Summary
Introducing components of algal carbon concentrating mechanisms (CCMs) into higher plant chloroplasts could increase photosynthetic productivity. A key component is the Rubisco‐containing pyrenoid that is needed to minimise CO 2 retro‐diffusion for CCM operating efficiency.
Rubisco in Arabidopsis was re‐engineered to incorporate sequence elements that are thought to be essential for recruitment of Rubisco to the pyrenoid, namely the algal Rubisco small subunit (SSU, encoded by rbcS) or only the surface‐exposed algal SSU α‐helices.
Leaves of Arabidopsis rbcs mutants expressing ‘pyrenoid‐competent’ chimeric Arabidopsis SSUs containing the SSU α‐helices from Chlamydomonas reinhardtii can form hybrid Rubisco complexes with catalytic properties similar to those of native Rubisco, suggesting that the α‐helices are catalytically neutral.
The growth and photosynthetic performance of complemented Arabidopsis rbcs mutants producing near wild‐type levels of the hybrid Rubisco were similar to those of wild‐type controls. Arabidopsis rbcs mutants expressing a Chlamydomonas SSU differed from wild‐type plants with respect to Rubisco catalysis, photosynthesis and growth. This confirms a role for the SSU in influencing Rubisco catalytic properties.
Keywords: Arabidopsis thaliana, carbon concentrating mechanism (CCM), Chlamydomonas reinhardtii, chloroplast, photosynthesis, pyrenoid, Rubisco, tobacco
Short abstract
See also the Commentary on this article by Sharwood, 214: 496–499.
Introduction
Rubisco (EC 4.1.1.39) catalyses net CO2 assimilation in all photosynthetic organisms. Despite this central role, Rubisco is an inefficient enzyme that limits photosynthetic productivity, particularly in plants with the C3 photosynthetic pathway. Rubisco has a slow carboxylation rate (k cat c) and a relatively low affinity for CO2, with a K m for CO2 at ambient O2 (K c air) close to the CO2 concentration in a C3 leaf mesophyll cell (Galmés et al., 2014). Rubisco also catalyses d‐ribulose‐1,5‐bisphosphate (RuBP) oxygenation, resulting in the energetically expensive photorespiratory pathway where previously fixed CO2 is lost (Sharkey, 1988). These features necessitate a large investment in the enzyme (up to 50% of leaf soluble protein) to support adequate rates of CO2 assimilation (Parry et al., 2013). Increasing the operating efficiency of Rubisco and reducing photorespiration are important approaches for improving yields in C3 crop plants (Whitney et al., 2011; Parry et al., 2013; Carmo‐Silva et al., 2015; Long et al., 2015; Ort et al., 2015).
The operating efficiency of Rubisco in C3 plants could be enhanced by elevating the CO2 concentration in the chloroplast by means of carbon concentrating mechanisms (CCMs). Possibilities include using components of biochemical CCMs (as in C4 and CAM photosynthesis) and/or the biophysical inorganic carbon accumulation mechanisms from cyanobacteria and eukaryotic algae (von Caemmerer et al., 2012; Price et al., 2013; Meyer et al., 2016). In algal CCMs, bicarbonate transporters and localisation of Rubisco and carbonic anhydrase within the chloroplast, and in most instances within the pyrenoid (a microcompartment commonly present in chloroplasts of microalgae), result in saturating CO2 concentrations around Rubisco (Morita et al., 1998; Giordano et al., 2005; Wang et al., 2015). Modelling approaches suggest that algal CCMs with a pyrenoid are likely to be more effective in maintaining elevated CO2 concentrations around Rubisco than those without (Badger et al., 1998). Modelling also reveals that the confinement of Rubisco to a microcompartment would be required for effective operation of a biophysical CCM in a higher plant (Price et al., 2013; McGrath & Long, 2014). Recent work has shown that algal CCM components, including carbonic anhydrases and bicarbonate transporters, can be expressed in appropriate subcellular locations in angiosperms (Atkinson et al., 2016). To achieve a functional algal CCM in an angiosperm it will also be necessary to introduce a Rubisco capable of assembling into a pyrenoid‐like structure.
Pyrenoid formation in the model green alga Chlamydomonas reinhardtii (hereafter Chlamydomonas) depends on the amino acid sequences of the small subunit of Rubisco (SSU, encoded by the rbcS nuclear gene family) and, more specifically, on two surface‐exposed α‐helices, which differ markedly between Chlamydomonas and higher plants (Meyer et al., 2012). In Chlamydomonas, rbcS deletion mutants can be rescued with a SSU variant from angiosperms (Arabidopsis, spinach or sunflower) without compromising in vitro Rubisco catalysis (Genkov et al., 2010). However, these hybrid Rubisco no longer assembled into a pyrenoid. Accordingly, lines expressing the hybrid Rubisco lacked a functional CCM, resulting in growth only at high CO2. Pyrenoid formation and CCM function were restored by expression of a chimeric SSU, where a higher plant SSU was modified with the algal SSU α‐helices (Meyer et al., 2012). Thus, assembling a pyrenoid‐like microcompartment in chloroplasts would probably require the incorporation of Chlamydomonas‐like α‐helical sequence into the native angiosperm SSU, in addition to other proteins involved in pyrenoid formation such as the Rubisco‐associated protein EPYC1 (Mackinder et al., 2016).
Here we examine how the incorporation of SSUs with α‐helices from Chlamydomonas SSU influences the biogenesis and catalysis of Rubisco in Arabidopsis leaves. The Rubisco large subunits (LSUs, encoded by rbcL) harbour the catalytic sites and are highly conserved between algae and angiosperms (Arabidopsis and Chlamydomonas LSUs are 88% identical at the level of amino acid sequences). By contrast, the SSU isoforms of Arabidopsis and Chlamydomonas are only 40–43% identical, even though their tertiary structures are extremely similar, including the positions of the α‐helices (Spreitzer, 2003). Although located on the distal ends of the octameric LSU core of Rubisco and distant from the catalytic sites, the amino acid sequence of the SSUs can affect the catalytic properties of the enzyme (Genkov & Spreitzer, 2009).
In Arabidopsis the SSUs are encoded by four genes. rbcS1A on chromosome 1 accounts for ~ 50% of SSU transcript, the remainder being contributed by the rbcS1B, rbcS2B and rbcS3B genes located contiguously on chromosome 5 (Yoon et al., 2001). An Arabidopsis double mutant lacking expression of rbcS1A and with strongly reduced expression of rbcS3B (the 1a3b mutant) has a low Rubisco content (30% of wild‐type plants) and slow growth (Izumi et al., 2012). In this study the 1a3b mutant was complemented with either the Arabidopsis rbcS1A (control), an rbcS1A variant encoding the Chlamydomonas α‐helix sequences or the native rbcS2 gene from Chlamydomonas. We compared the Rubisco content, catalytic properties, leaf photosynthesis and growth of multiple lines for each genotype produced. Our results show that the 1a3b mutant is a valuable background for attempts to assemble an algal CCM in an angiosperm chloroplast, and for wider examination of the contribution made by SSU genetic diversity to Rubisco properties.
Materials and Methods
Plant material and growth conditions
Arabidopsis (Arabidopsis thaliana (L.) Heynh. Col‐0) seeds were sown on compost, stratified for 3 d at 4°C and grown at 20°C, ambient CO2, 70% relative humidity and 150 μmol photons m−2 s−1 in 12 : 12 h light : dark. For comparisons of different genotypes, plants were grown from seeds of the same age and storage history, harvested from plants grown in the same environmental conditions. Tobacco (Nicotiana benthamiana L.) was cultivated in a glasshouse (minimum 20°C, natural light supplemented to give light periods of at least 12 h). An Arabidopsis rbcs1a rbcs2b mutant (double mutant 1a2b) was generated by crossing T‐DNA insertion lines GABI_608F01 (At1g67090) and GABI_324A03 (At5g38420). The 1a3b mutant (GABI_608F01 (At1g67090); SALK_117835 (At5g38410)) was provided by Hiroyuki Ishida, Department of Applied Plant Science, Tohoku University, Japan.
DNA and RNA extraction, PCR and RT‐qPCR
Genomic DNA was extracted from rosettes according to Li & Chory (1998). PCRs were performed as in McCormick & Kruger (2015) using gene‐specific primers (Supporting Information Table S1). Insertion copy numbers were obtained by quantification of 35S promoter copies (performed by iDNA Genetics, www.idnagenetics.com). mRNA was isolated from the sixth and seventh leaves of 28‐d‐old rosettes and complementary DNA was synthesized with oligo(dT) primers. Reverse transcription quantitative PCR (RT‐qPCR) was carried out as in Andriotis et al. (2010). Primers to test for expression of SSU genes were designed to amplify the unique 3′ region of the transcripts (Table S1). Amplification efficiency was determined with a calibration curve for each primer set. Three reference genes (At4g05320 (UBQ10), At1g13320 (PP2A) and At4g26410 (RHIP1) (Czechowski et al., 2005)) were used for normalisation. Calculations of relative expression ratios were performed according to Pfaffl (2001).
