Abstract
Umbilical cord blood is a traditional and convenient source of cells for hematopoietic stem cell transplantation. Thymic regulatory T cells (Tregs) are also present in cord blood, and there is growing interest in the use of autologous Tregs to provide a low-risk, fully human leukocyte antigen (HLA)-matched cell product for treating autoimmune diseases, such as type 1 diabetes. Here, we describe a good manufacturing practice (GMP)-compatible Treg expansion protocol using fluorescence-activated cell sorting, resulting in a mean 2,092-fold expansion of Tregs over a 16-day culture for a median yield of 1.26 × 109 Tregs from single-donor cryopreserved units. The resulting Tregs passed prior clinical trial release criteria for Treg purity and sterility, including additional rigorous assessments of FOXP3 and Helios expression and epigenetic analysis of the FOXP3 Treg-specific demethylated region (TSDR). Compared with expanded adult peripheral blood Tregs, expanded cord blood Tregs remained more naive, as assessed by continued expression of CD45RA, produced reduced IFN-γ following activation, and effectively inhibited responder T cell proliferation. Immunosequencing of the T cell receptor revealed a remarkably diverse receptor repertoire within cord blood Tregs that was maintained following in vitro expansion. These data support the feasibility of generating GMP-compliant Tregs from cord blood for adoptive cell transfer therapies and highlight potential advantages in terms of safety, phenotypic stability, autoantigen specificity, and tissue distribution.
Keywords: autoimmunity, cord blood, adoptive cell transfer, regulatory T cell, type 1 diabetes, T cell receptor, good manufacturing practices
Introduction
The inability of a host to control immune inflammation is a key pathological feature for the development of autoimmune diseases, including type 1 diabetes (T1D).1, 2 There is growing interest in the use of adoptive cell transfer (ACT) therapies involving regulatory T cells (Tregs) as a means to control host responses, given their essential role in limiting both innate and adaptive immune activation.3 A key component of the therapeutic activity of Tregs resides in their capacity to exert their function(s) as “living drugs.”4, 5 Specifically, Tregs function to maintain dominant peripheral tolerance by consuming growth factors of pathogenic T cells and exerting their tissue-directed suppressive activities through an array of anti-inflammatory biochemical pathways, expression of co-inhibitory receptors, and production of immunoregulatory cytokines.6 These seemingly redundant mechanisms allow Tregs to track to various tissues to suppress host inflammation in a tissue- and context-dependent manner. More recent studies have also suggested that Tregs may play an important role in tissue repair following damage,7, 8, 9 extending their potential benefits to the field of regenerative medicine.
T1D is a tissue-specific autoimmune disease characterized by the targeting of insulin-producing β cells in the pancreatic islets, primarily through the activity of autoreactive T effector cells (Teffs). A number of immunotherapeutic interventions have been conducted in patients with T1D, with the ultimate goal of altering the effective balance of cells to restore regulation by Tregs. At least two major approaches have been taken in this regard. First, agents have been used to target and selectively deplete autoreactive Teffs, with notable therapeutic responses observed following treatment with alefacept (a fusion protein disrupting the CD2:LFA3 interaction)10 and tepilizumab (anti-CD3)11 as monotherapies and anti-thymocyte globulin (ATG) for leukocyte depletion in combination with the stem cell mobilizing agent granulocyte colony-stimulating factor (G-CSF).12 One of the most potent interventions to date employed the combination therapy of autologous nonmyeloablative stem cell transplant plus preconditioning with cyclophosphamide and G-CSF along with high doses of ATG13, 14, 15, 16 or the chemotherapeutic agent fludarabine.17 T1D patients who received these therapies experienced a significant remission, some for 4 years or longer, and were able to stop exogenous insulin. However, significant side effects limit the use of such aggressive protocols, particularly in the setting of pediatric patients. The second major approach involves the administration of therapies to bolster the regulatory arm of the T cell compartment. In this regard, efforts have been made to treat subjects with T1D with exogenous low-dose interleukin-2 (IL-2), a selective Treg growth factor.18 Alternatively, autologous polyclonal Tregs can be isolated from peripheral blood and can also be bolstered by ex vivo expansion prior to ACT therapy. Initial phase I dose escalation trials conducted in subjects with graft versus host disease (GvHD)19, 20, 21 and subjects with recent-onset T1D22, 23 have demonstrated both initial persistence and safety. Although not powered to assess efficacy, these initial T1D trials did observe subjects who maintained relatively stable C-peptide responses when assessed by mixed-meal tolerance tests. Moreover, preliminary data from these efforts suggested that Treg persistence correlated with preservation of C-peptide in a number of subjects.
Umbilical cord blood (CB) has been used as a source of hematopoietic stem cells (HSCs) for transplantation since 1988.24, 25 HSC therapies can restore function in patients suffering from malignancies, bone marrow failure disorders, inborn errors of metabolism, and immunological disorders.26 Harvested cord blood units (CBUs) can also be used as a source of naive Tregs that have been established as a potential treatment to control xeno-GvHD,27 and multiple phase I clinical trials evaluating the safety of Treg ACT therapy in GvHD have reported that ex vivo-expanded, partially human leukocyte antigen (HLA)-matched Tregs from non-autologous CBUs are well tolerated.28, 29 We have previously demonstrated that the infusion of autologous, cryopreserved CB (cryoCB) alone30 or in combination with oral docosahexaenoic acid and vitamin D is safe in subjects with T1D.31 However, this unmanipulated CB product contains only a minor fraction of Tregs, which may explain the inability of these therapies to preserve C-peptide production.32 We hypothesized that a pure population of naive Tregs with high proliferative capacity could be isolated from cryoCB units (cryoCBUs) and expanded to therapeutically efficacious cell numbers. This approach may have several potential advantages over peripheral blood Tregs. CB is increasingly stored at birth, and acquisition of this material does not require a large-volume blood draw or leukapheresis procedure that may be onerous in pediatric patient populations.3 In addition, CB yields a highly naive repertoire of Tregs, presumably prior to the expansion of any pathogenic autoreactive Teff populations that could potentially contaminate a Treg preparation when isolated from T1D patients at the time of disease onset. Moreover, evidence from both animal models and human in vitro studies suggests that naive Tregs exhibit increased long-term phenotypic stability compared with memory Tregs, and this notion is also reflected in the epigenetic profile of these cells.33, 34
In this study, we developed a protocol to isolate CB-derived CD4+CD25+CD127lo/− Tregs (CB Tregs) and expand them for potential autologous Treg ACT therapeutic applications. By utilizing fluorescence-activated cell sorting (FACS) for isolation, an extremely pure population of Tregs can be isolated and induced to proliferate to therapeutic doses consistent with prior trial efforts.23 Our resulting protocol was then validated under good manufacturing practices (GMP) conditions and demonstrated to meet both clinical and sterility release criteria consistent with a prior Treg trial.23 Our GMP protocol resulted in a highly pure and potent population of Tregs that maintained their canonical phenotypic and functional qualities. Our data demonstrate that Tregs derived from cryoCB (cryoCB Tregs) can be effectively isolated and expanded with a purity and potency comparable with Tregs isolated from CB or adult peripheral blood (APB). Overall, these results support additional testing of cryoCB Tregs as a potential therapy for the treatment of T1D and other inflammatory and autoimmune diseases.
Results
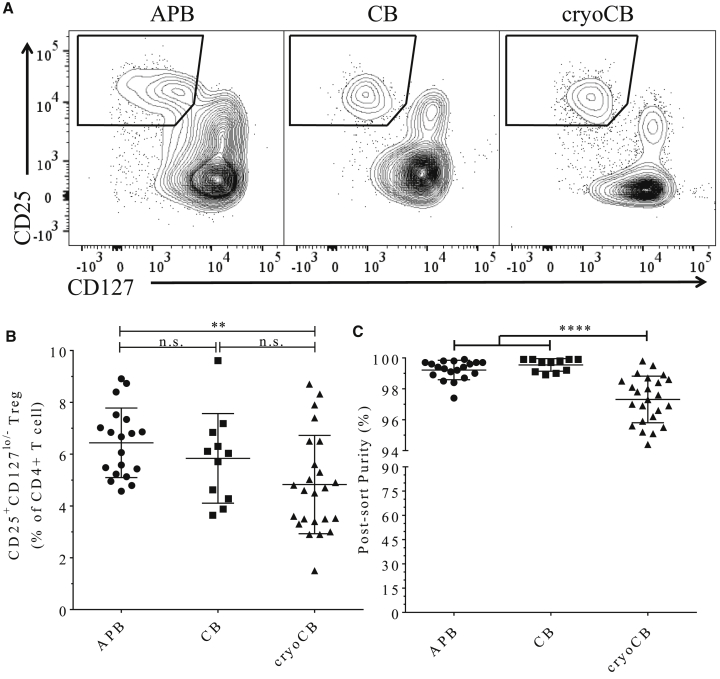
Identification and Isolation of CD4+CD25+CD127lo/− Tregs from CryoCBUs
Methods for the cryopreservation and handling of CBUs have been optimized for the recovery of CD34+ HSCs. We sought to determine whether cryoCB Tregs could survive and sustain sufficient surface marker expression to facilitate isolation by FACS and to enable in vitro expansion.35 Prior efforts utilizing CB for isolation of Tregs have relied primarily on microbead- and column-based positive selection of CD4+CD25+ T cells. In a 2008 study, microbead isolation of CB Tregs produced an average product of about 48% Tregs (CD25+FOXP3+ or CD127−FOXP3+) prior to expansion,27 and, in a 2011 study, only 64% of cells (range, 31%–96%) were considered Tregs (CD127−FOXP3+) at the end of expansion.28 Our FACS plots (Figure 1A) indicate that, although the Treg population was clearly more distinct in CB (whether freshly isolated or post-cryopreservation) over APB, it still contains a minor fraction of CD4+CD127+CD25intermediate T cells that would not have been eliminated by microbead isolation, which likely contributed to the non-FOXP3-expressing cells found contaminating the cultures in those previous efforts.27 Hence, we found FACS isolation of CD4+CD25+CD127lo/− Tregs to be sufficient and necessary, even in the setting of CB, for producing a highly pure end FOXP3+Helios+ Treg without the need for additional immunosuppressive agents (e.g., rapamycin) that negatively affect expansion kinetics.36 We observed variance in the Treg staining profiles in APB compared with fresh and cryoCB; however, the CD4+CD25+CD127lo/− Treg population was readily distinguishable from conventional CD4+ T cells (Tconvs) for all samples. Treg recovery was comparable across all three sources (Figure 1B). We did note that the post-sorting purity of CD4+CD25+CD127lo/− Tregs (by extracellular markers, attained by taking part of the sorted product and rerunning it through the sorter) was modestly reduced immediately post-sorting in cryopreserved versus fresh CB Tregs (APB Tregs = 99.0% ± 0.82%, CB Tregs = 99.47% ± 0.41%, cryoCB Tregs = 96.91% ± 1.49%, ****p < 0.0001; Figure 1C). However, we note that this reduction in post-sorting marker stability did not negatively affect the final purity of the Treg population post-expansion, as demonstrated below.
