of TCR surface expression impacts in vitro functional responses of the CAR T cells, we subjected CAR+ cells generated by the two methods to several tests of CAR function. Both cell products exhibited efficient activation and killing in response to CD19+ target cells. Importantly, we found no differences in the ability of TRAC HDR versus LV-generated CAR T cells to produce pro-inflammatory cytokines (interleukin 2 [IL-2], interferon [IFN]γ, and tumor necrosis factor α [TNF-α]) in response to the presence of CD19+ targets (Figure 4A). We also found no differences in the expression of exhaustion markers after prolonged (3 days) co-culture with the CD19+ Nalm-6 cells (Figure 4B).
Figure 4.
Equivalent In Vitro Function of CD19-Specific CAR Delivered via HDR or Lentiviral Platforms
Comparison of LV versus HDR-generated CAR T cell responses after co-culture with CD19+ Nalm6-GFP cells. (A) Cytometric bead array quantitation of cytokine production in cell culture supernatants 24 hr after co-incubation. (B) Immune checkpoint surface marker expression on mock or CAR T cells 3 days post-co-culture with Nalm6-GFP cells, assessed by flow cytometry and gating on GFP− T cells. The flow cytometry analysis from a single donor is shown. The data are representative of three unique donors. The error bars represent SEM.
As an additional proof-of-concept, we tested HDR insertion of a different CAR at the TRAC locus. We performed HDR editing of CD3+ PBMCs using the TRAC megaTAL nuclease, with AAV TRAC-anti-BCMA CAR as a donor template, to generate TRAC[BCMACAR] T cells. Of note, we have recently reported that delivery of mRNA encoding wild-type (WT) adenoviral helper proteins, Ad5 E4orf6/E1b55k WT, can enhance AAV transduction.35 Further, co-delivery of mRNA encoding specific mutant helper proteins in association with nuclease mRNA and AAV donor can improve HDR rates in cases where donor template delivery may be rate limiting35 (Y.H., unpublished data). Therefore, we used this combined approach to promote HDR within the TRAC locus. Using Ad5 E4orf6 and mutant E1b55 H354, we achieved CAR-expression rates of 40% at day 10, most (89%–94%) of which were CD3− (Figures 5A and 5B). Presence of the BCMACAR at the TRAC locus was confirmed by PCR and sequencing (Figure S4). IFNγ, IL-2, and TNF-α were detected by intracellular cytokine staining of CD4+ TRAC[BCMACAR] T cells after stimulation with the multiple myeloma cell line, RPMI8226. This response was specific to anti-BCMA CAR expression, as these cytokines were not upregulated in control TRAC[BFP] CD4+ T cells in this assay (Figure 5C). As a more rigorous in vitro test of the ability of TRAC[BCMACAR] cells to specifically target primary plasma cells, we cultured autologous B cells with soluble factors that supported a fraction of these cells (23%–35%) to differentiate into BCMA+ plasmablasts. After addition of TRAC[BCMACAR] cells, this population dropped to 0%–2% of CD4−/CD8− cells (a >90% decrease) (Figure 5D). The addition of TRAC[BFP] cells decreased the % BCMA+ only slightly (from 25% to 17%–20%, a <20% decrease).
Figure 5.
TRAC[BCMACAR] T Cells Produce Cytokine in the Presence of a Multiple Myeloma Cell Line and Eliminate BCMA+ Cells from In Vitro Plasmablast Cultures
CD3 and Protein L staining or BFP expression in AAV-alone and AAV plus TRAC megaTAL (AAV+MT) treated cells for AAV TRAC BCMACAR or AAV TRAC BFP at 12 days post-gene editing displayed as: (A) representative flow plots and (B) combined data from three donors. The bar graphs show mean percentage of cells in each quadrant by color. The error bars represent SEM. (C) Intracellular cytokine staining of TRAC[BCMACAR] and TRAC[BFP] T cells gated on CD4+ cells plated with the BCMA+ multiple myeloma cell line, RPMI8226. (D) In vitro generated plasma cells 48 hr post-mixing with T cell media alone, TCR-BCMACAR, or TCR-BFP T cells, gated on CD3−CD4−CD8− live lymphocytes. The representative flow plots from n = 3 experiments using autologous CAR T cell and plasma cell populations generated from independent PBMC donors are shown.
Discussion
Here, we demonstrate the use of megaTAL nucleases and AAV6 donor templates to generate CAR T cells at high-efficiency by HDR at two loci with potential clinical applications: the genes coding for the HIV co-receptor CCR5 and TCRα. CAR T cells produced using this method have possible utility for use in HIV+ lymphoma patients, where simultaneous CCR5 disruption could protect therapeutic cells from HIV infection and as an off-the-shelf therapy, where CAR+TCR− cells could be produced from an allogeneic donor. This work demonstrates proof of principle for HDR-delivered CARs to improve immunotherapies for specific patient populations.
