Abstract
Lentiviral vectors (LVs) are a highly valuable tool for gene transfer currently exploited in basic, applied, and clinical studies. Their optimization is therefore very important for the field of vectorology and gene therapy. A key molecule for LV function is the envelope because it guides cell entry. The most commonly used in transiently produced LVs is the vesicular stomatitis virus glycoprotein (VSV-G) envelope, whose continuous expression is, however, toxic for stable LV producer cells. In contrast, the feline endogenous retroviral RD114-TR envelope is suitable for stable LV manufacturing, being well tolerated by producer cells under constitutive expression. We have previously reported successful, transient and stable production of LVs pseudotyped with RD114-TR for good transduction of T lymphocytes and CD34+ cells. To further improve RD114-TR-pseudotyped LV cell entry by increasing envelope expression, we codon-optimized the RD114-TR open reading frame (ORF). Here we show that, despite the RD114-TRco precursor being produced at a higher level than the wild-type counterpart, it is unexpectedly not duly glycosylated, exported to the cytosol, and processed. Correct cleavage of the precursor in the functional surface and transmembrane subunits is prevented in vivo, and, consequently, the unprocessed precursor is incorporated into LVs, making them inactive.
Keywords: gene vector engineering, gene vector delivery, gene therapy, envelope glycoprotein, RD114-TR, codon optimization, lentiviral vector, RNA structure, packaging cell line
Introduction
Pseudotyping envelopes of viral vectors are heterologous glycoproteins with the key role of mediating vector entry into target cells. Thus, their nature, function, and density on the vector surface may deeply influence the transduction ability of the vectors.1 A powerful strategy to increase the expression of heterologous proteins in eukaryotic cells is codon optimization (co), which is an artificial process through which DNA sequences are modified by the introduction of silent mutations, generating synonymous codons. By degeneracy of the genetic code, all amino acids (aa) except Met and Trp are encoded by more than one codon; i.e., synonymous codons. Genetic code redundancy makes DNA triplets tolerant for point mutations, which do not result in corresponding aa mutations (silent mutations). Codon optimization is exploited to overcome species-specific codon usage bias and ultimately improve heterologous protein production. The frequency of codon distribution within the genome (codon usage bias) is variable and differs depending on species. It follows that tRNAs corresponding to synonymous codons are not equally abundant in different cell types and species. Therefore, for a certain aa, there are synonymous codons more often used, influencing the timing and efficiency of protein translation.2, 3, 4 The codon adaptation index (CAI) technique measures synonymous codon usage bias in a given species. The CAI uses a range (between 0 and 1, where 1 is the maximum translational efficiency) of high-rate expression genes (i.e., ribosomal proteins and elongation factors) to assess the relative contribution of each codon in a specific organism, allowing comparison with the nucleotide sequence of interest.5 Thus, it is possible to increase the expression of a certain gene in a specific organism/cell type by simply changing rare codons with more frequent ones, resulting in modification of the CAI. Codon optimization has been extensively used to increase the production of either recombinant proteins or viral vectors.6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17
RD114-TR is a chimeric mutant deriving from the feline endogenous retrovirus RD114 envelope, in which the TR domain of the gamma retroviral vector (γ-RV) Moloney leukemia virus (MLV) amphotropic 4070-A envelope, fused at the C-terminal end of RD114, increases envelope incorporation into lentiviral vector (LV) particles.18 RD114-TR is first translated in a non-functional precursor (PR) that is then processed by the membrane-associated endoprotease furin in the surface (SU) and transmembrane (TM) active subunits. RD114-TR processing occurs either in furin-rich compartments of the trans-Golgi network, where the PR accumulates during its way to the plasma membrane or in the recycling endosomes close to the plasma membrane.19 The cleavage and post-translational glycosylation of RD114-TR are crucial for trafficking to the plasma membrane and for incorporation into nascent virion coats. The TM subunit mediates plasma membrane anchoring of the SU subunit. Upon recognition and engagement of functional subunits to specific receptors, fusion between viral and cell membranes mediates the entry of the vector into target cells. RD114-TR-pseudotyped retroviral vectors are suitable for both ex vivo and in vivo gene therapy applications because they can be concentrated by centrifugation and are resistant to human serum complement inactivation.20, 21, 22, 23
To improve and simplify the expression of the RD114-TR envelope during development of the RD-MolPack packaging technology for stable and constitutive manufacturing of LVs,21, 23 we codon-optimized the entire RD114-TR open reading frame (ORF). This idea stemmed from our previous observation that RD114-TR expression is achieved only when the β-globin intron (BGI) is inserted between the promoter and the RD114-TR cDNA of the expression cassette of many different expression plasmids tested.23 To explain this constraint, we hypothesized that BGI may attenuate the negative effect of interfering sequences existing in the RD114-TR cDNA. To eliminate these sequences and to simplify the vector design, we decided to codon-optimize the entire RD114-TR ORF. In fact, the elimination of the interfering sequences would have avoided using the BGI, therefore reducing the size of the vector. Unexpectedly, we found that, despite the high level of transcription/translation and cytosol export, RD114-TRco is functionally dead. Our data strengthen the conclusion, also supported by other studies,24 that codon optimization may not always lead to functional improvement of the gene of interest.
Results
Expression of RD114-TR on RD3-MolPack-GFP Producer Cells and Their Derived LVs
We initially analyzed the expression of the RD114-TRWT envelope in RD3-MolPack-GFP producer cells and in their derived LVs23 to confirm previous studies describing proper processing and trafficking to the plasma membrane of the wild-type (WT) envelope.19 RD3-MolPack-GFP cells contain 12 copies of the integrated self inactivating (SIN)-RD114-TRWT-IN-rev-responsive element (RRE) transfer vector (TV) (Figure 1A, scheme 2), and the originated RD114-TRWT pseudo-typed LVs are proficient in cell transduction, as reported previously.23 We used two specific antibodies (Abs), each recognizing either the PR and SU (anti-SU) subunits or the PR and TM (anti-TM) subunits, respectively (Figure 1B). To visualize the expression of RD114-TRWT at the RD3-MolPack-GFP plasma membrane, we carried out pull-down (PD) of biotinylated and de-glycosylated total cell extracts. Because, in SDS-PAGE, glycosylated PR and SU molecules co-migrate,19 we first pulled down membrane proteins, which were first biotinylated in vivo and then deglycosylated by peptide N-glycosidase F (PNGaseF) treatment in vitro. PNGaseF cleaves the link between asparagine and N-acetylglucosamine residues (complex oligosaccharides) that are added in the endoplasmic reticulum (ER) and the Golgi stack. We here confirmed the results previously reported by Sandrin et al.,19 showing that both TM and SU subunits are correctly localized at the plasma membrane, whereas the PR does not reach and/or accumulate on it (Figure 2A, lane 8). The very low level of PR detected in the PNGaseF-treated sample likely derives from contamination of the endoplasmic reticulum or other membranes (Figure 2A, anti-SU, top right panel, lane 8). To further characterize RD114-TR glycosylation and trafficking to the plasma membrane, we treated in vitro producer cellular and derived vector extracts not only with PNGaseF but also with endoglycosidase H (EndoH) enzyme. The latter is active on N-linked high-mannose oligosaccharides (simplex oligosaccharides), added in the ER compartment, but not on high-glucose residues attached later during glycosylation in the Golgi apparatus. It follows that glycoproteins carrying complex oligosaccharides become resistant to the attack of EndoH (EndoH-resistant proteins). Of note, we observed that, in both cells and derived LVs, PR and TM subunits are EndoH-sensitive (Figure 2B, lanes 3 and 6, anti-TM). On the contrary, the SU subunit is EndoH-resistant because it carries complex oligosaccharides (Figure 2B, lanes 3 and 6, anti-SU). The TM contains one putative N-linked glycosylation site (NxS and NxT, where x is any aa), whereas SU contains 11 sites (Figure S1). It is possible that this unique N-linked site in TM is glycosylated with simplex and not complex oligosaccharides and that the TM subunit is transported to the plasma membrane linked to the SU. Furthermore, the average titer of RD3-MolPack-GFP LVs tested in this study is 1.6 × 106 ± 4.7 × 105 SEM transducing units (TU)/mL (n = 5), in line with our previous collective data.21, 23 Overall, these findings demonstrate that expression of RD114-TRWT in RD3-MolPack-GFP producer cells and stemmed LVs is correctly achieved.
