Abstract
Molecular changes during aging have been studied to understand the mechanism of aging progress. Herein, changes in microRNA (miRNA) expression in the whole blood of mice were studied to systemically reverse aging and propose them as non-invasive biomarkers. Through next-generation sequencing analysis, we selected 27 differentially expressed miRNAs during aging. The most recognized function involved was liver steatosis, a type of non-alcoholic fatty liver disease (NAFLD). Among 27 miRNAs, six were predicted to be involved in NAFLD, miR-16-5p, miR-17-5p, miR-21a-5p, miR-30c-5p, miR-103-3p, and miR-130a-3p; alterations in their blood and liver levels were confirmed by real-time qPCR. The expression of the genes associated in the network of these miRNAs, Bcl2, Ppara, E2f1, E2f2, Akt, Ccnd1, and Smad2/3, also was altered in the liver of aged mice. Following transfection of these miRNAs into 18-month-old mice, levels of miR-21a-5p, miR-103-3p, and miR-30c-5p increased, and their related genes exhibited a reversed expression in the liver. Expression of Mre11a, p16INK4a, and Mtor, reported to be aging-associated molecules, also was reversed in the livers of miRNA-transfected mice. These miRNAs could be non-invasive biomarkers for aging, and they might induce a reverse regulation of aging-associated pathways. This study provides preliminary data on reverse aging, which could be applied further for treatments of adult diseases.
Keywords: reverse aging, circulating miRNA, aging biomarker, non-alcoholic fatty liver disease, telomerase
Introduction
Recently, studies on molecular changes during the aging progress have been performed to understand the mechanisms and pathology of adult diseases, such as cancer, Alzheimer’s disease, and diabetes.1, 2, 3 Molecules such as telomerase, p16Ink4A, MTOR, NFKB1, and SIRT1 have been found to be involved in the aging process, and they have been used to predict longevity.4, 5, 6, 7 In addition, microRNAs (miRNAs) have been investigated to identify how they modulate aging-associated signaling pathways by binding to specific promoter regions of target mRNAs.8, 9 To this end, differentially expressed miRNAs (DERs), originating from a specific fraction of blood, such as cells, serum, or microvesicles (e.g., exosomes), have been identified in adult blood compared to that of infancy,10, 11, 12 which could provide key molecules to elucidate the aging mechanism.
In addition, transfection of miRNAs enables the modulation of biological processes.13, 14, 15 Specific miRNAs can be delivered to target tissues via the circulatory system, to modulate cellular pathways related to disease pathology in specific tissues.16, 17, 18 According to these studies, reprograming of gene expression could be used for disease therapy by introducing specific miRNAs into the blood that could eventually be delivered to target tissues. Such strategies are expected to have the capacity to modulate age-related genes, permitting reversal of cellular senescence and aging. However, there have not been reports on the reverse-aging effect in the aging body by injecting miRNAs into the circulatory system.
In the delivery of miRNAs from tissue to tissue, there are several routes for penetration through cell membranes, including complexation with proteins and lipids or encapsulation in vesicles, such as exosomes.19, 20 miRNAs also can be transferred directly from cell to cell; for example, miRNAs in macrophages were shown to be transferred via inclusion into the cellular membrane of hepatocarcinoma cells.21 These vesicular or cellular compartments in the blood would be a more stable source of miRNAs, and miRNAs isolated from them would have longer half-lives, than to free miRNAs in the blood. Therefore, if miRNAs are profiled in the whole blood of the aging body, the expanded pool of circulating miRNAs, including those from peripheral blood cells and vesicular fractions as well as cell-free forms, could aid in discovering important regulators of the aging process. They also have been proposed as non-invasive or minimally invasive biomarkers for aging.22
In this study, we analyzed whole blood as a source of miRNAs, including vesicular, cellular, and free miRNAs. We identified aging-related miRNAs in the whole blood of mice, and we analyzed their potency in differentially regulating the expression of well-known aging-related molecules in tissues. For this, we selected candidate miRNAs based on high-throughput screening of DERs through deep sequencing of RNA from whole blood. Then, the DERs were introduced into aged mice following encapsulation in a liposomal vehicle. Finally, we showed an acute response in the expression of aging-associated molecules after transfection of these DERs into aged mice.
Results
Age-Related miRNA Expression in Whole Blood
For preliminary screening of aging-related biomarkers in whole blood, including peripheral blood mononuclear cells (PBMCs), total RNA from 3-, 8-, and 12-month-old mice was analyzed by next-generation sequencing (NGS) analysis. When a 2-fold change was applied as the cutoff for selection of DERs, 155 miRNAs were found to be altered in 8-month-old mice compared to 3-month-old mice, and 196 genes were changed at 12 months compared to those at 8 months. Of the miRNAs, 46 were found to show >2-fold synchronous changes (step-by-step increase or decrease) between 8- and 12-month-old mice and 3- and 8-month-old mice. To select reliable biomarkers for aging, we limited DERs to those with a >4-fold synchronous alteration between 8- and 12-month-old mice and 3- and 8-month-old mice.
Table 1 shows the final 27 DERs selected, from the whole-blood RNA library, using this criteria. Among the 27 DEGs, only miR-3077-3p and miR-5107-5p increased with aging, whereas the other 25 miRNAs decreased over time.
Table 1.