Expression of rbcS genes in N. benthamiana and Arabidopsis 1a3b mutants
The α‐helices of rbcS1A (At1g67090) were replaced with those from the Chlamydomonas rbcS family (Fig. 1) using overlapping PCR with Phusion® High‐Fidelity DNA polymerase (as per the manufacturer's instructions; New England BioLabs). The promoter region (2 kb) upstream of rbcS1A was fused to the complete cDNA sequences of native or modified SSUs. The rbcS1A chloroplast transit peptide (TP) sequence was fused to the mature Chlamydomonas rbcS2 (Cre02.g120150) (Goldschmidt‐Clermont & Rahire, 1986) cDNA before promoter addition. Promoter–cDNA fusions were cloned into Gateway Entry vectors (pCR®8/GW/TOPO®TA Cloning® Kit; Thermo Fisher Scientific), then into the binary destination vector pGWB4 (Nakagawa et al., 2009) or pB7WG (Karimi et al., 2002) (Notes S1). Stop codons were removed to allow in‐frame C‐terminal fusion to a sequence encoding green fluorescent protein (GFP) in pGWB4. Binary vectors were transformed into Agrobacterium tumefaciens (AGL1) for transient gene expression in tobacco (Schöb et al., 1997) or stable insertion in Arabidopsis plants by floral dipping (Clough & Bent, 1998). Homozygous insertion lines were identified in the T3 generation by seedling segregation ratios on Murashige & Skoog (MS) medium (half‐strength) plates containing phosphinothricin (BASTA®, final concentration 10 ng μl−1) as a selectable marker. Lines used for subsequent analysis were checked for the presence of T‐DNA insertions at the rbcS1A and rbcS3B loci.
Figure 1.
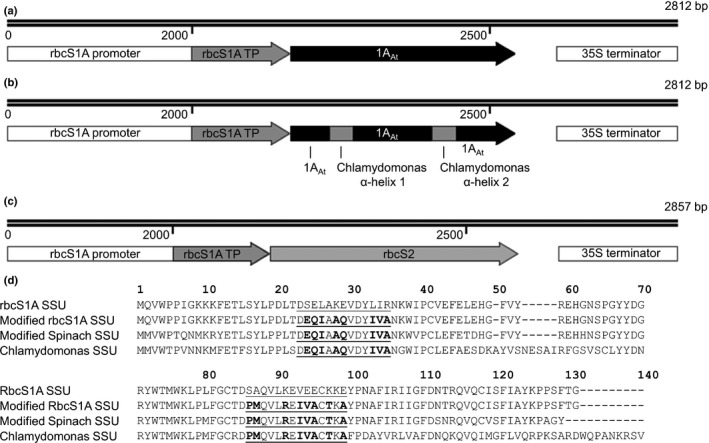
Gene expression cassettes for native and heterologous Rubisco small subunits. rbcS1A from Arabidopsis thaliana (1AA t) (a), rbcS1A with α‐helices from the Chlamydomonas reinhardtii rbcS family (1AA t MOD) (b), and mature rbcS2 from Chlamydomonas (S2Cr) (c) were expressed using the rbcS1A promoter (not drawn to scale) and 35S terminator. For S2Cr, the chloroplast transit peptide (TP) of Chlamydomonas rbcS2 (45 amino acids) was replaced with the rbcS1A TP (55 amino acids) from Arabidopsis to facilitate localisation of the mature rbcS2 to the chloroplast. (d) Alignments of the mature SSU peptides generated in this study. Numbering is relative to the Chlamydomonas rbcS2 sequence. Residues that comprise the two α‐helixes A and B are underlined, and those different from rbcS1A are in bold. For comparison with 1AA t MOD, the modified spinach SSU generated by Meyer et al. (2012) is included.
Protein quantification and Rubisco content
For determination of leaf protein and Rubisco contents on an area basis, soluble protein was extracted from 2 cm2 of snap frozen leaf material from 32‐d‐old plants (sixth and seventh leaf) in 500 μl of 50 mM Tricine‐NaOH (pH 8.0), 10 mM EDTA, 1% (w/v) PVP40, 20 mM 2‐mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride and 10 μM leupeptin. Following centrifugation at 2380 g for 5 min at 4°C, soluble protein was quantified using a Bradford‐based assay (Bio‐Rad) against BSA standards (Thermo Fisher Scientific). Rubisco content was determined in an aliquot of the extract via 14C‐CABP (carboxy‐d‐arabinitol 1,5‐bisphosphate) binding following incubation with 10 mM NaHCO3, 20 mM MgCl2 and the addition of 3 μl 12 mM 14C‐CABP (37 MBq mmol−1) for 25 min at room temperature (Whitney et al., 1999).
Subunit ratios were estimated by immunoblotting. Extracts were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) on a 4–12% (w/v) polyacrylamide gel (Bolt® Bis‐Tris Plus Gel; Thermo Fisher Scientific), transferred to polyvinylidene fluoride (PVDF) membrane then probed with rabbit serum raised against wheat Rubisco at 1 : 10 000 dilution (Howe et al., 1982) followed by Li‐Cor IRDye® 800CW goat anti‐rabbit IgG (Li‐Cor Inc.) at 1 : 10 000 dilution, then viewed on an Li‐Cor Odyssey CLx Imager. The contributions of LSU and SSUs were estimated from a five‐point standard curve of a wild‐type sample of known Rubisco content (0.1–2.4 μg Rubisco).
Rubisco catalytic properties
Whole 45‐d‐old rosettes (20–30 cm2) were rapidly frozen in liquid nitrogen (N2) and Rubisco was extracted as described by Prins et al. (2016), then activated for 45 min on ice before assays were conducted at 25°C. Catalytic properties of Rubisco from wild‐type and transgenic lines were determined from 14CO2 consumption, essentially as described by Prins et al. (2016) with alterations as per Orr et al. (2016), using 40 μl of extract. Six CO2 concentrations were used with O2 concentrations of 0 and 21%.
Rubisco specificity factor was determined on Rubisco purified from each genotype from c. 300 cm2 rosette tissue using the method described by Prins et al. (2016), with the omission of the final Sephacryl S‐200 step, which was found to be unnecessary for obtaining a clean extract (Orr et al., 2016). Rubisco CO2/O2 specificity (S C/O) was determined using the method of Parry et al. (1989). At least 10 measurements were made on the Rubisco purified from each genotype. Values were normalised based on measurements made in the same experiment on purified wheat (Triticum aestivum) Rubisco, which has an established S C/O of 100 (Parry et al., 1989).
Chlorophyll quantification
Leaf discs (c. 10 mg fresh weight) were frozen in liquid N2, powdered, and then mixed with 100 volumes of ice‐cold 80% (v/v) acetone, 10 mM Tris–HCl. Following centrifugation at 17 200 g for 10 min, chlorophyll was quantified according to Porra et al. (1989).
Measurement of photosynthetic parameters
Gas exchange and chlorophyll fluorescence were determined using a Li‐Cor LI‐6400 portable infra‐red gas analyser with a 6400‐40 leaf chamber on either the sixth or the seventh leaf of 35‐ to 45‐d‐old nonflowering rosettes grown in large pots to generate leaf area sufficient for gas exchange measurements (Flexas et al., 2007). For all gas exchange experiments, leaf temperature and chamber relative humidity were 20°C and c. 70%, respectively. Gas exchange data were corrected for CO2 diffusion from the measuring chamber as in Bellasio et al. (2016). Light response curves for net photosynthetic CO2 assimilation (A) were generated at ambient CO2 (400 μmol mol−1). A nonrectangular hyperbola was fitted to the light response (Marshall & Biscoe, 1980; Thornley, 1998). The response of A to varying sub‐stomatal CO2 concentration (C i) was measured at 1500 μmol photons m−2 s−1. To calculate the maximum rate of Rubisco carboxylation (V c,max) and the maximum photosynthetic electron transport rate (J max), the A/C i data were fitted to the C3 photosynthesis model as in Ethier & Livingston (2004) using the catalytic parameters K c air and affinity for O2 (K o) values for wild‐type Arabidopsis Rubisco at 20°C as reported in Walker et al. (2013). For estimates of the ratio of Rubisco oxygenase to carboxylase activity (V o/V c), leaves were measured under photorespiratory (ambient oxygen (O2), 21% (v/v)) or low‐photorespiratory (low O2, 2% (v/v)) conditions (Bellasio et al., 2014).
Maximum quantum yield of photosystem II (PSII) (F v/F m) was measured using a Hansatech Handy PEA continuous excitation chlorophyll fluorimeter (Hansatech Instruments) (Maxwell & Johnson, 2000). Nonphotochemical quenching (NPQ) analyses were performed using a Hansatech FMS1 pulse‐modulated chlorophyll fluorimeter. Rapid light response curves were generated by measuring the fluorescence response to a saturating pulse (applied every 30 s) under increasing levels of actinic light (0–1500 μmol photons m−2 s−1). Quenching parameters, including NPQs and NPQf, were derived as in Griffiths & Maxwell (1999).
Confocal laser scanning microscopy
Leaves were imaged with a Leica TCS SP2 laser scanning confocal microscope (Leica Microsystems) as in Atkinson et al. (2016).
Results
The 1a3b mutant of Arabidopsis provided a suitable genotype for examining the influence of heterologous SSUs on leaf photosynthesis and growth. Some aspects of the 1a3b mutant phenotype may reflect loss of distinct Rubisco isoforms (i.e. forms with different SSU compositions), as well as loss of total Rubisco activity. As a first step to evaluate this possibility, a second mutant, lacking expression of rbcS1A and a different minor SSU, rbcS2B (the 1a2b mutant) was included in some of the analyses. Quantification of T‐DNA copy numbers indicated that neither double mutant contained T‐DNA insertions other than those at their respective rbcS loci.