Figure 1.
Isolation of Tregs from APB, CB, and CryoCB by FACS Produces a Highly Pure Population
(A) CD4+ gated scatterplots showing gating for CD25+CD127lo/− Tregs from APB, CB, and cryoCB. Although all Treg recovery and purity values reached target ranges, a statistically significant sacrifice is made for cryoCB Tregs. (B) CD25+CD127lo/− Tregs as a percentage of CD4+ T cells were significantly less during FACS purification of cryoCB compared with APB (**p < 0.001, Tukey's multiple comparisons test). (C) Post-sorting purity of CD25+CD127lo/− Treg (attained by re-examining a small portion of the collected sample) was comparable for cells isolated from APB and CB but lower for cryoCB Tregs (Tukey's multiple comparisons test, ****p < 0.0001).
The number of cells that can be initially isolated has the potential to dictate therapeutic dosing capacity and even subsequent clinical efficacy. We observed that the Treg yield was comparable in CBUs versus cryoCBUs (Figure S1A). When the precise volume of CB collected after child birth could be determined from the CBU documentation, we observed the average number of cryoCB Tregs recovered per milliliter of CB to be 3.22 × 103 (SD = 1.65 × 103, n = 9) compared with a higher mean value from CB of 7.19 × 103 (SD = 2.96 × 103, n = 11, **p < 0.01; Figure S1B). Because storage and processing procedures vary for any CB bank, we compared the yield of Tregs from both academic research and private industry sources. Overall, the private banks did yield a higher average number of Tregs (Figure S1C); however, we noted that the samples were selected from a large number of available research units, and this pre-selection bias will not be feasible for autologous patient units. We therefore sought to identify a surrogate indicator of potential Treg cell yield from information uniformly recorded. When we analyzed three sources of cryoCB (an academic bank at the University of Florida [UF, a biorepository of cryopreserved research cords not deemed eligible for banking, obtained from the LifeCord Public Cord Blood Bank] and two private banks, ViaCord and Cord Blood Registry) for factors potentially predicting Treg yield, we found that the CD34+ cell count prior to cryopreservation was positively correlated with post-thaw Treg recovery in CB obtained from the UF biorepository (r2 = 0.823; **p < 0.01), but such correlation was not observed in CB obtained from the private banks (r2 = 0.028, p = 0.75; r2 = 0.006, p = 0.84; Figure S1C). We did not see any significant correlation between pre-cryopreservation total nucleated cell (TNC) count and Treg yield (Figure S1D), nor did we observe any effect of CB storage duration on Treg recovery (Figure S1E).
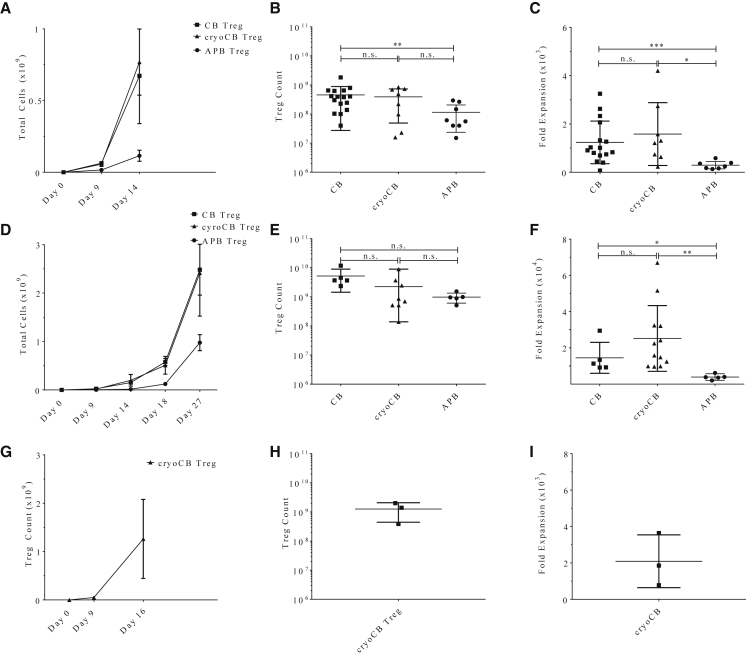
We also tested the feasibility of isolating cryoCB Tregs from the partitioned 20% fraction of cryoCBUs (a divided chamber of the cryoCBU consisting of about 20% of the total volume) to leave the remaining 80%-fraction of the unit for future treatments. Following the same methods stated previously for thawing and cell isolation, we found that, on average, 30,044 ± 6,035 cryoCB Tregs could be FACS-purified from the 20%-fraction (n = 5; Table 1). Following stimulation and expansion using protocol 2, we found that 1.98 × 108 ± 7.02 × 107 cryoCB Tregs could be obtained and used for potential autologous ACT therapy (Table 1). Based on these results, it would appear that isolation and expansion of even a minor fraction of a cryoCBU is feasible, but the downstream utility of this will require dose-finding studies for determining the necessary cell numbers for ACT.
Table 1.
The 20% Fraction of a cryoCB Can Produce a Substantial Number of Expanded cryoCB Tregs for ACT Therapy
| Unit No. | Day 0: cryoCB Tregs Isolated | Day 27 of Expansion: Cell Count | Fold Expansion |
|---|---|---|---|
| 1 | 26,568 | 176,000,000 | 6,625 |
| 2 | 42,158 | 298,000,000 | 7,069 |
| 3 | 39,145 | 221,000,000 | 5,646 |
| 4 | 29,456 | 105,000,000 | 3,565 |
| 5 | 27,894 | 189,000,000 | 6,776 |
| Average | 33,044 | 198,000,000 | 5,992 |
A benefit of using the 20% fraction (a divided chamber of the cryoCBU consisting of about 20% of the total volume) is that the remaining 80% can be used for future autologous therapies when needed. To determine the feasibility of using a 20% fraction to derive cryoCB Tregs from a cryoCBU, we isolated and expanded cryoCB Tregs using protocol 2 (Materials and Methods). Using FACS, 3.3044 × 104 ± 6.035 × 103 cryoCB Tregs were isolated (n = 5) and expanded to 1.98 × 108 ± 6.28 × 107 cells.
Tregs Derived from CryoCBUs Retain Their Lineage Characteristics and Suppressive Phenotype
We developed and optimized methods for the expansion of cryoCB Tregs utilizing methods adapted from Putnam et al.36 Tregs were isolated from both fresh and cryoCBUs, and these were compared with Tregs derived from APB. Tregs were expanded in vitro according to three unique protocols (from this point forward referred to as protocols 1–3), as described under Materials and Methods and depicted in Figure S2. Briefly, all three protocols included T cell stimulation on days 0 and 9 along with the presence of exogenous IL-2 in the culture medium. Protocol 1 was a 14-day expansion schedule that utilized an anti-CD3 (clone OKT3) loaded human K562 erythroleukemia-type artificial antigen-presenting cell (aAPC) line (KT/64/86; aAPCs expressing CD64 and CD86) on day 0, followed by anti-CD3/anti-CD28 bead restimulation on day 9. Because beads offer the advantage of off-the-shelf GMP compliance, protocols 2 and 3 did not include aAPCs but, rather, utilized anti-CD3/anti-CD28 beads for each round of T cell stimulation. For protocol 2, analyses were performed on day 27, whereas protocol 3 initially involved an expected 23-day expansion schedule but was subsequently shortened when we observed a sufficient Treg yield on day 16 for a targeted therapeutic dose for ACT (as shown below).
We first compared expansion from CB Tregs versus cryoCB Tregs and found that neither cell yield nor fold expansion were diminished by prior cryopreservation with expansion by protocol 1 (Figures 2A–2C) or protocol 2 (Figures 2D–2F). CB-derived Tregs, whether cryopreserved or freshly isolated, had increased proliferative capacity compared with APB Tregs (**p < 0.01 and *p < 0.05, respectively; Figure 2C). When protocol 1 and protocol 2 were directly compared, we found that CB and cryoCB Treg counts were similar on day 14 (the final day of protocol 1; p = 0.65 and p = 0.42, respectively). However, protocol 2 offered a higher mean Treg yield, although we observed greater variation across runs, likely because of the third round of stimulation with anti-CD3/anti-CD28 GMP beads and extended duration of culture. Using protocol 3, we found that Tregs expanded efficiently (Figures 2A, 2D, and 2G), yielding numbers sufficient for a therapeutic dose (estimating a 1.2 × 109 cell requirement to yield 30 × 106 cells/kg for an average 40-kg pediatric patient by day 16 (Figure 3H). We thus determined that protocol 3 offers the optimal GMP-compliant approach for CB Treg expansion based on off-the-shelf reagent availability and expansion parameters.
Figure 2.