While we found no significant differences in function between LV and HDR delivery of CAR gene cassettes at two loci, CAR T cells produced by the two methods are fundamentally different in several important ways. In HDR-generated CAR cells, gene disruption and CAR delivery occur concurrently at a single, known target site, allowing strict copy-number control as well as reducing the theoretical risk of insertional mutagenesis over randomly integrating retroviruses and transposons.14, 36 Further, enrichment for CAR+ cells simultaneously enriches for disruption of the gene, potentially simplifying production and purification of a desired engineered T cell population. Several groups have published two-step methods for generating TCR− CAR T cells that involve CAR delivery by gene addition (sleeping beauty transposons or lentivirus), followed by TCR-gene disruption using zinc-finger28 or TAL-effector nucleases (TALEN).32 Initial clinical application of TALEN-based TCR− CAR T cells33 speaks to the potential translational relevance of the one-step HDR-based methods for generating TCR− CAR T cells demonstrated here.
Refinement of gene editing methods as the field advances, including methods to further optimize AAV transduction and cellular processing, are likely to progressively increase the efficiency of HDR. Expanding on previous work, we explored several methods for increasing the efficiency of HDR. For example, the use of adenoviral helper mRNAs35 increased HDR with lower efficiency donor templates in our experiments with the anti-BCMA CAR and BFP donor templates at the TRAC locus (Figures 5, S6E, and S6F). Additionally, we confirmed the importance of removing the nuclease target site within the AAV donor (Figure S5) and found that reduction of viral packaging size by reducing homology arm length to 300 bp within candidate AAV donor constructs did not significantly decrease HDR (Figure S6).
We observed that the MFI of CAR surface expression was lower in HDR-generated cells versus LV, especially at the CCR5 locus and where a CAR expression cassette contained a downstream cis-linked fluorophore. Despite having fewer CARs on the cell surface by protein L staining, no difference was observed in function versus LV in in vitro assays using these cells (Figures 1 and 4). Interestingly, while this difference was evident at input in CCR5[CD19CAR] and LV CD19CAR T cells used in our in vivo experiments, output CAR T cells isolated from the spleen of tumor-bearing mice showed no significant difference in MFI (Figures 2E, S2B, and S2C). Whether this is due to increased expression from the MND promoter in activated cells via NF-κB signaling29, 37, 38 or whether the in vivo tumor challenge enriches for CAR T cells within a defined range of surface expression is not explored here.
While we have used exogenous promoters to drive expression in order to allow for direct comparison between LV and HDR-delivery, donor templates for HDR can be designed such that seamless integration at a target site puts CAR expression under the regulation of an endogenous gene.17 This may be a useful application of HDR for delivery of therapeutic cassettes that was not explored here.
In summary, we have demonstrated the use of HDR to generate gene targeted CAR T cells. Potential direct clinical applications of our process include, but are not limited to: the generation of CCR5-deficient CD19 CAR T cells for therapy of HIV-associated lymphoma; TCR-deficient CAR T cells to allow use of an allogeneic cell product in patients where generation of autologous T cells is impractical; autoimmune applications where the endogenous TCR repertoire may be pathogenic; and the use of endogenous promoters or regulatory elements to control CAR expression to facilitate more refined regulation of candidate CAR expression. Efficient gene targeting via HDR represents a valuable addition to the toolkit for engineering cellular therapeutics.
Materials and Methods
Plasmid Constructs
All CCR5 homology arms (HA) used in the pAAV shown in this manuscript are ∼0.6 kb length each (1.2 kb of CCR5 homology total) with a T to G mutation to render the homology arms resistant to CCR5 megaTAL binding and cleavage. The T to G mutation is at DNA matching chr3 nucleotide 46,373,690 of the December 2013 UCSC human genome sequence assembly. pAAV CCR5.MND.CD19CAR.2A.BFP was described previously18 and was also used to generate: pAAV CCR5.MND.CD19CAR (by deletion of coding sequence downstream of the CD9CAR by PCR and In-Fusion cloning into the CCR5.MND.CD19CAR.2A.BFP backbone after digestion by SalI and XhoI), and pAAV CCR5.MND.BCMACAR.2A.BFP (by replacing CD19CAR with synthesized BCMACAR coding sequence,30 GenScript). pAAV CCR5 MND.BFP and lentiviral plasmid pRRL MND.CD19CAR.2A.BFP.WPRE were described previously (M.H., unpublished data). A version of the latter vector without the 2A.BFP coding sequence (pRRL MND.CD19CAR.WPRE) was made by cloning a PCR amplification of the MND-CD19.CAR into the identical pRRL WPRE backbone.