Design and targeting of native and heterologous SSUs
Binary vectors were generated to express either the full‐length native Arabidopsis rbcS1A (1AAt), the mature Chlamydomonas rbcS2 N‐terminally fused to the chloroplast TP sequence from Arabidopsis rbcS1A (S2Cr), or the full‐length Arabidopsis rbcS1A modified to contain α‐helices matching the amino acid sequence as those of the Chlamydomonas SSU family (1AAtMOD) (Fig. 1; Notes S1). Chlamydomonas and Arabidopsis SSU α‐helices have the same number of amino acids, but differ in terms of chemical composition. Expression of the introduced proteins was driven by the promoter of Arabidopsis rbcS1A, which has the highest expression level of the Arabidopsis rbcS genes (Izumi et al., 2012).
To check the subcellular locations of introduced SSUs, they were initially generated as C‐terminal fusions to GFP and transiently expressed in leaves of N. benthamiana. Fluorescence microscopy revealed that all three fusion proteins were located in the chloroplast stroma (Fig. S1). Untagged SSUs were then stably expressed in the Arabidopsis 1a3b mutant.
Expression levels, leaf protein and Rubisco content of native and heterologous SSU isoforms
In wild‐type plants, rbcS1A transcripts were the most abundant (43% of the rbcS pool), followed by rbcS3B (28%), rbcS2B (21%) and rbcS1B (8%) (Fig. 2; Table S2). The 1a3b mutant had no detectable transcript for rbcS1A and much reduced levels of transcript for rbcS3B (c. 10% of wild‐type levels). Both rbcS1A and rbcS2B transcripts were below the level of detection in the 1a2b mutant. In the 1a3b mutant, transcript levels for the two undisrupted rbcS genes, rbcS1B and rbcS2B, were 50 and 170% of those in wild‐type plants, respectively. In the 1a2b mutant, rbcS1B and rbcS3B transcript levels were 120 and 140% of those in wild‐type plants, respectively. For both mutants, transcript levels for rbcL (ATCG00490) and for the overall rbcS pool were 50% of those in wild‐type plants.
Figure 2.
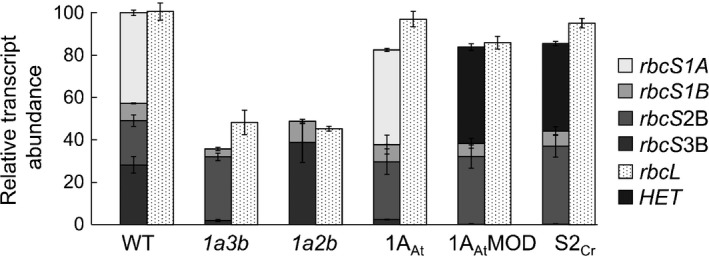
Transcript abundances of the Rubisco gene family in rbcs mutants and transgenic lines of Arabidopsis thaliana. Abundances of rbcS1A (At1g67090), rbcS1B (At5g38430), rbcS2B (At5g38420), rbcS3B (At5g38410) and rbcL (Atcg00490) transcripts were quantified relative to wild‐type levels (set at 100) from 28‐d‐old rosettes using RT‐qPCR with gene‐specific primers (Supporting Information Table S1). For wild‐type, 1a3b and 1a2b values are the means ± SE of measurements made on three individual 28‐d‐old rosettes. For transgenic lines values are means ± SE of measurements made on nine rosettes, three from each of the three lines. Full expression data are shown in Table S2. HET, heterologous rbcS.
For each of the three transgenic genotypes expressing native or heterologous SSUs in the 1a3b mutant background, at least six independent lines segregated in the T2 generation. Transgenic plants were screened for faster growth rates and maximum quantum yield of PSII (measured by dark‐adapted leaf fluorescence; F v/F m) compared to the 1a3b mutant (Fig. S2). For further analysis, homozygous T3 lines for each genotype were selected from the three best‐performing T2 segregating lines.
For each line of each transgenic genotype, transcript levels for the inserted transgene were comparable to those of the native rbcS1A gene in wild‐type plants (Fig. 2; Table S2). Levels of transcript of the undisrupted native Rubisco genes were altered in these lines relative to wild‐type plants. For rbcL, transcript levels were higher in transgenic than in 1a3b mutant plants, and in at least one independent line for each construct they were as high as in wild‐type plants. As in 1a3b mutants, transcript levels for rbcS2B in transgenic plants were generally higher than those in wild‐type plants (Fig. 2; Table S2).
The leaf Rubisco content in the 1a3b and 1a2b mutants was reduced by 70 and 50%, respectively, relative to wild‐type plants (Fig. 3a). Total soluble protein content in leaves of the mutants was also c. 60% of wild‐type values in both cases. This reduction was larger than could be accounted for by the reduction in Rubisco content alone (Fig. 3b; Table S3).
Figure 3.
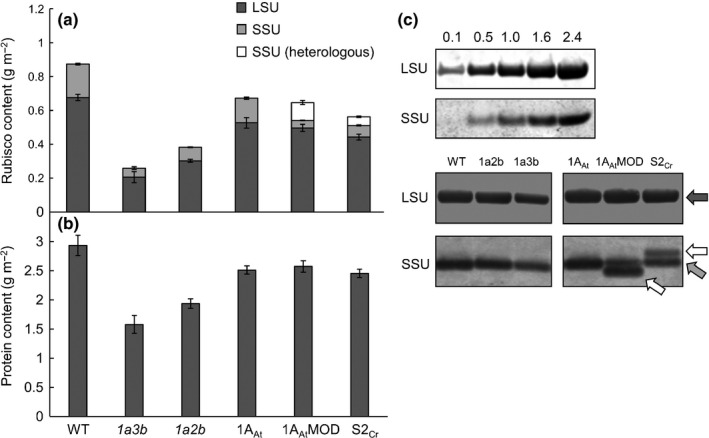
Rubisco and protein contents in rbcs mutants and transgenic lines of Arabidopsis thaliana. Rubisco (a) and total protein contents (b) are shown for 32‐d‐old plants. Rubisco content was determined via 14C‐CABP binding, and subunit ratios were estimated by immunoblotting. For wild‐type, 1a3b and 1a2b values are the means ± SE of measurements made on three individual rosettes. For transgenic lines values are means ± SE of measurements made on nine rosettes, three from each of the three lines. (c) Representative immunoblots for wild‐type plants and transgenic lines, probed with a serum containing polyclonal antibodies against Rubisco. Standard curves (0.1–2.4 μg Rubisco) are shown for wild‐type large subunit (LSU, 55 kDa) and small subunits (SSUs, 14.8 kDa), followed by protein amounts in different lines. Native LSU, SSU and heterologous SSUs (15.5 and 14 kDa, respectively) are indicated by dark grey, light grey and white arrows, respectively. Quantification of soluble protein and Rubisco is shown in Supporting Information Table S3.
Complementation of the 1a3b mutant restored total Rubisco to 75% of wild‐type levels for 1AAt and 1AAtMOD lines, and to 65% of wild‐type levels for S2Cr lines. Immunoblotting revealed that the heterologous SSUs 1AAtMOD and S2Cr had different mobilities on SDS‐PAGE gels from the native SSUs (Fig. 3c). This enabled quantification of the relative contributions of the LSU, the native SSUs and the heterologous SSUs to total Rubisco content (Fig. 3a). There were no significant differences in the ratio of LSU to SSU protein between any of the lines tested (Table S3). The 1AAtMOD and S2Cr transgenic lines retained the same amount of native SSU (i.e. products of the rbcS1B and rbcS2B genes) as the 1a3b mutant. Heterologous SSU levels were 2.4‐fold higher than native SSU levels in 1AAtMOD. By contrast, heterologous SSU levels were 1.4‐fold lower than native SSU levels in S2Cr lines.
Rubisco activity in mutant and transgenic plants
The in vitro catalytic properties of Rubisco from wild‐type plants (Table 1) were in good agreement with those of Galmés et al. (2014). The catalytic properties of Rubisco from 1a3b and 1a2b mutants were comparable to values for Rubisco from wild‐type plants. Rubisco from 1AAt lines had the same catalytic properties as Rubisco from wild‐type plants. This was also true for 1AAtMOD lines, despite the modification to the Rubisco SSU in these plants. However, k cat c and S C/O values were significantly lower for Rubisco from S2Cr lines than for Rubisco from wild‐type plants.
Table 1.
Catalytic parameters of Rubisco in rbcs mutants and transgenic lines of Arabidopsis thaliana
| Wild‐type | 1a3b | 1a2b | 1AAt | 1AAtMOD | S2Cr | |
|---|---|---|---|---|---|---|
| k cat c (s−1) | 4.1 ± 0.1 | 4.2 ± 0.1 | 4.1 ± 0.2 | 4.0 ± 0.1 | 4.1 ± 0.1 | 3.6 ± 0.1a |
| K c (μM) | 10.7 ± 0.7 | 9.5 ± 0.7 | 9.4 ± 1.1 | 10.4 ± 1.1 | 11.5 ± 0.9 | 9.6 ± 1.0 |
| K c air (μM) | 15.8 ± 1.0 | 14.3 ± 0.5 | 15.4 ± 1.5 | 16.9 ± 1.8 | 17.1 ± 1.0 | 16.4 ± 1.2 |
| k cat c/K c air | 0.25 ± 0.01 | 0.3 ± 0.02 | 0.27 ± 0.02 | 0.25 ± 0.03 | 0.24 ± 0.02 | 0.22 ± 0.03 |
| S C/O | 92.5 ± 1.0 (27) | 96.3 ± 1.7 (11) | 93.4 ± 1.7 (10) | 91.8 ± 1.0 (17) | 92.7 ± 0.8 (18) | 87.8 ± 0.9a (14) |
Rubisco specificity was determined from at least 10 replicate measurements for the enzyme purified from each line. Other catalytic parameters are calculated using the Michaelis–Menten model as described in Prins et al. (2016). The table shows mean ± SD values for three biological replicates, except for Rubisco specificity, which is the mean ± SD of the numbers of technical replicates shown in parentheses. All values were measured at 25°C. K c, K m for CO2 at 0% O2; K c air, K m for CO2 at 21% O2; k cat c, turnover number (mol carboxylation product mol−1 active site s−1); k cat c/K c air, Rubisco carboxylation efficiency at 21% O2; S C/O, Rubisco specificity factor.
