Abstract
Nonhuman primate (NHP) induced pluripotent stem cells (iPSCs) offer the opportunity to investigate the safety, feasibility, and efficacy of proposed iPSC-derived cellular delivery in clinically relevant in vivo models. However, there is need for stable, robust, and safe labeling methods for NHP iPSCs and their differentiated lineages to study survival, proliferation, tissue integration, and biodistribution following transplantation. Here we investigate the utility of the adeno-associated virus integration site 1 (AAVS1) as a safe harbor for the addition of transgenes in our rhesus macaque iPSC (RhiPSC) model. A clinically relevant marker gene, human truncated CD19 (hΔCD19), or GFP was inserted into the AAVS1 site in RhiPSCs using the CRISPR/Cas9 system. Genetically modified RhiPSCs maintained normal karyotype and pluripotency, and these clones were able to further differentiate into all three germ layers in vitro and in vivo. In contrast to transgene delivery using randomly integrating viral vectors, AAVS1 targeting allowed stable transgene expression following differentiation. Off-target mutations were observed in some edited clones, highlighting the importance of careful characterization of these cells prior to downstream applications. Genetically marked RhiPSCs will be useful to further advance clinically relevant models for iPSC-based cell therapies.
Keywords: nonhuman primate, induced pluripotent stem cells, gene editing, CRISPR/Cas9, AAVS1, rhesus macaque, off-target mutations
Graphical Abstract
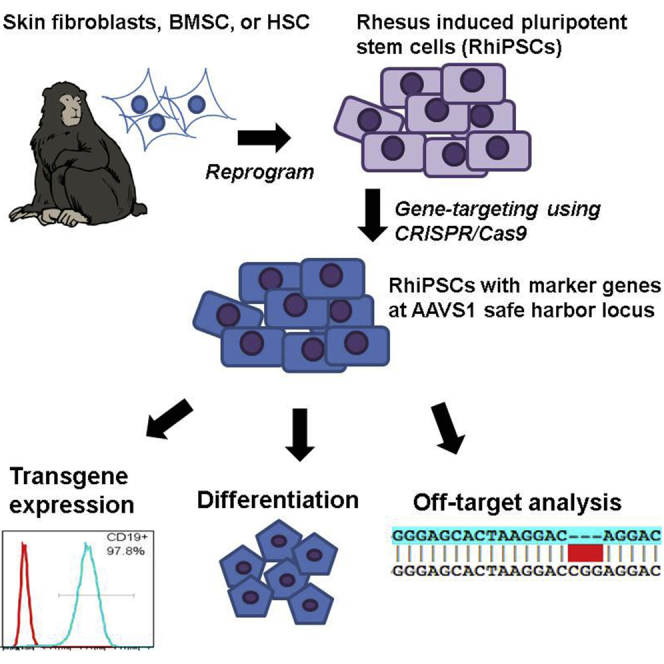
Dunbar and colleagues investigated the utility of the AAVS1 locus as a safe harbor for the addition of transgenes in rhesus macaque iPSCs using the CRISPR/Cas9 system. AAVS1 targeting supported stable transgene expression in differentiated cells of all three germ layers both in vitro and in vivo.
Introduction
The combination of somatic cell reprogramming and powerful new gene editing methods is poised to revolutionize personalized cell-based therapies for inherited and acquired diseases. However, preclinical investigation of safety and efficacy is required before moving these treatment modalities into human clinical trials. Nonhuman primate (NHP) induced pluripotent stem cells (iPSCs) have been valuable resources to model iPSC-based cell therapies in clinically relevant settings.1, 2, 3, 4 The close phylogenetic relationship of NHPs to humans, as well as their size, longevity, and the well-developed understanding of NHP physiology, makes these models very well suited to preclinical iPSC development. The ability to track derivation of tissues from iPSCs is necessary; thus, engineering NHP iPSCs to strongly and stably express reporter genes is desirable for monitoring of autologous or allogeneic transplanted cells in vivo. Additionally, development of these methodologies with reporter genes potentially paves the way for therapeutic gene addition or correction in iPSCs and their progeny.
Traditional methods of non-targeted gene transfer using viral vectors often suffer from transgene silencing during differentiation of PSCs.5, 6, 7 Genomic safe harbors (GSHs) refer to regions in the genome that permit sufficient expression of newly integrated DNA without adverse effects on the host cell or organism.8 Although specific loci or general criteria identifying prospective GSHs have been proposed,8, 9 no GSH has yet been fully validated in a large animal model or a clinical setting.10 Adeno-associated virus integration site 1 (AAVS1) locus in the first intron of PPP1R12C gene is one of most commonly used GSH sites in human cell studies. The AAVS1 locus has an open chromatin configuration in a variety of human cell types, including iPSCs, making this site readily accessible to site-specific endonucleases.11 Genes introduced into the AAVS1 site in human PSCs showed stable expression after in vitro and in vivo differentiation.5, 12, 13, 14, 15 The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9) system is a highly efficient RNA-guided DNA nuclease system modified from an adaptive immune system of bacteria and archaea to allow sequence-specific genome editing in human cells.16, 17, 18 Recent reports have indicated that the CRISPR/Cas9 system can be used for genetic modification in NHPs; however, this approach has not yet been applied to insert genes of interest into the AAVS1 safe harbor in NHP PSCs.19, 20, 21
We have identified a CRISPR/Cas9 target sequence in the rhesus macaque PPP1R12C gene, generated safe harbor-targeted rhesus macaque iPSC (RhiPSC) lines with two relevant marker genes, and documented the stability of expression both in iPSCs and after differentiation to tissues of interest. Genes encoding human truncated CD19 (hΔCD19) or GFP were chosen as reporters. GFP provides convenient and reliable in vivo imaging, but it is highly immunogenic and can result in a substantial loss of gene expression in immunocompetent recipients. The hΔCD19 is desirable for preclinical studies because it has not induced immune rejection in humans or NHPs.22, 23 To evaluate transgene stability in clinically relevant cells, we assessed expression of the transgene in directed differentiated cells representing all three germ layers. The AAVS1-targeting strategy combined with differentiation protocols described in this manuscript provides a foundation for the development of preclinical models for iPSC-based cell therapies.
Results
Generation of AAVS1-Targeted RhiPSC Clones via CRISPR/Cas9
The human AAVS1 locus is located in the first intron of the PPP1R12C gene on chromosome 19, and it is actively expressed in human pluripotent as well as differentiated cells, making it an attractive target for the introduction of a marker or other transgenes.8, 12 The rhesus PPP1R12C gene also is located on chromosome 19, and it has a comparable gene structure to the human ortholog, consisting of 22 exons, with 95% amino acid homology (Figure S1). We confirmed that rhesus PPP1R12C mRNA (Ensembl: ENSMMUT00000030906) was expressed in various rhesus cell types, including iPSCs, bone marrow stromal cells, and primary hepatocytes (Figure S1).