Figure 1.
Schematic of the Constructs and Abs Used in This Study
(A) Drawings of the plasmids in their linearized forms. (B) Scheme of the RD114-TR protein, where specific anti-TM and anti-SU Ab recognition and furin cleavage site are indicated. SA, splice acceptor; SD, splice donor; cPPT, central polypurine tract; hPGK, human phosphoglycerate kinase promoter; A, poly A sequence; T, cytoplasmic tail; R, R peptide.
Figure 2.
RD114-TRWT Expression on RD3-MolPack-GFP Producer Cells
(A) PD assay of cell surface-biotinylated proteins followed by western blot analysis. Membrane biotinylation was performed on intact cells. 1 mg of total cell extracts was incubated with 60 μL of streptavidin-conjugated Dynabeads for PD. The PD deglycosylated TM subunit migrated slightly higher than expected because of erroneous migration of the proteins in SDS-PAGE. (B) PNGaseF and EndoH deglycosylations were performed on cellular (30 μg whole-cell extracts) and viral (50 ng p24Gag equivalent) proteins obtained from RD3-MolPack-GFP cells, and their derived viral vectors were collected after 48 hr from cell seeding. Anti-calnexin and anti-ERK staining are internal controls for nonspecific biotinylation and PD; anti-p24Gag staining is for internal loading control. A long exposure of the film is included to better visualize the absence of the EndoH-resistant band of the TM subunit (lane 3) in the vectors. * indicates non-specific staining of the anti-TM Abs recognizing BSA present in the supernatants. ‡ refers to EndoH-resistant SU and TM proteins.
Functional Inactivation of the RD114-TR Envelope by Codon Optimization of the Entire ORF
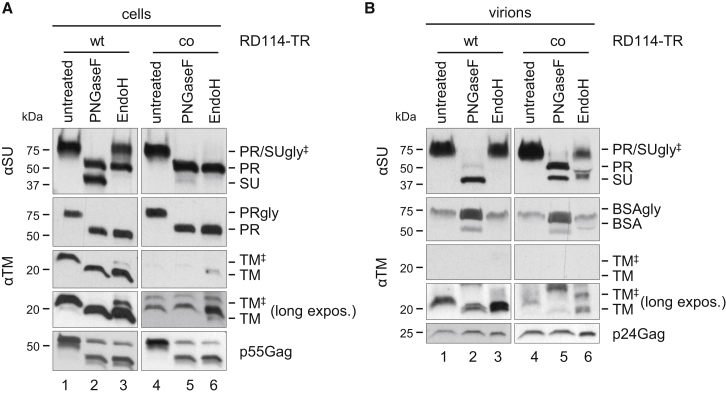
In an attempt to enhance the transduction efficiency of RD3-MolPack-derived LVs by increasing the expression and stability of RD114-TR glycoprotein, we codon-optimized its complete cDNA. After recoding, the CAI of the RD114-TR ORF shifted from 0.64 to 0.98, and the average GC content increased from 48% (WT) to 61% (co), resulting in 73% identity between the WT and co sequences (Figures S2 and S3). To test the function of RD114-TRco, the new ORF, cloned into the pIRES-puro3 expression vector, was transiently co-transfected in PK-7 cells together with the SIN-GFP TV to produce RD114-TRco-expressing LVs. RD114-TRWT-pseudotyped LVs were produced for comparison. We analyzed the expression of RD114-TR proteins by western blot, treating cell and virion extracts with or without PNGaseF and EndoH (Figure 3). Surprisingly, the pattern of RD114-TRco subunits greatly differed from that of the WT counterparts. In fact, both cell and LV protein extracts showed very high levels of PRco and very low levels or even absence of processed SUco and TMco subunits (Figures 3A and 3B). In contrast, the expression profile of RD114-TRWT in cell and vector extracts was identical to that of RD3-MolPack-GFP producer cells and the LVs shown in Figure 2. In agreement with these data, the viral titer of RD114-TRWT pseudotyped LVs calculated on CEM A3.01 cells was 3.9 × 104 ± 7.1 × 103 SEM TU/mL (n = 3), whereas that of RD114-TRco-pseudotyped LVs was consistently undetectable.
Figure 3.
RD114-TRWT and RD114-TRco Expression in Transiently Transfected PK-7 Cells and Generated LVs
(A) Western blot analysis of whole-cell proteins (30 μg) obtained from PK-7 cells after transfection of SIN-EGFP TV and envelope plasmids encoding either RD114-TRWT or RD114-TRco, treated or not treated with either PNGaseF or EndoH enzymes and then probed with anti-SU and anti-TM Abs. (B) Vector proteins (160 ng of p24Gag equivalent) obtained from the virions produced by the transfected cells shown in (A), treated or not treated with either PNGaseF or EndoH enzymes, and then probed with anti-SU and anti-TM Abs. Anti-HIV protein staining (p55Gag and p24Gag) was used as an internal control. A long exposure of the films is included to better visualize the absence of the EndoH-resistant band of the TM subunit (lanes 3 and 6). ‡ refers to EndoH-resistant SU and TM proteins.
RD114-TRco Is Correctly Cleaved by Furin In Vitro
To better understand the difference between PRWT and PRco processing, we tested whether codon optimization might have somehow compromised furin-mediated cleavage of RD114-TRco. To this purpose, we treated cell extracts derived from PK-7 cells transfected with either the RD114-TRWT or RD114-TRco plasmid with recombinant furin overnight at 16°C. Untreated and treated extracts were then analyzed by western blot using the anti-TM Ab (Figure 4A). We observed that, after furin treatment in vitro, the level of TMco subunit clearly increases (Figure 4A, lane 4), even though it is difficult to appreciate the corresponding decrement of PRco because of its high level of expression. On the contrary, the amount of PRWT is clearly decreased, although it is difficult to appreciate the corresponding increase of TMWT because the wild-type protein is already abundantly cleaved before cell protein extraction. Overall, these results support the idea that codon optimization does not compromise furin-mediated cleavage of the envelope, at least in vitro. Based on this notion, we then tried to understand why the PRco is not correctly processed in vivo. One possible explanation was that a large amount of PRco could trigger the phenomenon known as excess substrate inhibition. To exclude this possibility, we transfected HEK293T cells with a scalar amount of RD114-TRco plasmid and tested the corresponding cell extracts in a western blot to find the lowest possible dose of PRco substrate not inhibiting endogenous furin action (Figure 4B). We observed that, even at the lowest amount of plasmid generating detectable PRco, the TMco subunit was not visible, indicating that in vivo PRco is not processed (Figure 4B, lane 3). Further analyses are required to explain the defect underlying this obscure phenomenon.
Figure 4.
