Abstract
Integration site profiling and clonality analysis of viral vector distribution in gene therapy is a key factor to monitor the fate of gene-corrected cells, assess the risk of malignant transformation, and establish vector biosafety. We developed the Genome Integration Site Analysis Pipeline (GENE-IS) for highly time-efficient and accurate detection of next-generation sequencing (NGS)-based viral vector integration sites (ISs) in gene therapy data. It is the first available tool with dual analysis mode that allows IS analysis both in data generated by PCR-based methods, such as linear amplification method PCR (LAM-PCR), and by rapidly evolving targeted sequencing (e.g., Agilent SureSelect) technologies. GENE-IS makes use of trimming strategies, customized reference genome, and soft-clipped information with sequential filtering steps to provide annotated IS with clonality information. It is a scalable, robust, precise, and reliable tool for large-scale pre-clinical and clinical data analysis that provides users complete flexibility and control over analysis with a broad range of configurable parameters. GENE-IS is available at https://github.com/G100DKFZ/gene-is.
Keywords: gene therapy, bioinformatical analysis, next-generation sequencing, viral integration sites, LAM-PCR, targeted sequencing
Introduction
Viral vector gene therapy has demonstrated its tremendous potential and proved its efficacy in clinical trials.1, 2, 3, 4 However, the integration of viral vectors at certain genomic locations can not only alter gene expression but also can lead to malignant transformation, referred to as insertional mutagenesis.5 Vector position in each modified cell acts as a unique identifier that can be used to track and monitor integration points of the vector and unravel the likelihood of potential genotoxic and mutagenic events. Therefore, viral integration site (IS) profiling is of vital importance to illuminate and ensure the safety of the therapeutic vector system.
Traditionally, PCR-based methods are part of the experimental strategy used for IS analysis, such as linear amplification method (LAM)-PCR, while new targeted sequencing technologies also offer an efficient alternative approach for IS studies6, 7 (Figure S1). The advent of high-throughput sequencing technologies has revolutionized the possibilities of in-depth genome analyses. However, computational tools available for LAM IS analysis, like QuickMap,8 HISAP,9 and VISA,10 have issues with time efficiency and are not offering targeted sequencing IS analysis. Besides, these tools admit a limited set of reference genomes, do not expose multi-threading, and have limits on the amount of input data. The targeted sequencing analysis tools available, however (i.e., Virus-Clip11 and ViralFusionSeq12), have their primary focus on cancer genome data analysis. Also, the mechanism used to detect integration site is dissimilar from the objective of gene therapy safety studies, and it is not satisfactory and adequate.
Here we present the Genome Integration Site Analysis Pipeline (GENE-IS), a pipeline to analyze high-throughput sequencing data with a highly computationally efficient, automated, and reliable approach with various user-adjustable features. According to the best of our knowledge, this is the first available tool that allows the analysis of LAM-PCR and targeted sequencing-based gene therapy data. GENE-IS allows the analysis with any reference genome, provides a multi-threading option, has no limitation regarding input data, and provides an extensive range of user-customizable options. GENE-IS encompasses a broad analysis spectrum and is suitable for any viral (and non-viral) vector and reference host genome. Here we describe its applicability on unidirectional LAM-PCR sequence reads and targeted paired-end sequencing based on Illumina sequencing data.
Results
Comparison and Performance Evaluation
LAM-PCR Mode
To evaluate the reliability and computational performance of LAM-PCR analysis mode of GENE-IS, we performed several tests and comparisons with other tools, including QuickMap, HISAP, and VISA. VISPA13 was not included in this comparison for the reasons mentioned in the Discussion.
We generated two in silico datasets consisting of 500 (DS1.1) and 5,750 (DS1.2) sequences containing pre-located genomic positions. The number of sequences in the first dataset was kept low so that all tools were able to manage to complete the analysis within practical time. In addition, we used one in silico dataset of 1,000 (DS1.3) sequences provided by VISA authors along with their paper. DS1.1 was analyzed with all the tools, whereas DS1.2 was evaluated with GENE-IS, VISA, and HISAP while the QuickMap web service was suspended (Data S1). We evaluated DS1.3 by GENE-IS and HISAP; however, HISAP did not provide the results within the time limit for both DS1.2 and DS1.3. The VISA output available at the VISA server site was used to evaluate the tool (Data S2). Estimation of performance metrics for DS1.1 showed that GENE-IS and VISA had the highest sensitivity, specificity, precision, and accuracy, followed by HISAP and QuickMap. GENE-IS and VISA also showed the equally best performance in DS1.2 and DS1.3 (Table 1).