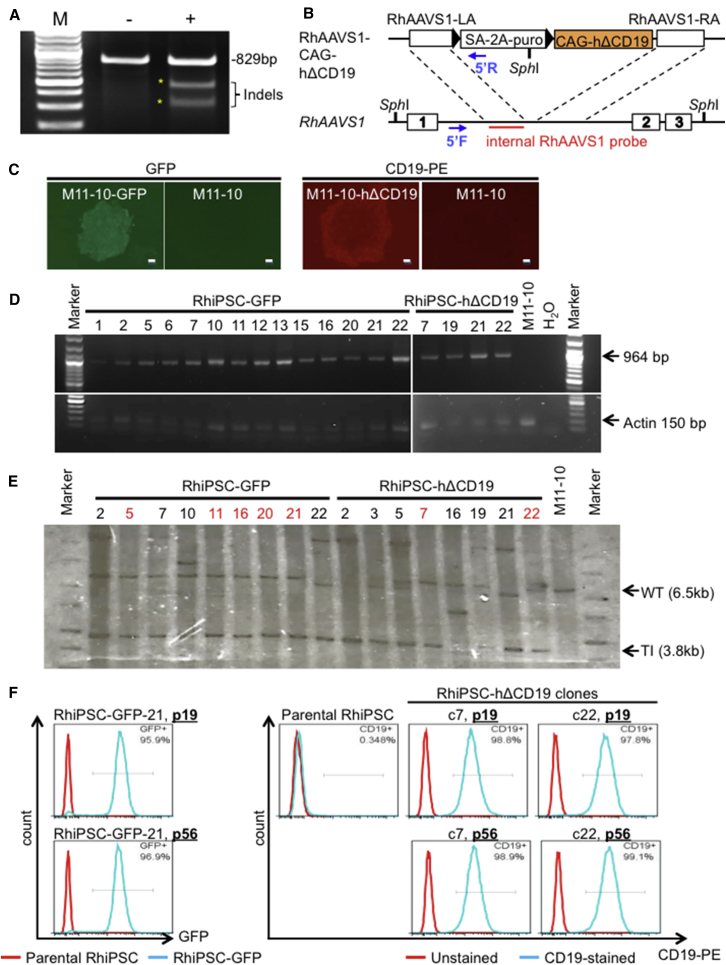
Next, we identified a CRISPR/Cas9 guide RNA (gRNA) intronic target sequence (ggggccactagggacaggaCtgg), which differs only at a single base compared to a highly efficient target sequence (ggggccactagggacaggaTtgg) found at the human AAVS1 site.17 We subsequently cloned this sequence into an all-in-one CRISPR/Cas9 plasmid expression vector. The cutting efficiency of this gRNA was 46%, as determined by the T7 endonuclease I (T7E1) assay performed in a fetal rhesus monkey kidney cell line, FRhK-4 (Figure 1A). A donor plasmid containing the hΔCD19 marker gene was constructed (Figure 1B), and CD19 expression from this plasmid was confirmed in 293T cells (Figure S1). The CRISPR/Cas9/gRNA and donor plasmids (GFP or hΔCD19) were delivered via nucleofection to three RhiPSC lines derived from three independent animals. After puromycin selection, GFP- or hΔCD19-positive colonies were manually picked and expanded for further characterization (Figure 1C). All RhiPSC clones tested had targeted integration of GFP or hΔCD19 at the intended AAVS1 site, confirmed by PCR (Figure 1D; Table 1). A total of 30 targeted clones were screened by Southern blot analysis. Among them, 12 clones had the desired monoallelic transgene insertion, without any additional random genomic insertions (Figure 1E; Table 1). The AAVS1 locus of the non-targeted allele was sequenced in each clone to detect any on-target mutations. The majority of clones (eight of nine tested) had various insertions or deletions (indels) at the non-targeted allele, leaving one of nine clones with an intact wild-type allele (Figure S2). The targeted clones maintained a normal karyotype (Figure S3), and they showed stable transgene expression during 7 months of in vitro culture (Figure 1F).
Figure 1.
Generation of Targeted Rhesus iPSC Clones Using CRISPR/Cas9 System
(A) Cutting efficiency of the rhesus AAVS1 gRNA was 46% in the rhesus FRhK-4 cell line by the T7E1 assay. (B) The rhesus AAVS1-CAG-human truncated CD19 (hΔCD19) donor plasmid was modified from human AAVS1-CAG-copGFP vector by replacing the human AAVS1 homology arms with orthologous rhesus AAVS1 sequences and the GFP cDNA with hΔCD19. (C) GFP- or hΔCD19-expressing RhiPSC colonies following nucleofection and puromycin selection. The GFP clone is shown under UV light, and the hΔCD19 clone is shown with anti-CD19 live staining. M11-10, unedited parental clone. Scale bars, 200 μm. (D) Puromycin-resistant and GFP- or hΔCD19-expressing CRISPR/Cas9-edited clones were plucked, isolated, and subjected to PCR screening for targeted integration, using one primer within the cassette and one outside in the predicted remaining AAVS1 genomic sequences. All puromycin-resistant and GFP- or hΔCD19-expressing clones tested had integration at the AAVS1 site. (E) Southern blot was performed to exclude RhiPSC clones with additional transgene integrations (labeled with black numbers). Red numbers indicate clones had single targeted integrations at the AAVS1 site. WT, wild-type; TI, targeted integration. (F) Transgene expression was maintained after long-term culture.
Table 1.
Summary of CRISPR/Cas9-Mediated Gene Editing in RhiPSCs
| Parental iPSC Clone | Reporter Gene | Targeted Insertiona | Single-Copy Integrationb | Off-Target Mutationsc |
|---|---|---|---|---|
| ZG15-M11-10 | hΔCD19 | 8/8 | 2/8 | 1/2 |
| GFP | 14/14 | 5/9 | 1/4 | |
| ZG32-3-4 | hΔCD19 | 4/4 | 2/4 | 1/1 |
| GFP | 4/4 | 1/4 | 1/1 | |
| ZH26-HS41 | hΔCD19 | 5/5 | 2/5 | 1/2 |
| Total | 35/35 (100%) | 12/30 (40%) | 5/10 (50%) |
Clones with targeted integration/transgene-positive clones, based on PCR analysis.
Monoallelic clones without random integration/clones with targeted integration, based on Southern blot analysis. The rest of colonies had additional random integration event or events, as well as the targeted integration event.
Clones with off-target mutations/clones with single-copy integration, based on Sanger sequencing.