Significant difference (P < 0.05) as determined by ANOVA followed by Tukey's HSD tests.
Growth phenotypes
Growth of transgenic lines was compared with that of (1) wild‐type plants, (2) the parental 1a3b mutant and (3) representative nontransgenic 1a3b mutant lines selected as out‐segregants from the T2 populations (Fig. 4). Fresh and dry weights of the out‐segregant mutant lines were the same as those of the parental 1a3b mutant at 28 d (Fig. 4c; Table S4). Out‐segregant lines had lower rates of rosette expansion than the parental 1a3b mutant (Fig. 4b), but this did not affect interpretation of the effects of the transgenes on growth.
Figure 4.

Growth analysis of rbcs mutants and transgenic lines of Arabidopsis thaliana. (a) Representative examples of 28‐d‐old rosettes (T3) for mutants and transgenic genotypes. (b) Rosette expansion of homozygous transgenic and 1a3b out‐segregant plants compared to that of wild‐type and 1a3b mutant plants. (c) Fresh and dry weights were compared after 28 d. For wild‐type (WT), 1a3b and 1a2b values are the means ± SE of measurements made on 10 individual rosettes. For transgenic lines values are means ± SE of measurements made on 30 rosettes, 10 from each of the three lines. See Supporting Information Table S4 for full dataset. seg, segregating T3 wild‐type.
As reported previously, 1a3b mutants had very low growth rates (Izumi et al., 2012). All three transgenic genotypes had greater rates of rosette expansion than 1a3b lines, with 1AAt and 1AAtMOD having higher expansion rates than S2Cr (Fig. 4b). The dry weight of 1AAt rosettes at 28 d was on average 84% of that of wild‐type plants, and was not significantly different from the wild‐type for two of the three lines. For 1AAtMOD and S2Cr lines, dry weight was on average 75 and 56%, respectively, of that of wild‐type plants. There was no significant difference in the ratio of dry weight to fresh weight between wild‐type plants and transgenic lines. All three transgenic genotypes had higher leaf area to weight ratios (rosette area per unit fresh or dry weight) than 1a3b mutants, and were not significantly different in this respect from wild‐type plants (Table S4).
Rosette expansion rates and fresh and dry weights in the 1a2b mutant were greater than in the 1a3b mutant, but lower than those of wild‐type and transgenic lines (Fig. 4c). The 1a2b mutant had a lower ratio of fresh to dry weight than the 1a3b mutant (Table S4). Although the specific leaf areas (rosette area per unit dry weight) of 1a2b and 1a3b mutants were comparable, rosette area per unit fresh weight was significantly higher in 1a2b than in 1a3b mutants.
Photosynthetic characteristics
At ambient CO2 and saturating light, all three transgenic genotypes had much higher rates of CO2 assimilation (A max) than the 1a3b mutant (A/photosynthetically active radiation (PAR) curves, Fig. 5a). A max was similar to that of wild‐type plants in 1AAt and 1AAtMOD lines but lower in S2Cr lines (Table 2). A max was higher in the 1a2b than in the 1a3b mutant, and was comparable in 1a2b and S2Cr lines. The apparent quantum efficiency (Φ) for all three transgenic lines was higher than in the 1a3b mutant and comparable with the wild‐type value. Light compensation point and respiration rate in the dark (R d) were the same in all lines.
Figure 5.
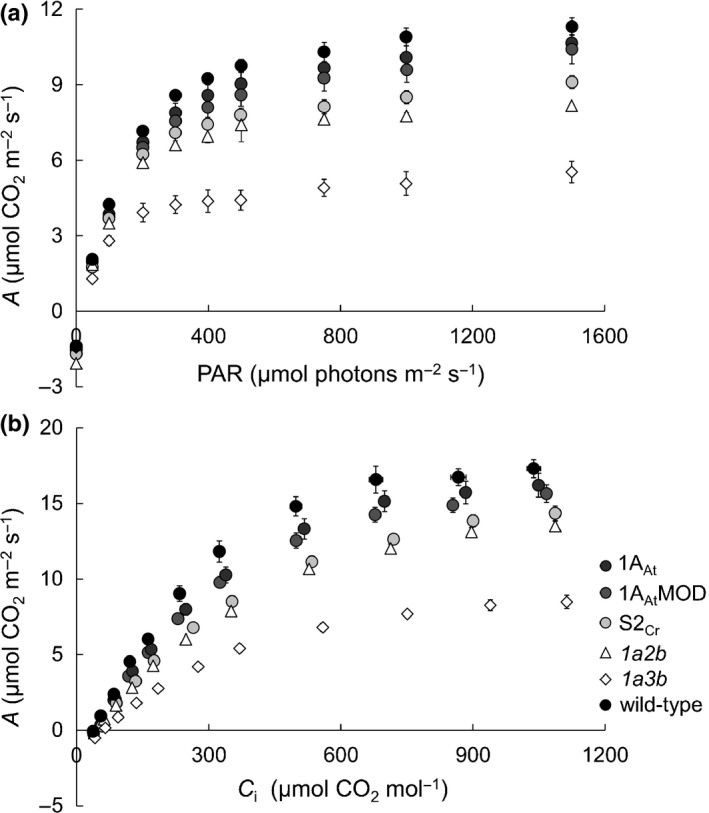
Photosynthesis response curves of rbcs mutants and transgenic lines of Arabidopsis thaliana. Measurements were made on the sixth or seventh leaf of 35‐ to 45‐d‐old nonflowering rosettes. (a) A/PAR curves show the response of CO 2 assimilation rates to different light levels at ambient CO 2 levels of 400 μmol mol−1. (b) A/C i curves showing the response of net CO 2 assimilation to different sub‐stomatal concentrations of CO 2 (C i) under saturating light (1500 μmol photons m−2 s−1). For wild‐type, 1a3b and 1a2b values are the means ± SE of measurements made on individual leaves from four different rosettes. For transgenic lines values are means ± SE of measurements made on 12 rosettes, four from each of the three lines.
Table 2.
Variables derived from photosynthetic response curves, based on gas exchange analysis of 35‐ to 45‐d‐old Arabidopsis thaliana plants
| Wild‐type | 1a3b | 1a2b | 1AAt | 1AAtMOD | S2Cr | |
|---|---|---|---|---|---|---|
| A amb (μmol CO2 m−2 s−1) | 5.7 ± 0.1a | 3.4 ± 0.3c | 4.7 ± 0.1b | 5.3 ± 0.2ab | 5.1 ± 0.2ab | 5.0 ± 0.1ab |
| A max (μmol CO2 m−2 s−1) | 13.5 ± 0.5a | 6.8 ± 0.5c | 10.4 ± 0.2b | 12.8 ± 0.6ab | 12.3 ± 0.7ab | 10.9 ± 0.4bc |
| g s (mol CO2 m−2 s−1) | 0.34 ± 0.06a | 0.42 ± 0.06a | 0.33 ± 0.02a | 0.3 ± 0.02a | 0.36 ± 0.03a | 0.41 ± 0.03a |
| Φ (mmol CO2 mol−1 photons) | 55.9 ± 1.9a | 42.2 ± 2.3b | 55.3 ± 1.4a | 53.5 ± 3.3a | 51.5 ± 1.8a | 53.6 ± 0.4a |
| LCP (μmol CO2 m−2 s−1) | 16.6 ± 1.3a | 18.8 ± 0.7a | 22.7 ± 0.8a | 18.0 ± 2.6a | 17.0 ± 1.5a | 20.9 ± 1.6a |
| V c,max (μmol CO2 m−2 s−1) | 31.4 ± 1.4a | 14.6 ± 0.4d | 20.9 ± 0.4c | 27.1 ± 1.3ab | 26.1 ± 1.1ab | 22.2 ± 0.6bc |
| J max (mmol e− m−2 s−1) | 73.3 ± 2.8a | 32.8 ± 1.3d | 53.7 ± 0.9c | 66.6 ± 3.0ab | 63.7 ± 2.1ab | 56.3 ± 1.6bc |
| Γ (μmol CO2 mol−1) | 39.4 ± 2.2b | 60.0 ± 3.9a | 42.8 ± 1.6b | 39.1 ± 0.9b | 41.4 ± 0.5b | 43.2 ± 0.8b |
| R d (μmol CO2 m−2 s−1) | 1.9 ± 0.2a | 1.8 ± 0.1a | 2.0 ± 0.1a | 1.8 ± 0.1a | 1.9 ± 0.1a | 1.8 ± 0.1a |
| Initial slope (A/C i) | 0.055 ± 0.003a | 0.024 ± 0.007d | 0.034 ± 0.006c | 0.045 ± 0.002b | 0.044 ± 0.002b | 0.036 ± 0.001bc |
For measurements of net photosynthetic CO2 assimilation (A)/photosynthetically active radiation (PAR), relative humidity was maintained at 68 ± 4% and ambient CO2 levels at 400 μmol mol−1. For measurements of A/sub‐stomatal CO2 concentration (C i), relative humidity was maintained at 73 ± 1% under a constant illumination of 1500 μmol photons m−2 s−1. All measurements were performed at 20°C. Values are the mean ± SE of measurements made on four leaves, each from a different plant (as shown in Fig. 5) followed by letters indicating significant differences (P < 0.05) as determined by ANOVA followed by Tukey's HSD tests. Values followed by the same letter are not significantly different. A amb, net photosynthesis measured at ambient CO2 and growth chamber light levels; A max, light‐saturated CO2 assimilation rate at ambient CO2; g s, stomatal conductance to CO2 (at ambient CO2); Φ, apparent quantum efficiency; LCP, light compensation point; V c,max, maximum rate of Rubisco carboxylation; J max, maximum electron transport rate; Γ, CO2 compensation point (C i–A); R d, respiration in the dark.