Fold Changes in the Expression of miRNAs Selected as DERs from the Whole Blood of Aged Mice
| miRNA | Log2(Fold Change) (3 Months versus 8 Months) | Log2(Fold Change) (8 Months versus 12 Months) |
|---|---|---|
| miR-6960-5p | −3.500 | −6.000 |
| miR-340-5p | −3.824 | −5.368 |
| miR-16-5p | −3.800 | −5.000 |
| miR-130a-3p | −2.284 | −4.786 |
| miR-17-5p | −2.937 | −4.250 |
| miR-132-3p | −2.720 | −4.000 |
| miR-92a-1-5p | −2.750 | −4.000 |
| miR-29b-3p | −7.500 | −3.500 |
| miR-103-3p | −2.546 | −3.412 |
| miR-194-5p | −2.765 | −3.400 |
| miR-181b-5p | −2.415 | −3.188 |
| miR-21a-5p | −2.231 | −3.075 |
| miR-30c-5p | −2.207 | −3.029 |
| miR-24-1-5p | −2.733 | −3.000 |
| miR-148a-3p | −2.158 | −2.806 |
| miR-144-5p | −2.310 | −2.682 |
| miR-5113 | −2.875 | −2.667 |
| miR-451b | −2.522 | −2.556 |
| miR-10b-5p | −3.321 | −2.545 |
| miR-3082-3p | −2.300 | −2.500 |
| miR-421-3p | −2.148 | −2.426 |
| miR-7688-5p | −2.000 | −2.308 |
| miR-1839-5p | −2.367 | −2.143 |
| miR-3074-1-3p | −2.733 | −2.143 |
| miR-505-5p | −2.625 | −2.000 |
| miR-3077-3p | 2.333 | 2.143 |
| miR-5107-5p | 12.000 | 2.833 |
Selection of Candidate Biomarkers Based on Network Analysis of DERs
To identify the molecular network of the selected DERs, the Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com) was used. Using 27 DERs, the molecular network, regarding cancer, organismal injury and abnormalities, and reproductive system disease, was proposed to be representative of aging in the whole blood. The molecular connections between miRNAs and proteins in this network are shown in Figure 1. In this network, ten of 27 DERs were connected to each other. The involvement of five miRNAs, miR-17-5p, miR-103-3p, miR-130a-3p, miR-148a-3p, and miR-30c-5p, in Ppara expression, miR-21a-5p in Akt and Pik3r1 expression, and miR-16-5p in Bcl2 and Ccnd1 expression also was proposed in this network.
Figure 1.
The Top Network Associated with the DERs Selected by the Criteria of 4-fold Change in Whole Blood
The functions related to this network are defined as cancer, organismal injury and abnormalities, and reproductive system disease. miRNAs, indicated as green half circles, represent the selected DERs. The connection with Ppara is shown using pink arrows. The analysis was performed using the Ingenuity Pathway Analysis (IPA) program at http://www.ingenuity.com. Error bars show SD.
Figure 2A shows the functional changes caused by 27 DERs in aged mice, analyzed by IPA. Pathway analysis with the IPA database can be used to explore the functionality of the critical genes at the system level.23 Using IPA core analysis, we also identified canonical pathways associated with the function of these genes. The five top canonical pathways were liver steatosis, heart failure, decreased level of albumin, renal necrosis, and cardiac dilation. Liver steatosis, a type of non-alcoholic fatty liver disease (NAFLD), showed the highest significance (p = 8.29 × 10−10) among these functional changes (p < 0.05). Histological analysis of liver tissue from 3-month-old to 24-month-old mice showed that fatty liver became more severe with age, which is a key sign of liver steatosis (Figure 2B). This supports the result that selected DERs from whole blood are closely related to liver aging, which appeared in the form of fatty liver. Of 27 DERs, miR-17-5p, miR-103-3p, miR-130a-3p, miR-30c-5p, miR-21a-5p, and miR-16-5p are related to NAFLD, and we selected these six miRNAs as candidate biomarkers from the whole blood of aged mice. Table 2 summarizes the six miRNAs related to NAFLD and their associated genes in the network shown in Figure 1. Bcl2, Ccnd1, E2f1, E2f2, E2f3, Ppara, and Smad2 are involved in this network, and they were predicted to be the targets of these miRNAs by the miRTarBase,24 starBase 2.0,25 and TargetScan databases.26
Figure 2.
Functional Changes Caused by DERs Identified in the Whole Blood of Aged Mice
(A) Disease states are expressed as probability of significance, calculated as p < 0.05 by IPA analysis. (B) Histological analysis of fatty liver according to age in mice. The deposited fats were observed in empty vacuoles (red arrows); thus, an elevated number of vacuoles implies the development of fatty liver. Error bars show SD.
Table 2.
The miRNAs Associated with NAFLD and Their Predicted Target Genes
| miRNA | Target Genea |
|---|---|
| miR-16-5p | Bcl2, Smad2, Akt, and E2f3 |
| miR-130a-3p | E2f2 |
| miR-17-5p | Akt, E2f2, E2f3, and Ppara |
| miR-103-3p | Akt, Bcl2, and E2f3 |
| miR-21a-5p | E2f2, Bcl2, E2f3, and Ppara |
| miR-30c-5p | Bcl2 |
Searched by miRTarBase, starBase 2.0, and TargetScan databases.
Confirmation of Expression of the Six Selected miRNAs in Whole Blood and Liver Tissue
Expression of the six miRNAs, miR-16-5p, miR-17-5p, miR-21a-5p, miR-30c-5p, miR-103-3p, and miR-130a-3p, was confirmed in the whole blood of aged mice by real-time qPCR. As shown in Figure 3A, all six miRNAs selected as candidate biomarkers were significantly downregulated at 24 months compared to their expression at 8 months of age. Although the expression of miR-30c-5p, miR-103-3p, and miR-130a-3p was significantly increased at 8 months compared to that at 3 months, it was obvious that the overall pattern of expression decreased with age. These results indicated that the selected miRNAs eventually decreased over 24 months, but there were some fluctuations during the initial process of aging, from 3 to 8 months.