Table 1.
Comparison of Sensitivity, Specificity, Precision, and Accuracy of LAM-PCR Mode of GENE-IS, VISA, HISAP, and QuickMap by Using In Silico Datasets of 500, 1,000, and 5,750 Sequences
| Tools | GENE-IS | VISA | HISAP | QuickMap |
|---|---|---|---|---|
| 500-Read In Silico Dataset | ||||
| Sensitivity | 1 | 1 | 1 | 0.99 |
| Specificity | 1 | 1 | 0.97 | 0.96 |
| Precision | 1 | 1 | 1 | 0.99 |
| Accuracy | 1 | 1 | 0.99 | 0.99 |
| 5,750-Read In Silico Dataset | ||||
| Sensitivity | 1 | 1 | –a |
–b |
| Specificity | 1 | 1 | ||
| Precision | 1 | 1 | ||
| Accuracy | 1 | 1 | ||
| 1,000-Read In Silico Dataset (Taken from VISA Web Server) | ||||
| Sensitivity | 1 | 1 | –a | –b |
| Specificity | 1 | 1 | ||
| Precision | 1 | 1 | ||
| Accuracy | 1 | 1 | ||
Analysis did not complete within 5 days.
Server suspended.
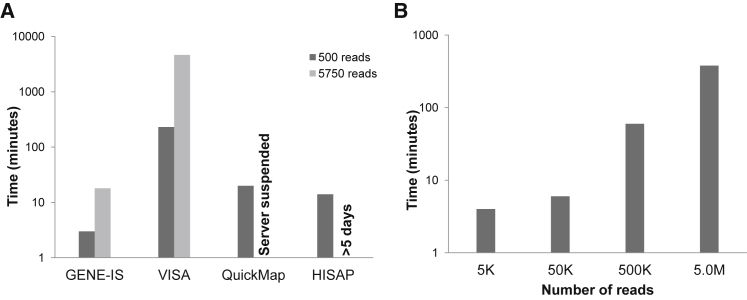
The time taken by each of these tools for analyzing DS1.1 and DS1.2 (Figure 1A) was compared to measure the performances. For the analysis of DS1.1, GENE-IS, VISA, HISAP, and QuickMap consumed 3, 230, 20, and 14 min, respectively. In the case of DS1.2, GENE-IS finished the analysis in 18 min and VISA took 76 hr and 59 min. An additional in silico dataset, consisting of ∼550,000 sequences (DS1.4), was generated for performing the computational efficiency comparison on large datasets between GENE-IS and VISA. At the time of writing this paper, it was not possible to upload more than 100,000 sequences on HISAP. After 240 hr (10 days), VISA did not complete the analysis, and the job was terminated. We also have analyzed previously published large pre-clinical datasets of a gene therapy study14 to establish the computational efficiency of GENE-IS (Figure S3). We were not able to evaluate this dataset with other pipelines because the number of reads widely exceeded the limit. Four in silico datasets, consisting of informative (IS-containing) and non-informative (non-IS-containing) reads of ∼5,000 (DS1.5), 50,000 (DS1.6), 500,000 (DS1.7), and 5.0 M (DS1.8) sequences, were used to depict GENE-IS time efficiency for varying number of reads (Figure 1B). In the time comparison of VISA, HISAP, and QuickMap tools, we have not taken into account the time to upload or download the data on these web servers.
Figure 1.
Comparison of Analysis Time of LAM-PCR Mode of GENE-IS
(A) Comparison with VISA, QuickMap, and HISAP for analyzing an in silico dataset of 500 and 5,750 sequences. In both datasets, all reads were informative (with IS). (B) Comparison between different datasets consisting of 5K, 50K, 500K, and 5.0M sequences (K, thousand; M, million). In these datasets, both kind of reads, i.e., informative (with IS) as well as non-informative (without IS) sequences, were present. This was done to depict computational efficiency in original settings, as experimental datasets always contain both types of reads.