There were substantial differences between the 1a3b mutant and the transgenic genotypes in the response of CO2 assimilation to changing external CO2 concentrations under saturating light (A/C i curves, Fig. 5b). Several photosynthetic parameters can be derived from A/C i curves (Table 2). The maximum rate of Rubisco carboxylation (V c,max) and maximum photosynthetic electron transport rate (J max) were not significantly different between wild‐type, 1AAt and 1AAtMOD plants, but were lower in S2Cr plants than in wild‐type plants. The initial linear slope of the A/C i curve (a measure of the carboxylation efficiency and activation state of Rubisco) was lower for transgenic genotypes than for wild‐type plants due to reduced Rubisco content in the transgenic lines (Fig. 3a). In the 1a2b mutant, V c,max, J max, the sub‐stomatal CO2 compensation point (Γ) and the initial slope of the A/C i curve were different from those of the 1a3b mutant, but similar to values for the S2Cr lines.
Gas exchange rates and chlorophyll fluorescence measurements under photorespiratory (ambient O2 (21%)) and nonphotorespiratory (low O2 (2%)) conditions were used to derive information about photorespiration (Table 3). Gross CO2 assimilation rates (GA, CO2 assimilation in the absence of respiration) and NADPH production (estimated from the photosynthetic electron transport rate, J NADPH) can together be used to estimate the ratio of Rubisco oxygenase to carboxylase activity (V o/V c) (Bellasio et al., 2014).
Table 3.
Estimates of in vivo Rubisco oxygenase and carboxylase activities made from measurements of gas exchange and chlorophyll fluorescence under ambient (21%) or low (2%) O2
| Wild‐type | 1a3b | 1a2b | 1AAt | 1AAtMOD | S2Cr | |
|---|---|---|---|---|---|---|
| GA Low (μmol m−2 s−1) | 9.48 ± 0.56a | 4.58 ± 0.4c | 5.69 ± 0.53c | 8.73 ± 0.09ab | 8.57 ± 0.64ab | 6.72 ± 0.49bc |
| GA amb (μmol m−2 s−1) | 6.17 ± 0.36a | 2.98 ± 0.25d | 3.67 ± 0.36 cd | 5.6 ± 0.15ab | 5.49 ± 0.42ab | 4.36 ± 0.38bc |
| J NADPHlow (μmol m−2 s−1) | 18.9 ± 1.1a | 9.1 ± 0.8c | 11.4 ± 1.1c | 17.5 ± 0.2ab | 17.1 ± 1.3ab | 13.4 ± 0.9bc |
| J NADPHamb (μmol m−2 s−1) | 19.9 ± 0.9a | 9.2 ± 0.7c | 11.9 ± 0.9bc | 18.4 ± 0.2a | 18.2 ± 1.4a | 14.1 ± 1b |
| V o (μmol m−2 s−1) | 2.21 ± 0.13a | 1.06 ± 0.11b | 1.34 ± 0.12b | 2.09 ± 0.06a | 2.06 ± 0.17a | 1.57 ± 0.07ab |
| V c (μmol m−2 s−1) | 7.27 ± 0.43a | 3.52 ± 0.29d | 4.35 ± 0.41c | 6.64 ± 0.12ab | 6.52 ± 0.49ab | 5.15 ± 0.42bc |
| V o/V c | 0.304 ± 0.002a | 0.302 ± 0.013a | 0.307 ± 0.008a | 0.313 ± 0.015a | 0.316 ± 0.012a | 0.307 ± 0.011a |
Arabidopsis thaliana plants (35–40 d old) were measured under 300 μmol photons m−2 s−1, and ambient CO2 of 300 μmol mol−1 as in Bellasio et al. (2014). For wild‐type, 1a3b and 1a2b values are the means ± SE of measurements made on individual leaves from five different rosettes. For transgenic lines, values are means ± SE of measurements made on 15 rosettes, five from each of the three lines. Values are followed by letters indicating significant difference (P < 0.05), as determined by ANOVA followed by Tukey's HSD tests. Values followed by the same letter are not significantly different. GA low, gross photosynthetic rate (A+R d) under 2% O2 (2%); GA amb, gross photosynthetic rate under 21% O2; J NADPHlow, NADPH produced for photosynthesis (derived from electron transport rate) under 2% O2; J NADPHamb, NADPH produced for photosynthesis under 21% O2; V o, Rubisco oxygenation rate; V c, Rubisco carboxylation rate.
The transgenic genotypes had higher GA and J NADPH values than the 1a3b mutant. Values for 1AAt and 1AAtMOD lines were similar to those of wild‐type plants, but values for S2Cr lines were lower. GA and J NADPH in the 1a2b mutant were higher than in the 1a3b mutant, and comparable with values for the S2Cr lines. There were no significant differences in V o/V c values between any of the lines, indicating that relative photorespiratory rates were similar across genotypes under the conditions used.
Chlorophyll content and dark‐adapted F v/F m values in the transgenic lines and the 1a2b mutant were higher than in the 1a3b mutant, and were not significantly different from those of wild‐type plants (Table S5). The 1a3b mutants had higher levels of NPQ than wild‐type plants, but NPQ in transgenic genotypes was comparable with that of wild‐type plants (Fig. 6). By contrast, the NPQ value for the 1a2b mutant was lower than that of wild‐type plants. NPQ has two components: fast relaxing quenching (qE: NPQfast) associated with photoprotection, and slow relaxing quenching (qI: NPQslow) associated with chronic photoinhibition (Walters & Horton, 1991). To calculate the contribution of these components in the mutant lines, NPQ was tracked following a period of high light (600 μmol photons m2 s−1 for 1 h) and subsequent recovery in darkness (1 h). qI was lower in both transgenic genotypes that in wild‐type plants, but qE was elevated in the 1a3b mutant and reduced in the 1a2b mutant. The qE : qI ratio was higher in the 1a3b mutant but lower in the 1a2b mutant than in wild‐type plants (Table S6).
Figure 6.
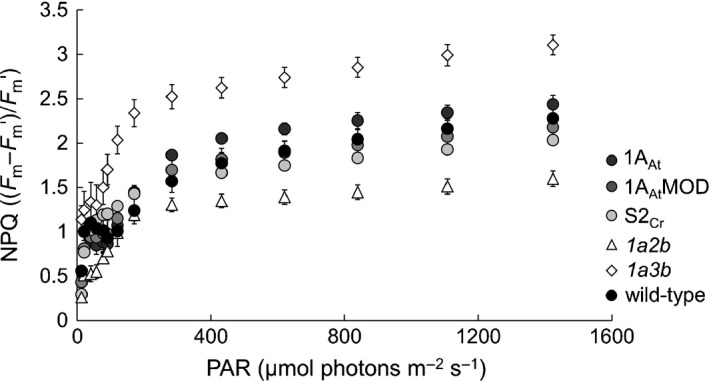
Nonphotochemical quenching response to light in leaves of rbcs mutants and transgenic lines of Arabidopsis thaliana. All plants were 28 d old. For wild‐type, 1a3b and 1a2b values are the means ± SE of measurements made on individual leaves from four different rosettes. For transgenic lines values are means ± SE of measurements on leaves from 12 plants, four from each of the three lines.
Discussion
Our results illustrate the impact of varying Rubisco content and native SSU composition on plant performance in Arabidopsis. Furthermore, we have shown that heterologous, pyrenoid competent SSUs assemble with the native LSU to produce a functional hybrid Rubisco with catalytic properties similar to the native Rubisco. This is a significant step towards the introduction of a functional algal CCM into higher plants.
Differences in native SSU composition of Rubisco have only minor implications for plant performance in Arabidopsis
The data presented here suggest that the four native SSUs in Arabidopsis are largely equivalent in the properties they convey to the Rubisco enzyme under the growth conditions tested. Four genotypes provided data that lead to this conclusion: (1) wild‐type plants, with the highest Rubisco content and with Rubisco containing almost exclusively rbcS1A, rbcS2B and rbcS3B SSUs (because of its very low transcript levels it is assumed that rbcS1B makes a very minor contribution to the SSU population); (2) 1AAt plants, with c. 78% of wild‐type Rubisco content and with Rubisco containing mainly rbcS1A and rbcS2B; (3) the 1a2b mutant, with 45% of wild‐type Rubisco content and with Rubisco containing mainly rbcS3B; and (4) the 1a3b mutant, with 30% of wild‐type Rubisco content and with Rubisco containing rbcS2B. The catalytic properties k cat c, K c air and S C/O of Rubisco at 25°C were similar in these four genotypes (Table 1), and thus they are largely independent of the native SSU composition of Rubisco in Arabidopsis.