In Vitro and In Vivo Furin Cleavage of RD114-TRco
(A) Western blot analysis of whole-cell proteins (30 μg) extracted from PK-7 cells, transfected with either RD114-TRWT or RD114-TRco plasmid and treated overnight at 16°C with 4 U/sample of recombinant furin. The filter was probed with the anti-TM Ab and, after stripping, with anti-HIV serum as an internal control. (B) Western blot analysis of whole-cell proteins (30 μg) extracted from HEK293T cells transfected with the indicated amounts of plasmid DNA encoding either RD114-TRWT or RD114-TRco. The filter was stained with anti-TM Ab and, after stripping, with anti-β actin Ab as an internal control.
Chimeric RD114-TR5′co and RD114-TR3′co Are Not Functional
We next evaluated whether partial recoding of the ORF could restore the function of the RD114-TRco envelope. To this aim, we generated two cDNAs recoded only in the 5′ or 3′ half of the cDNA sequence. We transiently transfected either the RD114-TR5′co or RD114-TR3′co chimera, cloned into the pIRES-puro3 plasmid, into PK-7 cells together with the SIN-EGFP TV. We then tested cellular and LV extracts in a western blot and LV titer in CEM A3.01 cells. Immunoblot analysis demonstrated that, for both chimeric RD114-TR glycoproteins, PRco processing was impaired (Figure S4). Furthermore, although we transfected equal amounts of RD114-TR, RD114-TR5′co, and RD114-TR3′co plasmid DNA, the expression of RD114-TR3′co was lower than that of RD114-TR5′co and RD114-TRWT (Figures S4A and S4C, lanes 3 and 4). The TM3′co and TM5′co subunits were not detectable in the respective LVs, whereas, after PNGaseF treatment, SU3′co and SU5′co were barely visible and visible, respectively. We explain the difference between anti-TM and anti-SU staining with an intrinsic difference in the specific affinity of the two Abs. In agreement, the titer of LVs pseudotyped with half-recoded envelopes was negative. Altogether, these results suggest that neither partial recoding restores the function of RD114-TRco.
Intracellular Localization of RD114-TRWT and RD114-TRco
To see whether RD114-TRco differs from RD114-TRWT in its subcellular localization, we carried out confocal microscope imaging in COS-7 cells transfected with pIRES-RD114-TR plasmids. Forty-eight hours after transfection, RD114-TR expression was visualized together with that of calnexin and VAMP8/Endobrevin, which are ER and early and late endosomal markers, respectively. As shown previously by Sandrin et al.,19 RD114-TRWT is expressed in the cytosol and perinuclear region and is co-localized mostly with calnexin and very poorly with Endobrevin/VAMP8. A similar staining pattern and subcellular localization was observed for RD114-TRco in either the COS-7 (Figure 5) or PK-7 cell experimental setting (Figure S5), indicating that ER and early and late endosome trafficking of RD114-TR is not affected by codon optimization.
Figure 5.
Subcellular Localization of RD114-TRWT and RD114-TRco
(A–D) COS-7 cells transfected with pIRES-RD114-TR plasmids were fixed, permeabilized, and stained at room temperature with the Abs as indicated. Endobrevin/VAMP8 is an early and late endosomal marker, and calnexin is an ER marker. Nuclei were stained with DAPI. Scale bar, 40 μm.
Analysis of the Splicing and Metabolism of RD114-TRWT and RD114-TRco mRNA
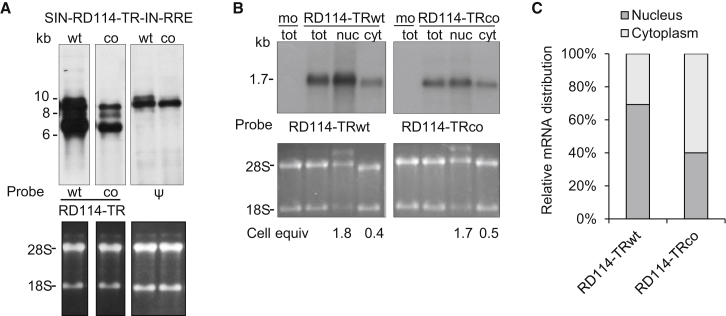
Because many groups have demonstrated that silent mutations affect correct pre-mRNA splicing by introducing cryptic splice sites or altering splicing control elements (i.e., exonic splicing enhancers and silencers),3, 25, 26 we and two service provider companies analyzed RD114-TRco mRNA both in silico and in vitro for the presence of potential cryptic splicing sites. The first in silico service-provided analysis identified one consensus (cryptic) splice donor site that was nullified by codon optimization, whereas the second service-provided analysis recognized no cryptic sites (Figures S2, S3, S6, and S7). We also examined the RD114-TRWT and RD114-TRco ORFs in silico using the NetGene2 server, which calculates the probability of cryptic splicing sites in pre-mRNA sequences. We did not pinpoint any differences between wild-type and codon-optimized ORFs. To further confirm these results, we assessed RD114-TRWT and RD114-TRco mRNA transcripts derived from PK-7 cells transiently transfected with the SIN-RD114-TRWT/co-IN-RRE constructs (Figure 1A, schemes 2 and 3) by northern blot (Figure 6A). Two sequence-specific probes targeting RD114-TRWT and RD114-TRco, respectively, recognized qualitatively comparable RD114-TR mRNA transcript patterns (Figure 6A). Similar results were obtained by using a probe directed against the packaging signal (ψ), which is a sequence common to both constructs. The overall steady-state level of RD114-TRco RNA detected by the ψ probe was only slightly reduced compared with the wild-type counterpart, but no extra spliced bands were observed. These results indicate that the two lentiviral vector plasmids were equally transfected and correctly expressed from the 5′ long terminal repeat (LTR)-cytomegalovirus (CMV) vector promoter. These findings indicate that no cryptic splicing sites are present either in the ORF or in the vector backbone (Figure 6A).
Figure 6.
Northern Blot Analysis of RD114-TRWT and RD114-TRco mRNA
(A) Total RNA (5 μg) extracted from PK-7 cells transiently transfected with SIN-eGFP TV and either SIN-RD114-TRWT or SIN-RD114-TRco plasmids was tested with two sequence-specific RD114-TRWT and RD114-TRco probes, respectively, and the common ψ probe as an internal control. Three bands were detected for both mRNAs, corresponding to the full-length (ψ and specific RD114-TR probes), the spliced, and the internal cassette transcripts, respectively (specific RD114-TR probes). Bottom: ethidium bromide (EtBr) staining of the agarose gel showing 28S and 18S RNAs. (B) Total, nuclear, and cytosolic mRNA (5 μg) extracted from PK-7 cells transiently mock-transfected (mo) and transfected with the pIRES-puro3-based plasmids encoding either RD114-TRWT or RD114-TRco were tested with two sequence-specific RD114-TRWT and RD114-TRco probes, respectively. A single band derived from the expression cassette was detected for both samples. Bottom: EtBr staining of the agarose gel showing 28S and 18S RNAs. (C) qRT-PCR analysis of the nuclear and cytoplasmic distribution of RD114-TRWT and RD114-TRco mRNA. Nuclear and cytoplasmic mRNA was retro-transcribed, and then qPCR was carried out on the corresponding cDNA using specific primers for RD114-TR WT and co genes and, as internal normalizers, specific primers for the U6 and GAPDH genes. The data were derived from a single qRT-PCR experiment in which each sample was run in sestuplicate.