Off-Target Analysis
To investigate off-target (OT) effects of our CRISPR/Cas9 system in RhiPSCs, potential OT sites were identified based on a recently published algorithm24 (Table S1). Among those, the top ten potential off-target sites were sequenced in ten edited RhiPSC clones. Based on BLAT online analysis, off-target site 2 (OT2), OT5, and OT8 located in the intron of CADPS2, ENSMMUG00000003164, and ITGA11, respectively, while other loci did not overlap with any gene. Sequencing analysis revealed that five of ten clones had mutations in OT5 and/or OT8, suggesting higher frequency of off-target events at gene-related sites among the top ten off-target sites (Table 1; Figure S4). Precise sequences of indels were confirmed by subcloning of PCR fragments (Figure S4). Indels ranged from 5- to 162-bp deletions; however, we also confirmed a large insertion (212 bp) into the OT8 site of one clone, which matched the sequence near the left homology arm of the donor plasmid DNA.
Stable Expression of Transgene in Edited RhiPSC Clones following In Vitro Spontaneous Differentiation
Among clones that were confirmed to have monoallelic integration of transgenes by Southern blot, two CD19 clones (clones 7 and 22 derived from parental ZG15-M11-10 iPSC) and two GFP clones (clones 5 and 21 derived from ZG15-M11-10 iPSC) were chosen for further in vitro and in vivo differentiation. Of note, subsequent analysis revealed that two of these clones (CD19-22 and GFP-5) had off-target mutations (Figure S4). Embryoid body (EB) formation was induced in the edited RhiPSC clones to monitor expression of the transgenes following spontaneous in vitro differentiation. After 15 days of culture, EB cells maintained strong expression of GFP or hΔCD19 (Figure S5).
Mesodermal Differentiation from Edited RhiPSC Clones
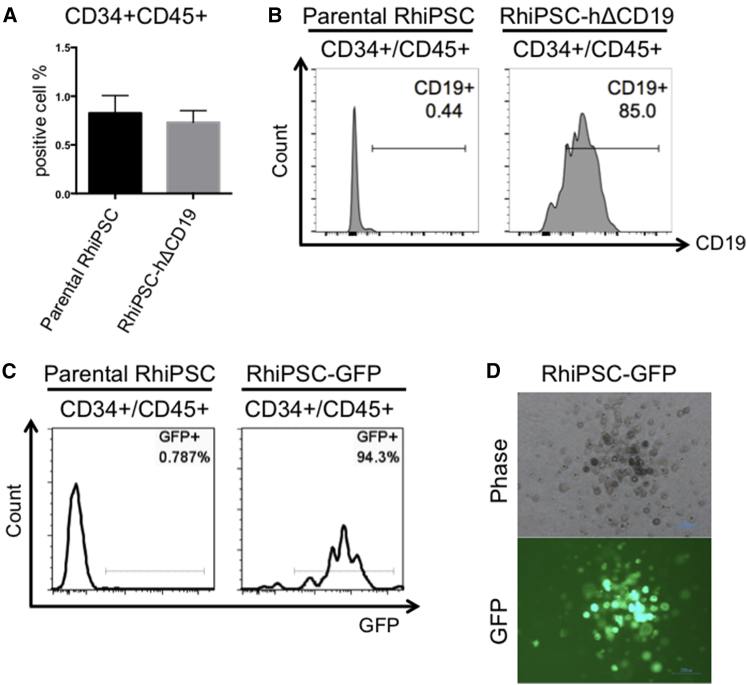
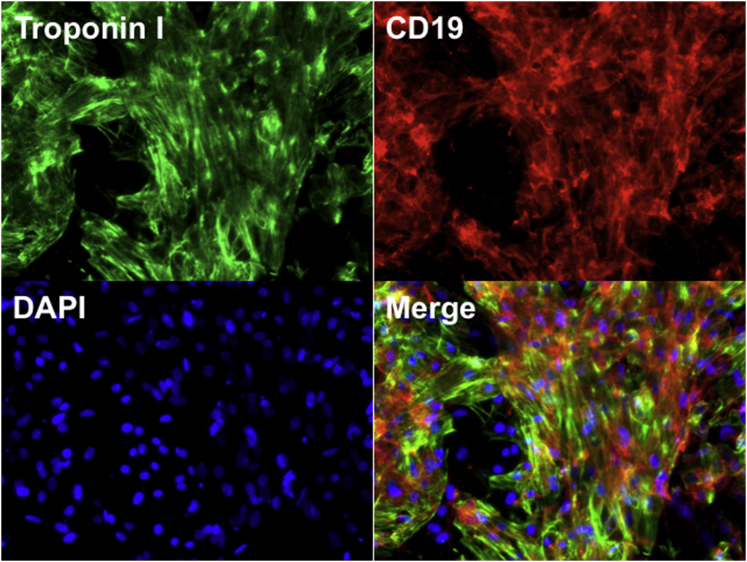
We then investigated whether genetically modified RhiPSCs could generate clinically relevant target tissues while maintaining stable expression of transgenes. Differentiation of rhesus PSCs into mesodermally derived hematopoietic stem/progenitor cells (HSPCs) using human protocols has been known to be extremely inefficient.25 We speculated that inefficient rhesus receptor activation by some human cytokines, particularly IL-3, could be a principal reason for the relatively low differentiation efficiency, given significant protein sequence differences between rhesus and human IL-3 and documented inefficient activity of human IL-3 on rhesus hematopoietic cells.26 We compared the effect of human IL-3 versus rhesus IL-3 during hematopoietic differentiation from RhiPSCs. Rhesus IL-3 significantly increased the number of CD34/45 (0.7% versus 5.4%, respectively) cells measured by flow cytometry on day 20 of differentiation as well as the output of colony-forming units (CFUs, Figure S6). The HSPC differentiation capacity of genetically modified RhiPSCs was comparable to the parental RhiPSC, with the majority of differentiated cells expressing transgenes (Figures 2A–2D). Similarly RhiPSC-derived cardiomyocytes, defined by positive troponin I staining, co-expressed the hΔCD19 (Figure 3).
Figure 2.
Hematopoietic Differentiation from Genetically Modified RhiPSCs
(A) A summary graph of flow cytometry analysis comparing hematopoietic differentiation from RhiPSC-hΔCD19 versus unedited clones. There was no significant difference between groups (Student’s t tests, p = 0.6620, n = 3). Error bars indicate SEM. (B and C) CD34+CD45+ cells derived from RhiPSC-hΔCD19 (B) and RhiPSC-GFP (C) clones maintained transgene expression after differentiation. (D) A representative image of transgene (GFP) expression from a CFU-M derived from hematopoietic differentiation of the RhiPSC-GFP-21 clone. A total of 24 colonies were produced in this differentiation experiment and 24 of 24 were GFP positive.
Figure 3.
Cardiac Differentiation from Genetically Modified RhiPSCs
After 30 days, terminally differentiated cardiomyocytes derived from RhiPSC-hΔCD19 co-expressed transgene (hΔCD19) and a cardiac-specific marker, troponin I. Images were taken at 200×. Representative images from the RhiPSC-hΔCD19-7 clone are shown.