Tregs Isolated from CB or CryoCB Were Expanded In Vitro According to Three Protocols
(A–I) Treg expansion efficiency is shown for (A–C) protocol 1, (D–F) protocol 2, and (G–I) protocol 3. (A, D, and G) The total Treg count over time was comparable for CB versus cryoCB (two-way ANOVA, NS = p ≥ 0.05). (B, E, and H) The Treg fold expansion from baseline on the final day of culture was not significantly different for CB versus cryoCB (Tukey's multiple comparisons test, NS = p ≥ 0.05). (C, F, and I) The Treg count on the final day of culture was not significantly different for CB versus cryoCB (Tukey's multiple comparisons test, NS = p ≥ 0.05). Protocol methodologies are outlined in Figure S1. p < 0.05 was considered statistically significant. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
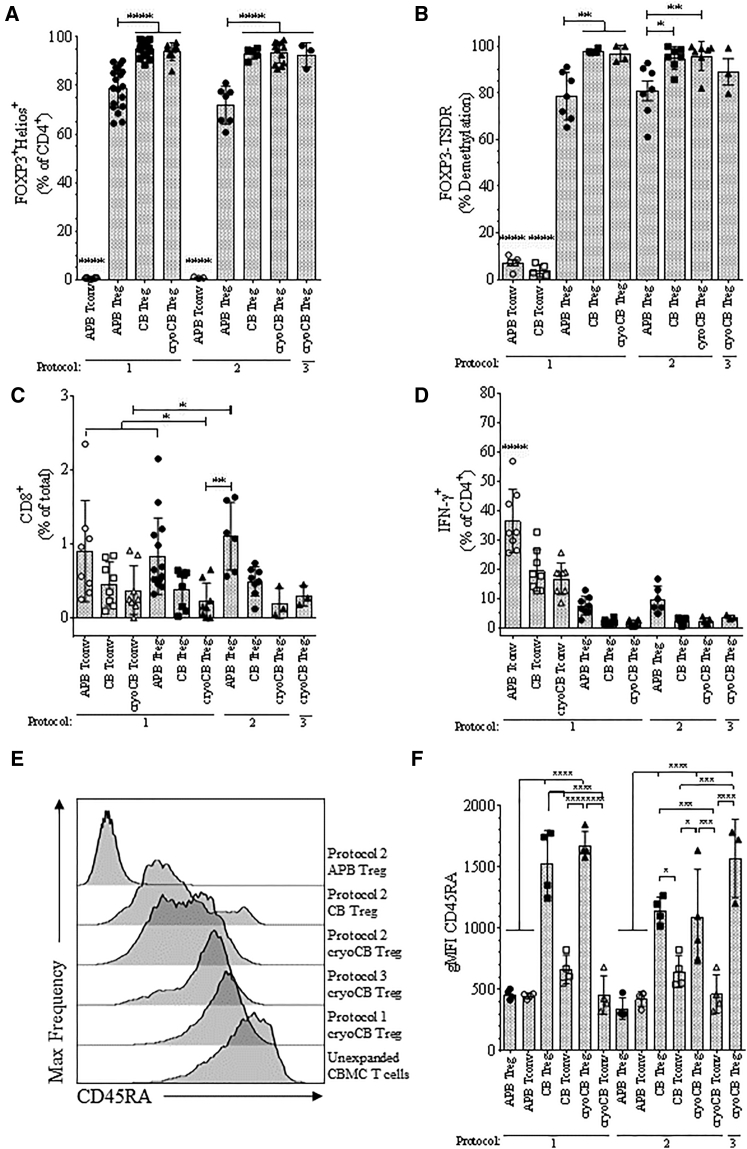
Figure 3.
Tregs Isolated and Expanded from CB, CryoCB, and APB Were Evaluated for Post-expansion Purity, Stability, and Naivety
(A) FOXP3+Helios+ Treg frequencies among CD4+ cells were significantly higher for Tregs expanded from CB or cryoCB compared with APB. Tregs were expanded via protocol 1 (ANOVA, **p < 0.001, NS = p ≥ 0.05). (B) The percentage of cells with demethylated FOXP3-TSDR was significantly greater among Tregs expanded from CB or cryoCB by protocols 1, 2, and 3 compared with Tregs expanded from APB by protocol 1 (**p < 0.01, ***p < 0.001). As expected, low FOXP3-TSDR percent demethylation was observed for CD4+ Tconv controls isolated and expanded from CB. (C and D) The percentages of (C) CD8+ cells and (D) IFN-γ+ cells were significantly lower for Tregs expanded from CB or cryoCB versus APB (ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, NS = p ≥ 0.05). Tregs or Tconvs were expanded via protocol 1. (C) CD8+ frequencies were comparable among CD4+ Tconvs isolated and expanded from APB, CB, or cryoCB. (D) As expected, IFN-γ+ cell frequencies were higher among Tconvs isolated and expanded from APB, CB, or cryoCB. (E and F) Representative histograms showing CD45RA expression by geometric mean fluorescence intensity (gMFI) in CB T cells and in some expanded populations (E) and mean gMFI for CD45RA expression on expanded Treg and Tconv populations (F), separated by expansion protocol.
We next examined APB, CB, and cryoCB for post-expansion Treg purity and CD8+ T cell contamination according to the University of California, San Francisco (UCSF) phase 1 trial clinical release criteria for polyclonal Tregs expanded from APB.23 Additionally, we assessed post-expansion suppressive function from cryopreserved expanded Treg preparations. Clinical microbiology release criteria were evaluated from culture aliquots collected at multiple time points throughout the expansion period, and all tests for endotoxin as well as bacterial and fungal contamination were found to be negative.
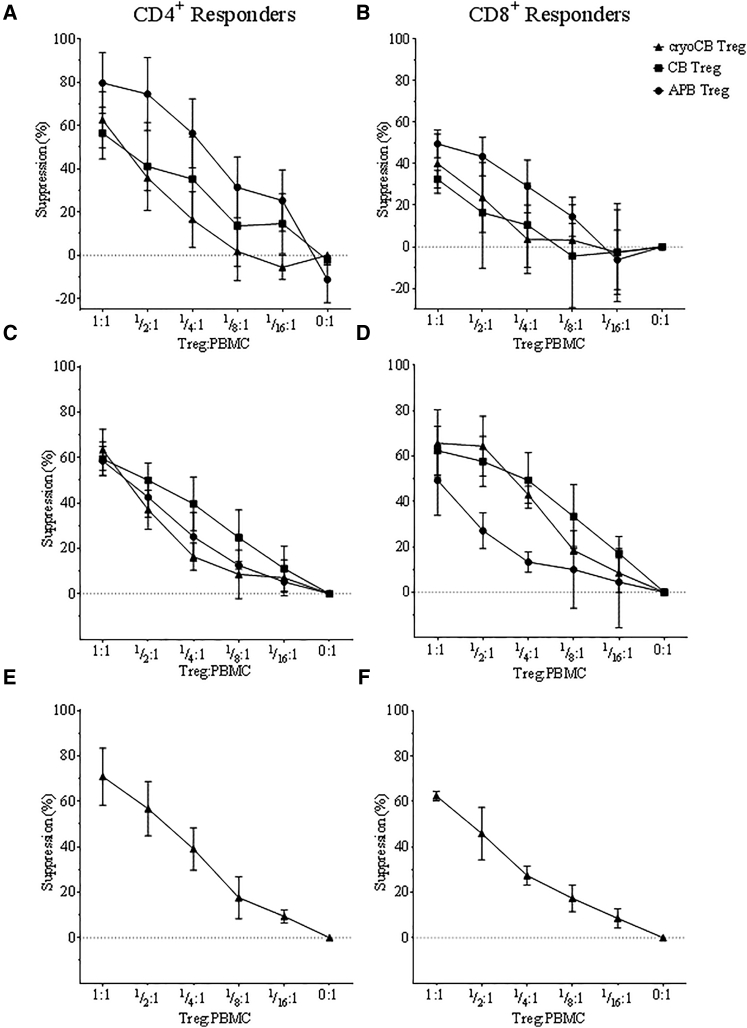
As expected, the CD4+ T cell population was almost entirely composed of FOXP3+Helios+ Tregs in both CB and cryoCB Treg expansions, whereas Treg purity was significantly lower in cells expanded from APB (protocols 1 and 2, ****p < 0.0001; Figure 3A). Indeed, all expanded CB and cryoCB Treg preparations greatly surpassed the release requirement for ≥60% FOXP3+.23 Epigenetic analysis of the Treg-specific demethylated region (TSDR) within the FOXP3 conserved non-coding sequence 2 (CNS2) locus confirmed that thymic Treg purity was greatest among Tregs isolated and expanded from fresh or cryopreserved CB (protocol 1: CB = 97.8% ± 1.0%, cryoCB = 96.9% ± 3.5%; protocol 2: CB = 92.1% ± 4.6%, cryoCB = 93.9% ± 8.2%; protocol 3: cryoCB = 89.0% ± 9.8%). APB Tregs demonstrated significantly less demethylation at the TSDR compared with cryoCB Tregs (protocol 1: mean = 78.5% ± 10.8%, **p < 0.01; protocol 2: mean = 80.9% ± 11.2%, **p < 0.01; Figure 3B). As expected, CB Tconv control cells exhibited nearly complete methylation of the TSDR (3.8% ± 2.6% demethylated, n = 5; Figure 3B). CD8+ T cell contamination was minimal, particularly in cells expanded from CB (protocol 1: APB Tregs = 0.8% ± 0.4%, CB Tregs = 0.4% ± 0.3%, cryoCB Tregs = 0.5% ± 0.3%; Figure 3C), presumably from the lower frequency of CD8+ T cell in CB.37 Again, these values were well below the clinical release criteria of ≤5% CD4−CD8+ contamination. Correspondingly, for each protocol, >99% of expanded cryoCB Tregs were CD4+, in accordance with the polyclonal APB Treg release criteria.23 Notably, interferon γ (IFN-γ) production was significantly higher among Tregs isolated and expanded from APB (protocol 1, 7.5% ± 3.2%; protocol 2, 9.7 ± 4.4%) compared with both fresh and cryopreserved CB preparations (protocol 1: CB = 1.8% ± 0.9%, **p < 0.01; cryoCB = 1.7% ± 0.9%, **p < 0.01; protocol 2: CB = 2.2% ± 1.2%, **p < 0.01; cryoCB = 2.2% ± 1.2%, **p < 0.01; Figure 3D). CD4+ T cells from CB, as expected, have nearly uniform expression of the CD45RA isoform characteristic of naive T cells (Figure 3E). Importantly, we observed that Tregs expanded from CB retained high levels of CD45RA expression, even following in vitro expansion (Figure 3F), in contrast to expanded APB Tregs that convert to the CD45RO isoform.38 Finally, Tregs were evaluated for functional suppressive capacity after expansion. Importantly, Tregs expanded from cryoCB, CB, and APB all demonstrated the ability to suppress both polyclonal CD4+ and CD8+ T cell responses (Figure 4).
Figure 4.