Nearly all the phenotypic differences between the four genotypes with different native SSU compositions can be explained by the differences in total Rubisco content alone. Across these four genotypes, parameters including leaf protein content (Fig. 3; Table S3), the response of photosynthesis to light and to CO2 (Fig. 5), Γ and J max (Table 2), and the rates of biomass accumulation and rosette expansion (Fig. 4; Table S4) responded to decreasing Rubisco activity in the manner expected for a single enzyme exercising a moderate degree of control over CO2 assimilation (Stitt & Schulze, 1994). Additionally, the responses were broadly in line with those observed for tobacco plants with varying amounts of Rubisco activity of probably constant SSU composition (Quick et al., 1991; Fichtner et al., 1993; Lauerer et al., 1993; Stitt & Schulze, 1994), and Arabidopsis plants with strong suppression of expression of all four SSU genes (Zhan et al., 2014).
Some features of the four genotypes did not vary consistently with Rubisco content. For example, chlorophyll content, F v/F m and Φ were strongly affected only in the genotype with the lowest levels of Rubisco, 1a3b (Tables 2, S5). Other parameters including leaf soluble protein content and specific leaf area were affected only in genotypes with less than 50% of wild‐type Rubisco levels (Fig. 3; Table S4). Our data in these respects are reminiscent of those obtained for tobacco under limiting light in which Rubisco activity was varied by expression of antisense RNA that targeted all of the SSUs (Quick et al., 1991; Fichtner et al., 1993; Lauerer et al., 1993; Stitt & Schulze, 1994). Reductions of c. 40% or less in Rubisco activity in tobacco plants under limiting light (as in our experiments) had relatively little effect on the rate of photosynthesis and few pleiotropic consequences. Greater reductions progressively affected photosynthesis and downstream processes, different processes being affected at different levels of Rubisco reduction (Quick et al., 1991; Stitt & Schulze, 1994). Future experiments will investigate which phenotypic differences between the lines are exaggerated when plants are grown in saturating light.
For processes associated with photoprotection, qualitatively different phenotypes were observed in the 1a3b and 1a2b mutants. NPQ was elevated in the 1a3b mutant. NPQ was also elevated in tobacco and rice with reduced levels of Rubisco (Quick et al., 1991; Lauerer et al., 1993; Ruuska et al., 2000; Ushio et al., 2003; von Caemmerer et al., 2004): this effect may result from reduced ATP consumption for CO2 assimilation, and hence a higher ΔpH across the thylakoid membrane. Lumen acidification promotes activity of the energy‐dissipating xanthophyll cycle (Ruuska et al., 2000; Johnson et al., 2009; Zaks et al., 2012). By contrast, with 1a3b plants and other plant species with reduced Rubisco, NPQ was reduced in 1a2b mutants. In particular, 1a2b plants had a much reduced rate of relaxation of NPQ immediately following the onset of darkness (the qE or fast component of NPQ). The exact mechanism underlying qE is not known (e.g. Johnson et al., 2009; Zaks et al., 2012). However, as mature rbcS2B and rbcS3B have identical amino acid sequences, the difference in NPQ between the 1a2b and 1a3b mutants is likely to stem from the pleiotropic effects of the different degrees of reduction of Rubisco activity in the two mutants, rather than from the different SSU compositions of their Rubiscos.
It is clear from previous work that SSUs can influence Rubisco catalysis. For example, overexpression of specific native or heterologous SSU proteins altered the catalysis of Rubisco in rice leaves, resulting in properties that are more like those of C4 plants (i.e. higher k cat c, but also higher K c (lower CO2 affinity) than for native rice Rubisco) (Ishikawa et al., 2011; Morita et al., 2014). Over‐expression in Arabidopsis of a pea SSU, differing from Arabidopsis SSUs by 40 amino acids, resulted in Rubisco with slightly reduced carboxylase activity and capacity for activation (Getzoff et al., 1998). Similarly, Rubisco properties were changed by introduction of a sorghum SSU into rice (Ishikawa et al., 2011). However, except in the case of the rice SSU above, little is known about the functional importance of sequence variation between SSUs within a species. SSU isoforms in a single species are typically very similar. In Chlamydomonas, for example, the two SSUs differ by only four amino acid residues (all outside the α‐helices) and appear to be functionally equivalent (Rodermel et al., 1996; Genkov et al., 2010). In Arabidopsis, the mature rbcS1A differs from rbcS1B, rbcS2B and rbcS3B by only eight amino acids, six of which are conserved between the three B‐class SSUs (Fig. S3). Two of these are located in the first α‐helix.
Chlamydomonas‐like SSUs generate a functional hybrid Rubisco in Arabidopsis
Introduction of either a Chlamydomonas SSU (S2Cr) or a modified version of rbcS1A (1AAtMOD) into the Arabidopsis 1a3b mutant substantially complemented several aspects of the 1a3b phenotype. In a previous study, a Chlamydomonas SSU introduced into pea chloroplasts was not processed to the mature, active form, probably due to differences in chloroplast import machinery between Chlamydomonas and higher plants (Su & Boschetti, 1994). In this study, replacing the Chlamydomonas SSU TP with the rbcS1A TP directed the mature protein to the chloroplast stroma (Fig. S1). Expression of S2Cr or 1AAtMOD increased Rubisco content in the 1a3b mutant without significantly enhancing levels of the remaining native SSUs, and thus both introduced proteins promoted expression of the native LSU and assembled into catalytically active hybrid Rubiscos. These results are consistent with the idea that rbcL transcription and LSU synthesis adjust according to the availability of SSU (Wollman et al., 1999; Wostrikoff & Stern, 2007; Wostrikoff et al., 2012; Zhan et al., 2014).
Photosynthesis was restored almost to wild‐type levels in 1AAtMOD (Fig. 5). Furthermore, the catalytic characteristics of Rubisco in 1AAtMOD plants, where c. 70% of the SSU pool was heterologous, were comparable to those of 1AAt and wild‐type plants (Table 1). This suggests that the SSU α‐helix regions alone do not affect Rubisco biogenesis or catalysis, and that Rubisco in higher plants can be made compatible with the requirements of the algal CCM without affecting enzyme performance.
Rubisco in S2Cr plants had lower k cat c and S C/O values than those of wild‐type and 1AAtMOD Rubisco, even though the S2Cr SSU pool contained only c.40% Chlamydomonas SSU. S2Cr lines generally performed less well than 1AAtMOD lines. Neither S2Cr nor 1AAtMOD lines are likely to be Rubisco‐limited because they both have c. 70% of the Rubisco content of wild‐type plants (Quick et al., 1991). Differences in photosynthesis and growth between S2Cr and 1AAtMOD lines are thus likely to result largely from SSU‐dependent differences in Rubisco catalytic properties. In Chlamydomonas, expression of a higher plant SSU can impart improved catalysis and S C/O (Genkov et al., 2010). The data shown here demonstrate that the reverse is also true: an algal SSU can negatively affect catalytic properties of the hybrid Rubisco in a higher plant. Since the 1AAtMOD and S2Cr SSUs have the same α‐helices, differences in catalytic properties of the hybrid enzyme must arise from sequence differences in regions of the SSU outside of these helices.
The Chlamydomonas SSU protein differs in several respects from the Arabidopsis SSUs, including the presence of additional amino acid residues at the C‐terminus and in the loop between β‐strands A and B (Spreitzer, 2003). The latter forms the entrance of the solvent channel and may be important for carboxylation rates and S C/O (Karkehabadi et al., 1995; Esquivel et al., 2013). Hybrid Rubisco enzymes with SSUs that diverge significantly in amino acid sequence from the native SSU frequently have altered stability and properties, and a lower capacity for assembly with the native LSU. The poor complementation of Arabidopsis Rubisco in S2Cr warrants further study to expand upon existing knowledge in this area, including the functional capacity of the chaperone Rubisco activase when presented with hybrid Rubiscos.
rbcs mutants of Arabidopsis are a useful platform for Rubisco analyses and the assembly of an algal CCM
This study shows that Arabidopsis mutants lacking SSU isoforms are a useful platform for attempts to assemble a functional algal CCM in higher plants. Introduction of 1AAtMOD, containing α‐helices believed to be necessary for pyrenoid assembly, had no apparent effect on Rubisco function and assembly, and plant performance was generally close to wild‐type levels under our growth conditions.
For aggregation of Rubisco into a pyrenoid, additional algal CCM components will be required. Cryo‐electron tomography of Chlamydomonas pyrenoids showed that Rubisco proteins are not randomly arranged, and periodicity is consistent with hexagonal close packing, with a space of 2–4.5 nm between each protein depending on their relative orientations (Engel et al., 2015). Other factors, such as linker proteins, are probably needed. Recently, a multiple repeat linker‐protein, EPYC1 (formerly known as LCI5), has been identified in Chlamydomonas that is associated with Rubisco during aggregation within the pyrenoid (Mackinder et al., 2016). The 1AAtMOD and S2Cr Arabidopsis lines are ideal backgrounds in which to test candidates for these other factors as they emerge, to clarify the nature of SSU‐associated interactions, and to integrate other essential algal CCM components (Atkinson et al., 2016).