Targeted Sequencing Mode
We decided to compare GENE-IS targeted sequencing analysis mode with the accessible virus IS analysis pipelines, including Virus-Clip and ViralFusionSeq. VirusFinder2.015, 16 and Vy-PER17 were not taken into the comparison for the reasons examined in the Discussion.
To perform the comparison, we generated an in silico dataset of 5,600 (DS2.1) sequences containing pre-located genomic and vector positions that were analyzed by GENE-IS, Virus-Clip, and ViralFusionSeq with default parameters. GENE-IS detected all the DS2.1 integration sites and showed the highest sensitivity, specificity, accuracy, and precision, whereas Virus-Clip reported only one genomic IS with a sequence count of 5,600 (Table 2). The output of ViralFusionSeq did not include the exact positions of genomic integration sites. Therefore, it was not possible to compare its output with GENE-IS. The detailed comparison between GENE-IS and Virus-Clip IS along with ViralFusionSeq output is provided in Data S3.
Table 2.
Comparison of Targeted Sequencing Data Analysis by GENE-IS Targeted Sequencing Mode and Virus-Clip by Using an In Silico Dataset of 5,600 Reads and a Control Sample, Lentiviral Vector-Transduced HeLa Cells, with Three Pre-determined Integration Sites
| Tools | GENE-IS | Virus-Clip |
|---|---|---|
| 5,600-Read In Silico Dataset | ||
| Sensitivity | 1 | –a |
| Specificity | 1 | |
| Precision | 1 | |
| Accuracy | 1 | |
| Real Control HeLa Sample, ∼37.5 Million Reads | ||
| With sequence count threshold per integration site >1 | ||
| Sensitivity | 1 | 1 |
| Specificity | 1 | 0.063 |
| Precision | 1 | 0.038 |
| Accuracy | 1 | 0.097 |
| Without sequence count threshold per integration site | ||
| Sensitivity | 1 | 1 |
| Specificity | 0.28 | 0.059 |
| Precision | 0.19 | 0.036 |
| Accuracy | 0.38 | 0.091 |
Statistical measures were estimated without using any sequence count threshold per integration site and by using a threshold greater than 1, i.e., in this case only those integration sites were counted that were supported by at least two independent reads.
Analysis didn’t report each individual genomic IS.
Additionally, we have evaluated a control lentiviral vector-transduced HeLa cell sample containing three pre-determined ISs (DS2.2). It consists of about 37 million reads of 100-bp paired-end sequencing reads (HiSeq; Section S6). ViralFusionSeq did not complete the analysis in less than 5 days, and it was excluded from this comparison. GENE-IS and Virus-Clip tools were able to detect all three known ISs, but Virus-Clip detected significantly higher false-positive ISs. We estimated sensitivity, specificity, precision, and accuracy (1) directly on the results of the pipelines and (2) using the value of the IS sequence count as threshold parameter. In the second case, imposing the threshold of having at least two reads to support an IS, GENE-IS detected no false-positive ISs and all four performance indices converged to 1. However, Virus-Clip still detected 74 false-positive ISs (Table 2). The full comparison between GENE-IS and Virus-Clip is provided in Data S4.
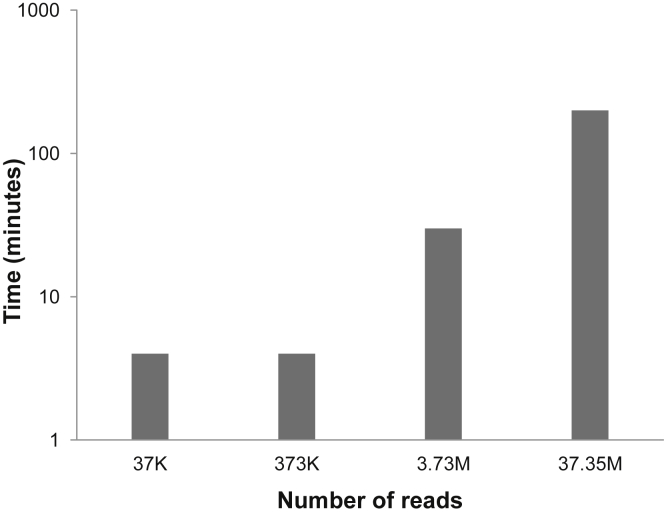
To estimate the time efficiency of GENE-IS targeted sequencing mode, we also generated large simulated in silico datasets. Four datasets comprising 37,200 (DS2.3), 373,200 (DS2.4), 3,735,000 (DS2.5), and 37,350,000 (DS2.6) raw read pairs were subjected to analysis with targeted sequencing paired-end mode. The time consumed by GENE-IS for analysis of DS2.3 and DS2.4 was 4 min, whereas for DS2.5 and DS2.6 it took 29 and 200 min, respectively (Figure 2).