Suppressive Function of CB, CryoCB, and APB Tregs
(A–F) T cells (responders) were isolated from PBMCs, labeled with CTV, and plated in increasing proportions with expanded Tregs (suppressors) from protocol 1 (A and B), protocol 2 (C and D), and protocol 3 (E and F) that were labeled with the cell proliferation dye eFluor 670, in ratios as indicated along the x axis. The cells were activated in vitro with soluble anti-CD3 and anti-CD28, and the proliferation of CD4+ or CD8+ responder cells (Tresp) was measured via flow cytometry after labeling with fluorescently conjugated anti-CD4 and anti-CD8 antibodies to distinguish the populations. Percent suppression was calculated as: 100 − [100 × (percentage of proliferating cells with Treg present)/(percentage of proliferating cells without Tresp present)]. Suppressive capacity was not significantly different for Tregs expanded from cryoCB, CB, or APB (two-way ANOVA, p = n.s., all).
CB Tregs Exhibit a Highly Diverse Receptor Repertoire that Is Maintained following Expansion
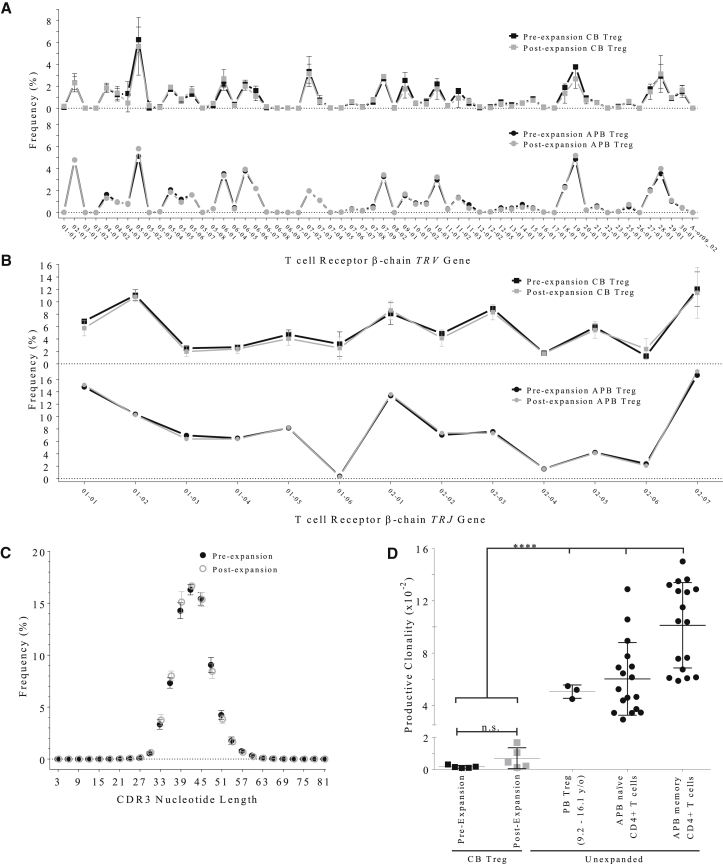
Treg T cell receptor (TCR) diversity has been demonstrated to be beneficial in maintaining self-tolerance.39 Moreover, a report by Yang et al.40 demonstrated a distinctive murine TCR repertoire among Tregs generated early in development during the perinatal period, which exhibit less clonal expansion and are uniquely capable of defending tissues against autoimmune destruction compared with Tregs derived from adult mice. Therefore, we sought to determine the relative diversity of the polyclonal Treg populations derived from CB relative to those observed in APB Tregs. For this analysis, we conducted immunosequencing of the complementarity-determining region 3 (CDR3) β chain loop of the TCR (TCRβ), a highly variable region formed as a result of TCR V(D)J gene segment recombination that serves to engage antigen peptides presented by HLA molecules.41 We compared Treg TCRβ V-gene (TRBV) and J-gene (TRBJ) frequencies pre- and post-expansion from either CB or APB and found that V-gene (Figure 5A) and J-gene (Figure 5B) distributions were not noticeably changed as a result of expansion for either Treg source. Similarly, upon evaluating the frequency of CDR3 sequence lengths, for which a non-Gaussian distribution would indicate a monoclonal expansion, we observed a normal distribution with a mean length of 42 amino acids (aa) in both pre- and post-expansion Tregs (Figure 5C).
Figure 5.
Immunosequencing of the TCR β Chain Demonstrates Increased Receptor Diversity of CB Tregs
Immunosequencing of the TCR β chain was performed for Tregs isolated from fresh CB prior to and after in vitro expansion according to protocol 1. (A and B) TRBV gene usage (A) and TRBJ gene usage (B) were not different among pre-expansion (black) and post-expansion (gray) CB Treg samples. (C) The distribution of CDR3 nucleotide lengths was comparable in pre-expansion (closed circles) and post-expansion (open circles) CB Treg samples. (D) Analysis of productive clonality (an inverse indicator of TCR clonal diversity) did not show a significant difference between post-expansion CB Tregs (gray squares) and pre-expansion samples (black squares). Compared with pre- or post-expansion CB Tregs, productive clonality was significantly higher among unexpanded Tregs isolated from peripheral blood (PB) samples obtained from young donors (age 9.2–16.1 years), adult PB naive CD4+ T cells, or adult PB memory CD4+ T cells (Tukey's multiple comparisons test, ****p < 0.0001).
In assessing TCR diversity, we quantified CB Treg productive clonality, a score of the sample productive diversity normalized to sample productive entropy. A productive clonality score near 1 would indicate very few productive rearrangements (monoclonal) and little TCR diversity, whereas a score near 0 would indicate a more polyclonal sample. For CB Tregs, productive clonality did not significantly change during expansion (n = 5; NS, p = 0.15, two-tailed paired Student’s t test; Figure 5D). A pre-expansion productive clonality of 1.64 × 10−3 ± 8.82 × 10−4 increased to 2.60 × 10−3 ± 1.70 × 10−3 post-expansion, indicating that CB Tregs express a very diverse receptor repertoire both before and after expansion. For perspective, previous studies have observed the mean productive clonality of unexpanded peripheral blood Tregs obtained from three children (9.2–16.1 years old) to be 5.05 × 10−2 ± 5.09 × 10−3 (n = 3),42 and, similarly, we discovered unexpanded naive T cells obtained from APB to be 6.03 × 10−2 ± 2.78 × 10−2 (Figure 5D). Additionally, APB Tregs were FACS-purified from a single T1D patient sample from clinical trial NCT01210664,23 and the productive clonality value was found to be 6.10 × 10−2 (n = 1; J.A.B., unpublished data). Taken together, these data imply a substantial reduction in Treg TCR repertoire diversity from infancy to adulthood (*p < 0.0001).
Discussion
T cells for ACT therapies have been applied for treatment of certain cancers and infections (reviewed by Busch et al.43), and clinical trials of autologous Tregs, harvested by leukapheresis and expanded from peripheral blood, have demonstrated safety in adults with immune-mediated disorders, including GvHD and T1D.22, 23 Although peripheral blood offers a large pool of available cells, an increasing number of peripheral blood T cells adopt a terminally differentiated memory phenotype with subject age, reducing expansion capacity and the likelihood of successful engraftment and survival.43 Conversely, CB-derived Tregs are far less likely to adopt a memory phenotype given the lack of central and effector memory subsets. As such, CB Tregs may offer an optimal population of CD28+CD27+ naive Tregs with improved purity following FACS isolation from CB versus peripheral blood and capacity for greater stability during extended expansions, facilitating long-term engraftment.44 CryoCB Tregs, therefore, are a potentially optimal source for autologous Tregs for ACT therapies.
Infusion of autologous whole CBUs was shown to be safe but did not preserve C-peptide in pediatric T1D patients,30 suggesting a potential need for higher doses of Tregs. This prompted the development of a GMP-compliant protocol to expand Tregs from cryoCBUs to produce a highly pure and functionally suppressive cell population for autologous ACT therapy. Here we compared three expansion protocols using Tregs isolated from fresh and cryoCB as well as APB. Although protocol 1 is well validated, it utilizes aAPCs and, therefore, presents reagent and GMP production challenges. Protocol 2 is GMP-compliant and utilizes multiple microbead restimulations to yield the greatest number of expanded Tregs. Unfortunately, the microbeads, which bind more effectively to cells, are difficult to remove from cultures, and, at the time of these experiments, had not yet been validated for clinical release criteria (although the microbeads have since been used in the ThRIL trial45). Protocol 3 utilized GMP-approved materials for expanded T cell products suitable for testing in humans that have been successfully utilized in a phase I trial of APB Tregs.23
The post-sorting purity was lower from cryopreserved versus fresh CB, but expansion curves were nearly identical from the two sources of CB. Meanwhile, both CB and cryoCB Tregs expanded more efficiently than Tregs from APB. A critical requirement for any effective Treg ACT therapy is that it must produce suppressive, lineage-stable Tregs that do not produce inflammatory cytokines. CryoCB Treg expansion products from the three unique protocols performed comparably in an analysis of Treg-suppressive capacity and stability (defined as FOXP3 and Helios co-expression and demethylation of the FOXP3 TSDR). Importantly, expanded cryoCB Tregs met previously determined clinical release criteria pertaining to the percentage of cells that maintain FOXP3 positivity, low CD8+ T cell contamination, and sterility.23 The target dose is not yet determined, but a dose escalation trial using Tregs expanded from ABP has demonstrated safety with doses as high as 2.9 × 109 infused Tregs.23 We were able to expand cryoCB Tregs to numbers near and even above this value. Additional clinical studies are needed to definitively identify the target dose for patients with T1D. As an initial phase I trial, we propose to move forward, specifically, with the goal of treating pediatric subjects with T1D in a dose escalation safety trial (at 10, 20, and 30 × 106 Treg/kg body weight). These doses are in line with prior clinical trials using peripheral blood Tregs.22, 46 Current expansions were performed using Tregs isolated from the complete CBU, and ongoing efforts are needed to generate Treg expansions from a fraction of the CBU to allow for potential repeat dosing.