Author contributions
A.J.M. and N.A. planned and designed the research and wrote the manuscript. A.M.S., D.J.O., M.T.M., H.G. and E.C‐S. assisted in experimental design, data analysis and writing of the manuscript. A.J.M., N.A. and N.L. performed the research, data analysis, collection, and assisted with data interpretation and writing.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Transient expression of Rubisco small subunit–GFP fusion proteins in tobacco.
Fig. S2 Impact of native and heterologous SSUs on photosynthesis and growth in the Arabidopsis mutant 1a3b background.
Fig. S3 Alignments of the mature Arabidopsis SSU amino acid sequences.
Table S1 Sequences of synthetic oligonucleotides used in this study
Table S2 Transcript abundances of the Rubisco gene family in rbcs mutants and transgenic lines
Table S3 Rubisco and soluble protein contents for rbcs mutants and transgenic lines
Table S4 Rosette area and biomass for rbcs mutants and transgenic lines
Table S5 Chlorophyll characteristics and maximum quantum yield of PSII (F v/F m) for rbcs mutants and transgenic lines
Table S6 Photosynthetic nonphotochemical quenching capacity for rbcs mutants
Notes S1 Expression vectors for Rubisco small subunit (rbcS) cassettes.
Acknowledgements
This work was funded by the UK Biotechnology and Biological Sciences Research Council (Institute Strategic Programme Grant BB/J004561/1 to the John Innes Centre, grants BB/I024453/1 and BB/M006468/1 to A.M.S. and A.J.M., respectively, and a PhD studentship from the John Innes Foundation to N.L.). E.C‐S. acknowledges financial support from the Lancaster Environment Centre. We thank Hiroyuki Ishida (Tohoku University) for seeds of the 1a3b mutant and the reviewers for their helpful comments.
See also the Commentary on this article by Sharwood, 214: 496–499.
References
- Andriotis VM, Pike MJ, Bunnewell S, Hills MJ, Smith AM. 2010. The plastidial glucose‐6‐phosphate/phosphate antiporter GPT1 is essential for morphogenesis in Arabidopsis embryos. Plant Journal 64: 128–139. [DOI] [PubMed] [Google Scholar]
- Atkinson N, Feike D, Mackinder LC, Meyer MT, Griffiths H, Jonikas MC, Smith AM, McCormick AJ. 2016. Introducing an algal carbon‐concentrating mechanism into higher plants: location and incorporation of key components. Plant Biotechnology Journal 14: 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, Leggat W, Price GD. 1998. The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast‐based CO2‐concentrating mechanisms in algae. Canadian Journal of Botany 76: 1052–1071. [Google Scholar]
- Bellasio C, Beerling DJ, Griffiths H. 2016. An Excel tool for deriving key photosynthetic parameters from combined gas exchange and chlorophyll fluorescence: theory and practice. Plant, Cell & Environment 39: 1180–1197. [DOI] [PubMed] [Google Scholar]
- Bellasio C, Burgess SJ, Griffiths H, Hibberd JM. 2014. A high throughput gas exchange screen for determining rates of photorespiration or regulation of C4 activity. Journal of Experimental Botany 65: 3769–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer S, Lawson T, Oxborough K, Baker NR, Andrews TJ, Raines CA. 2004. Stomatal conductance does not correlate with photosynthetic capacity in transgenic tobacco with reduced amounts of Rubisco. Journal of Experimental Botany 55: 1157–1166. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Quick WP, Furbank RT. 2012. The development of C4 rice: current progress and future challenges. Science 336: 1671–1672. [DOI] [PubMed] [Google Scholar]
- Carmo‐Silva E, Scales JC, Madgwick PJ, Parry MA. 2015. Optimizing Rubisco and its regulation for greater resource use efficiency. Plant, Cell & Environment 38: 1817–1832. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Schaffer M, Kuhn Cuellar L, Villa E, Plitzko JM, Baumeister W. 2015. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo‐electron tomography. eLife 4: e04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquivel MG, Genkov T, Nogueira AS, Salvucci ME, Spreitzer RJ. 2013. Substitutions at the opening of the Rubisco central solvent channel affect holoenzyme stability and CO2/O2 specificity but not activation by Rubisco activase. Photosynthesis Research 118: 209–218. [DOI] [PubMed] [Google Scholar]
- Ethier GJ, Livingston NJ. 2004. On the need to incorporate sensitivity to CO2 transfer conductance into the Farquhar‐von Caemmerer‐Berry leaf photosynthesis model. Plant, Cell & Environment 27: 137–153. [Google Scholar]
- Fichtner K, Quick WP, Schulze ED, Mooney HA, Rodermel SR, Bogorad L, Stitt M. 1993. Decreased ribulose‐1,5‐bisphosphate carboxylase‐oxygenase in transgenic tobacco transformed with ‘antisense’ rbcS. V. Relationship between photosynthetic rate, storage strategy, biomass allocation and vegetative plant growth at three different nitrogen supplies. Planta 190: 332–345. [Google Scholar]
- Flexas J, Ortuno MF, Ribas‐Carbo M, Diaz‐Espejo A, Florez‐Sarasa ID, Medrano H. 2007. Mesophyll conductance to CO2 in Arabidopsis thaliana . New Phytologist 175: 501–511. [DOI] [PubMed] [Google Scholar]
- Galmés J, Kapralov MV, Andralojc PJ, Conesa MÀ, Keys AJ, Parry MAJ, Flexas J. 2014. Expanding knowledge of the Rubisco kinetics variability in plant species: environmental and evolutionary trends. Plant, Cell & Environment 37: 1989–2001. [DOI] [PubMed] [Google Scholar]
- Genkov T, Meyer M, Griffiths H, Spreitzer RJ. 2010. Functional hybrid Rubisco enzymes with plant small subunits and algal large subunits: engineered rbcS cDNA for expression in Chlamydomonas. Journal of Biological Chemistry 285: 19833–19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genkov T, Spreitzer RJ. 2009. Highly conserved small subunit residues influence Rubisco large subunit catalysis. Journal of Biological Chemistry 284: 30105–30112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getzoff TP, Zhu GH, Bohnert HJ, Jensen RG. 1998. Chimeric Arabidopsis thaliana ribulose‐1,5‐bisphosphate carboxylase/oxygenase containing a pea small subunit protein is compromised in carbamylation. Plant Physiology 116: 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M, Beardall J, Raven JA. 2005. CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annual Review of Plant Biology 56: 99–131. [DOI] [PubMed] [Google Scholar]
- Goldschmidt‐Clermont M, Rahire M. 1986. Sequence, evolution and differential expression of the two genes encoding variant small subunits of ribulose bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii . Journal of Molecular Biology 191: 421–432. [DOI] [PubMed] [Google Scholar]
- Griffiths H, Maxwell K. 1999. In memory of C. S. Pittendrigh: does exposure in forest canopies relate to photoprotective strategies in epiphytic bromeliads? Functional Ecology 13: 15–23. [Google Scholar]
- Howe CJ, Auffret AD, Doherty A, Bowman CM, Dyer TA, Gray JC. 1982. Location and nucleotide sequence of the gene for the proton‐translocating subunit of wheat chloroplast ATP synthase. Proceedings of the National Academy of Sciences, USA 79: 6903–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa C, Hatanaka T, Misoo S, Miyake C, Fukayama H. 2011. Functional incorporation of sorghum small subunit increases the catalytic turnover rate of Rubisco in transgenic rice. Plant Physiology 156: 1603–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M, Tsunoda H, Suzuki Y, Makino A, Ishida H. 2012. RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity. Journal of Experimetnal Botany 63: 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MP, Perez‐Bueno ML, Zia A, Horton P, Ruban AV. 2009. The zeaxanthin‐independent and zeaxanthin‐dependent qE components of nonphotochemical quenching involve common conformational changes within the photosystem II antenna in Arabidopsis. Plant Physiology 149: 1061–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. 2002. GATEWAY vectors for Agrobacterium‐mediated plant transformation. Trends in Plant Science 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Karkehabadi S, Peddi SR, Anwaruzzaman M, Taylor TC, Cederlund A, Genkov T, Andersson I, Spreitzer RJ. 1995. Chimeric small subunits influence catalysis without causing global conformational changes in the crystal structure of ribulose‐1,5‐bisphosphate carboxylase/oxygenase. Biochemistry 44: 9851–9861. [DOI] [PubMed] [Google Scholar]
- Lauerer M, Saftic D, Quick WP, Labate C, Fichtner K, Schulze ED, Rodermel SR, Bogorad L, Stitt M. 1993. Decreased ribulose‐1,5‐bisphosphate carboxylase/oxygenase in transgenic tobacco transformed with ‘antisense’ rbcS. VI. Effect on photosynthesis in plants grown at different irradiance. Planta 190: 332–345. [Google Scholar]
- Li J, Chory J. 1998. Preparation of DNA from Arabidopsis. Methods in Molecular Biology 82: 55–60. [DOI] [PubMed] [Google Scholar]
- Long SP, Marshall‐Colon A, Zhu XG. 2015. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161: 56–66. [DOI] [PubMed] [Google Scholar]
- Mackinder LCM, Meyer MT, Mettler‐Altmann T, Chen VK, Mitchell MC, Caspari O, Rosenzweig ESF, Pallesen L, Reeves G, Itakura A et al 2016. A repeat protein links Rubisco to form the eukaryotic carbon‐concentrating organelle. Proceedings of the National Academy of Sciences, USA 113: 5958–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B, Biscoe PV. 1980. A model for C3 leaves describing the dependence of net photosynthesis on irradiance. Journal of Experimental Botany 31: 29–39. [Google Scholar]