To assess whether mRNA metabolism differs between RD114-TRWT and RD114-TRco, we studied mRNA nuclear-cytoplasm export in the PK-7 cell setting using the pIRES-puro3-based expression vectors, which generate only one mRNA transcript. Northern blot analysis of total, nuclear (nucl), and cytoplasmic (cyt) mRNAs and quantification by Typhoon Phosphorimager of the band intensity normalized by cellular equivalents loaded revealed that the unique codon-optimized mRNA is exported 1.4-fold more (WT cyt/nucl band intensity = 1.1 and co cyt/nucl band intensity = 1.6; 1.6/1.1 = 1.4) than wild-type mRNA (Figure 6B). qRT-PCR analysis, using the expression of nuclear U6 and cytosolic/total GAPDH genes as an internal normalizer, revealed that RD114-TRco mRNA is exported 3.6-fold more than RD114-TRWT mRNA (Figure 6C). Overall, these data establish that recoding affects nuclear export but not transcription and splicing processes.
Analysis of the Secondary Structures of the RD114-TRWT and RD114-TRco mRNA
We then investigated whether codon optimization could influence mRNA secondary structure and, thereafter, protein translation, as reported recently by several groups.3, 4, 27 Thus, we examined RD114-TRWT and RD114-TRco mRNA sequences by MFOLD software (Figures 7A and 7B). This computational analysis predicts the most thermodynamically stable RNA configurations (up to 50) based on the free energy value (ΔG) of the molecules, where a lower ΔG indicates a higher stability. We retrieved 33 different configurations for RD114-TRWT and 37 for RD114-TRco (Figure 7E). As expected, wild-type structures are very different from codon-optimized ones; the average ΔG for RD114-TRWT mRNA is −462.25 (where ΔG = −468.70 is the most stable configuration), whereas the average ΔG for RD114-TRco mRNA is −679.39 (where ΔG = −687.60 is the most stable configuration). This finding indicates that recoded mRNA molecules are more stable than wild-type counterparts (p < 0.0001) (Figure 7E).
Figure 7.
Prediction of RD114-TRWT and RD114-TRco mRNA Secondary Structures
(A and B) Schematics of the most thermodynamically stable (lowest free energy value) secondary structures of RD114-TRWT (A) and RD114-TRco (B) mRNA, predicted by the MFOLD software. Data are annotated using color-based characters, indicating the probability of base-pairing from 0 (light blue) to 1 (red), where 1 is the maximum pairing level. (C and D) The 5′ end substructures of 320 bp (C) and 330 bp (D) of RD114-TRWT and RD114-TRco mRNA, respectively, are shown in the insets. (E) Boxplot of RD114-TRWT and RD114-TRco mRNA secondary structure ΔG, calculated by MFOLD software. Statistical analysis was performed by Wilcoxon rank-sum test. Statistical significance was set as ***p < 0.001.
Because 5′ end and 3′ end substructures are fundamentally important for translational dynamics and protein folding,28 we scanned the 5′ and 3′ ends of all wild-type and recoded mRNAs to identify any possible structural conserved domains. Over the 33 conformations of RD114-TRWT mRNA, we identified a conserved domain at both the 5′ end (nucleotides [nt] 1–320) and 3′ end (nt 1,308–1,677) (Figure 7C). These 5′ end and 3′ end domains are also conserved in the corresponding region of the RD114-TRco mRNA structure. Above the 37 structures calculated by MFOLD for RD114-TRco mRNA, we identified nt 1–330 at the 5′ end and nt 1,390–1,677 at the 3′ end (Figure 7D). To evaluate the similarity between identified domains, we studied RD114-TRWT and RD114-TRco mRNA 5′ end and 3′ end substructures with SimTree software. This software compares each node complexity (branch-loop) of two structures that eventually grades in a similarity score (between 0–1, where 1 is the maximum). The score is normalized by the number of nucleotides of the substructures from two RNA structures showing the lowest ΔG in MFOLD (Table S1). At the 5′ end of the RD114-TRWT mRNA substructure, 26 complexities were identified, which corresponded to only ten complexities in RD114-TRco (normalized symmetric similarity [nss] = 0.5213) (Figure 7A). At the 3′ end mRNA substructures, 42 complexities were found in RD114-TRWT and only 18 in RD114-TRco (nss = 0.6178) (Figure 7B).
These results point out that codon optimization of the RD114-TR gene introduced significant alterations at both the 5′ end and 3′ end of the RD114-TRco mRNA secondary structure. Such alterations might affect mRNA processes and eventually alter protein structure and function.
Discussion
Codon optimization and de-optimization have been used extensively for a lot of different biotechnological practices, primarily in heterologous systems to increase recombinant protein yield and as an adaptive response to environmental conditions and natural host selection in bacteria, yeasts and viruses.29, 30, 31, 32, 33, 34 In some eukaryotes (i.e., C. elegans and Drosophila melanogaster), it has been exploited to control intracellular tRNA to modulate translational efficiency.35, 36, 37, 38 In autologous hosts, such as the mammalian Chinese hamster ovary (CHO) or HEK293 cell lines, which are the most widespread systems for manufacturing pharmaceuticals,7 codon optimization is a valuable strategy to prevent transcriptional silencing, mRNA destabilization, or inefficient translation other than being a powerful tool to increase immunogenicity in DNA vaccinology applications.9, 10, 16, 17, 39 Finally, HIV-derived LV production has also benefited from codon optimization by enhancing production of structural and functional viral proteins (i.e., gag and pol);9, 11, 12 neutralizing cis-repressive sequences present in gag/pol genes, thereby making the expression of these genes Rev-independent;13, 14 and eliminating homology between packaging gag-pol genes and the cis-regulatory packaging (ψ) sequence contained in the packaging construct and TV, respectively, therefore reducing the risk of generating a replication-competent lentivirus (RCL).15
Based on these premises, we analyzed DNA codon optimization of the RD114-TR gene with the aim of improving envelope translation in RD3-MolPack producer cells. In fact, codon optimization would have neutralized the interfering sequences contained in the RD114-TR ORF, thereby sparing the use of the BGI in vector design and, at the same time, increasing the production and density of RD114-TR on the cellular plasma membrane and, consequently, on virion coats. The increased envelope density on the LVs would have eventually enhanced their functional titer. We asked for two recoding analyses by two independent companies, which provided similar results (Figure S8). Therefore, we believe that the quality of the analysis could not have affected the final output. The consistently negative results obtained with the chimeric RD114-TR3′co and 5′co support this idea.
In this study, we show that RD114-TRWT expressed either stably (RD3-MolPack-24 cells) or transiently (PK-7 cells) naturally traffics from the ER through the Golgi network to reach the plasma membrane. PRWT is processed in SUWT and TMWT subunits, which are eventually embedded into nascent LVs. In contrast, RD114-TRco reaches the plasma membrane mainly as unprocessed PRco, and maturation into SUco and TMco functional subunits is drastically reduced or even absent. As a consequence, a high level of PRco is erroneously incorporated into budded viral particles, which become defective vectors.
Because N-linked glycosylation is crucial for maturation of different envelope (Env) proteins, such as the hepatitis C virus glycoprotein E2 and human T cell leukemia virus type I (HTLV-I) envelopes,39, 40, 41 we studied the glycosylation status of RD114-TRWT and RD114-TRco. Here we confirm the conclusions reached by Sandrin et al.,19 showing that PRWT, SUWT, and TMWT are deglycosylated by PNGaseF. Furthermore, we also expanded glycosylation studies showing that, in contrast to SUWT, TMWT is EndoH-sensitive. This result suggests that TMWT either reaches the plasma membrane anchored to SU or loses complex oligosaccharides when on the membrane, or, alternatively, does not need complex oligosaccharides for its function. The analysis of PRco glycosylation demonstrates sensitivity to both PNGaseF and EndoH enzymes, suggesting defective glycosylation of the recoded protein. Further studies will clarify whether a correlation between the observed defective glycosylation and maturation of RD114-TRco does exist.