Tissue distribution of T cells is controlled by the specificity of the TCR.47 Moreover, activation through the TCR is required for Treg-suppressive activity. Hence, we asked how the diversity of CB Treg TCRs compared pre- and post-expansion by conducting next-generation sequencing of the TCR β-chain (TRB) locus. The data support the notion that our in vitro expansion protocol induces broad expansion of all clones without significant deviations toward oligoclonality. The β-chain V-gene frequency and the sample clonality indicate that, although the TCR diversity decreases slightly during in vitro expansion, post-expansion CB Tregs remain far more polyclonal than APB Tregs.42 This has important implications when considering the use of APB or CB as a source of Treg material, either fresh or following TCR or chimeric antigen receptor (CAR) gene transfer approaches, which are increasingly in development, particularly given the successes in animal models using TCR-transgenic Tregs.48 This notion of an optimal Treg cell population will provide for optimal suppression of tissue-reactive Teff populations while also minimizing the potential for untoward immunosuppression of pathogens and potential tumor antigens. We argue that due to their uniquely thymic origin, CB Tregs contain receptors capable of seeding a wide variety of peripheral tissues early in life, would lack any enriched clones of pathogens that may become enriched through chronic infection in life (e.g., to cytomegalovirus [CMV] or Epstein-Barr virus [EBV]), and would lack Tregs arising to neo-antigens that suppress protective tumor immunity.49 Additionally, CB Tregs will remain subject to the normal homeostatic controls of a host, including the ability to attenuate suppression in the instance of acute viral infection.50 As a whole, these data suggest that our new GMP-compliant protocol yields cells suitable in number, phenotype, and function for testing as a potential treatment in humans with autoimmune disease, including T1D.
These expanded cryoCB Tregs would provide a practical alternative to the need for a large-volume blood draw or leukapheresis procedure currently needed for autologous ACT therapy, a benefit of particular importance for pediatric T1D patients. That said, unlike peripheral blood collection, the singular opportunity to store autologous CB creates a situation in which efforts to utilize fractions of stored CBUs, facilitated by multi-compartment storage bags, are needed to allow for longitudinal treatment regimens. Given the high safety profile observed with whole CB infusion30 as well as peripheral blood Treg ACT therapy,5, 23 we expect autologous CB Treg ACT therapy to be well tolerated. Moreover, expanded CB Tregs may offer an allogeneic cell source when autologous cell therapy is demonstrated to be safe. Indeed, allogeneic cells (when sufficiently major histocompatibility complex [MHC]-matched) from CB donors without T1D or associated risk alleles might offer improved therapeutic benefits given the potential advantages of non-autoimmune background genetics and intrinsic function as well as greater availability from a broader patient base.
In summary, Tregs can be consistently isolated from cryoCBUs using standard methods and reagents and stimulated to expand into a number of cells that can be considered within therapeutic dose ranges of prior trials. Therefore, the findings reported here support continued testing of Tregs expanded from cryoCB for safety and efficacy as a potential treatment to prevent or cure T1D.
Materials and Methods
Sample Collection and Mononuclear Cell Isolation
“Fresh” CB (fresh is defined as processed within 48 hr of cord blood draw) was collected by New York Blood Center’s National Cord Blood Program into CBUs containing 35 mL of citrate phosphate dextrose (CPD) anticoagulant.51 CBUs (n = 11) were then shipped to the University of Florida Diabetes Institute laboratories and immediately processed for isolation of cord blood mononuclear cells (CBMCs) by density gradient centrifugation (Ficoll-Paque PLUS, GE Healthcare).
CryoCBUs (n = 19) were obtained from private cord blood banks (Cbr Systems and ViaCord, a subsidiary of PerkinElmer) or through anonymous donation to a UF biorepository of cryopreserved research CBUs obtained from LifeCord, a public cord blood bank. CryoCB was collected and processed according to the standard operating procedures of the individual banks from units meeting NetCord-FACT International Standards for Cord Blood Collection, Banking, and Release for Administration, most notably at a minimum gestational age of 34 weeks, but with additional exclusion criteria including infectious diseases in the mother, severe pregnancy complications, or premature delivery with a birth weight of less than 1,500 g or when perinatal asphyxia is present in the fetus, and cell counts.52 All samples were obtained under Institutional Review Board (IRB)-exempt approved protocols at UF (non-human exempt protocol IRB201300072). CryoCBUs were thawed as described previously,53 resuspended in 60 mL of dextran-HSA wash solution (divided into two 50-mL conical tubes), allowed to warm to room temperature, and underlaid with 15 mL Ficoll-Paque PLUS per conical tube for density gradient centrifugation and CBMC isolation.
Peripheral blood was collected after informed consent, in accordance with the IRB at UF (protocol IRB201400703), from healthy control subjects (mean age, 28.8 ± 5.4 years; range, 22.5–38.7 years; n = 19) in sodium-heparinized Vacutainer tubes (Becton Dickinson) and processed within 24 hr for isolation of peripheral blood mononuclear cells (PBMCs) by density gradient centrifugation (Ficoll-Paque PLUS, GE Healthcare).
Sample Processing and Isolation of T Cells by FACS
Unless otherwise noted, cell isolation and culture guidelines, including cell numbers, volumes, and respective culture flasks, have been described previously.36 For research-grade Treg expansions, T cells subsets were isolated on a FACSAria III high-speed cell sorter (BD Biosciences) with the following antibodies: CD4-Pacific Blue (clone RPA-T4), CD127-PE (clone hIL-7R-M21), and CD25-APC (clone 2A3). CD4+CD25+CD127lo/− Tregs and CD4+CD127+ Tconvs were sorted into 400 μL of fetal bovine serum (FBS, United States Department of Agriculture [USDA]-approved origin, Atlanta Biologicals) using an aseptic technique. For protocol 3, Treg isolation was described previously.23
In Vitro Expansion Procedures
Given the naive and highly refractory nature of CB Tregs, we tested a number of stimulation conditions and expansion durations to optimize final cell yield and purity. This entailed the use of activating aAPCs and/or activation with commercially available antibody-conjugated microbeads. Cell culture volume and flask recommendations are listed in Table S1. The conditions for these three protocols can be visualized in Figure S2 and are outlined below.
Protocol 1: 14-Day aAPC Stimulation and Anti-CD3- and Anti-CD28-Coated Microbead Restimulation
FACS-isolated cells were plated according to Putnam et al.36 aAPCs were prepared from KT64/86, a K562-derived cell line constitutively expressing high-affinity Fc receptor, CD64, and CD86 for co-stimulation (a kind gift from Drs. James Riley and Bruce Levine, University of Pennsylvania). aAPCs were generated, cultured, and prepared for co-culture as described previously.54, 55 Briefly, Fc-binding receptors on KT64/86 were pre-cleared of serum immunoglobulins by culture in serum-free medium (SFM) overnight and then irradiated at 10,000 rad. Anti-CD3 (clone OKT-3, Miltenyi Biotec) monoclonal antibody (mAb) was loaded on KT64/86 at 1 μg/106 cells at 4°C for 30 min, washed twice with SFM, and cryopreserved in CryoStor CS10 (BioLife Solutions). After Treg FACS purification (described above), KT64/86 aAPCs were added to culture at a 1:1 aAPC-to-Treg cell ratio. CB Tregs and cryoCB Tregs were expanded in complete RPMI 1640 (cRPMI)—consisting of RPMI 1640 (Life Technologies) supplemented with 10% FBS (Atlanta Biologicals), 1 M HEPES, 1 mM sodium pyruvate, 100× minimum essential medium (MEM) non-essential amino acid solution, 50 mM 2-mercaptoethanol (2-ME), and 100 U penicillin/streptomycin (Gibco)—plus 600 IU/mL Proleukin (human recombinant IL-2 [hrIL-2], Prometheus Laboratories). APB Tregs were expanded in cRPMI plus 300 IU/mL Proleukin (hrIL-2, Prometheus Laboratories). On day 2, the culture volume was doubled, and IL-2 was added (at the aforementioned concentrations, assuming consumption). Cells were resuspended, and fresh medium and IL-2 were added on days 4, 6, 8, 11, and 13. On day 9, cells were restimulated with Dynabeads (human T-activator CD3/CD28 for T cell expansion and activation, Dynal Invitrogen) at a 1:1 ratio.
Protocol 2: 27-Day GMP-Compliant Anti-CD3-/Anti-CD28-Coated Microbead Stimulation and Multiple Restimulations
FACS-isolated cells were plated evenly at 2.5 × 104 to 5.0 × 104 Tregs/well in a 96-well flat-bottom plate (Costar) and activated with anti-CD3/anti-CD28-coated microbeads (MACS GMP ExpAct Treg kit for research use, Miltenyi Biotec) at a 4:1 bead-to-cell ratio. CB Tregs and cryoCB Tregs were expanded in cRPMI plus 600 IU/mL Proleukin (hrIL-2, Prometheus Laboratories). APB Tregs were expanded in cRPMI plus 300 IU/mL Proleukin (hrIL-2, Prometheus Laboratories). On day 2, the culture volume was doubled, and fresh IL-2 was added (at the aforementioned concentrations, assuming consumption). Cells were resuspended, and fresh medium and IL-2 were added on days 4, 6, 8, 11, 13, 15, 17, 20, 22, 24, and 26, assuming consumption. On days 9 and 18, cells were restimulated with fresh anti-CD3/anti-CD28-coated beads at a 1:1 ratio.
Protocol 3: 16-Day GMP Anti-CD3-/Anti-CD28-Coated Microbead Stimulation and Restimulation
Except where noted, protocol 3 followed the previously published protocol.38 Briefly, FACS-isolated cells were plated and activated with Dynabeads at a 4:1 bead-to-cell ratio. Cells were cultured either in X-VIVO 15 or in X-VIVO 15 customized by Lonza by substituting 100% of the glucose in the base medium with D-glucose (6,6-2H2, 99%) (Cambridge Isotope Laboratories, catalog no. DLM-349-MPT) supplemented with 10% human heat-inactivated pooled APB serum. On day 2, the culture volume was doubled, and IL-2 was added (300 IU/mL, Proleukin). Cells were resuspended, and fresh medium and IL-2 were added (600 IU/mL, Proleukin) on days 5, 7, 12, and 14, assuming consumption. On day 9, cells were restimulated with additional Dynabeads at a 1:1 ratio.