Figure 2.
Histogram of the Analysis Time Consumed by GENE-IS Targeted Sequencing Mode
Time required for processing four in silico datasets of 37K, 373K, 3.73M, and 37M reads by GENE-IS is shown.
Discussion
GENE-IS is an efficient, flexible, and easily scalable pipeline for analyses of large-scale high-throughput sequence data generated by different experimental approaches, including PCR and targeted sequencing methods. It showed marked improvements in minimizing the execution time as compared to the available tools and reliability in IS detection to a single-base resolution. Further time reduction in analysis process can be achieved by parallelizing the demultiplexing and the mapping steps. A primary intent of the present work is to achieve time efficiency without compromising sensitivity and specificity measures. For this reason, we have used a combination of mapping tools, including Burrows-Wheeler Aligner (BWA) MEM algorithm18 for initial alignment followed by BLAT19 (Section S7). It is worthwhile to specify that the statistical metrics of GENE-IS do not vary with read lengths, although a significant effect of read length on computational efficiency was observed (Section S8).
The web-based nature of previously published LAM tools VISA, HISAP, and QuickMap for PCR-based IS data analyses has certain limitations regarding (1) amount of input data, (2) analysis time, and (3) pre-defined reference genomes available. Most importantly, none of these tools allowed the analysis of paired-end reads (generated, e.g., by targeted sequencing or whole-genome sequencing). QuickMap and HISAP are not compatible for analysis of large datasets. Although the VISA tool allows large-scale analysis, it is restricted in the input reads. Besides, it had a substantial limitation regarding reference genome, as only the hg38 version could be used for the study. GENE-IS provides an option for incorporating any known reference or vector genome, which is a crucial factor for flexible investigation. We also consider here that VISPA was not tested in the present study because it requires the installation of Galaxy20 and Apache Hadoop21 as the data processing framework. In our opinion, the hardware and software requirements needed to install and use VISPA are a substantial limitation and far beyond the possibilities of a routine laboratory.
The tools available for whole-genome or targeted sequencing data analyses, like VirusFinder2.0, ViralFusionSeq, Vy-PER, and Virus-Clip, are tailored for specific analysis requirements (with the focus on virus detection in cancer genome sequence data), which differ from our objective. VirusFinder2.0 requires an elaborate installation (as it has a very high number of external dependencies), which includes installation of ten primary tools that have further dependencies with secondary dependencies that also have to be installed. In addition, it reports those ISs that show high clonal expansion, but it fails to detect clones existing at a lower frequency. As an example from clinical gene therapy studies, it is necessary to track the clones over the years, to control the clonal dynamics in the patient and detect adverse events. The usage of BLAST222 in ViralFusionSeq suffers from two disadvantages: (1) it requires excessive execution time, and (2) after the release of the newer BLAST+ version,23 the authors do not recommend BLAST2 use anymore. Moreover, it makes use of BWA backtrack algorithm, which is slower and designed for short Illumina sequencing reads (up to 100 bp). Vy-PER and Virus-Clip are comparatively recently published tools. The focus of Vy-PER is significantly different from our study, as its principal aim is to detect different candidate viruses integrated into the host genome; consequently, their primary emphasis lies on the filtering techniques to detect potential viruses followed by IS detection. All these above-mentioned available tools have their own merits and are suitable for the purposes for which they have been designed, but they differ significantly from our analysis aims. In general, these tools offer reduced flexibility to the end users, as it is not possible to adjust key procedures such as quality filtering, adapter trimming, and many others.