Although PRco is cleaved in vitro by recombinant furin both under reducing and non-reducing conditions (data not shown), it is not cleaved in vivo. To explain this result, we reasoned that cleavage in vivo could be prevented by an excess of substrate: PRco is, in fact, much more abundant compared with PRWT. Alternatively, the deficit of PRco processing could be secondary to a deficit of retrograde transport of PRco from the cell membrane to the endosomes, where the active form of furin is accumulated.42 We ruled out the first hypothesis because furin is not active in vivo, even with a very low amount of PRco substrate, whereas the second hypothesis requires further analysis to be formally accepted.
Synonymous mutations have been considered ineffective for a long time, and, for this reason, they are also named silent mutations. However, their nature has been recently re-evaluated because evidence has shown that these mutations have a great effect on pre-mRNA splicing and mRNA secondary structure formation, therefore affecting protein translational efficiency and folding.4, 43 Even a single synonymous codon substitution can have a significant effect on protein folding and function. Protein dysfunction can be caused either by disruption (or introduction) of splicing enhancers, by altering mRNA stability at the global and local level, or by altering the kinetic of protein production, the ribosomal pausing sites, and co-translational folding.27, 44 Our results exclude that codon optimization has introduced aberrant pre-mRNA splicing sites. Rather, they establish that RD114-TRco mRNA is exported more efficiently into the cytosol than RD114-TRWT mRNA and support the theory that some alterations occurred at the mRNA secondary structure, thereby influencing protein translation. We focused our study on the mRNA 5′ end and 3′ end because previous findings from others demonstrated that these two domains crucially influence translation dynamics, such as translation initiation and RNA global and local stability.45, 46, 47 The in silico MFOLD and SinTree software analyses highlighted that the secondary structures of RD114-TRWT and RD114-TRco mRNA are significantly different. Especially some conserved domains at the 5′ end and the 3′ end of RD114-TRWT mRNA are lost in the RD114-TRco isoform. Interestingly, the generation of chimeric RD114-TR5co′ and RD114-TR3co′ led to even worse functional impairment. These findings suggest that RD114-TRco inactivity is not due to single mutations clustered at the 5′ end or 3′ end but, more likely, due to conformational modifications distributed along the mRNA molecule that affect global mRNA stability and, thereby, protein folding and processing. Sandrin et al.19 demonstrated that modifications in the cytoplasmic tail of RD114 and RD114-TR alter PR subunit transport from the cell membrane to the trans-Golgi network. In particular, transport of the envelopes associated with core protein (i.e., Gag) to the endosomal compartment, where active furin accumulates, is important because it affects cleavage efficiency.19 We observed, by confocal microscope imaging, that both RD114-TRWT and RD114-TRco are localized mostly in the ER compartment when assessed either in the presence (PK-7 setting) or in the absence (COS-7) of Gag protein. To this extent, secondary structure modifications identified in RD114-TRco mRNA might result in alteration of protein folding, which, in turn, is responsible for protein dysfunction. However, further investigations are necessary to validate this hypothesis.
Altogether, this study suggests that RD114-TR is not suitable for codon optimization and that this strategy cannot be applied to improve its performance.
Materials and Methods
Plasmids
The phCMV-RD114-TR plasmid, obtained by F.-L. Cosset (INSERM), encodes the chimeric RD114-TR envelope that derives from fusion of the extracellular and transmembrane domains of the feline endogenous retrovirus RD114 envelope and the cytoplasmic tail (TR) of the amphotropic (A)-MLV 4070 envelope18 (Figure 1A, scheme 1). The pIRES-RD114-TRWT-internal ribosome entry site (IRES)-puro-woodchuck hepatitis post-transcriptional regulatory element (WPRE) plasmid was obtained by excising the CMV-RD114-TRWT cassette from the phCMV-RD114-TR plasmid and cloning it into the pIRES-puro 3 plasmid (Clontech Laboratories, a Takara Bio Company) (Figure 1A, scheme 7). The generation of the SIN-RD114-TRWT and SIN-RD114-TRco vectors (Figure 1A, schemes 2 and 3) as well as the constructs encoding the HIV gag, pol, and rev genes (Figure 1A, schemes 4 and 5, respectively) have been described previously.21 The SIN-GFP TV encoding the EGFP gene was kindly provided by L. Naldini (Tiget, OSR) (Figure 1A, scheme 6). The RD114-TR ORF was codon-optimized, synthesized, and cloned in either the pMK-RQ or pMS-RQ plasmid by GENEART. We further cloned the RD114-TRco ORF into either the pIRES-puro3 or SIN-LV plasmid. Four different molecules were generated: pIRES-CMV-RD114-TR-FLco, in which full-length (FL) cDNA was codon-optimized and cloned into the EcoRV and NsiI sites of the pIRES-puro3 plasmid by excising the ORF from the pMK-RQ-RD114-TR-FLco plasmid; pIRES-CMV-RD114-TR-5′co, obtained by recoding only the 5′-half sequence (789 bp) of the RD114-TRWT ORF (RD114-TR5′co was modified by adding Eco47III and NsiI restriction sites at the 5′ end and 3′ end of the gene sequence, respectively, and the ORF was then cloned into the Eco47III and NsiI sites of pIRES-puro3 plasmid); pIRES-CMV-RD114-TR-3′co, obtained by recoding only the 3′-half sequence (789 bp) and cloning into the Eco47III and SphI restriction sites of the pIRES-puro3 plasmid by excising the ORF from pMK-RQ-RD114-TR-3′co (Figure 1A, scheme 7); and SIN-RD114-TRco FL (Figure 1A, scheme 3), generated by inserting RD114-TRco into a SIN-LV through a three-step cloning strategy. First, the RD114-TRWT-IRES-puro-WPRE fragment was excised from the SIN-RD114-TRWT-IN-RRE vector (Figure 1A, scheme 2) and cloned into the EcoRI site of a pGEM-T plasmid, generating the pGEM-RD114-TRWT-IRES-puro-WPRE plasmid. Then, RD114-TRWT-IRES-puro was excised from pGEM-RD114-TRWT-IRES-puro-WPRE (obtaining the pGEM-WPRE intermediate) using BamHI, and the RD114-TRco-IRES-puro ORF was excised from the pIRES-CMV-RD114-TR-FL-co using EcoRV/XbaI enzymes. RD114-TRco-IRES-puro was then cloned into the pGEM-WPRE intermediate through blunt ligation, obtaining the pGEM-RD114-TRco-IRES-puro-WPRE plasmid. Finally, the RD114-TRco-IRES-WPRE ORF was cut out from pGEM-RD114-TRco-IRES-WPRE and cloned into the EcoRI site of SIN-RD114-TRWT-IN-RRE, generating the SIN-RD114-TRco-IN-RRE vector.
Cells
HEK293T cells and their derivative PK-7 clone, which constitutively expresses the HIV gag-pol-rev genes,21 were propagated in Iscove’s modified Dulbecco’s medium (IMDM) (BioWhittaker, Lonza Group) supplemented with 10% Australian fetal calf serum (FCS) (BioWhittaker) and a combination of 1% penicillin-streptomycin and glutamine (PSG) (Lonza). The CEM A3.01 T cell line48 was grown in RPMI 1640 medium (BioWhittaker) supplemented with 10% FCS and 1% PSG. COS-7 cells were grown in DMEM (BioWhittaker) supplemented with 10% FCS and 1% PSG.