Figure 3.
Expression Level of miRNAs and Proteins in Whole Blood and Liver According to Aging Stage
(A and B) Expression is shown of differentially expressed miRNAs in (A) whole blood and (B) liver tissue of mice, assessed by qPCR and according to age. (C) Expression of proteins involved in the miRNA network was analyzed by western blotting. Red asterisk(s), comparison to 8-month-old mice; black asterisk(s), comparison to 3-month-old mice (*p < 0.05, **p < 0.01, and ***p < 0.001). Error bars show SD.
To determine the biological relevance of miRNAs as biomarkers for tissue aging, expression in the liver was analyzed by qPCR. Figure 3B shows that the expression of all six miRNAs was significantly decreased at 12 months of age compared to that at 8 months of age. The mean increased expression from 3 to 8 months of age was similar for all six miRNAs, and it was significant for miR-16-5p, miR-21a-5p, and miR-30c-5p. The selected six miRNAs showed aging-dependent decreases in liver expression at 12 months of age, but they showed some fluctuation in the initial stage of aging, from 3 to 8 months, similar to that observed in whole-blood samples.
To confirm the decreased miRNA levels in aged livers, the expression of genes involved in the network (Figure 1; Table 2) was analyzed by western blotting. Figure 3C shows that proteins such as BCL2, PPARA, CCND1, and SMAD2/3 were significantly decreased with age, and the expression of E2F1, E2F2, and AKT significantly increased with aging.
Thus, decreased expression of six miRNAs, selected from high-throughput screening of whole-blood samples, in the liver as well as the whole blood of aged mice was confirmed by qPCR. In addition, expression of the associated genes was found to be significantly altered, which supported the hypothesis that the selected miRNAs changed during the aging process.
Transfection of miRNAs into Aged Mouse Livers via the Circulatory System
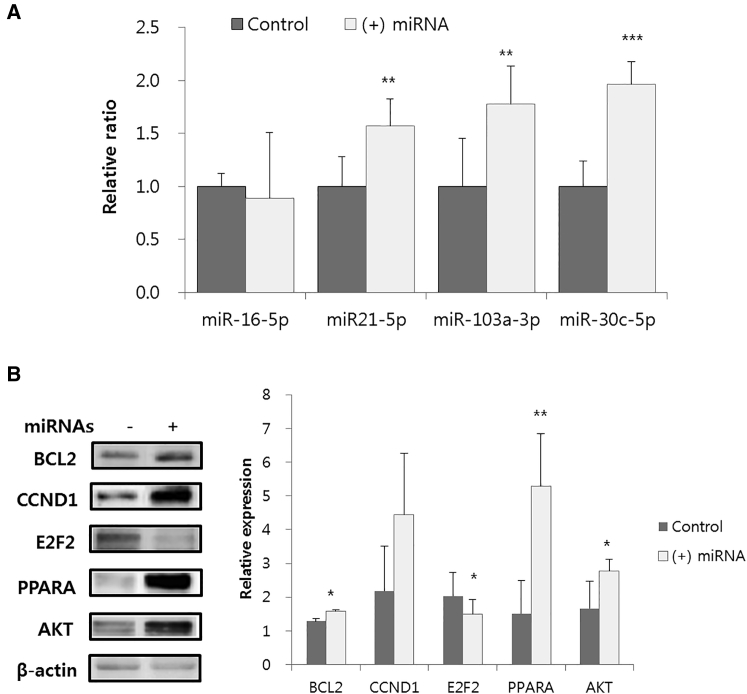
Mice (18 months old, n = 8) were divided into two groups, one of which was injected with vehicle only (control group), while the other was injected with the six miRNAs (miR-16-5p, miR-130a-3p, miR-17-5p, miR-103-3p, miR-30c-5p, and miR-21a-5p) encapsulated in a vehicle ([+] miRNA group). Then 12 hr after intraperitoneal injection of the six miRNAs (twice, every 12 hr), the level of miRNAs in the liver was measured by qPCR. We detected a significant increase in three miRNAs, miR-21a-5p, miR-103-3p, and miR-30c-5p (Figure 4A). miR-16-5p was detected in the control and (+) miRNA groups, but the expression was not significantly different.
Figure 4.
Alteration of miRNA and Protein Level Induced by Six miRNAs in Liver
(A and B) Changes in (A) miRNA and (B) protein levels in the liver of 18-month-old mice after intraperitoneal injection of six miRNAs (*p < 0.05, **p < 0.01, and ***p < 0.001). (+) miRNA indicates the expression level in mice in the group injected with the six miRNAs. Error bars show SD.
miR-17-5p and miR-130a-3p were not detected in either the control or (+) miRNA group. These results were interpreted to indicate differences in delivery efficiency and degradation rate in liver tissue or circulatory system, following injection of the miRNA encapsulated in the liposomal vehicles.
As shown in Figure 4B, we also analyzed protein levels following transfection of miRNA into the liver. BCL2, CCND1, and PPARA were observed to decrease with aging (Figure 3C) but to increase in the livers of mice in the (+) miRNA group (Figure 4B). E2F2, which was increased in aged mice, showed decreased expression following miRNA transfection. AKT was increased with aging in liver tissues and also increased by the transfection of the six miRNAs.