Virus-Clip has an easier installation procedure and is more comparable to our approach. However, it cannot be directly applied in gene therapy data analysis because it does not report each IS position. It clusters all those reads that have the same vector sequence integrating into the genome and reports only one respective genomic IS. This was shown in the analysis of DS2.1, where it failed to detect 5,600 individual ISs and instead reported only one genomic IS with a sequence count of 5,600. It also listed a number of false-positive ISs, as was remarked in the analysis of DS2.2. GENE-IS showed a reduction of the false-positive rate using a sequence count threshold (Table 2). No significant reduction was observed in Virus-Clip with the same threshold. According to our analysis, the main reason for this noise is the tendency of Virus-Clip to pick vector fragments that align with the genome with similar homology score. Concerning computational efficiency, Virus-Clip is faster compared to the targeted sequencing GENE-IS mode. Virus-Clip performs the initial alignment with vector only and then a subset of sequences is processed further. GENE-IS completes the initial mapping step with a customized reference genome composed by the organism and viral genome. Also, we have employed various extra steps that consume additional time. These activities include paired-end quality filtering, unique and multiple hits IS distinction, re-mapping of individual sequences, and retrieving each genomic IS per vector IS. ViralFusionSeq is the most time-consuming tool and is not optimal for large-scale analysis, as it took ∼20 hr for the analysis of DS2.1, whereas for Virus-Clip and GENE-IS it was finished within few minutes.
The dependence of GENE-IS on the Linux platform can be a limitation for general users, as web-based tools have the advantage of a simple user interface. However, to meet the growing analysis demands and the requirements of standard analysis tools like BWA and SAMtools,24 most of the next-generation sequencing (NGS) analysis pipelines (i.e., Virus-Finder2.0, ViralFusionSeq, Virus-Clip, and Vy-Per) are established on Linux-based architectures. It is worth mentioning that GENE-IS needs a setup that requires approximately half an hour on a standard workstation. The speed depends on the number of the raw reads to be analyzed and the amount of the parallelism imposed (e.g., an i7 architecture with four cores and 16 gigabytes (GB) random access memory (RAM) is sufficient to analyze a MiSeq run in a few hours). Keeping in mind the installation and usage limitations for inexperienced users, we have provided the detailed instruction manual to instruct the user to download, install, and use the pipeline. It is worthwhile to consider that, after the installation, the parameters in a text file, referred to as a configuration file, have to be changed to fit the analysis requirements.
GENE-IS outperformed the available tools by providing a dual analysis mode for LAM-PCR- and targeted sequencing-based large-scale NGS datasets with remarkable computational efficiency and flexibility to use any reference genome. Also, GENE-IS will be under active development in the future to further improve computational efficiency and offer enhanced analysis options, providing additional user-customizable features.
Materials and Methods
Synthetic Datasets
To generate DS1.1, we randomly extracted 500 sequences of 200-bp length from the hg38 build of the human genome. Barcode, megaprimer, and long terminal repeat (LTR) sequences were added at the beginning of each genomic sequence; linker cassette (LC) was added at the end of the sequence. We generated this dataset in FASTQ and FASTA formats, as VISA, HISAP, and QuickMap only accept FASTA input format. HISAP and QuickMap support at latest the hg19 build; therefore, genomic coordinates of the test dataset were converted to the hg38 build by UCSC liftover25 for comparison and adjusted for differences in sequences between hg19 and hg38 versions by manual inspection. Similarly, DS1.2 and DS1.4 were generated as mentioned above; however, sequences of 250-bp length were produced. DS1.5, DS1.6, DS1.7, and DS1.8 were generated with IS- as well as non-IS-containing sequences.
In the case of targeted sequencing mode, for DS2.1 we randomly extracted 5,600 sequences from the hg38 build of the human genome to generate the paired-end FASTQ dataset of 250-bp length. A vector sequence of 50 bp was added at the end of each sequence in one file. We additionally generated four in silico datasets (DS2.3, DS2.4, DS2.5, and DS2.6) by extracting 1,000 random sequences of 1 kb from the hg38 build. The vector was inserted at known genomic locations in the middle of each fragment. This library was used with a freely available program, profile-based Illumina pair-end Reads Simulator (pIRS),26 with default parameters to simulate four 100-bp sequencing datasets. Each dataset samples the library at four different coverage values: 1, 10, 100, and 1,000. DS2.3, DS2.4, DS2.5, and DS2.6 contain 37,200, 373,200, 3,735,000, and 37,350,000 sequences, respectively.
