Abstract
Monogenic disorders (MGDs), which are caused by single gene mutations, have a serious effect on human health. Among these, β-thalassemia (β-thal) represents one of the most common hereditary hematological diseases caused by mutations in the human hemoglobin β (HBB) gene. The technologies of induced pluripotent stem cells (iPSCs) and genetic correction provide insights into the treatments for MGDs, including β-thal. However, traditional approaches for correcting mutations have a low efficiency and leave a residual footprint, which leads to some safety concerns in clinical applications. As a proof of concept, we utilized single-strand oligodeoxynucleotides (ssODNs), high-fidelity CRISPR/Cas9 nuclease, and small molecules to achieve a seamless correction of the β-41/42 (TCTT) deletion mutation in β thalassemia patient-specific iPSCs with remarkable efficiency. Additionally, off-target analysis and whole-exome sequencing results revealed that corrected cells exhibited a minimal mutational load and no off-target mutagenesis. When differentiated into hematopoietic progenitor cells (HPCs) and then further to erythroblasts, the genetically corrected cells expressed normal β-globin transcripts. Our studies provide the most efficient and safe approach for the genetic correction of the β-41/42 (TCTT) deletion in iPSCs for further potential cell therapy of β-thal, which represents a potential therapeutic avenue for the gene correction of MGD-associated mutants in patient-specific iPSCs.
Keywords: β thalassemia, induced pluripotent stem cells, CRISPR/Cas9, ssODNs
Introduction
β thalassemia (β-thal) is a monogenic blood disease that is caused by a lack of or reduction in β-globin chain synthesis. Patients with β-thal major, also known as Cooley’s anemia, have severe anemia and usually require frequent transfusions and iron chelation therapy.1 However, complications related to iron overload cannot be completely managed through chelation therapy, and compliance with a chronic transfusion regimen is difficult to maintain throughout a patient’s lifetime. To date, the only available cure for β-thal is allogeneic hematopoietic stem cell (HSC) transplantation.2 However, fully matched donors are rare for β-thalassemia patients, and most of those who receive mismatched transplants suffer from immune complications such as graft rejection or graft versus host disease.3 Gene therapy has recently provided an alternative approach for β-thalassemia by permanent expression of the β-globin gene in patient-derived HSCs using a lentiviral vector, and these patients require no further blood transfusions.4 The use of a viral vector has safety concerns for patients in that it may cause random integrations in multiple sites of the host genome, which would carry risks of insertional oncogenesis; this has already been observed in gene therapy for other diseases.5
Induced pluripotent stem cell (iPSC) technology presents new concepts of regenerative therapy by replacing impaired tissues and cells with stem cells or functional cells differentiated from patient- specific iPSCs.6 However, for inherited diseases (such as β-thal, which is caused by genetic mutations), the mutations in iPSCs must be corrected prior to use in regeneration therapy.7 The tremendously low efficiency of traditional homology-directed repair (HDR) in iPSCs has restricted its use in treatment. New gene-editing tools, such as zinc finger nuclease (ZFN),8 transcription activator-like effector nuclease (TALEN),9 and clustered regulatory interspaced short palindromic repeat (CRISPR)/Cas9 endonuclease,10 can greatly improve the efficiency of gene targeting by inducing site-specific double-strand breaks (DSBs). Those technologies have previously been applied to the genetic correction of differentiated mutations of human hemoglobin β (HBB) caused in β-thalassemia in combination with a gene-targeting vector, which required a drug-resistant gene for selection-positive colonies.11 The Cre/Loxp (or Flp/flipase recognition target [FRT]) recombination systems and the PiggyBac transposon system are often used to remove the selection markers; nevertheless, this leaves an indelible footprint in the genome, uses a targeting construct, and needs two drug selection steps.10 Single-stranded DNA oligonucleotides (ssODNs) were recently applied as a repair template to generate precise point mutations in human cells combined with ZFN, TALEN, and CRISPR/Cas9 through HDR.12 Compared with gene-targeting vectors, which require several weeks to construct, ssODNs can be synthesized in just a few days and achieve precise and scarless genome engineering in one step without any selection markers. Thus, for the sake of simplicity and to address safety concerns, ssODN-mediated seamless genome editing is highly desirable in the genetic correction of inherited mutations in patient-specific iPSCs for stem cell therapies.