LV Production, Titration, and Purification
LVs were produced from either HEK293T or PK-7 cells by transient transfection.21 Briefly, 3–5 × 106 cells were plated on 100-mm2 tissue culture dishes (Becton Dickinson). After 24 hr of culture, the EGFP TV, rev, packaging, and envelope constructs were co-transfected at a 4:1:0.88:0.48 ratio using either Profection mammalian transfection system calcium phosphate (Promega) or FuGENE 6 transfection reagent (Roche Diagnostics) according to the manufacturer’s instructions. Transfection efficiency was calculated 48 hr later by analyzing the percentage of EGFP-positive cells by fluorescence-activated cell sorting (FACS) analysis.
Transduction
The CEM A3.01 cell line was transduced by spinoculation at 1,024 × g for 2 hr at 37°C in the presence of Polybrene (8 μg/ml) (Sigma-Aldrich). Transduction efficiency was obtained by FACS analysis of EGFP expression. Physical titer was evaluated by measuring the level of p24Gag released in the culture supernatant with the Alliance HIV-1 p24 antigen ELISA kit (PerkinElmer) according to the manufacturer’s instructions.
Northern Blot Assay
PK-7 cells were transfected with the pIRES-RD114-TR or SIN-RD114-TR TV plasmid encoding either RD114-TRWT or RD114-TRco. Forty-eight hours after transfection, total, nuclear, and cytoplasmic RNAs were extracted by Trizol reagent (Life Technologies) following the manufacturer’s instructions and analyzed by northern blot assay. Five micrograms RNA/sample was run on 0.8% agarose-formaldehyde gel, transferred onto a Hybond-N membrane by capillary transfer, and finally probed with 1 × 106 dpm/mL of a 32P-labeled 550-bp RD114-TRWT or RD114-TRco probe in PerfectHyb Plus hybridization buffer (Sigma-Aldrich). Membranes were extensively washed and then exposed to X-ray films at –80°C or to a Typhoon Phosphorimager 9000 (GE Healthcare) for direct quantification of the radioactive signal. After stripping, membranes were re-hybridized with an internal control probe encompassing the packaging sequence (ψ) to detect full-length mRNAs.
qRT-PCR
Total, nuclear, and cytoplasmic RNAs, obtained as described above, were retrotranscribed with the SuperScript first-strand synthesis system kit for RT-PCR (Invitrogen). The cDNA (1.25 ng) was quantified by qPCR SYBR Green technology with the following specific primers: RD114-TRWT (for 5′ aac ggg tca gtc ttc ctc tg; rev 5′ atc aat ggc agg aat ggg ga), RD114-TRco (for 5′ ccg tgc agt tca ttc ctc tg; rev 5′ ctc agc ttg gtg tac tgg gt), U6 (for 5′ ctc gct tcg gca gca ca; rev aac gct tca cga att tgc gt 5′), and GAPDH (for 5′ tgc acc aca act gct tag c; rev 5′ ggc atg gac tgt ggt cat gag). Normalization was calculated using GAPDH for total and cytosolic mRNA and U6 for nuclear mRNA.
Western Blot Assay
Cellular extracts and viral proteins derived from isolated cell-free virus-like particles (VLPs) or LVs were prepared as described previously.21, 49 Briefly, LV supernatants were concentrated by centrifugation at 15,000 × g for 90 min at 4°C. Then, the liquid phase was gently removed, and pelleted virions were directly lysed by adding 5 μL of PBS/0.5% NP-40 (Calbiochem, Merck-Millipore, #492016). Proteins were size-fractionated on 8%, 12%, or 4%–15% gradient SDS-PAGE (Mini-Protean TGX gels, #456-1084, Bio-Rad). Then, proteins were electroblotted on either Hybond enhanced chemiluminescence (ECL) nitrocellulose membranes (GE Healthcare) or Transblot turbo transfer pack membranes (Bio-Rad, #170-4159). Membranes were blocked in 5% low-fat dry milk Tris-buffered saline (TBS), 1% Tween 20 (TBS-T) and then incubated with the appropriate primary antibody diluted in 5% BSA and TBS-T. The anti-TM RD114-TR rabbit serum, kindly provided by F.-L. Cosset (INSERM),19 was diluted 1:1,000. The anti-SU RD114-TR mAb, generated by Areta International, was diluted 1:50. The mouse anti-p24Gag Ab (Acris Antibodies) was diluted 1:500. The anti-extracellular signal-related kinase-1 (ERK) rabbit Ab was diluted 1:1,000 (Cell Signaling Technology, #16). The anti-calnexin rabbit Ab was diluted 1:2,000 (Santa Cruz Biotechnology, G1910). The extravidin horseradish peroxidase (HRP) Ab was diluted 1:2,000 (Sigma-Aldrich, #E2886). The anti-HIV human serum, obtained from an AIDS patient, was kindly donated by G. Poli (OSR) and diluted 1:1,000. The secondary HRP-linked Abs anti-human (#NA933V) and anti-rabbit (NA934V) (GE Healthcare) were diluted 1:5,000. The anti-mouse (#A2066) Ab (Sigma-Aldrich) was diluted 1:10,000. ECL western blotting detection reagent (GE Healthcare, RPMN2106) was used for the chemiluminescence reaction.
Immunofluorescence
PK-7 and COS-7 cells were transfected with pIRES-RD114-TR plasmids encoding either RD114-TRWT or RD114-TRco, seeded on poly-L-lysine-coated glass slides (Thermo Fisher Scientific). Forty-eight hours after transfection, cells were fixed with PBS and 3% paraformaldehyde (PFA)/0.1 mM CaCl2/0.1 mM MgCl2, permeabilized with PBS and 0.1% Triton X-100, and then stained with the following Abs: rabbit anti-VAMP8 (Endobrevin Synaptic Systems, catalog no. 1047 302) at 1:200 dilution, rabbit anti-calnexin (Santa Cruz, H-70, catalog no. sc-11397) at 1:50 dilution, and mouse anti-SU RD114-TR mAb at 1:300 dilution. The secondary Abs were Alexa Fluor goat anti-rabbit A488 (Invitrogen, catalog no. A11034) and Alexa Fluor goat anti-mouse A568 (Invitrogen, catalog no. A11031). Nuclei were stained with PBS/0.1 μg/mL DAPI. Slides were mounted with fluorescence mounting medium (Dako), and images were captured with a laser-scanning confocal microscope (Leica TCS SP5) with an HCX PL APO λblue 63× (NA 1.4) objective in oil immersion. Images were acquired using the Leica Application Suite (LAS) Advanced Fluorescence (AF) software (Leica Microsystems) and processed by the public domain ImageJ image processing and analysis software (http://rsb.info.nih.gov/ij/) (Image Processing and Analysis in Java).
In Vitro Deglycosylation Assays and Furin Digestion
Protein extracts from either cells or virions were treated with PNGaseF and EndoH enzymes according to the manufacturer’s instructions (New England Biolabs, #P0704S and #P0702S, respectively). Briefly, proteins were first denatured for 10 min at 99°C and then digested for 1 hr at 37°C with 250–500 U of PNGaseF or EndoH enzyme. 4× loading buffer containing β-mercaptoethanol was added to samples that were then boiled for 5 min at 99°C and finally loaded onto 4%–15% SDS-PAGE precast gels (Mini-Protean TGX gels, #456-1084, Bio-Rad) for western blot analysis. In vitro furin digestion was carried out by treating 35 μg cellular extracts with 4 U of recombinant furin (NEB, #P8077S) for 16 hr at 16°C following the manufacturer’s instructions.