The analysis with all the Linux-based tools was performed on a 10.04 LTE Linux machine with 48 GB RAM and an eight-core Xeon central processing unit (CPU). The detail of criterion employed for statistical measure estimation is available in Section S4.
Pipeline
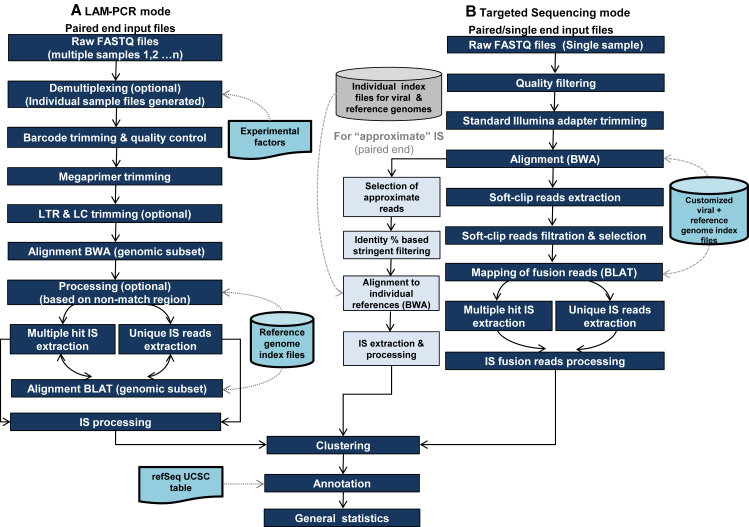
GENE-IS consists of two main analysis modes specific for LAM-PCR and targeted sequencing IS analysis (Figure 3). In LAM-PCR, several samples are commonly pooled together for simultaneous sequencing to enhance time and cost efficiency of the process. To reduce potential cross contamination, each of the samples is tagged with a unique combination of barcodes. As a first step, we decided to implement a demultiplexing strategy based on double barcodes (Section S2). In the next step, experimental sequences as sequencing adapters and vector-specific fragments are removed from the reads to obtain only the genomic portion. The megaprimer is searched and trimmed without mismatches. A set of configurable trimming parameters can be employed to improve the quality of the reads (Figure S2). After this pre-processing of sequences, the complete set of virus vector flanking genomic sequences is mapped to the reference genome by BWA MEM algorithm. The selection of potential IS sequences is evaluated and controlled by stringent filtering based on quality of reads and alignment score. A configurable threshold is applied for allowing unmapped bases at the 5′ end of mapped sequences (Figure S2). In the final steps, the subset of candidate sequences is re-aligned by BLAT aligner and processed to extract the final subset of IS-contributing sequences.
Figure 3.
Overview of GENE-IS Framework
(A and B) The LAM-PCR (A) and targeted sequencing modes (B). In case of paired-end targeted sequencing, the candidate reads for exact IS are those that contain fusion events of vector and genome in one read and the other read supports the fusion event read. For approximate IS candidate reads, one read of the pair has 100% matching identity with vector and the other with genome region.
In targeted sequencing data, the sequence reads can contain any sub-region of viral vector and genomic portion within the entire read. The individual targeted sequencing FASTQ sample, paired-end or single-end reads, is subjected to quality filtering and trimming of standard Illumina adapters. The union of the organism and vector genomes creates a customized reference genome that is used by BWA MEM to map the paired (single)-end reads. In the resulting set of mapped reads, the fusion reads are identified since they contain the aligned and the not-aligned part within the same read. The soft-clipped (SC) region represents the not-aligned part of the sequence and can be a genome, vector, or an unspecified sequence. The IS identification involves extraction of all the SC reads longer than a configurable threshold (20 bp by default). In paired-end reads, each SC-containing read is evaluated on the basis of the support from the second read of pair by taking into account read pairing, mapping quality, and alignment score. This stringent control and subsequent filtering is the critical step that strictly reduces the detection of false-positive ISs. At the next stage, the high-quality IS-containing subset of reads is re-mapped and processed in a number of steps to obtain a final set of viral-genome true IS reads, which are further analyzed for identification of exact breakpoints at base level. In the case of paired-end targeted sequencing data, the reads that do not contain fused viral-genome junctions within a single read are referred to as approximate reads, and they are processed independently by an approximate IS module (optional). In this case, one read aligns to the vector and the other to the genome. GENE-IS uses the stringent alignment identity (100%) to ensure the best possible removal of artifact IS by processing only read sequences that possess 100% sequence identity with vector and genome.