Plasmid pAAV TRAC CD19CAR was cloned by ligation of the pAAV backbone with a gene synthesized fragment (GENEWIZ) containing an MND promoter-CD19CAR cDNA-SV40 polyA signal sequence cassette (as above) within TRAC HA. The TRAC HA used in the body of this manuscript is comprised of a total of 1,281 nucleotides: 22,546,933-22,547,576 (643 bp; 5′HA) and 22,547,577-22,548,215 (638 bp; 3′HA) of NC_000014.9 chromosome 14 Reference GRCh38.p7 primary assembly, located proximal to (but not including) the TRAC megaTAL cleavage site. AAV with varying sizes of TRAC HA are used and described in the Supplemental Information. To generate pAAV TRAC BCMACAR, the BCMACAR coding sequence was PCR amplified from the pAAV CCR5.MND.BCMACAR.2A.BFP construct with addition of XhoI site at the 5′ end and stop codon and NotI at the 3′ end. The PCR fragment was subcloned between TRAC homology arms in the above AAV vector via XhoI and Not I sites, between the MND promoter and polyA signal sequence. As a control, BFP coding sequence was PCR amplified and subcloned between the MND promoter and poly A signal sequence. The resulting constructs were verified by sequencing.
Plasmids used for in vitro production of mRNA encoding CCR5 megaTAL18 and Ad5 helper proteins E4orf6 and E1b55k-H354 were described previously.35 The TRAC megaTAL coding sequence34 was cloned into pEVL20039 for in vitro transcription.
Production of Recombinant AAV, Lentivirus, and mRNA
Recombinant AAVs used in Figures 1, 2, 5, S1–S4, S6D, and S6E were generated and titered using the triple transfection method and serotype 6 helper plasmid described previously.18 Recombinant AAVs used in Figures 3, 4, S5, and S6A–S6C were prepared using a two plasmid transfection system. Semi-confluent 293T cells were transfected with pDP6rs (Plasmid Factory) and a pAAV plasmid containing the transgene of interest flanked by AAV2-derived ITR elements. For each transfection, a semi-confluent 15 cm plate of 293T cells were transfected with 6 μg of pAAV plasmid and 18 μg of pDP6rs plasmid using PEIpro transfection reagent (VWR) according to manufacturer’s instructions. Cells were collected 72 hr post-transfection and AAV was harvested and purified as described,18 then titered using the AAVpro titration kit (Clontech). VSV-G pseudotyped LVs were made as described.40 For in vitro production of mRNA, plasmids containing cDNA cassette of interest downstream of a T7 promoter were linearized 3′ of their stop codon (CCR5 megaTAL, E4orf6, and E1b55k-H354) or 3′ of the encoded polyA tail (TRAC megaTAL) by restriction digest. mRNAs were synthesized using the T7 mScript Standard mRNA Production Kit (CellScript) with modifications described previously,18 with the exception that the TRAC megaTAL, having an encoded 3′ poly A tail, was not subjected to additional poly-adenylation. mRNA used in Figures 3 and 4 was produced using a linearized plasmid template and the T7 HiScribe ARCA mRNA Kit (New England Biolabs) and purified with the RNeasy Mini Kit (QIAGEN).
Isolation and Culture of Primary Human Cells and Cell Lines
Primary human cells were obtained from healthy adult donors at Seattle Children’s Research Institute (SCRI) and the Fred Hutchinson Cancer Research Center (FHCRC) using protocols approved by the respective Institutional Review Boards. Primary human cells used in Figures 3 and 4 were purchased from Key Biologics. For most of the experiments presented here, T cells were enriched directly from whole peripheral blood using a negative selection kit (RosetteSep Human T Cell Enrichment Cocktail, STEMCELL Technologies), either freshly prepared or thawed from stocks frozen at 2–4 × 107 cells/mL in culture media with 10% DMSO. Whole PBMCs were used to generate cells used in Figures 3 and 4. For tests of plasma cell killing by BCMACAR T cells in vitro, autologous B and T cells were purified using EasySep Human Pan-B or T Cell Enrichment Kits (StemCell Technologies) from PBMCs obtained from either: CD34-depleted apheresis product from non-mobilized donors by the FHCRC Hematopoietic Cell Processing and Repository Core or from PBMCs collected at SCRI as described.18
Standard primary human T cell culturing conditions were incubation in a humidified environment at 37°C with 5% CO2, maintaining a density of ∼1 × 106 cells/mL by replenishing media every 2–3 days. T cell media was: RPMI-1640 with 20% fetal calf serum (Omega Scientific), 1 × GlutaMAX, 20 mM N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid (HEPES), 55 μm 2-mercaptoethanol (2ME; GIBCO), and recombinant human interleukin-2 (IL-2; 50 ng/mL), IL-7 (5 ng/mL), and IL-15 (5 ng/mL) from PeproTech as described.18 Nalm-6, K562, and RPMI8226 cells (ATCC) and Nalm6-GFP (Nalm-6 transduced with LV MND GFP; Sather et al.18) were maintained in T cell media without cytokines; T cell cytokines were added for mixed cultures (below). Culturing of primary B cells for assays using in vitro generated plasma B cells is described below.