Cell Surface Biotinylation
RD3-MolPack-SIN-GFP producer cells were plated in 60-mm2 culture dishes at 1 × 106 cells/cm2 density. Forty-eight hours after cell seeding, RD3-MolPack-SIN-GFP cells reached about 90% confluency. LV supernatant was harvested and filtered with a 0.45-μm filter (Merck Millipore). Cellular monolayers were gently washed in PBS supplemented with 1 mM MgCl2 and 0.1 mM CaCl2 to keep epithelial junctions tight and impermeable to molecules. Cells were then incubated on ice for 30 min with 0.5 mg/ml EZ-Link Sulfo-NHS-LC-Biotin (Thermo Scientific) in PBS/1 mM MgCl2/0.1 mM CaCl2 and gently shaken. After biotinylation, cell monolayers were washed for 5 min with PBS/100 mM Glycine/1 mM MgCl2/0.1 mM CaCl2 for quenching the biotin excess. Cells were finally lysed as described previously,21, 49 and protein extracts were quantified by protein assay (Bio-Rad, #500-0006). One milligram of proteins was incubated with 50–60 μl of biotin binder magnetic beads (Dynabeads MyOne Streptavidin T1, #65602, Invitrogen) for 1 hr at room temperature by gentle rocking. Beads were washed four to five times with 1 mL of PBS/0.1% BSA, and then protein/bead complexes were processed with PNGaseF. After addition of 4× loading dye and boiling for 5 min at 99°C, proteins were separated by SDS-PAGE on 4%–15% precast gels (Mini-Protean TGX gels, #456-1084, Bio-Rad) and analyzed by western blot assay.
Virion PD Assay
Matched p24Gag equivalents of vector particles were incubated with 5 μL of anti-SU Ab (0.9 mg/mL) for 3 hr at 4°C under rocking conditions. The PD was performed by washing three times 100 μL of Dynabeads (sheep anti-mouse immunoglobulin G [IgG] Dynabeads, Invitrogen, #422.01) with PBS/0.5% BSA/2mM EDTA and then rocking them for 30 min at room temperature in the presence of LV particles and the anti-SU Ab. After virion PD, the Dynabeads were washed several times, 4× loading dye was added, and proteins were separated by SDS-PAGE on 4%–15% precast gels for western blot analysis.
RNA Splicing and Structure Prediction Web Servers
Prediction of RNA splice sites was generated by the software made available by the NetGene2 Server (http://www.cbs.dtu.dk/services/NetGene2/) and prediction of mRNA structure by the software MFOLD (http://mfold.rit.albany.edu/?q=mfold/RNA-Folding-Form) using the following parameters: linear RNA sequence; 37°C folding temperature; 1 M NaCl ionic condition: number of calculated folding; differences between the calculated foldings = default parameters; maximum extension of the calculated loops = 30; maximum asymmetry between the calculated loops = 30; and no limit in base pairing distance. The comparison between different structures was obtained by the SimTree software (http://bioinfo.cs.technion.ac.il/SimTree/).
Statistical Methods
Statistical analysis was performed using JMP statistical software and by running the Wilcoxon-Mann-Whitney ranked-sum non-parametric test. The significance level was set as p < 0.001.
Author Contributions
E.Z. and M.P. contributed to the conception, acquisition, analysis, and interpretation of data and drafting the article. C.P. and A.S. contributed to the conception, acquisition and analysis of data. C.S., E.G., S.B., C.A., and S.C. contributed to the acquisition and analysis of data. C. Bordignon and G.P.R. contributed to the final approval of the version to be published. C. Bovolenta contributed to the conception, design, analysis, and interpretation of the data, drafting and approving the final version to be published.
Conflicts of Interest
All authors except E.Z. are employees of MolMed S.p.A.
Acknowledgments
We wish to thank Niek Van Til (Erasmus MC) for his help with the recoding process and silent mutations; Eugenio Montini and Stefania Merella (Tiget, OSR) for their consulting on splicing analysis; Davide Cittaro (Bioinformatics Facility, OSR) for his help with in silico analysis of RNA; Massimo Alessio (Proteome Biochemistry, OSR) for discussing results; and Cesare Covino (Alembic, OSR) for assistance with confocal microscopy.
Footnotes
Supplemental Information includes eight figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtm.2017.01.002.
Supplemental Information
References
- 1.Moulard M., Decroly E. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta. 2000;1469:121–132. doi: 10.1016/s0304-4157(00)00014-9. [DOI] [PubMed] [Google Scholar]
- 2.Chamary J.V., Hurst L.D. The price of silent mutations. Sci. Am. 2009;300:46–53. doi: 10.1038/scientificamerican0609-46. [DOI] [PubMed] [Google Scholar]
- 3.Chamary J.V., Parmley J.L., Hurst L.D. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 4.Sauna Z.E., Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 5.Sharp P.M., Li W.H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maertens B., Spriestersbach A., von Groll U., Roth U., Kubicek J., Gerrits M., Graf M., Liss M., Daubert D., Wagner R., Schäffer F. Gene optimization mechanisms: a multi-gene study reveals a high success rate of full-length human proteins expressed in Escherichia coli. Protein Sci. 2010;19:1312–1326. doi: 10.1002/pro.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wurm F.M. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 2004;22:1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 8.Fath S., Bauer A.P., Liss M., Spriestersbach A., Maertens B., Hahn P., Ludwig C., Schäfer F., Graf M., Wagner R. Multiparameter RNA and codon optimization: a standardized tool to assess and enhance autologous mammalian gene expression. PLoS ONE. 2011;6:e17596. doi: 10.1371/journal.pone.0017596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ngumbela K.C., Ryan K.P., Sivamurthy R., Brockman M.A., Gandhi R.T., Bhardwaj N., Kavanagh D.G. Quantitative effect of suboptimal codon usage on translational efficiency of mRNA encoding HIV-1 gag in intact T cells. PLoS ONE. 2008;3:e2356. doi: 10.1371/journal.pone.0002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frelin L., Ahlén G., Alheim M., Weiland O., Barnfield C., Liljeström P., Sällberg M. Codon optimization and mRNA amplification effectively enhances the immunogenicity of the hepatitis C virus nonstructural 3/4A gene. Gene Ther. 2004;11:522–533. doi: 10.1038/sj.gt.3302184. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda Y., Takeuchi Y., Martin F., Cosset F.L., Mitrophanous K., Collins M. Continuous high-titer HIV-1 vector production. Nat. Biotechnol. 2003;21:569–572. doi: 10.1038/nbt815. [DOI] [PubMed] [Google Scholar]
- 12.Meintjes P.L., Rodrigo A.G. Evolution of relative synonymous codon usage in Human Immunodeficiency Virus type-1. J. Bioinform. Comput. Biol. 2005;3:157–168. doi: 10.1142/s0219720005000953. [DOI] [PubMed] [Google Scholar]
- 13.Wagner R., Graf M., Bieler K., Wolf H., Grunwald T., Foley P., Uberla K. Rev-independent expression of synthetic gag-pol genes of human immunodeficiency virus type 1 and simian immunodeficiency virus: implications for the safety of lentiviral vectors. Hum. Gene Ther. 2000;11:2403–2413. doi: 10.1089/104303400750038507. [DOI] [PubMed] [Google Scholar]
- 14.Kuate S., Wagner R., Uberla K. Development and characterization of a minimal inducible packaging cell line for simian immunodeficiency virus-based lentiviral vectors. J. Gene Med. 2002;4:347–355. doi: 10.1002/jgm.290. [DOI] [PubMed] [Google Scholar]
- 15.Delenda C. Lentiviral vectors: optimization of packaging, transduction and gene expression. J. Gene Med. 2004;6(Suppl 1):S125–S138. doi: 10.1002/jgm.501. [DOI] [PubMed] [Google Scholar]