Additionally, the final release criteria for this protocol require that the final cell product be evaluated for purity (≤5% CD8+ cells, <100 beads/3 × 106 cells, and endotoxin ≤3.5 endotoxin units [EU]/mL), phenotype (≥95% CD4+ cells and ≥60% FOXP3+), sterility (negative for mycoplasma, anaerobic and aerobic bacteria, gram stain, fungal culture, potassium hydroxide [KOH] exam), and viability (≥85%).23
Analysis of Populations Pre- and Post-expansion
Phenotypic analyses and suppression analysis were performed in batches on rested cells 1 day after thawing post-cryogenic storage. Following in vitro expansion, Tregs were evaluated for continued expression of CD4, CD8, CD25, CD127, FOXP3, Helios, and IFN-γ. On day 14 (protocol 1), day 27 (protocol 2), or day 16 (protocol 3, Figure S2), 2 × 106 cells were removed from culture and activated for 4 hr with phorbol myristate acetate (PMA; 10 μg/mL) and ionomycin (500 nM) in the presence of GolgiStop (4 μL/6 mL of culture, BD Biosciences). Cells were then stained for surface markers (CD4-PE-Cy7, clone RPA-T4; CD8-APC-H7, clone SK1; CD25-BB515, clone 2A3; CD127-PE, clone hIL-7R-M21; 3 μL/test). Intracellular staining was achieved with the FOXP3 staining kit (BioLegend) according to the manufacturer’s instructions and modified as follows. Cells were washed and fixed for 30 min at 23°C using fixation/permeabilization buffer. Cells were washed, resuspended in permeabilization buffer, and incubated overnight at 4°C. The next day, the samples were washed in permeabilization buffer. Cells were subsequently blocked with human immunoglobulin G (IgG) (5 μg/test) for 5 min and stained with anti-human FOXP3-PE (clone 259D, 5 μl/test) and Helios-Pacific Blue (clone 22F6, 5 μL/test). Flow cytometric data were collected on an LSRFortessa cytometer (BD Biosciences) and analyzed with FlowJo software (version 10.0.0.7r2, Tree Star).
Bisulfite Sequencing and TSDR Real-Time PCR
Thymic Treg lineage commitment and stability are subject to epigenetic control at the conserved non-coding sequence 2 (CNS2) of the FOXP3 gene,56 commonly referred to as the TSDR.57 We assessed the degree of CpG demethylation at the TSDR of cultures following expansion by real-time PCR TSDR as described previously.58 Genomic DNA was isolated from purified T cells using the DNeasy tissue kit (QIAGEN) following the supplier’s recommendations. Bisulfite treatment of genomic DNA was performed with the EZ DNA methylation kit (Zymo Research). Briefly, 500 ng genomic DNA was sodium bisulfite-treated overnight at 50°C in the dark, followed by 10-min incubation on ice. After washing and desulphonation, bisulfite-converted DNA was eluted with 16 μL elution buffer.
Bisulfite-converted DNA samples were used for PCR amplification of FOXP3 TSDR fragments in a reaction volume of 50 μL containing 25 μL of ZymoTaq PreMix buffer (Zymo Research) and 0.5 μM each of the primers FOXP3_TSDRfw (ATATTTTTAGATAGGGATATGGAGATGATTTGTTTGG) and FOXP3_TSDRrev (AATAAACATCACCTACCACATCCACCAACAC). After activation at 95°C for 10 min, amplification was performed as follows: 50 cycles at 95°C for 30 s, at 55°C for 30 s, and at 72°C for 1 min. Amplified PCR products were purified with the QIAquick PCR purification kit (QIAGEN) and sequenced in both directions, applying the PCR primers and ABI Big Dye Terminator v1.1 cycle sequencing chemistry (Applied Biosystems), followed by capillary electrophoresis on an ABI 3100 genetic analyzer. Trace files were interpreted using ESME, a Linux system-based software that normalizes sequence traces, corrects for incomplete bisulfite conversion, and allows for quantification of methylation signals. For each sample, both PCR amplification and sequencing were repeated once. The primers used for sequencing reactions are the same as PCR amplification of bisulfite-converted genomic DNA.
TSDR real-time PCR was performed in 96-well white trays with a LightCycler 480 system (Roche Diagnostics) with a final reaction volume of 20 μL, containing 10 μL LightCycler 480 Probes Master Mix (Roche Diagnostics), 3 μL bisulfite-converted DNA sample or standards, and 1 μM of each primer (TSDR_Forward GGTTTGTATTTGGGTTTTGTTGTTATAGT, TSDR_Reverse CTATAAAATAAAATATCTACCCTCTTCTCTTCCT). The probes (6-carboxyfluorescein [FAM]-labeled methylated FOXP3, CGGTCGGATGCGTC; VIC-labeled unmethylated FOXP3, TGGTGGTTGGATGTGTTG) were added to a final concentration of 150 nM. All samples were analyzed in duplicate. After initial denaturation at 95°C for 10 min, the samples were subjected to 50 cycles at 95°C for 15 s and at 61°C for 1 min.
Various ratios of methylated and unmethylated FOXP3 TSDR template DNA were mixed and used to generate a standard curve. Genomic DNA extracted from sorted T lymphocytes, which are either fully methylated or unmethylated, were bisulfite-converted and used for PCR amplification of the FOXP3 TSDR fragment with the same pairs of primers and protocol as bisulfite-converted genomic DNA (described above).
Amplified PCR products were purified with the QIAquick gel extraction kit (QIAGEN). The concentration of purified DNA was determined with a GE NanoVue spectrophotometer (GE Healthcare). 17 differing ratios of methylated and unmethylated bisulfite-converted DNA mixtures were prepared and used as real-time standards to generate a standard curve.
In Vitro Suppression Assays
The in vitro-suppressive capacity of expanded Treg populations was determined based on their ability to suppress the proliferation of allogeneic CD4+ and CD8+ responder T cells derived from a single PBMC set from a healthy adult donor (standardized responder). PBMCs were harvested from whole blood by density gradient centrifugation (Ficoll-Paque PLUS, GE Healthcare) and cryopreserved in multiple aliquots (∼10 × 106 cells/vial). For each assay, an individual vial of PBMCs was thawed and labeled with Cell Trace Violet (CTV, 2.5 μM, Life Technologies), whereas cryoCB and CB Tregs were freshly collected on either day 14 or day 21 of expansion and labeled with the cell proliferation dye eFluor 670 (1.25 μM, eBioscience) and plated at ratios of 1:1, :1, :1, :1, :1, and 1:0 Tregs to responder PBMCs (5 × 104 Tregs at 1:0 condition) in 96-well round-bottom tissue culture plates (CellTreat) in 200 μL/well. All in vitro suppression assays were cultured in cRPMI. The cells were stimulated with soluble anti-CD3 (2 μg/mL, clone OKT3, BD Biosciences) and anti-CD28 (1 μg/mL, clone CD28.2, BD Biosciences) and incubated for 4 days at 37°C in a 5% CO2 incubator. On day 4, the cells were harvested and stained with Live/Dead Yellow according to the manufacturer’s protocols (Life Technologies), CD4-PE-Cy7 (clone RPA-T4, 3 μL/test), CD8-APC-H7 (clone SK1, 3 μL/test), CD25-BB515 (clone 2A3, 3 μL/test), and CD127-PE (clone hIL-7R-M21, 3 μL/test). CD4+ and CD8+ responder proliferation was assessed via flow cytometry. The responder populations were gated by forward scatter area (FSC-A) and side scatter area (SSC-A), and live cells were gated for CD4+ and CD8+. Percent suppression was calculated as: 100 − [100 × (percentage of proliferating cells with Treg present)/(percentage of proliferating cells without Tresp present)]. Analysis was completed using FlowJo software (Tree Star, v10.2), and proliferation was determined based on the division index of live-gated responder populations.
T Cell Receptor β Chain Sequencing
Samples were sequenced using the immunoSEQ assay (Adaptive Biotechnologies) utilizing the deep-level resolution to identify and quantitate the TCR β chain (TRB). In brief, the somatically rearranged TRB was amplified from 159.36–1,200 ng genomic DNA using a two-step, amplification bias-controlled multiplex PCR approach, and libraries were sequenced with raw Illumina sequence reads demultiplexed and processed as described previously.59, 60 Clonality was calculated according to the equation provided by Adaptive Technologies and their immunoSEQ software: 1 − (entropy) / log2(# of productive unique reads).
Additional data from peripheral blood samples used for TCR productive clonality comparisons were taken from the immuneACCESS open access database (Adaptive Biotechnologies). Henderson et al.42 isolated, by FACS, CD4+CD25+CD127low Tregs from the peripheral blood (PB) of three young donors, whose ages ranged from 9.2–16.1 years. In the second study included, Emerson et al.61 FACS-isolated CD4+CD45RA+CD62L+ (naive) and CD4+CD45RA−CD45RO+ (memory) T cells from 17 APB samples. Each study also used the immunoSEQ assay (Adaptive Biotechnologies) for TRB sequencing.
Data Analysis
Statistical analyses were performed using Prism (v7.01, GraphPad). Unless otherwise noted, statistical analysis was performed using two-tailed unpaired Student’s t test with Welsh’s correction, one-way ANOVA with Geisser-Greenhouse correction and Tukey’s multiple comparisons test, or two-way ANOVA with Tukey’s multiple comparisons test, as noted in the figures legends (where p < 0.05 was considered statistically significant; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Author Contributions
H.R.S. researched the data and wrote the manuscript. A.L. Putnam and J.C. researched the data and reviewed/edited the manuscript. A.L. Posgai contributed to discussions and wrote the manuscript. E.H.R., J.R.W., K.F.G., M.K., A.L., H.L.B., K.S.B., K.T.B., L.P., A.B., M.A.A., and J.A.B. contributed to discussions and reviewed/edited the manuscript. M.J.H. conceived the study and reviewed/edited the manuscript. T.M.B. conceived the study and wrote the manuscript.
Conflicts of Interest
K.F.G. and M.K. are employees of ViaCord, LLC, a subsidiary of PerkinElmer, and, as such, are PerkinElmer stock shareholders. H.L.B. and K.S.B. are employees of Cbr Systems, Inc. (Cord Blood Registry), a subsidiary of AMAG Pharmaceuticals, and, as such, are AMAG Pharmaceuticals stock shareholders. J.A.B. and A.L. Putnam are co-inventors on patents (US 20080131445 A1 and US 7722862 B2) filed in connection with the manufacturing of the Treg product. J.A.B. has received funding from Caladrius Biosciences and other in-kind contributions from BD Biosciences. The remaining authors declare that they have no competing interests.
Acknowledgments
We would like to thank Drs. James Riley and Bruce Levine (University of Pennsylvania) for kindly providing the KT64/86 (aAPC) cell line. We would also like to thank Michael R. Lee (UCSF) for his technical contributions toward this effort. These efforts were supported by non-restricted research grants and kind gifts (of CB units) from ViaCord, LLC (a subsidiary of Perkin Elmer) and CBR Systems, Inc. (a subsidiary of AMAG Pharmaceuticals) (to M.J.H. and T.M.B). Additional project support was provided by research grants from JDRF in the form of Cord Blood Center Grant 4-2007-1065 (to M.A.A.) and Career Development Award 2-2012-280 (to T.M.B.), a grant from the McJunkin Family Foundation (to M.J.H.), and NIH Grants P01 AI42288 (to M.A.A. and T.M.B.) and R01 DK106191 (to T.M.B).