Endodermal Differentiation from Edited RhiPSCs
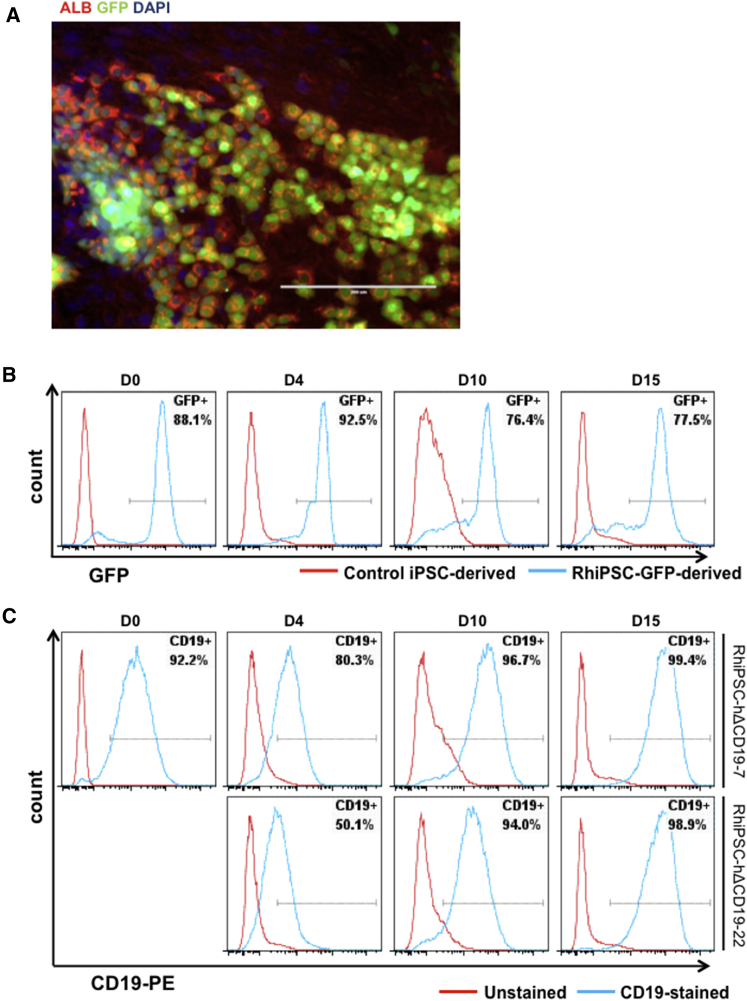
We also investigated whether edited clones could be differentiated into endoderm-derived cells. We adapted a recently published protocol that described the derivation of functional hepatocyte-like cells from human embryonic stem cells (ESCs) and iPSCs.27, 28 Although the kinetics of hepatic differentiation of RhiPSCs was similar to that of human iPSCs, we found that hypoxic culture (5% O2) during definitive endoderm induction was necessary in RhiPSC differentiation (Figure S7). After 4 days of differentiation, pluripotent genes were downregulated and RhiPSCs generated homogeneous definitive endoderm, which expressed SOX17 and FOXA2. Hepatoblasts, expressing alpha-fetoprotein (AFP) but no albumin (ALB), were present by day 12, and, following the addition of dexamethasone, ALB+ hepatocyte-like cells were observed by day 15. The efficiency of generation of hepatocyte-like cells was lower from all RhiPSC clones as compared to human iPSC clones, indicating some species differences and room for further optimization. However, edited RhiPSC clones generated hepatocyte-like cells co-expressing ALB and the knocked-in transgenes (Figure 4A). Marker genes were expressed throughout the differentiation procedure, indicating no significant silencing of the transgenes (Figures 4B and 4C). Of note, for some hΔCD19 clones, we measured lower mean fluorescence intensity of the marker during early stages of hepatic differentiation.
Figure 4.
Hepatic Differentiation from Genetically Modified RhiPSCs
(A) Imaging demonstrates co-expression of GFP and ALB after 15 days of differentiation of an RhiPSC-GFP clone. Scale bar, 200 μm. (B) GFP expression at days 0, 4, 10, and 15 of hepatic differentiation from an RhiPSC-GFP clone is shown. (C) hΔCD19 expression at days 0, 4, 10, and 15 of hepatic differentiation from two RhiPSC-hΔCD19 clones is shown.
Ectodermal Differentiation from Edited RhiPSCs
To study transgene expression during directed differentiation toward ectoderm, the RhiPSC-hΔCD19-7 line was differentiated toward neural lineages. At day 10, RhiPSC-derived neural stem cells, defined by positive PAX6 staining, co-expressed the transgene (hΔCD19) (Figure S8).
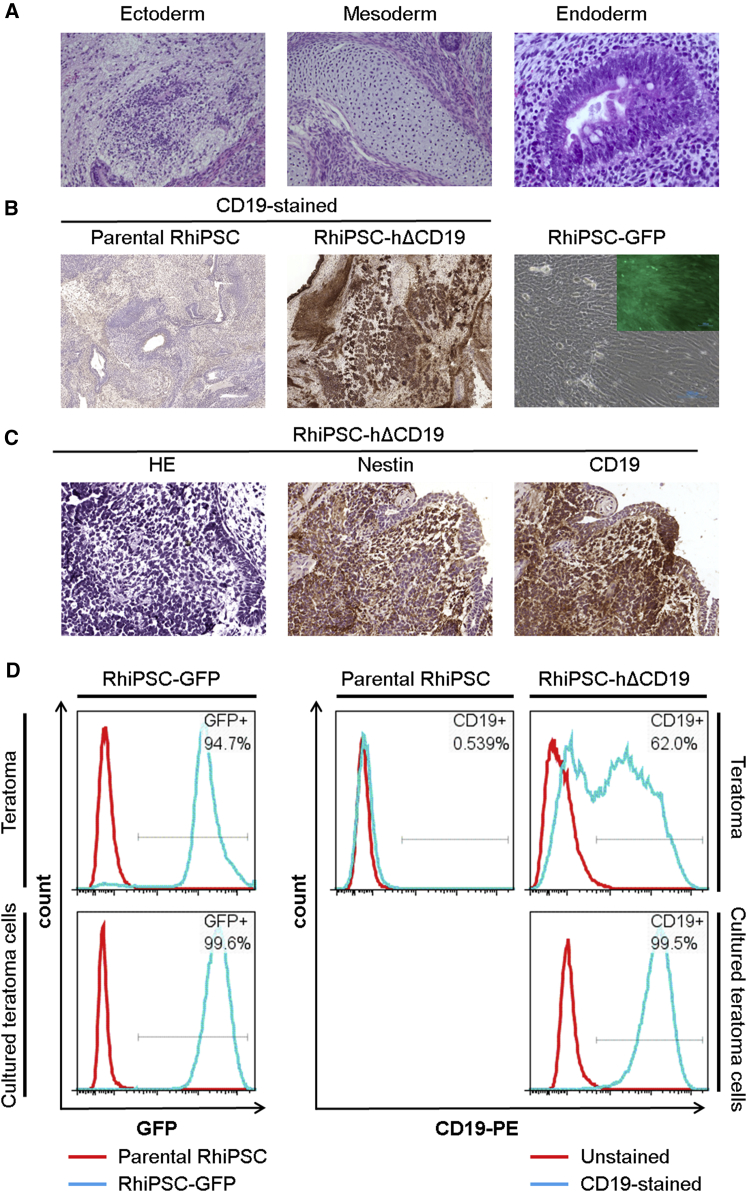
Stable Expression of Transgene in Edited RhiPSC Clones following Teratoma Formation
To monitor long-term in vivo gene expression stability, undifferentiated edited RhiPSC clones were injected subcutaneously into the NOD-scid IL2rγnull (NSG) mice and monitored for teratoma formation. All clones induced teratoma formation within 6–8 weeks (Figure 5A), a similar latency to parental clones. The majority of cells present in the teratomas expressed GFP or hΔCD19, including mature cells derived from all three germ layers (Figures 5B–5D).