- 16.Li L., Saade F., Petrovsky N. The future of human DNA vaccines. J. Biotechnol. 2012;162:171–182. doi: 10.1016/j.jbiotec.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niazi K.R., Ochoa M.T., Sieling P.A., Rooke N.E., Peter A.K., Mollahan P., Dickey M., Rabizadeh S., Rea T.H., Modlin R.L. Activation of human CD4+ T cells by targeting MHC class II epitopes to endosomal compartments using human CD1 tail sequences. Immunology. 2007;122:522–531. doi: 10.1111/j.1365-2567.2007.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandrin V., Boson B., Salmon P., Gay W., Nègre D., Le Grand R., Trono D., Cosset F.L. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood. 2002;100:823–832. doi: 10.1182/blood-2001-11-0042. [DOI] [PubMed] [Google Scholar]
- 19.Sandrin V., Muriaux D., Darlix J.L., Cosset F.L. Intracellular trafficking of Gag and Env proteins and their interactions modulate pseudotyping of retroviruses. J. Virol. 2004;78:7153–7164. doi: 10.1128/JVI.78.13.7153-7164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Nunzio F., Piovani B., Cosset F.L., Mavilio F., Stornaiuolo A. Transduction of human hematopoietic stem cells by lentiviral vectors pseudotyped with the RD114-TR chimeric envelope glycoprotein. Hum. Gene Ther. 2007;18:811–820. doi: 10.1089/hum.2006.138. [DOI] [PubMed] [Google Scholar]
- 21.Stornaiuolo A., Piovani B.M., Bossi S., Zucchelli E., Corna S., Salvatori F., Mavilio F., Bordignon C., Rizzardi G.P., Bovolenta C. RD2-MolPack-Chim3, a packaging cell line for stable production of lentiviral vectors for anti-HIV gene therapy. Hum. Gene Ther. Methods. 2013;24:228–240. doi: 10.1089/hgtb.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Relander T., Johansson M., Olsson K., Ikeda Y., Takeuchi Y., Collins M., Richter J. Gene transfer to repopulating human CD34+ cells using amphotropic-, GALV-, or RD114-pseudotyped HIV-1-based vectors from stable producer cells. Mol. Ther. 2005;11:452–459. doi: 10.1016/j.ymthe.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Marin V., Stornaiuolo A., Piovan C., Corna S., Bossi S., Pema M., Giuliani E., Scavullo C., Zucchelli E., Bordignon C. RD-MolPack technology for the constitutive production of self-inactivating lentiviral vectors pseudotyped with the nontoxic RD114-TR envelope. Mol. Ther. Methods Clin. Dev. 2016;3:16033. doi: 10.1038/mtm.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauro V.P., Chappell S.A. A critical analysis of codon optimization in human therapeutics. Trends Mol. Med. 2014;20:604–613. doi: 10.1016/j.molmed.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 26.Wang G.S., Cooper T.A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 27.Bartoszewski R.A., Jablonsky M., Bartoszewska S., Stevenson L., Dai Q., Kappes J., Collawn J.F., Bebok Z. A synonymous single nucleotide polymorphism in DeltaF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J. Biol. Chem. 2010;285:28741–28748. doi: 10.1074/jbc.M110.154575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 29.Frenkel-Morgenstern M., Danon T., Christian T., Igarashi T., Cohen L., Hou Y.M., Jensen L.J. Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels. Mol. Syst. Biol. 2012;8:572. doi: 10.1038/msb.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu D., Kazana E., Bellanger N., Singh T., Tuite M.F., von der Haar T. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 2014;33:21–34. doi: 10.1002/embj.201385651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauring A.S., Acevedo A., Cooper S.B., Andino R. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe. 2012;12:623–632. doi: 10.1016/j.chom.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlin S., Blaisdell B.E., Schachtel G.A. Contrasts in codon usage of latent versus productive genes of Epstein-Barr virus: data and hypotheses. J. Virol. 1990;64:4264–4273. doi: 10.1128/jvi.64.9.4264-4273.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong E.H., Smith D.K., Rabadan R., Peiris M., Poon L.L. Codon usage bias and the evolution of influenza A viruses. Codon Usage Biases of Influenza Virus. BMC Evol. Biol. 2010;10:253. doi: 10.1186/1471-2148-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J., Liu W.J., Peng S.W., Sun X.Y., Frazer I. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J. Virol. 1999;73:4972–4982. doi: 10.1128/jvi.73.6.4972-4982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angov E. Codon usage: nature’s roadmap to expression and folding of proteins. Biotechnol. J. 2011;6:650–659. doi: 10.1002/biot.201000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharp P.M., Emery L.R., Zeng K. Forces that influence the evolution of codon bias. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:1203–1212. doi: 10.1098/rstb.2009.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocha E.P. Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004;14:2279–2286. doi: 10.1101/gr.2896904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duret L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–289. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- 39.Pique C., Pham D., Tursz T., Dokhélar M.C. Human T-cell leukemia virus type I envelope protein maturation process: requirements for syncytium formation. J. Virol. 1992;66:906–913. doi: 10.1128/jvi.66.2.906-913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan-Fook C., Jiang W.R., Clarke B.E., Zitzmann N., Maidens C., McKeating J.A., Jones I.M. Hepatitis C virus glycoprotein E2 binding to CD81: the role of E1E2 cleavage and protein glycosylation in bioactivity. Virology. 2000;273:60–66. doi: 10.1006/viro.2000.0407. [DOI] [PubMed] [Google Scholar]
- 41.Goffard A., Dubuisson J. Glycosylation of hepatitis C virus envelope proteins. Biochimie. 2003;85:295–301. doi: 10.1016/s0300-9084(03)00004-x. [DOI] [PubMed] [Google Scholar]
- 42.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan J., Wainwright M.S., Comeron J.M., Saitou N., Sanders A.R., Gelernter J., Gejman P.V. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 44.Tsai C.J., Sauna Z.E., Kimchi-Sarfaty C., Ambudkar S.V., Gottesman M.M., Nussinov R. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J. Mol. Biol. 2008;383:281–291. doi: 10.1016/j.jmb.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nackley A.G., Shabalina S.A., Tchivileva I.E., Satterfield K., Korchynskyi O., Makarov S.S., Maixner W., Diatchenko L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 46.Kudla G., Murray A.W., Tollervey D., Plotkin J.B. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu W., Zhou T., Wilke C.O. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput. Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Folks T., Benn S., Rabson A., Theodore T., Hoggan M.D., Martin M., Lightfoote M., Sell K. Characterization of a continuous T-cell line susceptible to the cytopathic effects of the acquired immunodeficiency syndrome (AIDS)-associated retrovirus. Proc. Natl. Acad. Sci. USA. 1985;82:4539–4543. doi: 10.1073/pnas.82.13.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porcellini S., Gubinelli F., Alberici L., Piovani B.M., Rizzardi G.P., Bovolenta C. Chim3 confers survival advantage to CD4+ T cells upon HIV-1 infection by preventing HIV-1 DNA integration and HIV-1-induced G2 cell-cycle delay. Blood. 2010;115:4021–4029. doi: 10.1182/blood-2009-09-243030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.