T Cell Transduction and Gene Editing
Gene editing of CD3+ enriched PBMCs was performed as described.18 Briefly, 1 × 106 T cells were stimulated 1:1 with Dynabeads Human T-Activator CD3/CD28 Beads (Invitrogen) for 48 hr, then washed, and cultured in T cell media for 16 hr. There were 2.4 × 107 cells/mL that were electroporated using either the 100 μL or 10 μL tip of the Neon Transfection System (Invitrogen) as described (Sather et al.18), with 1 μg of nuclease-encoding mRNA per every ∼2.5 × 105 cells. For experiments where mRNAs encoding Ad5 helper proteins were co-transfected with the nuclease (Figures 5, S4, and S6), we added 0.025 μg each of mRNA encoding E4orf6 and E1b55k-H354 per 1 μg of nuclease mRNA (a 1:1:40 ratio). After electroporation cells were incubated at 30°C for 2 hr, followed by addition of 20% of the total culture volume of AAV (∼2–4 × 104 MOI), then returned to 30°C incubation for ∼20–24 hr. Cells were then returned to 37°C and standard T cell culturing conditions.
LV transduction of T cells was performed by addition of 10 μL (∼2 MOI) of LV and 4 μg/mL polybrene (Sigma-Aldrich) to ∼1 × 106 primary human T cells in 1 mL culture media. Cells were incubated at 37°C overnight, then 1 mL T cell media added, followed by standard culturing conditions.
After gene editing or LV transfer, fluorescent protein expressing cells were in some cases enriched using a BD FACSAria cell sorter (BD Biosciences).
CAR T cells shown in Figures 3 and 4 were generated directly from whole PBMCs as follows. Thawed PBMCs (1 × 106/mL) were activated with 50 ng/mL human anti-CD3 and anti-CD28 (OKT3 and 15E8, respectively, both Miltenyi Biotec) in X-VIVO 15 media (Lonza) supplemented with 5% human serum, type AB (Valley Biomedical), 2 mM GlutaMAX, 10 mM HEPES, and 250 IU/mL recombinant human IL-2 (CellGenix GmbH). Gene editing was performed as above, with the following differences: mRNA electroporation was performed after 3 days in culture; purified AAV was added within 1 hr following electroporation, with a maximum dose of 15% of total culture volume; and cells were expanded in culture for 5–6 days before phenotyping and functional assays. For LV transduction, 24 hr post-activation, LV was added to cells at a MOI of 20, followed by overnight incubation, then media was replenished, and cells cultured using standard conditions. At 5 to 6 hr post-LV or HDR, TCR and CAR expression and T cell phenotype were determined by flow cytometry after labeling with APC anti-CD62L (Dreg-56), AlexaFluor488 anti-CD3 (SK7), PE anti-CD45RA (HI100), all from BioLegend, and recombinant PE-CD19-His (Creative Biomart).
Flow Cytometry and Statistical Analyses
Fluorescent protein expressing or labeled cells were detected using either an LSR II (BD), or Attune NxT (Thermo Fisher) flow cytometer, and analyzed using FlowJo X software (Tree Star). GraphPad Prism 6 (GraphPad Software) was used to visualize data and to perform tests of statistical significance; specific tests used are noted in the figure legends. Data presented here show the mean ± SEM unless otherwise noted.
In Vitro Functional Assays of CD19CAR T Cells
K562 CD19+ and control target cell lines were previously described18 and consist of K562 cells (ATCC) stably transduced with LV carrying either MND.CD19.T2A.GFP (CD19pos K562) or MND.BCMA.T2A.iRFP (irrelevant antigen control; CD19neg K562). Cytotoxicity assays were performed as described.18 Briefly, equal numbers (5 × 104) of CD19pos and CD19neg K562 target cells (T) were co-cultured with control or CD19CAR T cells (E; generated either by HDR or LV) at various E:T ratios. At 48 hr, cytotoxicity was calculated as percent specific lysis: 100% × (1 − (%CD19pos/%CD19neg at noted E:T)/(%CD19pos/CD19neg at 0:1 E:T)) by flow cytometry.