These results indicate that miRNAs encapsulated in a vehicle can be transfected into the livers of aged mice. Of the injected miRNAs, three were increased in the liver, whereas the other three were not significantly changed or not detected in either group. However, the success of miRNA transfection was supported by changes in the expression of proteins involved in the miRNA target network.
Acute Reversal of Aging by Delivery of miRNA in Liver
Because miRNAs transfected into tissues could affect the expression of proteins in the network, it was expected that these miRNAs also could reverse known aging indicators. Telomere-related genes have been reported to be involved in the aging process; thus, we quantified the expression of telomere-related enzymes following miRNA transfection. The telomere-related genes evaluated in this study were Men1, Terf2, Tnks2, Tep1, Tert, and Mre11a. Expression of Mre11a (which encodes a double-strand break repair protein) was ∼40-fold higher in the livers of the (+) miRNA group than in control mice (Figure 5A). Decreased expression of Mre11a is associated with aging and has been reported to contribute to cellular senescence.27 Although expression of other telomere-related proteins did not change significantly, transfection of these six miRNAs into the liver was demonstrated to reverse the expression of aging-related genes.
Figure 5.
Expression of Aging-Associated Molecules in Livers Transfected with Six miRNAs
(A and B) The (A) mRNA levels of telomerase-related genes and (B) expression of proteins involved in aging pathways. (+) miRNA indicates the expression level in mice in the group injected with the six miRNAs. Error bars show SD.
Expression of other aging markers, such as p16INK4A, MTOR, and SIRT1, in the liver also was analyzed following miRNA transfection. Figure 5B shows that the expression of p16INK4A and MTOR significantly decreased after miRNA transfection, but SIRT1 was not significantly altered.
Discussion
Profiling of miRNAs has been performed in whole blood10 and PBMCs28 of humans; in these studies, the majority of miRNAs were downregulated with age. In our study, 25 of 27 miRNAs selected as DERs from the whole blood using a 4-fold change criterion were decreased, similar to previous results observed in humans (Table 1). However, the set of DERs identified in mice showed no overlaps with the set of DERs identified in humans; miRNAs that are deregulated in mouse serum also have been reported, but only two (miR-10-5p and miR-5107-5p) of 48 miRNAs were identical to the DERs identified here from whole blood.22
Using pathway analysis, we identified six miRNAs from whole blood as circulatory biomarkers of aging; liver steatosis was proposed to be the most critical change caused by the selected DERs (Figures 2A and 2B). The selected miRNAs were all associated with NAFLD and the network of these miRNAs is shown in Figure 1. In this network, BCL2, PPARA, CCND1, SMAD2/3, E2F1, E2F2, and AKT were involved. The genes for these proteins were predicted as target genes by miRNA databases (Table 2).
Even though we selected DERs that showed a linear relationship with aging through NGS analysis, four miRNAs (with the exception of miR-17-5p and miR-21a-5p), analyzed by qPCR, showed increased expression at 8 months of age compared to that at 3 months, which was the opposite of NGS results (Figure 3A). This result was attributed to the difference in the sample preparation methods and the sensitivity of instruments used in analysis. Before NGS analysis, total RNA samples were prepared and then the small RNA fraction was separated using a small RNA sample preparation kit, followed by cDNA synthesis. In contrast, the quantification of a specific miRNA by qPCR was performed with cDNA synthesized through the addition of a poly(A) tail to total RNA. In addition, some fluctuations in the small RNA proportion during aging have been reported in Caenorhabditis elegans29 and Bombyx mori,30 wherein the total number of miRNAs and small RNAs, respectively, showed a concave pattern during development. Therefore, it was interpreted that the proportion of small RNA (mainly composed of miRNA) to total RNA at 3 months was lower than that at 8 months; thus, the final concentration in the total RNA at 3 months by qPCR would be lower than that in the small RNA fraction by NGS analysis.
When we analyzed the selected six miRNAs in liver tissue, the expression levels were significantly decreased at 12 months of age (Figure 3B), even though they were significant as late as at 24 months of age in whole blood. This result indicates that the expression change in liver occurred before changes in the blood. Nevertheless, it revealed that deregulation of the selected six miRNAs occurred in both liver and blood, which implies that these aging-related miRNAs are common to liver and blood.
The expression of BCL2, PPARA, E2F1, E2F2, AKT, CCND1, and SMAD2/3, which were associated in the network of the six miRNAs, also was analyzed in 3-, 8-, and 12-month-old mice by western blotting. Among these proteins, BCL2, PPARA, CCND1, and SMAD2/3 were downregulated and E2F1, E2F2, and AKT were upregulated along with the decreased expression of the six miRNAs in aging. Various mechanisms that link miRNAs in miRNA-induced silencing complexes (miRISCs) to reduced expression of target proteins by the inhibition of translation or induction of mRNA degradation have been reported.31 However, many miRNAs that enhance gene expression also have been reported, mediated through the repression of negative transcriptional regulators.32 The capability of activating gene expression directly or indirectly has been revealed in response to different cell types and conditions.26, 33, 34 In this study, the downregulated genes also were predicted as the target genes of the miRNAs by several databases, such as miRTarBase, but the actual mechanism of regulation was assumed as indirect targeting that led to the positive regulation of gene expression.