- Maxwell K, Johnson GN. 2000. Chlorophyll fluorescence – a practical guide. Journal of Experimental Botany 51: 659–668. [DOI] [PubMed] [Google Scholar]
- McCormick AJ, Kruger NJ. 2015. Lack of fructose 2,6‐bisphosphate compromises photosynthesis and growth in Arabidopsis in fluctuating environments. Plant Journal 81: 670–683. [DOI] [PubMed] [Google Scholar]
- McGrath JM, Long SP. 2014. Can the cyanobacterial carbon‐concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiology 164: 2247–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MT, Genkov T, Skepper JN, Jouhet J, Mitchell MC, Spreitzer RJ, Griffiths H. 2012. Rubisco small‐subunit α‐helices control pyrenoid formation in Chlamydomonas. Proceedings of the National Academy of Sciences, USA 109: 19474–19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MT, McCormick AJ, Griffiths H. 2016. Will an algal CO2‐concentrating mechanism work in higher plants? Current Opinion in Plant Biology 31: 181–188. [DOI] [PubMed] [Google Scholar]
- Morita E, Abe T, Tsuzuki M, Fujiwara S, Sato N, Hirata A, Sonoike K, Nozaki H. 1998. Presence of the CO2‐concentrating mechanism in some species of the pyrenoid‐less free‐living algal genus Chloromonas (Volvocales, Chlorophyta). Planta 204: 269–276. [DOI] [PubMed] [Google Scholar]
- Morita K, Hatanaka T, Misoo S, Fukayama H. 2014. Unusual small subunit that is not expressed in photosynthetic cells alters the catalytic properties of Rubisco in rice. Plant Physiology 164: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Ishiguro S, Kimura T. 2009. Gateway vectors for plant transformation. Plant Biotechnology 26: 275–284. [Google Scholar]
- Orr DJ, Alcântara A, Kapralov MV, Andralojc PJ, Carmo‐Silva E, Parry MAJ. 2016. Surveying Rubisco diversity and temperature response to improve crop photosynthetic efficiency. Plant Physiology 172: 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP et al 2015. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proceedings of the National Academy of Sciences, USA 112: 8529–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MA, Andralojc PJ, Scales JC, Salvucci ME, Carmo‐Silva AE, Alonso H, Whitney SM. 2013. Rubisco activity and regulation as targets for crop improvement. Journal of Experimental Botany 64: 717–730. [DOI] [PubMed] [Google Scholar]
- Parry MAJ, Keys AJ, Gutteridge S. 1989. Variation in the specificity factor of C3 higher plant Rubiscos determined by the total consumption of Ribulose‐P2. Journal of Experimental Botany 40: 317–320. [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Research 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. 1989. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta (BBA) – Bioenergetics 975: 384–394. [Google Scholar]
- Price GD, Pengelly JJ, Forster B, Du J, Whitney SM, von Caemmerer S, Badger MR, Howitt SM, Evans JR. 2013. The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. Journal of Experimental Botany 64: 753–768. [DOI] [PubMed] [Google Scholar]
- Prins A, Orr DJ, Andralojc PJ, Reynolds MP, Carmo‐Silva E, Parry MAJ. 2016. Rubisco catalytic properties of wild and domesticated relatives provide scope for improving wheat photosynthesis. Journal of Experimental Botany 67: 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick WP, Schurr U, Fichtner K, Schulze ED, Rodermel SR, Bogorad L, Stitt M. 1991. The impact of decreased Rubisco on photosynthesis, growth, allocation and storage in tobacco plants which have been transformed with antisense rbcS. Plant Journal 1: 51–58. [Google Scholar]
- Rodermel S, Haley J, Jiang CZ, Tsai CH, Bogorad L. 1996. A mechanism for intergenomic integration: abundance of ribulose bisphosphate carboxylase small‐subunit protein influences the translation of the large‐subunit mRNA. Proceedings of the National Academy of Sciences, USA 93: 3881–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, von Caemmerer S, Badger MR, Andrews TJ, Price GD, Robinson SA. 2000. Xanthophyll cycle, light energy dissipation and electron transport in transgenic tobacco with reduced carbon assimilation capacity. Australian Journal of Plant Physiology 27: 289–300. [Google Scholar]
- Schöb H, Kunz C, Meins F Jr. 1997. Silencing of transgenes introduced into leaves by agroinfiltration: a simple, rapid method for investigating sequence requirements for gene silencing. Molecular and General Genetics 256: 581–585. [DOI] [PubMed] [Google Scholar]
- Sharkey TD. 1988. Estimating the rate of photorespiration in leaves. Physiologia Plantarum 73: 147–152. [Google Scholar]
- Spreitzer RJ. 2003. Role of the small subunit in ribulose‐1,5‐bisphosphate carboxylase/oxygenase. Archives of Biochemistry and Biophysics 414: 141–149. [DOI] [PubMed] [Google Scholar]
- Stitt M, Schulze D. 1994. Does Rubisco control the rate of photosynthesis and plant growth? An exercise in molecular ecophysiology. Plant, Cell & Environment 17: 465–487. [Google Scholar]
- Su Q, Boschetti A. 1994. Substrate‐ and species‐specific processing enzymes for chloroplast precursor proteins. Biochemical Journal 300: 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornley J. 1998. Dynamic model of leaf photosynthesis with acclimation to light and nitrogen. Annals of Botany 81: 421–430. [Google Scholar]
- Ushio A, Makino A, Yokota S, Hirotsu N, Mae T. 2003. Xanthophyll cycle pigments and water‐water cycle in transgenic rice with decreased amounts of ribulose‐1,5‐bisphosphate carboxylase and the wild‐type rice grown under different N levels. Soil Science and Plant Nutrition 49: 77–83. [Google Scholar]
- Walker B, Ariza LS, Kaines S, Badger MR, Cousins AB. 2013. Temperature response of in vivo Rubisco kinetics and mesophyll conductance in Arabidopsis thaliana: comparisons to Nicotiana tabacum . Plant, Cell & Environment 36: 2108–2119. [DOI] [PubMed] [Google Scholar]
- Walters RG, Horton P. 1991. Resolution of components of non‐photochemical chlorophyll fluorescence quenching in barley leaves. Photosynthesis Research 27: 121–133. [DOI] [PubMed] [Google Scholar]
- Wang Y, Stessman DJ, Spalding MH. 2015. The CO2 concentrating mechanism and photosynthetic carbon assimilation in limiting CO2: how Chlamydomonas works against the gradient. Plant Journal 82: 429–448. [DOI] [PubMed] [Google Scholar]
- Whitney SM, Houtz RL, Alonso H. 2011. Advancing our understanding and capacity to engineer nature's CO2‐sequestering enzyme, Rubisco. Plant Physiology 155: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, von Caemmerer S, Hudson GS, Andrews TJ. 1999. Directed mutation of the Rubisco large subunit of tobacco influences photorespiration and growth. Plant Physiology 121: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F‐A, Minai L, Nechushtai R. 1999. The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochimica et Biophysica Acta (BBA) – Bioenergetics 1411: 21–85. [DOI] [PubMed] [Google Scholar]
- Wostrikoff K, Clark A, Sato S, Clemente T, Stern D. 2012. Ectopic expression of Rubisco subunits in maize mesophyll cells does not overcome barriers to cell type‐specific accumulation. Plant Physiology 160: 419–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K, Stern D. 2007. Rubisco large‐subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proceedings of the National Academy of Sciences, USA 104: 6466–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M, Putterill JJ, Ross GS, Laing WA. 2001. Determination of the relative expression levels of Rubisco small subunit genes in Arabidopsis by rapid amplification of cDNA ends. Analytical Biochemistry 291: 237–244. [DOI] [PubMed] [Google Scholar]
- Zaks J, Amarnath K, Kramer DM, Niyogi KK, Fleming GR. 2012. A kinetic model of rapidly reversible nonphotochemical quenching. Proceedings of the National Academy of Sciences, USA 109: 15757–15762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan GM, Li RJ, Hu ZY, Liu J, Deng LB, Lu SY, Hua W. 2014. Cosuppression of RBCS3B in Arabidopsis leads to severe photoinhibition caused by ROS accumulation. Plant Cell Reports 33: 1091–1108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Transient expression of Rubisco small subunit–GFP fusion proteins in tobacco.
Fig. S2 Impact of native and heterologous SSUs on photosynthesis and growth in the Arabidopsis mutant 1a3b background.
Fig. S3 Alignments of the mature Arabidopsis SSU amino acid sequences.
Table S1 Sequences of synthetic oligonucleotides used in this study
Table S2 Transcript abundances of the Rubisco gene family in rbcs mutants and transgenic lines
Table S3 Rubisco and soluble protein contents for rbcs mutants and transgenic lines
Table S4 Rosette area and biomass for rbcs mutants and transgenic lines
Table S5 Chlorophyll characteristics and maximum quantum yield of PSII (F v/F m) for rbcs mutants and transgenic lines
Table S6 Photosynthetic nonphotochemical quenching capacity for rbcs mutants
Notes S1 Expression vectors for Rubisco small subunit (rbcS) cassettes.