Footnotes
Supplemental Information includes two figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtm.2016.12.003.
Supplemental Information
References
- 1.Bach J.F. Autoimmune diseases as the loss of active “self-control”. Ann. N Y Acad. Sci. 2003;998:161–177. doi: 10.1196/annals.1254.017. [DOI] [PubMed] [Google Scholar]
- 2.Thompson J.A., Perry D., Brusko T.M. Autologous regulatory T cells for the treatment of type 1 diabetes. Curr. Diab. Rep. 2012;12:623–632. doi: 10.1007/s11892-012-0304-5. [DOI] [PubMed] [Google Scholar]
- 3.Trzonkowski P., Bacchetta R., Battaglia M., Berglund D., Bohnenkamp H.R., ten Brinke A., Bushell A., Cools N., Geissler E.K., Gregori S. Hurdles in therapy with regulatory T cells. Sci. Transl. Med. 2015;7:304ps18. doi: 10.1126/scitranslmed.aaa7721. [DOI] [PubMed] [Google Scholar]
- 4.Riley J.L., June C.H., Blazar B.R. Human T regulatory cell therapy: take a billion or so and call me in the morning. Immunity. 2009;30:656–665. doi: 10.1016/j.immuni.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gitelman S.E., Bluestone J.A. Regulatory T cell therapy for type 1 diabetes: May the force be with you. J. Autoimmun. 2016;71:78–87. doi: 10.1016/j.jaut.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Tang Q., Bluestone J.A. The Foxp3+ regulatory T cell: a jack of all trades, master of regulation. Nat. Immunol. 2008;9:239–244. doi: 10.1038/ni1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nosbaum A., Prevel N., Truong H.A., Mehta P., Ettinger M., Scharschmidt T.C., Ali N.H., Pauli M.L., Abbas A.K., Rosenblum M.D. Cutting Edge: Regulatory T Cells Facilitate Cutaneous Wound Healing. J. Immunol. 2016;196:2010–2014. doi: 10.4049/jimmunol.1502139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu B., Yang M., Wang Q. Interleukin-33 in tumorigenesis, tumor immune evasion, and cancer immunotherapy. J. Mol. Med. (Berl.) 2016;94:535–543. doi: 10.1007/s00109-016-1397-0. [DOI] [PubMed] [Google Scholar]
- 9.Villalta S.A., Rosenthal W., Martinez L., Kaur A., Sparwasser T., Tidball J.G., Margeta M., Spencer M.J., Bluestone J.A. Regulatory T cells suppress muscle inflammation and injury in muscular dystrophy. Sci. Transl. Med. 2014;6:258ra142. doi: 10.1126/scitranslmed.3009925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rigby M.R., Harris K.M., Pinckney A., DiMeglio L.A., Rendell M.S., Felner E.I., Dostou J.M., Gitelman S.E., Griffin K.J., Tsalikian E. Alefacept provides sustained clinical and immunological effects in new-onset type 1 diabetes patients. J. Clin. Invest. 2015;125:3285–3296. doi: 10.1172/JCI81722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodle E.S., Bluestone J.A., Zivin R.A., Jolliffe L.K., Auger J., Xu D., Thistlethwaite J.R. Humanized, nonmitogenic OKT3 antibody, huOKT3 gamma(Ala-Ala): initial clinical experience. Transplant. Proc. 1998;30:1369–1370. doi: 10.1016/s0041-1345(98)00278-4. [DOI] [PubMed] [Google Scholar]
- 12.Haller M.J., Gitelman S.E., Gottlieb P.A., Michels A.W., Rosenthal S.M., Shuster J.J., Zou B., Brusko T.M., Hulme M.A., Wasserfall C.H. Anti-thymocyte globulin/G-CSF treatment preserves β cell function in patients with established type 1 diabetes. J. Clin. Invest. 2015;125:448–455. doi: 10.1172/JCI78492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voltarelli J.C., Couri C.E., Stracieri A.B., Oliveira M.C., Moraes D.A., Pieroni F., Coutinho M., Malmegrim K.C., Foss-Freitas M.C., Simões B.P. Autologous nonmyeloablative hematopoietic stem cell transplantation in newly diagnosed type 1 diabetes mellitus. JAMA. 2007;297:1568–1576. doi: 10.1001/jama.297.14.1568. [DOI] [PubMed] [Google Scholar]
- 14.Snarski E., Milczarczyk A., Torosian T., Paluszewska M., Urbanowska E., Król M., Boguradzki P., Jedynasty K., Franek E., Wiktor-Jedrzejczak W. Independence of exogenous insulin following immunoablation and stem cell reconstitution in newly diagnosed diabetes type I. Bone Marrow Transplant. 2011;46:562–566. doi: 10.1038/bmt.2010.147. [DOI] [PubMed] [Google Scholar]
- 15.Li L., Shen S., Ouyang J., Hu Y., Hu L., Cui W., Zhang N., Zhuge Y.Z., Chen B., Xu J., Zhu D. Autologous hematopoietic stem cell transplantation modulates immunocompetent cells and improves β-cell function in Chinese patients with new onset of type 1 diabetes. J. Clin. Endocrinol. Metab. 2012;97:1729–1736. doi: 10.1210/jc.2011-2188. [DOI] [PubMed] [Google Scholar]
- 16.D’Addio F., Valderrama Vasquez A., Ben Nasr M., Franek E., Zhu D., Li L., Ning G., Snarski E., Fiorina P. Autologous nonmyeloablative hematopoietic stem cell transplantation in new-onset type 1 diabetes: a multicenter analysis. Diabetes. 2014;63:3041–3046. doi: 10.2337/db14-0295. [DOI] [PubMed] [Google Scholar]
- 17.Cantú-Rodríguez O.G., Lavalle-González F., Herrera-Rojas M.A., Jaime-Pérez J.C., Hawing-Zárate J.A., Gutiérrez-Aguirre C.H., Mancias-Guerra C., González-Llano O., Zapata-Garrido A., Villarreal-Pérez J.Z., Gómez-Almaguer D. Long-Term Insulin Independence in Type 1 Diabetes Mellitus Using a Simplified Autologous Stem Cell Transplant. J. Clin. Endocrinol. Metab. 2016;101:2141–2148. doi: 10.1210/jc.2015-2776. [DOI] [PubMed] [Google Scholar]
- 18.Klatzmann D., Abbas A.K. The promise of low-dose interleukin-2 therapy for autoimmune and inflammatory diseases. Nat. Rev. Immunol. 2015;15:283–294. doi: 10.1038/nri3823. [DOI] [PubMed] [Google Scholar]
- 19.Hippen K.L., Merkel S.C., Schirm D.K., Nelson C., Tennis N.C., Riley J.L., June C.H., Miller J.S., Wagner J.E., Blazar B.R. Generation and large-scale expansion of human inducible regulatory T cells that suppress graft-versus-host disease. Am. J. Transplant. 2011;11:1148–1157. doi: 10.1111/j.1600-6143.2011.03558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trzonkowski P., Bieniaszewska M., Juścińska J., Dobyszuk A., Krzystyniak A., Marek N., Myśliwska J., Hellmann A. First-in-man clinical results of the treatment of patients with graft versus host disease with human ex vivo expanded CD4+CD25+CD127- T regulatory cells. Clin. Immunol. 2009;133:22–26. doi: 10.1016/j.clim.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Di Ianni M., Falzetti F., Carotti A., Terenzi A., Castellino F., Bonifacio E., Del Papa B., Zei T., Ostini R.I., Cecchini D. Tregs prevent GVHD and promote immune reconstitution in HLA-haploidentical transplantation. Blood. 2011;117:3921–3928. doi: 10.1182/blood-2010-10-311894. [DOI] [PubMed] [Google Scholar]
- 22.Marek-Trzonkowska N., Myśliwiec M., Dobyszuk A., Grabowska M., Derkowska I., Juścińska J., Owczuk R., Szadkowska A., Witkowski P., Młynarski W. Therapy of type 1 diabetes with CD4(+)CD25(high)CD127-regulatory T cells prolongs survival of pancreatic islets - results of one year follow-up. Clin. Immunol. 2014;153:23–30. doi: 10.1016/j.clim.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Bluestone J.A., Buckner J.H., Fitch M., Gitelman S.E., Gupta S., Hellerstein M.K., Herold K.C., Lares A., Lee M.R., Li K. Type 1 diabetes immunotherapy using polyclonal regulatory T cells. Sci. Transl. Med. 2015;7:315ra189. doi: 10.1126/scitranslmed.aad4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gluckman E., Broxmeyer H.A., Auerbach A.D., Friedman H.S., Douglas G.W., Devergie A., Esperou H., Thierry D., Socie G., Lehn P. Hematopoietic reconstitution in a patient with Fanconi’s anemia by means of umbilical-cord blood from an HLA-identical sibling. N. Engl. J. Med. 1989;321:1174–1178. doi: 10.1056/NEJM198910263211707. [DOI] [PubMed] [Google Scholar]
- 25.Wagner J.E., Kernan N.A., Steinbuch M., Broxmeyer H.E., Gluckman E. Allogeneic sibling umbilical-cord-blood transplantation in children with malignant and non-malignant disease. Lancet. 1995;346:214–219. doi: 10.1016/s0140-6736(95)91268-1. [DOI] [PubMed] [Google Scholar]
- 26.Warwick R., Armitage S. Cord blood banking. Best Pract. Res. Clin. Obstet. Gynaecol. 2004;18:995–1011. doi: 10.1016/j.bpobgyn.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Hippen K.L., Harker-Murray P., Porter S.B., Merkel S.C., Londer A., Taylor D.K., Bina M., Panoskaltsis-Mortari A., Rubinstein P., Van Rooijen N. Umbilical cord blood regulatory T-cell expansion and functional effects of tumor necrosis factor receptor family members OX40 and 4-1BB expressed on artificial antigen-presenting cells. Blood. 2008;112:2847–2857. doi: 10.1182/blood-2008-01-132951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunstein C.G., Miller J.S., Cao Q., McKenna D.H., Hippen K.L., Curtsinger J., Defor T., Levine B.L., June C.H., Rubinstein P. Infusion of ex vivo expanded T regulatory cells in adults transplanted with umbilical cord blood: safety profile and detection kinetics. Blood. 2011;117:1061–1070. doi: 10.1182/blood-2010-07-293795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunstein C.G., Miller J.S., McKenna D.H., Hippen K.L., DeFor T.E., Sumstad D., Curtsinger J., Verneris M.R., MacMillan M.L., Levine B.L. Umbilical cord blood-derived T regulatory cells to prevent GVHD: kinetics, toxicity profile, and clinical effect. Blood. 