For the subset of reads that give multiple hits in the genome (i.e., sequences reflecting repetitive elements of the host genome), a homology ratio is calculated between primary and secondary alignment score, and this value is compared with a defined threshold value. All the reads with an alignment score ratio higher than the user-provided threshold value (default 0.95) are flagged as ambiguous, and they are processed to generate a multiple aligned IS result file.
The clustering module performs topographical clustering of the resulting set of ISs, where we commonly use a window size of ±5 bp by default (user-customizable parameter) to cluster all ISs that are within this range to account for experimental PCR or sequencing biases. The ISs are reported along with the respective sequence count or frequency. The final set of ISs is annotated according to the nearby gene based on the UCSC refSeq Table,27 by employing BEDTools28 and our in-house Perl and Shell scripts.
GENE-IS has been tested at Ubuntu 10.10, 12.04, and 14.04 LTS 64-bit operating system. It was developed exploiting a combination of Perl (v5.10.1), Python (2.7.2), and shell programming languages in order to orchestrate the delivery of the intermediate results to the pipeline tools and to complete customized operations on tabular files. Skewer29 has been used for the trimming of adapters, and SAMtools is employed for the intermediate processing of SAM alignment files along with our Perl and Shell scripts. To make GENE-IS usable for a large audience without any profound computational expertise, we have written template configuration text files as an example for each GENE-IS mode (Figures S6 and S7). The CSV results and statistics files are generated at the end with detailed information (Section S10).
Author Contributions
M.S. and R.F. designed and supervised the study and revised the manuscript. M.S. and C.v.K. conceived the study and coordinated on the project. S.A. developed the software, performed benchmarking analyses, and wrote the manuscript. R.F. contributed in the development of the software and writing of the manuscript. S.W. conducted the laboratory experiments used for testing the software.
Conflicts of Interest
C.v.K. and M.S. are co-founders of GeneWerk GmbH.
Acknowledgments
We would like to gratefully acknowledge Gabor Veres and Olivier Negre (from Bluebird Bio Inc.) for providing pre-clinical datasets for analysis. We thank our colleagues at the Translational Oncology group for helpful feedback and the Genomics and Proteomics Core Facility group at German Cancer Research Center (DKFZ). There were no funding sources for this project.
Footnotes
Supplemental Information includes Supplemental Materials and Methods, seven figures, one table, and four data files and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2016.12.001.
Supplemental Information
This dataset was taken from VISA paper from VISA website.
An in silico data of 5600 sequences with pre-determined IS was used.