To assay upregulation of T cell activation markers, 1 × 105 CD19CAR T cells were plated at 1 × 106/mL in T cell media with 5 × 104 CD19pos or CD19neg K562 in a 96-well plate. Flow cytometry was performed at 6–8 or 24 hr time points after staining for PE anti-CD137 (4B4-1, BD Bioscience), APC anti-CD3 (BW264/56, Miltenyi Biotec), and Live/Dead Fixable Near-IR Dead Cell Stain Kit (Invitrogen).
Internalization of CD107a, a feature of cytotoxic T cell degranulation, was assessed after co-culture of 1.0 × 105 CD19CAR T cells with 5.0 × 104 CD19pos or CD19neg K562 cells (total cell density was 1 × 106 per mL) and 4 μL/mL PE anti-CD107a (H4A3, BD Bioscience). Following a 1 hr incubation, 1 μL GolgiStop (BD Bioscience) was added per milliliter culture and incubated an additional 5 hr. Cell mixture was washed with PBS and cells labeled with APC anti-CD3 (BW264/56, Miltenyi Biotec) and Live-Dead Near-IR viability dye (Life Technologies), followed by flow cytometry.
To assess cytokine production, 0.5 × 105 LV CD19CAR or TRAC[CD19CAR] T cells were cultured at a 1:1 ratio with Nalm-6-GFP (CD19+). After a 24 hr incubation period, 20 μL of supernatant was removed for cytokine analysis and T cells were cultured for 24 hr more. At the end of the 2 day period, T cells were stained with APC anti-CTLA4 (L3D10, BioLegend), PE anti-PD1 (J105, eBioscience), PE-Cy7 anti-TIM3 (F38-2E2, eBioscience), and APC anti-LAG3 (3DS223H, eBioscience). Cytokine secretion was quantified using the MultiCyt QBeads cytokine PlexScreen and analyzed with the iQue Plus Screener (Intellicyt).
Analysis of Cytokine Levels of Activated BCMACAR T Cells
There were 1.5 × 105 BCMACAR T cells that were incubated with 7.5 × 104 RPMI8226 (1 × 106/mL) and 1 μL/mL GolgiPlug for 5 hr at 37°C, then placed at 4°C overnight. Cells were then labeled with the following antibodies from BD Bioscience: FITC anti-CD4 (RPA-T4), PerCPCy5.5 anti-CD8 (RPA-T8), and Alexa700 anti-CD3 (SP34-2). Cells were subsequently permeabilized and fixed using BD Cytofix/Cytoperm Kit (BD Biosciences), followed by labeling with the following human cytokine antibodies: AlexaFluor647 anti-IFNγ (4S.B3, BioLegend), PE anti-IL-2 (MQ1-17H12, BD Biosciences), and APC-Cy7 anti-TNF-α (Mab11, BioLegend).
BCMACAR T Cell Killing of In Vitro-Generated Plasma Cells
B cells obtained from human PBMCs (described above) were seeded in 96-well plates at 2.5 × 106 cells/mL, then cultured at 37°C with 5% CO2 for 3 days in RPMI supplemented with 10% FBS, 100 U/mL Pen/Strep, 0.11 mg/mL sodium pyruvate, 10 mM HEPES, 2 mM glutamine, 55 μM 2ME with IL-21 (50 ng/mL), anti-CD40 (5 μg/mL, IC10 Southern Biotech), and R848 (Resiquimod, 3 μg/mL, Sigma-Aldrich). CAR T cells were added to B cells at a 1:1 cell ratio, keeping the total cell concentration between 0.5–1 × 106 in B cell media as above, but with the addition of IL-2 (50 ng/mL), IL-7 (5 ng/mL), and IL-15 (5 ng/mL). The relative proportion of CD4−CD8− BCMA+ plasma cells in each sample was then assessed 2 days post-addition of T cells by flow cytometry after staining with FITC anti-CD4 (RPA-T4, BD Bioscience), PerCP-Cy5.5 anti CD-8 (RPA-T8, BD Bioscience), PE anti-BCMA (19F2, BioLegend), Alexa700 anti-CD138 (MI15, Biolegend), and PE-Cy7 anti-CD19 (HIB19, eBioscience).