Figure 5.
Stable Transgene Expression in CRISPR/Cas9-Edited Clones after In Vivo Differentiation
(A) Representative images of a teratoma formed by an edited RhiPSC-hΔCD19-7 clone. On H&E staining, all three germ-line tissue types were found in each injected animal, demonstrating pluripotency of edited clones. Ectoderm and mesoderm images were taken at 200× and Endoderm was 400×. (B) Representative images show transgene expression in teratoma tissues. Left panel: original RhiPSC-derived teratoma (20×); middle panel: RhiPSC-hΔCD19-7-derived teratoma (20×); right: primary cells isolated from teratoma tissue derived from an RhiPSC-GFP-21 clone (100×). (C) Transgene (hΔCD19) expression in the nestin-positive neuroectoderm (800×). Serial sections of a teratoma derived from RhiPSC-hΔCD19-7 were used for staining. (D) Teratoma cells were dispersed using collagenase II, dispase, and trypsin for flow cytometry analysis. Edited clones exhibited strong expression of GFP (left panel) or hΔCD19 (right panel) after in vivo teratoma formation (top row). Teratoma-derived single cells were further cultured in vitro for 1 week and re-analyzed by flow cytometry (bottom row).
Discussion
The development of PSC-based systems for disease modeling and regenerative therapies of human diseases is promising. Gene targeting can be used to correct certain genetic mutations or to add therapeutic genes into safe harbor loci in PSC-derived therapeutic products. To monitor long-term cell survival, migration, and incorporation of these genetically modified PSCs into the recipient organs, engineered PSCs with proper reporter genes in large animal models are desirable. Traditionally, most genome editing and marker introduction have been done using integrating vectors, such as lentiviruses, due to reliable efficiency and the need to have the marker or therapeutic gene passed on to all daughter cells.29 However, random integration, particularly in stem cells, can be associated with either of two undesirable outcomes: ectopic activation of nearby proto-oncogenes or inactivation due to gene silencing.30, 31 In human cells, it is now feasible to achieve high gene-targeting efficiency with the CRISPR/Cas9 system. Recently, gene-modified cynomolgus monkeys were generated using CRISPR/Cas9, demonstrating that CRISPR/Cas9-mediated gene editing is also feasible in primates.21 It was confirmed in another study where authors performed targeted genome editing in a single rhesus ESC line at the hypoxanthine-guanine phosphoribosyl transferase (HPRT) locus.20
This is the first study reporting successful CRISPR/Cas9-mediated gene addition at the AAVS1 safe harbor locus in NHP PSCs. Modified cells retained their pluripotent phenotype and showed no abnormal karyotype. Importantly, we demonstrated stable expression of our transgenes in undifferentiated and terminally differentiated RhiPSCs. However, for some hΔCD19 clones, we measured heterogeneous transgene expression of the marker during intermediate stages of differentiation. Ordovás et al. recently reported that transgene expression inhibition at the AAVS1 site was observed in a lineage-dependent manner in human iPSCs due to DNA methylation and other unknown mechanisms.32 We did not observe transgene silencing in GFP clones that showed robust GFP expression not only in undifferentiated status but also throughout the differentiation toward all three germ layers. This result suggests that the AAVS1 site can indeed support stable transgene expression in rhesus macaque. On the other hand, tissue- and transgene-specific post-transcriptional regulation may explain the heterogeneity of hΔCD19 expression in some cell types.
In this study, we developed protocols for the efficient differentiation of RhiPSCs toward hematopoietic cells and hepatocytes, both under development in our rhesus models of in vivo tissue regeneration. These differentiation studies required some modifications from human differentiation approaches. Derivation of blood cells from rhesus PSCs has been found to be less efficient compared with humans using the identical protocols.25 Rhesus bone marrow cells have been shown to form in vitro hematopoietic colonies inefficiently in response to human IL-3, with 50-fold lower potency of human in comparison to rhesus IL-3, likely due to several important changes in the IL-3 amino acid sequence between the two species.26 We found that hematopoietic differentiation of rhesus PSCs can be greatly enhanced by replacing human with rhesus IL-3. Although NHPs are highly similar to humans, our data highlight the importance of careful optimization to increase differentiation efficiency.
While CRISPR/Cas9 shows great promise and is highly efficient, this approach also has been shown to result in off-target genomic changes.24, 33, 34, 35, 36 Previous studies on a limited number of human CRISPR/Cas9-edited PSC clones reported a low incidence of off-target mutagenesis as detected by whole-genome sequencing, but they concluded that these changes were most likely not related to CRISPR/Cas9 editing, due to low sequence similarity between the off-target indels and the intended target sites.37, 38 However, a recent study using deep sequencing to detect off-target editing of human AAVS1-T2 gRNA, the counterpart of our rhesus AAVS1 gRNA, found a significant number of indels at several endogenous loci in 293T cells and one site in a targeted human iPSC line.39 In the current study, we identified potential off-target sites in the rhesus genome using the same algorithm,24 and indeed we found indels at one or two sites in half the tested clones. Although we did not see any difference in reporter gene expression, differentiation efficiency, or teratoma formation between clones with or without off-target mutations, the long-term effects of these mutations on behavior of differentiated or undifferentiated iPSCs are unclear. These findings suggest that continued efforts to improve the specificity of CRISPR/Cas9 editing are desirable, for instance, optimization of gRNA choice to decrease off-target effects33 or the use of alternative Cas9 enzymes.36, 40 It is also prudent to screen edited iPSC clones for off-target mutations before utilization.