The results indicated that gene expression had been significantly altered, as reported in previous studies on aging mechanisms. For example, decreased expression of BCL2 in T cell subsets in humans35 and PPARA in rat spleens36 was reported with aging. Decreased CCND1 expression was reported to be caused by p16Ink4A37 and was related to embryonic stem cell senescence.38 Significant alteration in TGF-β-signaling pathways was shown to result in a loss of the protective SMAD2/3 pathway during the aging of mice.39 A previous report revealed that E2F1 enhanced cellular senescence in human fibroblast cells by negatively regulating FOXO3.40
Administration of blood plasma from young mice to aged mice was reported to induce a reversal of aging, resulting in recovery from cognitive and synaptic plasticity impairments.41 Thus, we hypothesized that transfection of blood constituents in aged mice would induce differential expression of aging-related molecules. When we transfected the selected six miRNAs into the liver of 18-month-old mice, the levels of three miRNAs (miR-21a-5p, miR-103-3p, and miR-30c-5p) were significantly increased (Figure 4A). Interestingly, expression of the associated proteins (BCL2, CCND1, E2F2, and PPARA) was significantly reversed with increased levels of these three miRNAs in liver, following the injection of six miRNAs (Figure 4B).
Telomere length is correlated with longevity and disease resistance during normal aging in animals; in mammalian somatic cells, short telomeres can trigger replicative senescence.42 Telomerase subunits include TERT, TEP1, TERC, and DKC1.43 Telomeric repeat-binding factor 2 (TERF2)44 and tankyrase-2 (TNKS2)45 are components of the shelterin nucleoprotein complex, which plays a key role in the protective activity of telomeres. Stabilization of telomeric DNA is assisted by the double-strand break repair protein MRE11A, which was shown to exhibit statistically significant age-dependent downregulation in human lymphocytes.27 It was suggested that maintenance of a higher expression of MRE11A might be responsible for the longer lifespan observed in the longevity group.
Figure 5A shows the expression of these telomere length-related genes, such as Men1, Tert, Tep1, Terf2, Tnks2, and Mre11a. Among these, Mre11a was dramatically increased in the livers of the (+) miRNA group. Elevated levels of Mre11a supported the hypothesis that miRNA transfection into liver could reverse the expression of aging-associated molecules in the liver. In our study, the selected six miRNAs clearly induced Mre11a, but they did not induce telomerase activity, as seen with Tert and Tep1 expression.
Other aging-associated proteins have been identified in animals, such as p16Ink4A,46, 47 MTOR,5, 48 and SIRT1.49 When the six miRNAs were transfected into the liver, p16Ink4A and MTOR decreased significantly; this indicated a reversal in the expression of aging-related molecules. The expression of SIRT1 has been reported to decline with aging in animals and human tissues, including lung, fat, heart, and blood vessels.50 In our study, SIRT1 was upregulated on average but this change was not statistically significant. This result indicates that the transfection of six miRNAs reversed aging-associated upregulation of p16Ink4A and MTOR but could not significantly reverse the downregulation of SIRT1.
In this study, the alteration in levels of aging-related molecules lasted for only 1 day; therefore, the observed effect was acute rather than chronic. We set out to detect an immediate response following transfection of miRNAs, before longer-term adaptation occurred. Overall, transfection of the six miRNAs selected in this study showed promise for reversing the expression of aging-associated pathways in animal models.
Conclusions
In this study, we investigated the effects of miRNA transfection on selected aging biomarkers. By analyzing the acute response in the target tissue, miRNA transfection was confirmed, and it was shown to induce a reversal in the expression of aging-related molecules. This is the first example of modulation of aging pathways through in vivo transfection of miRNAs. This result suggests that miRNA transfection via the circulatory system shows potential to induce changes in aging-related molecules, an important step toward the reversal of aging and development of therapeutics for aging-related diseases.
Materials and Methods
Animals
C57BL/6 male mice aged 2, 7, 11, and 23 months were purchased from Central Lab Animal. The animals were maintained after purchase at DGIST Animal Laboratory in accordance with the Institutional Animal Care Guidelines. The Animal Care and Use Committee of DGIST approved all animal protocols. After 1 month of acclimation, animals were sacrificed and tissues (blood, liver, and lung) were sampled immediately.
Small RNA Sequencing Method
For high-throughput screening of miRNAs differentially expressed in aged mice, NGS was performed with total RNA isolated from the whole blood of mice. Total RNA was isolated from whole blood using the Hybride-R RNA Kit (GeneAll), according to the manufacturer’s instructions. Total RNA integrity was verified using an Agilent 2100 Bioanalyzer (Agilent Technologies), with an RNA integrity number (RIN) value >8 as a cutoff value. Small RNA sequencing libraries were prepared using a TruSeq Small RNA Library Preparation kit (Illumina), according to the manufacturer’s instructions. A small RNA (1 μg) sample pooled with equal amount of each mouse (n = 3) was ligated with RNA 3′ and RNA 5′ adapters. Reverse transcription followed by PCR was used to create cDNA constructs based on small RNA ligated to 3′ and 5′ adapters. This process selectively enriched fragments with adapter molecules on both ends. The small RNA fraction was purified with the Pippin Prep electrophoresis platform (Sage Science). The quality of the libraries was verified by capillary electrophoresis using an Agilent 2100 Bioanalyzer (Agilent Technologies). The libraries were pooled in equimolar amounts and loaded on the flow cell of a HiSeq 2000 sequencing system (Illumina). Sequencing was performed to generate 1 × 50-bp length read.