Murine Xenograft Model of In Vivo Target Cell Killing
Murine studies were performed in a specific pathogen-free facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, in accordance with the NIH Guide for the Care and Use of Laboratory Animals, and the Seattle Children’s Research Institute’s Institutional Animal Care and Use Committee. NOD-Scid-IL2RγNULL mice at 8–10 weeks of age were injected intravenously (i.v.) with 0.5 × 106 Raji-ffluc (human Raji B cells stably expressing a firefly luciferase-eGFP fusion protein,41 gift from Michael Jensen). In vivo bioluminescence imaging of Raji-ffluc cells was performed following subcutaneous injection of 150 μL D-Luciferin monopotassium salt (28.57 mg/mL in sterile saline; Pierce) in a custom box that allowed simultaneous imaging of six mice (Ellard Instrumentation). Mice were imaged at the signal plateau (between 10 and 14 min post-D-Luciferin injection) using an IVIS Spectrum imager (PerkinElmer) under isoflurane anesthesia. The tumor burden was quantified as the total photon flux per second within a region of interest (ROI) that encompassed the head through pelvic region of the mouse; the ROIs were identically sized for all measurements. Sensitivity settings were adjusted at each time point to maintain 600–60,000 counts per pixel. Pseudocolor maps are scaled at each time point as follows: 1 × 104 − 1 × 105 (day 5), 5 × 105 − 5 × 106 (day 12), 1 × 106 − 1 × 107 (day 16), 5 × 106 − 5 × 107 (days 21 and 26), and 1 × 107 − 1 × 108 p/sec/cm2/sr (days 32 and 45). Imaging was first performed on day 5 after xenograft transfer. At this time, mice were divided into groups of approximately equal average and variance of tumor burden and injected with volumes of bulk edited or LV-transduced primary human T cells from frozen aliquots to achieve 2.4 × 106 CAR+ T cells per mouse (LV = 15 × 106 cells that were 16% CAR+; AAV CCR5-CD19CAR = 12 × 106 cells, 20% CAR+; and mock T cells = 15 × 106 human T cells). Mice were euthanized when pre-determined humane endpoints were observed (typically onset of hindlimb paralysis, >20% weight loss, or general malaise). Splenic lymphocytes obtained from euthanized mice were labeled with a viability dye (Near-IR Fixable Live/Dead, Thermo Fisher) and FITC anti-human CD45 (2D1, eBioscience), APC anti-mouse CD45 (30-F11, eBioscience), PE anti-human CD3 (OKT3, BioLegend), and biotin-Protein L (GenScript), followed by PE-streptavidin (BD Biosciences).
Author Contributions
Conceptualization: M.H., B.L., Y.H., W.-H.L., A.A., and D.J.R.; Methodology and Investigation: M.H., B.L., Y.H., W.-H.L., A.E.G., H.M.J., and J.S.; Writing - Original Draft: M.H. and K.S.; Writing - Review & Editing: M.H., B.L., W.-H.L., K.S., A.M.S., A.A., and D.J.R.; and Supervision: S.W.J., A.M.S., A.A., and D.J.R.
Conflicts of Interest
B.L., W.-H.L., and A.A. are full time employees of bluebird bio. A.M.S. is a consultant and shareholder in bluebird bio. All other authors declare no conflict of interest.
Acknowledgments
We thank Michael Jensen and Cindy Chang at the Ben Towne Center for Childhood Cancer Research for use of the IVIS instrument. This work was supported by Seattle Children’s Program for Cell and Gene Therapy and the Center for Immunity and Immunotherapies and bluebird bio. D.J.R. received support as a recipient of the Children’s Guild Association Endowed Chair in Pediatric Immunology.
Footnotes
Supplemental Information includes six figures and can be found with this article online at http://dx.doi.org/10.1016/j.omtm.2016.12.008.
Contributor Information
Alexander Astrakhan, Email: sastrakhan@bluebirdbio.com.
David J. Rawlings, Email: drawling@uw.edu.
Supplemental Information
References
- 1.Sadelain M., Brentjens R., Rivière I. The basic principles of chimeric antigen receptor design. Cancer Discov. 2013;3:388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fesnak A.D., June C.H., Levine B.L. Engineered T cells: the promise and challenges of cancer immunotherapy. Nat. Rev. Cancer. 2016;16:566–581. doi: 10.1038/nrc.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maus M.V., June C.H. Making better chimeric antigen receptors for adoptive T-cell therapy. Clin. Cancer Res. 2016;22:1875–1884. doi: 10.1158/1078-0432.CCR-15-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brentjens R.J., Davila M.L., Riviere I., Park J., Wang X., Cowell L.G., Bartido S., Stefanski J., Taylor C., Olszewska M. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med. 2013;5:177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kochenderfer J.N., Dudley M.E., Carpenter R.O., Kassim S.H., Rose J.J., Telford W.G., Hakim F.T., Halverson D.C., Fowler D.H., Hardy N.M. Donor-derived CD19-targeted T cells cause regression of malignancy persisting after allogeneic hematopoietic stem cell transplantation. Blood. 