In summary, we demonstrate that gene targeting into the safe harbor AAVS1 is feasible in rhesus macaque, and targeted RhiPSC clones maintain stable transgene expression without significant silencing during in vitro and in vivo differentiation. The AAVS1-targeting strategy in combination with efficient differentiation protocols described in this manuscript will be useful to further develop autologous preclinical models for iPSC-based cell therapy. For example, a reporter gene inserted at the AAVS1 locus could be used as a marker to closely track the iPSC-derived cells in vivo and to assess efficacy of the transplanted cells. Additionally, the introduction of suicide genes at this site might permit on-demand ablation of transplanted cells in the case of an adverse event.41, 42 Further exploration of NHP homologs of other human safe harbor loci, such as ROSA26 and CLYBL,41, 42 will allow multiplexed transgene knockin and cell marking.
Materials and Methods
Animal Use
All animals used in this study were housed and handled in accordance with protocols approved by the Animal Care and Use Committee of the National Heart, Lung and Blood Institute (H-0294).
Vector Construction and gRNA Screening
The all-in-one CRISPR/Cas9 plasmid PX458 (Addgene 48138), expressing Streptococcus pyogenes Cas9, EGFP, and U6 promoter-driven custom gRNA sequences, was modified to replace CBh with CAG promoter, which was known to drive strong and stable transgene expression in human PSCs,7, 43 and was deposited to Addgene (79144). A rhesus AAVS1 candidate gRNA sequence was identified, consisting of ggggccactagggacaggac (chromosome 19: 61115594–61115613), with a single base change from the corresponding human AAVS1 T2 gRNA sequence.17 To access its cutting efficiency, T7E1 assay was performed. Briefly, rhesus FRhK-4 cell line was transfected with the candidate gRNA, and the AAVS1 region of the target cells was amplified by PCR using Platinum PCR SuperMix (Thermo Fisher Scientific, Life Technologies). Sequences for the primers we utilized are listed in Table S2. The PCR products were denatured (95°C for 5 min) and reannealed (95°C–85°C, ramp rate of –2°C/s followed by 85°C–25°C, ramp rate of –0.3°C/s) to allow heteroduplex formation between wild-type DNA and CRISPR/Cas9-editied DNA. T7E1 (2.5 U/reaction) was added to heteroduplex and incubated at 37°C for 20 min. The resulting cleaved and full-length PCR products were visualized using 2.5% agarose gel. The donor plasmid used for transgene knockin was modified from the human AAVS1-CAG-copGFP vector41 (Addgene 66577) by replacing the human AAVS1 homology arms with rhesus homologous sequences. Rhesus AAVS1 5′ and 3′ homology arm sequences were amplified from RhiPSC genomic DNA, and they were cloned into the vector following digestion with NotI for the 5′ homology arm and PmeI for the 3′ homology arm using Gibson Assembly method. The rhesus AAVS1-CAG-copGFP donor plasmid was deposited at Addgene (73439). The copGFP marker gene was replaced with hΔCD196, 23 following digestion with BsrGI and MluI. The PCR primers used to produce each insert for Gibson Assembly are listed in Table S2.
RhiPSC Culture and EB Differentiation
RhiPSC lines derived from three independent rhesus macaques were used for gene targeting. ZG15-M11-10 and ZG32-3-4 lines were derived from bone marrow stromal cells and skin fibroblasts, respectively, using the Cre-excisableSTEMCCA vector.1 ZH26-HS41 was derived from bone marrow CD34+ cells using the CytoTune-iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific). RhiPSCs were cultured either on mouse embryonic fibroblasts (MEFs, GlobalStem) feeder or Matrigel (BD Biosciences) plates. RhiPSC medium was knockout (KO)/DMEM (Gibco, Life Technologies) supplemented with 20% knockout serum replacement (Life Technologies), 20 ng/ml human bFGF (PeproTech), 0.1 mM minimum essential medium (MEM) nonessential amino acids (Life Technologies), 1% penicillin-streptomycin-glutamine (Life Technologies), and 0.1 mM 2-mercaptoethanol (Sigma-Aldrich). For EB differentiation, day 3–4 undifferentiated RhiPSCs were harvested using a cell scraper and incubated in EB formation medium (RhiPSC medium without bFGF) overnight. The medium was replaced with fresh medium every 4–5 days.
Gene Targeting, Colony Isolation, and Screening
One day before the transfection, puromycin-resistant MEFs (puro-MEF, GlobalStem) were plated. On day 0, nucleofection of RhiPSCs was performed with the 4D-Nucleofector System (Lonza), and 1 × 106 cells were plated onto the puro-MEF plates in RhiPSC medium supplemented with 10 μM Rock inhibitor (Stemgent). Then 2 days following transfection, 0.5 μg/ml puromycin (Sigma-Aldrich) was added for 5 days, and the medium was changed daily until ESC-like colonies appeared. To select hΔCD19-positive clones, cells were live-stained with an anti-human CD19-PE (BD Biosciences). GFP- or hΔCD19-positive colonies were mechanically transferred onto new MEFs and expanded for further analyses. PCR and Southern blot analysis were performed as previously described12, 44 in order to screen for targeted integration and exclude clones with random integration. Sequences for the primers we utilized are listed in Table S2. An AAVS1 5′ homology arm probe was used for Southern blot.
Teratoma Formation
Teratoma formation by RhiPSCs was assessed as previously described.1 Frozen tissue sections were stained with anti-human CD19 (1:800, BD Biosciences) and Nestin (1:400, Neuromics) antibodies.
Off-Target Analyses
To perform off-target analyses, we developed an in-house Python script that calculated the scores for each off-target site, based on the previously published algorithm24 as well as the updated formula posted by the same group (http://crispr.mit.edu/about). The reference genome was downloaded from the Ensembl website (http://www.ensembl.org, release 77), and the unmasked version was used. The script searches for all possible guide sequences in a given region, and it lists off-target sites associated with guide sequences up to four mismatches. We validated our script by comparing results with the online tool for a test region within the human genome. Using this pipeline, 403 candidate off-target sites in the rhesus genome were computationally identified. The top ten potential off-target loci were amplified using the primer sets listed in Table S2. The Platinum PCR SuperMix High Fidelity was used as directed, with optimization of annealing temperature to 61°C for amplification of OT10. The purified PCR products were Sanger-sequenced, and each sequence chromatogram was analyzed with the online Tracking of In/dels by Decomposition (TIDE) software (https://tide.nki.nl). To confirm a precise sequence of indels in some clones, subcloning of PCR fragments was performed using the Zero Blunt TOPO cloning kit (Invitrogen).