References
- 1.Aiuti A., Slavin S., Aker M., Ficara F., Deola S., Mortellaro A., Morecki S., Andolfi G., Tabucchi A., Carlucci F. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 2.Bainbridge J.W.B., Smith A.J., Barker S.S., Robbie S., Henderson R., Balaggan K., Viswanathan A., Holder G.E., Stockman A., Tyler N. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 3.Boztug K., Schmidt M., Schwarzer A., Banerjee P.P., Díez I.A., Dewey R.A., Böhm M., Nowrouzi A., Ball C.R., Glimm H. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N. Engl. J. Med. 2010;363:1918–1927. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranzani M., Annunziato S., Adams D.J., Montini E. Cancer gene discovery: exploiting insertional mutagenesis. Mol. Cancer Res. 2013;11:1141–1158. doi: 10.1158/1541-7786.MCR-13-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt M., Schwarzwaelder K., Bartholomae C., Zaoui K., Ball C., Pilz I., Braun S., Glimm H., von Kalle C. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 7.Ustek D., Sirma S., Gumus E., Arikan M., Cakiris A., Abaci N., Mathew J., Emrence Z., Azakli H., Cosan F. A genome-wide analysis of lentivector integration sites using targeted sequence capture and next generation sequencing technology. Infect. Genet. Evol. 2012;12:1349–1354. doi: 10.1016/j.meegid.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Appelt J.U., Giordano F.A., Ecker M., Roeder I., Grund N., Hotz-Wagenblatt A., Opelz G., Zeller W.J., Allgayer H., Fruehauf S., Laufs S. QuickMap: a public tool for large-scale gene therapy vector insertion site mapping and analysis. Gene Ther. 2009;16:885–893. doi: 10.1038/gt.2009.37. [DOI] [PubMed] [Google Scholar]
- 9.Arens A., Appelt J.U., Bartholomae C.C., Gabriel R., Paruzynski A., Gustafson D., Cartier N., Aubourg P., Deichmann A., Glimm H. Bioinformatic clonality analysis of next-generation sequencing-derived viral vector integration sites. Hum. Gene Ther. Methods. 2012;23:111–118. doi: 10.1089/hgtb.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hocum J.D., Battrell L.R., Maynard R., Adair J.E., Beard B.C., Rawlings D.J., Kiem H.P., Miller D.G., Trobridge G.D. VISA--Vector Integration Site Analysis server: a web-based server to rapidly identify retroviral integration sites from next-generation sequencing. BMC Bioinformatics. 2015;16:212. doi: 10.1186/s12859-015-0653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho D.W., Sze K.M., Ng I.O. Virus-Clip: a fast and memory-efficient viral integration site detection tool at single-base resolution with annotation capability. Oncotarget. 2015;6:20959–20963. doi: 10.18632/oncotarget.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J.W., Wan R., Yu C.S., Co N.N., Wong N., Chan T.F. ViralFusionSeq: accurately discover viral integration events and reconstruct fusion transcripts at single-base resolution. Bioinformatics. 2013;29:649–651. doi: 10.1093/bioinformatics/btt011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calabria A., Leo S., Benedicenti F., Cesana D., Spinozzi G., Orsini M., Merella S., Stupka E., Zanetti G., Montini E. VISPA: a computational pipeline for the identification and analysis of genomic vector integration sites. Genome Med. 2014;6:67. doi: 10.1186/s13073-014-0067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Negre O., Bartholomae C., Beuzard Y., Cavazzana M., Christiansen L., Courne C., Deichmann A., Denaro M., de Dreuzy E., Finer M. Preclinical evaluation of efficacy and safety of an improved lentiviral vector for the treatment of β-thalassemia and sickle cell disease. Curr. Gene Ther. 2015;15:64–81. doi: 10.2174/1566523214666141127095336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q., Jia P., Zhao Z. VirusFinder: software for efficient and accurate detection of viruses and their integration sites in host genomes through next generation sequencing data. PLoS ONE. 2013;8:e64465. doi: 10.1371/journal.pone.0064465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q., Jia P., Zhao Z. VERSE: a novel approach to detect virus integration in host genomes through reference genome customization. Genome Med. 2015;7:2. doi: 10.1186/s13073-015-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forster M., Szymczak S., Ellinghaus D., Hemmrich G., Rühlemann M., Kraemer L., Mucha S., Wienbrandt L., Stanulla M., Franke A., UFO Sequencing Consortium within I-BFM Study Group Vy-PER: eliminating false positive detection of virus integration events in next generation sequencing data. Sci. Rep. 2015;5:11534. doi: 10.1038/srep11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent W.J. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apache Hadoop. [http://hadoop.apache.org].
- 22.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn R.M., Haussler D., Kent W.J. The UCSC genome browser and associated tools. Brief. Bioinform. 2013;14:144–161. doi: 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X., Yuan J., Shi Y., Lu J., Liu B., Li Z., Chen Y., Mu D., Zhang H., Li N. pIRS: Profile-based Illumina pair-end reads simulator. Bioinformatics. 2012;28:1533–1535. doi: 10.1093/bioinformatics/bts187. [DOI] [PubMed] [Google Scholar]
- 27.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang H., Lei R., Ding S.W., Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This dataset was taken from VISA paper from VISA website.
An in silico data of 5600 sequences with pre-determined IS was used.