Analysis of DERs and Pathways Related to Aging
DERs were selected according to two criteria as follows: (1) a 4-fold change in expression between mice of 3 and 8 months of age and 8 and 12 months of age, and (2) these changes occurred in the same direction, for example, they were both up- or downregulated at each aging step. Pathway analysis was performed to determine the most relevant signaling pathways associated with DERs, using the IPA (http://www.ingenuity.com) database. Briefly, the DERs and their extreme expression values in age-dependent samples were uploaded to the dataset files of IPA. Then, core analysis was performed and the resultant functional changes and the candidate crucial genes were identified.
Histological Analysis
Histological processing and embedding in wax were performed by conventional techniques. Briefly, the biopsies were fixed in 4% paraformaldehyde for 24 hr and embedded in paraffin. Sections ∼4 μm thick were stained with hematoxylin for 10 min, washed, and stained with eosin for 2 min. After washing with water, the slides were gradually dehydrated in 50%, 70%, 90%, and 100% ethanol. The stained sections were examined and images were captured with a light microscope (Leica ICC50 HD, Leica Microsystems).
Delivery of miRNA to Liver Tissue
C57BL/6 male mice (18 months old) were randomly divided into two groups (n = 4), which were administrated PBS (control group) or a six-miRNA mixture composed of miR-16-5p, miR-130a-3p, miR-17-5p, miR-103-3p, miR-30c-5p, and miR-21a-5p ([+] miRNA group). All miRNA mimics were purchased from Bioneer. The miRNAs were encapsulated by a liposome-based transfection reagent (Liver in vivo Transfection Kit, Altogen Biosystems) for the delivery of miRNAs. Then the encapsulated miRNAs were administered by intraperitoneal injection into each group of mice, according to the manufacturer’s instructions. The injection was performed twice, 12 hr apart, with 60 nmol/animal for each miRNA. The mice were sacrificed at 12 hr after the second injection.
miRNA and mRNA Expression Analysis by qPCR
Total RNA was isolated from blood and tissues using the Hybride-R RNA Kit (GeneAll), according to the manufacturer’s instructions. cDNA for miRNA analysis was synthesized from 5 ng total RNA using the Universal cDNA synthesis kit II (Exiqon). For the determination of miRNA expression, qPCR was performed using the miRCURY LNA Universal RT micro RNA PCR LNAPCR primer sets for miR-16-5p, miR-130a-3p, miR-17-5p, miR-103-3p, miR-30c-5p, and miR-21a-5p (Exiqon), and the SYBR green PCR kit (Exiqon) in an ABI7900HT Real-Time PCR System (Thermo Fisher Scientific). cDNA for mRNA analysis was synthesized from 1 μg total RNA using PrimeScript 1st strand cDNA Synthesis Kit (Exiqon). For the determination of mRNA expression, qPCR was performed using gene-specific primer pairs and the Roche SYBR-Green master mix in an ABI7900HT Real-Time PCR System (Thermo Fisher Scientific).
Protein Expression Analysis Western Blotting
Total protein from liver tissues was obtained after homogenizing tissues in 100–300 μL RIPA buffer (Sigma-Aldrich). After incubation on ice for 15 min, centrifugation was performed at 10,000 × g for 30 min, and the supernatant (protein) was collected for analysis. Protein concentrations were determined using a BCA protein assay kit (Bio-Rad). Samples (50 μg) were separated by 10% SDS-PAGE, and they were transferred to polyvinylidene difluoride (PVDF) membranes using a transfer apparatus (Trans-Blot SD semi-dry transfer cell, Bio-Rad), according to the manufacturer’s instructions. After blocking with 5% BSA in Tris-buffered saline with Tween 20 (TBST) for 1 hr at room temperature, membranes were incubated overnight at 4°C with primary antibodies against BCL2 (Cell Signaling Technology), PPARA (Santa Cruz Biotechnology), E2F1 (Santa Cruz Biotechnology), E2F2 (Santa Cruz Biotechnology), Akt (Cell Signaling Technology), CCND1 (Cell Signaling Technology), SMAD2/3 (Santa Cruz Biotechnology), p16Ink4A (Abcam), MTOR (Bioworld Technology), SIRT1 (Abcam), and β-actin (Santa Cruz Biotechnology), followed by horseradish peroxidase (HRP)-conjugated anti-mouse IgG at room temperature for 1 hr. Blots were washed with TBST three times and developed with an ECL system (GE Healthcare Life Sciences), according to the manufacturer’s protocols.
Statistical Analysis
For all experiments, data from three independent experiments were analyzed using a Student’s t test and are reported as mean ± SD. Sigma Plot version 12.3 was used (Systat Software) to determine the p values. A p value < 0.05 was considered statistically significant.
Author Contributions
J.-H.K. performed major experiments, such as animal treatment and molecular analysis, and wrote the manuscript. B.-R.L. and E.-S.C. maintained animals and performed staining of tissues for histological analysis. K.-M.L., S.-K.C., and J.H.C. contributed to the sacrifice of animals and preparation of tissue sections. W.B.J. performed network and pathway analysis of differentially expressed genes. E.K. designed the overall experiments and wrote the manuscript based on the integrated analysis of the whole data.
Conflicts of Interest
The authors declare no conflict of interest related to this manuscript.
Acknowledgments
This study was supported in part by the Ministry of Science, ICT, & Future Planning of the Republic of Korea (DGIST Basic Research Fund 16-BD-06 and 2017010021 and 2016R1A2B4014728).