Hematopoietic Differentiation
Undifferentiated iPSCs were harvested with Accutase (EMD Millipore), and they were either plated on Matrigel for monolayer differentiation or incubated in ultra-low-attachment 96-well plates (Corning) to generate EBs, in STEMdiff APEL Medium (STEMCELL Technologies) supplemented with 10 μM Rock inhibitor. After 1–2 days, medium was replaced with STEMdiff APEL Medium with 30 ng/mL VEGF (R&D Systems), 30 ng/mL BMP-4 (R&D Systems), 40 ng/mL SCF (Amgen), and 50 ng/mL Activin A (R&D Systems). On day 4, this medium was replaced with hematopoietic differentiation medium: STEMdiff APEL Medium supplemented with 300 ng/mL SCF, 300 ng/mL Flt-3 ligand (Miltenyi Biotec), 10 ng/mL IL-3 (human IL-3 from R&D Systems, rhesus IL-3 from ProSpec), 10 ng/mL IL-6 (R&D Systems), 50 ng/mL G-CSF (filgrastim, Amgen), and 25 ng/mL BMP-4. Medium was replaced with fresh hematopoietic differentiation medium every 3–4 days. After hematopoietic differentiation, cells were dissociated into single cells and analyzed by flow cytometry or plated in CFU assays. For flow cytometry, cells were stained for viability using LIVE/DEAD Fixable Violet Dead Cell Stain Kit (ViViD, Invitrogen), along with CD34 and CD45 (BD Biosciences). Data were acquired on a LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star). For CFU assays, 50,000 EB-derived cells were plated in MethoCult H4435 medium (STEMCELL Technologies) supplemented with 10 ng/mL rhesus IL-3 and incubated for 12–14 days.
Cardiac Differentiation
Cardiomyocytes were differentiated from RhiPSCs by treating with BMP-4, Activin A, and bFGF followed by the Wnt inhibitor IWP-2. At day 30 of differentiation, cells were stained with anti-human CD19-PE and anti-cardiac Troponin I (1:100, Santa Cruz Biotechnology) antibodies.
Hepatic Differentiation
RhiPSCs were first differentiated to SOX17- and FOXA2-positive definitive endoderm in STEMdiff Definitive Endoderm medium (STEMCELL Technologies) in 5% O2. After 4 days, cells were transferred to a normoxic incubator and further differentiated to hepatoblasts in medium with 100 ng/ml hepatocyte growth factor and 1% DMSO for 8 days, as previously described.27, 28 Finally, cells were pushed toward hepatocyte-like cells with the addition of 10−7 M dexamethasone for 3 additional days. At each time point, cells were analyzed by Taqman RT-qPCR and immunostaining as previously described.27
Neural Induction from RhiPSCs
RhiPSCs were differentiated toward neural stem cells using PSC Neural Induction Medium (Gibco). After 7 days of differentiation, cells were passaged according to the manufacturer’s protocol and cultured for 3 more days. Cells were stained with anti-human CD19-PE and anti-PAX6 (1:100, BioLegend) antibodies.
Statistical Analysis
Prism 6 software (GraphPad) was used in statistical analysis and graph creation. Student’s t tests were utilized to analyze flow cytometry and CFU data; p < 0.05 was considered statistically significant. Data are presented as mean ± SEM in all figure panels in which error bars are shown.
Author Contributions
S.H., J.Z., and C.E.D. conceived the study and designed the work. S.H., R.Y., K.C., M.A.F.C., M.J., Y.L., H.L., A.C., A.A.A., and M.P. collected and analyzed data. M.B., R.K.M., C.L.S., H.L.M., T.J.L., I.T., X.W., S.P., T.W., J.Z., and C.E.D performed data analysis and interpretation. S.H., R.Y., T.W., and C.E.D. wrote the manuscript.
Acknowledgments
This research was supported by funding from the Division of Intramural Research at the National Heart, Lung and Blood Institute (NHLBI) at the NIH. We thank Gianpietro Dotti for sharing the pCDH-iCasp9-2A-h ΔCD19 lentiviral vector. We thank the NHLBI Pathology core for tissue processing and Building 50 staff for provision of excellent animal care. We also thank Shiqin Judy Yu for assisting with the iPSC culture, Macallister Harris for his support during the Southern blot analysis, and Alec Nickolls for feedback on neural stem cell staining.
Footnotes
Supplemental Information includes eight figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2016.10.007.
Supplemental Information
References
- 1.Hong S.G., Winkler T., Wu C., Guo V., Pittaluga S., Nicolae A., Donahue R.E., Metzger M.E., Price S.D., Uchida N. Path to the clinic: assessment of iPSC-based cell therapies in vivo in a nonhuman primate model. Cell Rep. 2014;7:1298–1309. doi: 10.1016/j.celrep.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emborg M.E., Liu Y., Xi J., Zhang X., Yin Y., Lu J., Joers V., Swanson C., Holden J.E., Zhang S.C. Induced pluripotent stem cell-derived neural cells survive and mature in the nonhuman primate brain. Cell Rep. 2013;3:646–650. doi: 10.1016/j.celrep.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hallett P.J., Deleidi M., Astradsson A., Smith G.A., Cooper O., Osborn T.M., Sundberg M., Moore M.A., Perez-Torres E., Brownell A.L. Successful function of autologous iPSC-derived dopamine neurons following transplantation in a non-human primate model of Parkinson’s disease. Cell Stem Cell. 2015;16:269–274. doi: 10.1016/j.stem.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong S.G., Dunbar C.E., Winkler T. Assessing the risks of genotoxicity in the therapeutic development of induced pluripotent stem cells. Mol. Ther. 2013;21:272–281. doi: 10.1038/mt.2012.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith J.R., Maguire S., Davis L.A., Alexander M., Yang F., Chandran S., ffrench-Constant C., Pedersen R.A. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells. 2008;26:496–504. doi: 10.1634/stemcells.2007-0039. [DOI] [PubMed] [Google Scholar]
- 6.Wu C., Hong S.G., Winkler T., Spencer D.M., Jares A., Ichwan B., Nicolae A., Guo V., Larochelle A., Dunbar C.E. Development of an inducible caspase-9 safety switch for pluripotent stem cell-based therapies. Mol. Ther. Methods Clin. Dev. 2014;1:14053. doi: 10.1038/mtm.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou J., Sweeney C.L., Chou B.K., Choi U., Pan J., Wang H., Dowey S.N., Cheng L., Malech H.L. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells: functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood. 2011;117:5561–5572. doi: 10.1182/blood-2010-12-328161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2011;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 9.Papapetrou E.P., Lee G., Malani N., Setty M., Riviere I., Tirunagari L.M., Kadota K., Roth S.L., Giardina P., Viale A. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011;29:73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papapetrou E.P., Schambach A. Gene insertion into genomic safe harbors for human gene therapy. Mol. Ther. 2016;24:678–684. doi: 10.1038/mt.