2016;127:1044–1051. doi: 10.1182/blood-2015-06-653667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haller M.J., Wasserfall C.H., Hulme M.A., Cintron M., Brusko T.M., McGrail K.M., Sumrall T.M., Wingard J.R., Theriaque D.W., Shuster J.J. Autologous umbilical cord blood transfusion in young children with type 1 diabetes fails to preserve C-peptide. Diabetes Care. 2011;34:2567–2569. doi: 10.2337/dc11-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haller M.J., Wasserfall C.H., Hulme M.A., Cintron M., Brusko T.M., McGrail K.M., Wingard J.R., Theriaque D.W., Shuster J.J., Ferguson R.J. Autologous umbilical cord blood infusion followed by oral docosahexaenoic acid and vitamin D supplementation for C-peptide preservation in children with Type 1 diabetes. Biol. Blood Marrow Transplant. 2013;19:1126–1129. doi: 10.1016/j.bbmt.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haller M.J., Viener H.-L., Wasserfall C., Brusko T., Atkinson M.A., Schatz D.A. Autologous umbilical cord blood infusion for type 1 diabetes. Exp. Hematol. 2008;36:710–715. doi: 10.1016/j.exphem.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X., Bailey-Bucktrout S., Jeker L.T., Bluestone J.A. Plasticity of CD4(+) FoxP3(+) T cells. Curr. Opin. Immunol. 2009;21:281–285. doi: 10.1016/j.coi.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann P., Eder R., Boeld T.J., Doser K., Piseshka B., Andreesen R., Edinger M. Only the CD45RA+ subpopulation of CD4+CD25high T cells gives rise to homogeneous regulatory T-cell lines upon in vitro expansion. Blood. 2006;108:4260–4267. doi: 10.1182/blood-2006-06-027409. [DOI] [PubMed] [Google Scholar]
- 35.Woods E.J., Liu J., Pollok K., Hartwell J., Smith F.O., Williams D.A., Yoder M.C., Critser J.K. A theoretically optimized method for cord blood stem cell cryopreservation. J. Hematother. Stem Cell Res. 2003;12:341–350. doi: 10.1089/152581603322023070. [DOI] [PubMed] [Google Scholar]
- 36.Putnam A.L., Brusko T.M., Lee M.R., Liu W., Szot G.L., Ghosh T., Atkinson M.A., Bluestone J.A. Expansion of human regulatory T-cells from patients with type 1 diabetes. Diabetes. 2009;58:652–662. doi: 10.2337/db08-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong M.T., Ong D.E.H., Lim F.S.H., Teng K.W.W., McGovern N., Narayanan S., Ho W.Q., Cerny D., Tan H.K., Anicete R. A High-Dimensional Atlas of Human T Cell Diversity Reveals Tissue-Specific Trafficking and Cytokine Signatures. Immunity. 2016;45:442–456. doi: 10.1016/j.immuni.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Bluestone J.A., Buckner J.H., Fitch M., Gitelman S.E., Gupta S., Hellerstein M.K., Herold K.C., Lares A., Lee M.R., Li K. Type 1 diabetes immunotherapy using polyclonal regulatory T cells. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aad4134. 315ra189–315ra189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wing J.B., Sakaguchi S. TCR diversity and Treg cells, sometimes more is more. Eur. J. Immunol. 2011;41:3097–3100. doi: 10.1002/eji.201142115. [DOI] [PubMed] [Google Scholar]
- 40.Yang S., Fujikado N., Kolodin D., Benoist C., Mathis D. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 2015;348:589–594. doi: 10.1126/science.aaa7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes M.M., Yassai M., Sedy J.R., Wehrly T.D., Huang C.Y., Kanagawa O., Gorski J., Sleckman B.P. T cell receptor CDR3 loop length repertoire is determined primarily by features of the V(D)J recombination reaction. Eur. J. Immunol. 2003;33:1568–1575. doi: 10.1002/eji.200323961. [DOI] [PubMed] [Google Scholar]
- 42.Henderson L.A., Volpi S., Frugoni F., Janssen E., Kim S., Sundel R.P., Dedeoglu F., Lo M.S., Hazen M.M., Beth Son M. Next-Generation Sequencing Reveals Restriction and Clonotypic Expansion of Treg Cells in Juvenile Idiopathic Arthritis. Arthritis Rheumatol. 2016;68:1758–1768. doi: 10.1002/art.39606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Busch D.H., Fräßle S.P., Sommermeyer D., Buchholz V.R., Riddell S.R. Role of memory T cell subsets for adoptive immunotherapy. Semin. Immunol. 2016;28:28–34. doi: 10.1016/j.smim.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horwitz M.E., Chao N.J., Rizzieri D.A., Long G.D., Sullivan K.M., Gasparetto C., Chute J.P., Morris A., McDonald C., Waters-Pick B. Umbilical cord blood expansion with nicotinamide provides long-term multilineage engraftment. J. Clin. Invest. 2014;124:3121–3128. doi: 10.1172/JCI74556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Safinia N., Vaikunthanathan T., Fraser H., Thirkell S., Lowe K., Blackmore L., Whitehouse G., Martinez-Llordella M., Jassem W., Sanchez-Fueyo A. Successful expansion of functional and stable regulatory T cells for immunotherapy in liver transplantation. Oncotarget. 2016;7:7563–7577. doi: 10.18632/oncotarget.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marek-Trzonkowska N., Mysliwiec M., Dobyszuk A., Grabowska M., Techmanska I., Juscinska J., Wujtewicz M.A., Witkowski P., Mlynarski W., Balcerska A. Administration of CD4+CD25highCD127- regulatory T cells preserves β-cell function in type 1 diabetes in children. Diabetes Care. 2012;35:1817–1820. doi: 10.2337/dc12-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li M.O., Rudensky A.Y. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat. Rev. Immunol. 2016;16:220–233. doi: 10.1038/nri.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang Q., Henriksen K.J., Bi M., Finger E.B., Szot G., Ye J., Masteller E.L., McDevitt H., Bonyhadi M., Bluestone J.A. In vitro-expanded antigen-specific regulatory T cells suppress autoimmune diabetes. J. Exp. Med. 2004;199:1455–1465. doi: 10.1084/jem.20040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen C.J., Gartner J.J., Horovitz-Fried M., Shamalov K., Trebska-McGowan K., Bliskovsky V.V., Parkhurst M.R., Ankri C., Prickett T.D., Crystal J.S. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J. Clin. Invest. 2015;125:3981–3991. doi: 10.1172/JCI82416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baca Jones C., Pagni P.P., Fousteri G., Sachithanantham S., Dave A., Rodriguez-Calvo T., Miller J., von Herrath M. Regulatory T cells control diabetes without compromising acute anti-viral defense. Clin. Immunol. 2014;153:298–307. doi: 10.1016/j.clim.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butler M.G., Menitove J.E. Umbilical cord blood banking: an update. J. Assist. Reprod. Genet. 2011;28:669–676. doi: 10.1007/s10815-011-9577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lauber S., Latta M., Klüter H., Müller-Steinhardt M. The Mannheim cord blood bank: experiences and perspectives for the future. Transfus. Med. Hemother. 2010;37:90–97. doi: 10.1159/000289589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenau E.H., Sugrue M.W., Haller M., Fisk D., Kelly S.S., Chang M., Hou W., Eldjerou L., Slayton W., Cogle C.R., Wingard J.R. Characteristics of thawed autologous umbilical cord blood. Transfusion. 2012;52:2234–2242. doi: 10.1111/j.1537-2995.2011.03556.x. [DOI] [PubMed] [Google Scholar]
- 54.Maus M.V., Thomas A.K., Leonard D.G., Allman D., Addya K., Schlienger K., Riley J.L., June C.H. Ex vivo expansion of polyclonal and antigen-specific cytotoxic T lymphocytes by artificial APCs expressing ligands for the T-cell receptor, CD28 and 4-1BB. Nat. Biotechnol. 2002;20:143–148. doi: 10.1038/nbt0202-143. [DOI] [PubMed] [Google Scholar]
- 55.Suhoski M.M., Golovina T.N., Aqui N.A., Tai V.C., Varela-Rohena A., Milone M.C., Carroll R.G., Riley J.L., June C.H. Engineering artificial antigen-presenting cells to express a diverse array of co-stimulatory molecules. Mol. Ther. 2007;15:981–988. doi: 10.1038/mt.sj.6300134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klein L., Jovanovic K. Volume 23. Elsevier; 2011. pp. 401–409. (Regulatory T cell lineage commitment in the thymus. Seminars in immunology). [DOI] [PubMed] [Google Scholar]
- 57.Polansky J.K., Kretschmer K., Freyer J., Floess S., Garbe A., Baron U., Olek S., Hamann A., von Boehmer H., Huehn J. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 2008;38:1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 58.Fuhrman C.A., Yeh W.I., Seay H.R., Saikumar Lakshmi P., Chopra G., Zhang L., Perry D.J., McClymont S.A., Yadav M., Lopez M.C. Divergent Phenotypes of Human Regulatory T Cells Expressing the Receptors TIGIT and CD226. J. Immunol. 2015;195:145–155. doi: 10.4049/jimmunol.1402381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carlson C.S., Emerson R.O., Sherwood A.M., Desmarais C., Chung M.-W., Parsons J.M., Steen M.S., LaMadrid-Herrmannsfeldt M.A., Williamson D.W., Livingston R.J. Using synthetic templates to design an unbiased multiplex PCR assay. Nat. Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 60.Robins H.S., Campregher P.V., Srivastava S.K., Wacher A., Turtle C.J., Kahsai O., Riddell S.R., Warren E.H., Carlson C.S. Comprehensive assessment of T-cell receptor β-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emerson R., Sherwood A., Desmarais C., Malhotra S., Phippard D., Robins H. Estimating the ratio of CD4+ to CD8+ T cells using high-throughput sequence data. J. Immunol. Methods. 2013;391:14–21. doi: 10.1016/j.jim.2013.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.