References
- 1.Campisi J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawarabayashi T., Younkin L.H., Saido T.C., Shoji M., Ashe K.H., Younkin S.G. Age-dependent changes in brain, CSF, and plasma amyloid (β) protein in the Tg2576 transgenic mouse model of Alzheimer’s disease. J. Neurosci. 2001;21:372–381. doi: 10.1523/JNEUROSCI.21-02-00372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunasekaran U., Gannon M. Type 2 diabetes and the aging pancreatic beta cell. Aging (Albany NY) 2011;3:565–575. doi: 10.18632/aging.100350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ressler S., Bartkova J., Niederegger H., Bartek J., Scharffetter-Kochanek K., Jansen-Dürr P., Wlaschek M. p16INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell. 2006;5:379–389. doi: 10.1111/j.1474-9726.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- 5.Perl A. mTOR activation is a biomarker and a central pathway to autoimmune disorders, cancer, obesity, and aging. Ann. N Y Acad. Sci. 2015;1346:33–44. doi: 10.1111/nyas.12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balistreri C.R., Candore G., Accardi G., Colonna-Romano G., Lio D. NF-κB pathway activators as potential ageing biomarkers: targets for new therapeutic strategies. Immun. Ageing. 2013;10:24. doi: 10.1186/1742-4933-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho S.H., Chen J.A., Sayed F., Ward M.E., Gao F., Nguyen T.A., Krabbe G., Sohn P.D., Lo I., Minami S. SIRT1 deficiency in microglia contributes to cognitive decline in aging and neurodegeneration via epigenetic regulation of IL-1β. J. Neurosci. 2015;35:807–818. doi: 10.1523/JNEUROSCI.2939-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L.H., Chiou G.Y., Chen Y.W., Li H.Y., Chiou S.H. MicroRNA and aging: a novel modulator in regulating the aging network. Ageing Res. Rev. 2010;9(Suppl 1):S59–S66. doi: 10.1016/j.arr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Lai C.Y., Wu Y.T., Yu S.L., Yu Y.H., Lee S.Y., Liu C.M., Hsieh W.S., Hwu H.G., Chen P.C., Jeng S.F., Chen W.J. Modulated expression of human peripheral blood microRNAs from infancy to adulthood and its role in aging. Aging Cell. 2014;13:679–689. doi: 10.1111/acel.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu D., Tahara H. The role of exosomes and microRNAs in senescence and aging. Adv. Drug Deliv. Rev. 2013;65:368–375. doi: 10.1016/j.addr.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Noren Hooten N., Fitzpatrick M., Wood W.H., 3rd, De S., Ejiogu N., Zhang Y., Mattison J.A., Becker K.G., Zonderman A.B., Evans M.K. Age-related changes in microRNA levels in serum. Aging (Albany NY) 2013;5:725–740. doi: 10.18632/aging.100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Z., Jiang J., Kokkinaki M., Tang L., Zeng W., Gallicano I., Dobrinski I., Dym M. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cells. 2013;31:2205–2217. doi: 10.1002/stem.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo M., Weng Y., Tang J., Hu M., Liu Q., Jiang F., Yang D., Liu C., Zhan X., Song P. MicroRNA-450a-3p represses cell proliferation and regulates embryo development by regulating Bub1 expression in mouse. PLoS ONE. 2012;7:e47914. doi: 10.1371/journal.pone.0047914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi J., Qiao Y., Wang P., Li S., Zhao W., Gao C. microRNA-210 negatively regulates LPS-induced production of proinflammatory cytokines by targeting NF-κB1 in murine macrophages. FEBS Lett. 2012;586:1201–1207. doi: 10.1016/j.febslet.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Miniarikova J., Zanella I., Huseinovic A., van der Zon T., Hanemaaijer E., Martier R., Koornneef A., Southwell A.L., Hayden M.R., van Deventer S.J. Design, characterization, and lead selection of therapeutic miRNAs targeting Huntingtin for development of gene therapy for Huntington’s disease. Mol. Ther. Nucleic Acids. 2016;5:e297. doi: 10.1038/mtna.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W., Tian S.S., Hang P.Z., Sun C., Guo J., Du Z.M. Combination of microRNA-21 and microRNA-146a attenuates cardiac dysfunction and apoptosis during acute myocardial infarction in mice. Mol. Ther. Nucleic Acids. 2016;5:e296. doi: 10.1038/mtna.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiraki M., Nishimura J., Takahashi H., Wu X., Takahashi Y., Miyo M., Nishida N., Uemura M., Hata T., Takemasa I. Concurrent targeting of KRAS and AKT by miR-4689 is a novel treatment against mutant KRAS colorectal cancer. Mol. Ther. Nucleic Acids. 2015;4:e231. doi: 10.1038/mtna.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannafon B.N., Ding W.Q. Intercellular communication by exosome-derived microRNAs in cancer. Int. J. Mol. Sci. 2013;14:14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu Z., Jiang C., Wu J., Ding Y. Exosomes as potent regulators of HCC malignancy and potential bio-tools in clinical application. Int. J. Clin. Exp. Med. 2015;8:17088–17095. [PMC free article] [PubMed] [Google Scholar]