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rensburg R., Beyer I., Yao X.Y., Wang H., Denisenko O., Li Z.Y., Russell D.W., Miller D.G., Gregory P., Holmes M. Chromatin structure of two genomic sites for targeted transgene integration in induced pluripotent stem cells and hematopoietic stem cells. Gene Ther. 2013;20:201–214. doi: 10.1038/gt.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Y., Liu C., Cerbini T., San H., Lin Y., Chen G., Rao M.S., Zou J. Stable enhanced green fluorescent protein expression after differentiation and transplantation of reporter human induced pluripotent stem cells generated by AAVS1 transcription activator-like effector nucleases. Stem Cells Transl. Med. 2014;3:821–835. doi: 10.5966/sctm.2013-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekine K., Takebe T., Taniguchi H. Fluorescent labeling and visualization of human induced pluripotent stem cells with the use of transcription activator-like effector nucleases. Transplant. Proc. 2014;46:1205–1207. doi: 10.1016/j.transproceed.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Oceguera-Yanez F., Kim S.I., Matsumoto T., Tan G.W., Xiang L., Hatani T., Kondo T., Ikeya M., Yoshida Y., Inoue H., Woltjen K. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods. 2016;101:43–55. doi: 10.1016/j.ymeth.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Zhang P.W., Haidet-Phillips A.M., Pham J.T., Lee Y., Huo Y., Tienari P.J., Maragakis N.J., Sattler R., Rothstein J.D. Generation of GFAP:GFP astrocyte reporter lines from human adult fibroblast-derived iPS cells using zinc-finger nuclease technology. Glia. 2016;64:63–75. doi: 10.1002/glia.22903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeder M.L., Gersbach C.A. Genome-editing technologies for gene and cell therapy. Mol. Ther. 2016;24:430–446. doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y., Zheng Y., Kang Y., Yang W., Niu Y., Guo X., Tu Z., Si C., Wang H., Xing R. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum. Mol. Genet. 2015;24:3764–3774. doi: 10.1093/hmg/ddv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu S., Rong Z., Lu X., Xu Y., Fu X. Gene targeting through homologous recombination in monkey embryonic stem cells using CRISPR/Cas9 system. Stem Cells Dev. 2015;24:1147–1149. doi: 10.1089/scd.2014.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 22.Barese C.N., Felizardo T.C., Sellers S.E., Keyvanfar K., Di Stasi A., Metzger M.E., Krouse A.E., Donahue R.E., Spencer D.M., Dunbar C.E. Regulated apoptosis of genetically modified hematopoietic stem and progenitor cells via an inducible caspase-9 suicide gene in rhesus macaques. Stem Cells. 2015;33:91–100. doi: 10.1002/stem.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Stasi A., Tey S.K., Dotti G., Fujita Y., Kennedy-Nasser A., Martinez C., Straathof K., Liu E., Durett A.G., Grilley B. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med. 2011;365:1673–1683. doi: 10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajesh D., Chinnasamy N., Mitalipov S.M., Wolf D.P., Slukvin I., Thomson J.A., Shaaban A.F. Differential requirements for hematopoietic commitment between human and rhesus embryonic stem cells. Stem Cells. 2007;25:490–499. doi: 10.1634/stemcells.2006-0277. [DOI] [PubMed] [Google Scholar]
- 26.Burger H., van Leen R.W., Dorssers L.C., Persoon N.L., Lemson P.J., Wagemaker G. Species specificity of human interleukin-3 demonstrated by cloning and expression of the homologous rhesus monkey (Macaca mulatta) gene. Blood. 1990;76:2229–2234. [PubMed] [Google Scholar]
- 27.Carpentier A., Tesfaye A., Chu V., Nimgaonkar I., Zhang F., Lee S.B., Thorgeirsson S.S., Feinstone S.M., Liang T.J. Engrafted human stem cell-derived hepatocytes establish an infectious HCV murine model. J. Clin. Invest. 2014;124:4953–4964. doi: 10.1172/JCI75456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carpentier A., Nimgaonkar I., Chu V., Xia Y., Hu Z., Liang T.J. Hepatic differentiation of human pluripotent stem cells in miniaturized format suitable for high-throughput screen. Stem Cell Res. (Amst.) 2016;16:640–650. doi: 10.1016/j.scr.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wianny F., Bernat A., Huissoud C., Marcy G., Markossian S., Cortay V., Giroud P., Leviel V., Kennedy H., Savatier P., Dehay C. Derivation and cloning of a novel rhesus embryonic stem cell line stably expressing tau-green fluorescent protein. Stem Cells. 2008;26:1444–1453. doi: 10.1634/stemcells.2007-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivière I., Dunbar C.E., Sadelain M. Hematopoietic stem cell engineering at a crossroads. Blood. 2012;119:1107–1116. doi: 10.1182/blood-2011-09-349993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlesinger S., Goff S.P. Retroviral transcriptional regulation and embryonic stem cells: war and peace. Mol. Cell. Biol. 2015;35:770–777. doi: 10.1128/MCB.01293-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ordovás L., Boon R., Pistoni M., Chen Y., Wolfs E., Guo W., Sambathkumar R., Bobis-Wozowicz S., Helsen N., Vanhove J. Efficient recombinase-mediated cassette exchange in hPSCs to study the hepatocyte lineage reveals AAVS1 locus-mediated transgene inhibition. Stem Cell Reports. 2015;5:918–931. doi: 10.1016/j.stemcr.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cradick T.J., Fine E.J., Antico C.J., Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith C., Gore A., Yan W., Abalde-Atristain L., Li Z., He C., Wang Y., Brodsky R.A., Zhang K., Cheng L., Ye Z. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veres A., Gosis B.S., Ding Q., Collins R., Ragavendran A., Brand H., Erdin S., Cowan C.A., Talkowski M.E., Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith C., Abalde-Atristain L., He C., Brodsky B.R., Braunstein E.M., Chaudhari P., Jang Y.Y., Cheng L., Ye Z. Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol. Ther. 2015;23:570–577. doi: 10.1038/mt.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerbini T., Funahashi R., Luo Y., Liu C., Park K., Rao M., Malik N., Zou J. Transcription activator-like effector nuclease (TALEN)-mediated CLYBL targeting enables enhanced transgene expression and one-step generation of dual reporter human induced pluripotent stem cell (iPSC) and neural stem cell (NSC) lines. PLoS ONE. 2015;10:e0116032. doi: 10.1371/journal.pone.0116032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irion S., Luche H., Gadue P., Fehling H.J., Kennedy M., Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat. Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- 43.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winkler T., Hong S.G., Decker J.E., Morgan M.J., Wu C., Hughes W.M., 5th, Yang Y., Wangsa D., Padilla-Nash H.M., Ried T. Defective telomere elongation and hematopoiesis from telomerase-mutant aplastic anemia iPSCs. J. Clin. Invest. 2013;123:1952–1963. doi: 10.1172/JCI67146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.