2013;122:4129–4139. doi: 10.1182/blood-2013-08-519413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruz C.R., Micklethwaite K.P., Savoldo B., Ramos C.A., Lam S., Ku S., Diouf O., Liu E., Barrett A.J., Ito S. Infusion of donor-derived CD19-redirected virus-specific T cells for B-cell malignancies relapsed after allogeneic stem cell transplant: a phase 1 study. Blood. 2013;122:2965–2973. doi: 10.1182/blood-2013-06-506741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali S.A., Shi V., Maric I., Wang M., Stroncek D.F., Rose J.J., Brudno J.N., Stetler-Stevenson M., Feldman S.A., Hansen B.G. T cells expressing an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of multiple myeloma. Blood. 2016;128:1688–1700. doi: 10.1182/blood-2016-04-711903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grupp S.A., Kalos M., Barrett D., Aplenc R., Porter D.L., Rheingold S.R., Teachey D.T., Chew A., Hauck B., Wright J.F. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalos M., Levine B.L., Porter D.L., Katz S., Grupp S.A., Bagg A., June C.H. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med. 2011;3:95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh H., Figliola M.J., Dawson M.J., Olivares S., Zhang L., Yang G., Maiti S., Manuri P., Senyukov V., Jena B. Manufacture of clinical-grade CD19-specific T cells stably expressing chimeric antigen receptor using Sleeping Beauty system and artificial antigen presenting cells. PLoS ONE. 2013;8:e64138. doi: 10.1371/journal.pone.0064138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kebriaei P., Singh H., Huls M.H., Figliola M.J., Bassett R., Olivares S., Jena B., Dawson M.J., Kumaresan P.R., Su S. Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J. Clin. Invest. 2016;126:3363–3376. doi: 10.1172/JCI86721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beatty G.L., Haas A.R., Maus M.V., Torigian D.A., Soulen M.C., Plesa G., Chew A., Zhao Y., Levine B.L., Albelda S.M. Mesothelin-specific chimeric antigen receptor mRNA-engineered T cells induce anti-tumor activity in solid malignancies. Cancer Immunol. Res. 2014;2:112–120. doi: 10.1158/2326-6066.CIR-13-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan R.A., Kakarla S. Genetic modification of T cells. Cancer J. 2014;20:145–150. doi: 10.1097/PPO.0000000000000033. [DOI] [PubMed] [Google Scholar]
- 14.Porteus M.H. Towards a new era in medicine: therapeutic genome editing. Genome Biol. 2015;16:286. doi: 10.1186/s13059-015-0859-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoban M.D., Bauer D.E. A genome editing primer for the hematologist. Blood. 2016;127:2525–2535. doi: 10.1182/blood-2016-01-678151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubbard N., Hagin D., Sommer K., Song Y., Khan I., Clough C., Ochs H.D., Rawlings D.J., Scharenberg A.M., Torgerson T.R. Targeted gene editing restores regulated CD40L function in X-linked hyper-IgM syndrome. Blood. 2016;127:2513–2522. doi: 10.1182/blood-2015-11-683235. [DOI] [PubMed] [Google Scholar]
- 18.Sather B.D., Romano Ibarra G.S., Sommer K., Curinga G., Hale M., Khan I.F., Singh S., Song Y., Gwiazda K., Sahni J. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med. 2015;7:307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Exline C.M., DeClercq J.J., Llewellyn G.N., Hayward S.B., Li P.W., Shivak D.A., Surosky R.T., Gregory P.D., Holmes M.C., Cannon P.M. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunthel C.J., Lechowicz M.J. Human immunodeficiency virus lymphomas in the era of antiretrovirals: Is it finally time to change the discussion? Cancer. 2016;122:2621–2623. doi: 10.1002/cncr.30110. [DOI] [PubMed] [Google Scholar]
- 21.Dezube B.J., Aboulafia D.M., Pantanowitz L. Plasma cell disorders in HIV-infected patients: from benign gammopathy to multiple myeloma. AIDS Read. 2004;14:372–374. 377–379. [PubMed] [Google Scholar]
- 22.Castillo J., Pantanowitz L., Dezube B.J. HIV-associated plasmablastic lymphoma: lessons learned from 112 published cases. Am. J. Hematol. 2008;83:804–809. doi: 10.1002/ajh.21250. [DOI] [PubMed] [Google Scholar]
- 23.Lederman M.M., Penn-Nicholson A., Cho M., Mosier D. Biology of CCR5 and its role in HIV infection and treatment. JAMA. 2006;296:815–826. doi: 10.1001/jama.296.7.815. [DOI] [PubMed] [Google Scholar]