- 21.Aucher A., Rudnicka D., Davis D.M. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J. Immunol. 2013;191:6250–6260. doi: 10.4049/jimmunol.1301728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhahbi J.M., Spindler S.R., Atamna H., Yamakawa A., Guerrero N., Boffelli D., Mote P., Martin D.I. Deep sequencing identifies circulating mouse miRNAs that are functionally implicated in manifestations of aging and responsive to calorie restriction. Aging (Albany NY) 2013;5:130–141. doi: 10.18632/aging.100540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu S.D., Lin F.M., Wu W.Y., Liang C., Huang W.C., Chan W.L., Tsai W.T., Chen G.Z., Lee C.J., Chiu C.M. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J.H., Li J.H., Shao P., Zhou H., Chen Y.Q., Qu L.H. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ju Y.J., Lee K.H., Park J.E., Yi Y.S., Yun M.Y., Ham Y.H., Kim T.J., Choi H.M., Han G.J., Lee J.H. Decreased expression of DNA repair proteins Ku70 and Mre11 is associated with aging and may contribute to the cellular senescence. Exp. Mol. Med. 2006;38:686–693. doi: 10.1038/emm.2006.81. [DOI] [PubMed] [Google Scholar]
- 28.Noren Hooten N., Abdelmohsen K., Gorospe M., Ejiogu N., Zonderman A.B., Evans M.K. microRNA expression patterns reveal differential expression of target genes with age. PLoS ONE. 2010;5:e10724. doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato M., Chen X., Inukai S., Zhao H., Slack F.J. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA. 2011;17:1804–1820. doi: 10.1261/rna.2714411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jagadeeswaran G., Zheng Y., Sumathipala N., Jiang H., Arrese E.L., Soulages J.L., Zhang W., Sunkar R. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics. 2010;11:52. doi: 10.1186/1471-2164-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilczynska A., Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015;22:22–33. doi: 10.1038/cdd.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie W., Rajasekhar M., Flamant S., Rasko J.E.J. Conserved expression patterns predict microRNA targets. PLoS Comput. Biol. 2009;5:e1000513. doi: 10.1371/journal.pcbi.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valinezhad Orang A., Safaralizadeh R., Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int. J. Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Place R.F., Li L.-C., Pookot D., Noonan E.J., Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aggarwal S., Gupta S. Increased apoptosis of T cell subsets in aging humans: altered expression of Fas (CD95), Fas ligand, Bcl-2, and Bax. J. Immunol. 1998;160:1627–1637. [PubMed] [Google Scholar]
- 36.Gelinas D.S., McLaurin J. PPAR-α expression inversely correlates with inflammatory cytokines IL-1β and TNF-α in aging rats. Neurochem. Res. 2005;30:1369–1375. doi: 10.1007/s11064-005-8341-y. [DOI] [PubMed] [Google Scholar]
- 37.Kim W.Y., Sharpless N.E. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Yao M., Wang Y., Zhang P., Chen H., Xu Z., Jiao J., Yuan Z. BMP2-SMAD signaling represses the proliferation of embryonic neural stem cells through YAP. J. Neurosci. 2014;34:12039–12048. doi: 10.1523/JNEUROSCI.0486-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Kraan P.M. Age-related alterations in TGF beta signaling as a causal factor of cartilage degeneration in osteoarthritis. Biomed. Mater. Eng. 2014;24(1, Suppl):75–80. doi: 10.3233/BME-140976. [DOI] [PubMed] [Google Scholar]
- 40.Xie Q., Peng S., Tao L., Ruan H., Yang Y., Li T.M., Adams U., Meng S., Bi X., Dong M.Q., Yuan Z. E2F transcription factor 1 regulates cellular and organismal senescence by inhibiting Forkhead box O transcription factors. J. Biol. Chem. 2014;289:34205–34213. doi: 10.1074/jbc.M114.587170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villeda S.A., Plambeck K.E., Middeldorp J., Castellano J.M., Mosher K.I., Luo J., Smith L.K., Bieri G., Lin K., Berdnik D. Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat. Med. 2014;20:659–663. doi: 10.1038/nm.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou Y., Sfeir A., Gryaznov S.M., Shay J.W., Wright W.E. Does a sentinel or a subset of short telomeres determine replicative senescence? Mol. Biol. Cell. 2004;15:3709–3718. doi: 10.1091/mbc.E04-03-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gramatges M.M., Bertuch A.A. Short telomeres: from dyskeratosis congenita to sporadic aplastic anemia and malignancy. Transl. Res. 2013;162:353–363. doi: 10.1016/j.trsl.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakaguchi A.Y., Padalecki S.S., Mattern V., Rodriguez A., Leach R.J., McGill J.R., Chavez M., Giambernardi T.A. Chromosomal sublocalization of the transcribed human telomere repeat binding factor 2 gene and comparative mapping in the mouse. Somat. Cell Mol. Genet. 1998;24:157–163. doi: 10.1023/b:scam.0000007118.47691.d7. [DOI] [PubMed] [Google Scholar]
- 45.Hsiao S.J., Poitras M.F., Cook B.D., Liu Y., Smith S. Tankyrase 2 poly(ADP-ribose) polymerase domain-deleted mice exhibit growth defects but have normal telomere length and capping. Mol. Cell. Biol. 2006;26:2044–2054. doi: 10.1128/MCB.26.6.2044-2054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coppé J.P., Rodier F., Patil C.K., Freund A., Desprez P.Y., Campisi J. Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. J. Biol. Chem. 2011;286:36396–36403. doi: 10.1074/jbc.M111.257071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krishnamurthy J., Torrice C., Ramsey M.R., Kovalev G.I., Al-Regaiey K., Su L., Sharpless N.E. Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson S.C., Rabinovitch P.S., Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donmez G., Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;9:285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y., Liang Y., Vanhoutte P.M. SIRT1 and AMPK in regulating mammalian senescence: a critical review and a working model. FEBS Lett. 2011;585:986–994. doi: 10.1016/j.febslet.2010.11.047. [DOI] [PubMed] [Google Scholar]