- 24.Perez E.E., Wang J., Miller J.C., Jouvenot Y., Kim K.A., Liu O., Wang N., Lee G., Bartsevich V.V., Lee Y.L. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Didigu C.A., Wilen C.B., Wang J., Duong J., Secreto A.J., Danet-Desnoyers G.A., Riley J.L., Gregory P.D., June C.H., Holmes M.C., Doms R.W. Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood. 2014;123:61–69. doi: 10.1182/blood-2013-08-521229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osborn M.J., Webber B.R., Knipping F., Lonetree C.L., Tennis N., DeFeo A.P., McElroy A.N., Starker C.G., Lee C., Merkel S. Evaluation of TCR gene editing achieved by TALENs, CRISPR/Cas9, and megaTAL nucleases. Mol. Ther. 2016;24:570–581. doi: 10.1038/mt.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torikai H., Reik A., Liu P.Q., Zhou Y., Zhang L., Maiti S., Huls H., Miller J.C., Kebriaei P., Rabinovich B. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. 2012;119:5697–5705. doi: 10.1182/blood-2012-01-405365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Challita P.M., Skelton D., el-Khoueiry A., Yu X.J., Weinberg K., Kohn D.B. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J. Virol. 1995;69:748–755. doi: 10.1128/jvi.69.2.748-755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carpenter R.O., Evbuomwan M.O., Pittaluga S., Rose J.J., Raffeld M., Yang S., Gress R.E., Hakim F.T., Kochenderfer J.N. B-cell maturation antigen is a promising target for adoptive T-cell therapy of multiple myeloma. Clin. Cancer Res. 2013;19:2048–2060. doi: 10.1158/1078-0432.CCR-12-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torikai H., Cooper L.J. Translational implications for off-the-shelf immune cells expressing chimeric antigen receptors. Mol. Ther. 2016;24:1178–1186. doi: 10.1038/mt.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poirot L., Philip B., Schiffer-Mannioui C., Le Clerre D., Chion-Sotinel I., Derniame S., Bas C., Potrel P., Lemaire L., Duclert A. Multiplex genome-edited T-cell manufacturing platform for “off-the-shelf” adoptive T-cell immunotherapies. Cancer Res. 2015;75:3853–3864. doi: 10.1158/0008-5472.CAN-14-3321. [DOI] [PubMed] [Google Scholar]
- 33.Qasim W., Amrolia P.J., Samarasinghe S., Ghorashian S., Zhan H., Stafford S., Butler K., Ahsan G., Gilmour K., Adams S. First clinical application of talen engineered universal CAR19 T cells in B-ALL. Blood. 2015;126:2046. [Google Scholar]
- 34.Boissel S., Jarjour J., Astrakhan A., Adey A., Gouble A., Duchateau P., Shendure J., Stoddard B.L., Certo M.T., Baker D., Scharenberg A.M. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2014;42:2591–2601. doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gwiazda K.S., Grier A.E., Sahni J., Burleigh S.M., Martin U., Yang J.G., Popp N.A., Krutein M.C., Khan I.F., Jacoby K. High efficiency CRISPR/Cas9-mediated gene editing in primary human T-cells using mutant adenoviral E4orf6/E1b55k “helper” proteins. Mol. Ther. 2016;24:1570–1580. doi: 10.1038/mt.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.June C.H., Riddell S.R., Schumacher T.N. Adoptive cellular therapy: a race to the finish line. Sci. Transl. Med. 2015;7:280ps7. doi: 10.1126/scitranslmed.aaa3643. [DOI] [PubMed] [Google Scholar]
- 37.Griffin G.E., Leung K., Folks T.M., Kunkel S., Nabel G.J. Activation of HIV gene expression during monocyte differentiation by induction of NF-kappa B. Nature. 1989;339:70–73. doi: 10.1038/339070a0. [DOI] [PubMed] [Google Scholar]
- 38.Baeuerle P.A., Henkel T. Function and activation of NF-kappa B in the immune system. Annu. Rev. Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 39.Grier A.E., Burleigh S., Sahni J., Clough C.A., Cardot V., Choe D.C., Krutein M.C., Rawlings D.J., Jensen M.C., Scharenberg A.M., Jacoby K. pEVL: A linear plasmid for generating mRNA IVT templates with extended encoded poly(A) sequences. Mol. Ther. Nucleic Acids. 2016;5:e306. doi: 10.1038/mtna.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X., Shin S.C., Chiang A.F., Khan I., Pan D., Rawlings D.J., Miao C.H. Intraosseous delivery of lentiviral vectors targeting factor VIII expression in platelets corrects murine hemophilia A. Mol. Ther. 2015;23:617–626. doi: 10.1038/mt.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudecek M., Sommermeyer D., Kosasih P.L., Silva-Benedict A., Liu L., Rader C., Jensen M.C., Riddell S.R. The nonsignaling extracellular spacer domain of chimeric antigen receptors is decisive for in vivo antitumor activity. Cancer Immunol. Res. 2015;3:125–135. doi: 10.1158/2326-6066.CIR-14-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milone M.C., Fish J.D., Carpenito C., Carroll R.G., Binder G.K., Teachey D., Samanta M., Lakhal M., Gloss B., Danet-Desnoyers G. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol. Ther. 2009;17:1453–1464. doi: 10.1038/mt.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.