Because of the low efficiency of transfection and high fraction of non-homologous end joining (NHEJ) repair of DSBs caused by CRISPR/Cas9, ssODN-mediated gene editing produces a low number of desired iPSCs. Small-molecule compounds, such as L755507,13 were recently shown to significantly enhance CRISPR-mediated HDR. In this study, we investigated a highly efficient and seamless genetic correction of a common deletion mutant, β-41/42 (TCTT), in the HBB gene of β-thal patient-specific iPSCs. To optimize ssODN-mediated HDR with Cas9/guide RNA (gRNA), we first established a dual fluorescent reporter that harbored the deletion mutant β-41/42 (TCTT) of HBB. The gRNA and ssODN with the highest efficiencies were selected to genetically correct the deletion mutant β-41/42 (TCTT) of endogenous HBB in patient-specific iPSCs. To reduce non-specific DNA contacts, we used the newly developed high-fidelity CRISPR/Cas9 nucleases SpCas9-HF1.14 A fluorescence-linked Cas9- and gRNA-expressing vector was applied to target iPSCs, and fluorescence-activated cell sorting (FACS) was used to enrich the transfected cells. In combination with the small-molecule compound L755507, we achieved highly efficient (up to biallelic correction of 54%) genetic correction of the deletion mutant β-41/42 (TCTT) in β-thal patient-specific iPSCs with ssODNs. Whole-exome sequencing was applied to evaluate the off-targeting effect, and the sequencing results showed that the iPSCs were genetically corrected and exhibited a minimal mutational load. Furthermore, the functionality of genetically corrected iPSCs was assessed through in vitro differentiation and RT-PCR. HBB was correctly transcripted.
Our results showed that the use of ssODNs and small molecules can lead to CRISPR/Cas9-mediated gene targeting with efficient, seamless, and biallelic correction of the gene mutant in one step, which represents a more efficient and safer alternative to other current technologies. This strategy has important implications for personalized treatment in β-thal patients and holds great potential for the treatment of other monogenic disorders (MGDs).
Results
Design and Optimization of gRNAs and ssODNs with a Dual Fluorescent Reporter
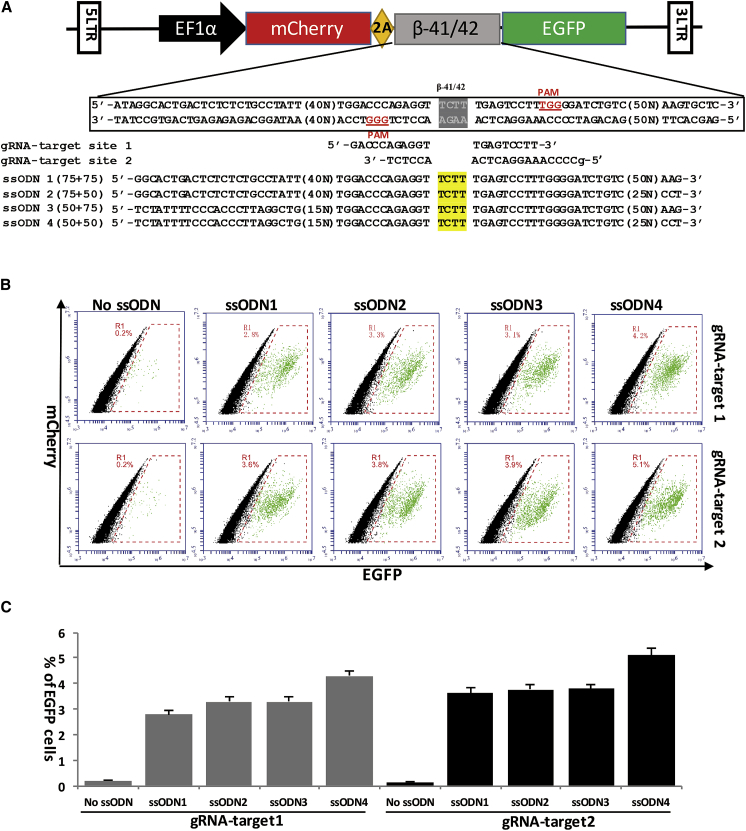
Although ssODNs have some advantages in precise gene editing combined with CRISPR/Cas9-mediated HDR compared with double-stranded DNA (dsDNA) donors, its length was limited to less than 200 nucleotides (nt) by oligonucleotide synthesis technology.15 In light of the characterization of gRNA-guided Cas9 cleavage,16 gRNAs were designed to target the mutation sites that are adjacent to the protospacer-adjacent motif (PAM) site. Based on this principle, we designed two gRNAs that directly target the deletion mutant site (Figure 1A). We previously found that ssODNs with a 100-nt homologous sequence can achieve the highest efficiency of Cas9/gRNA-mediated HDR in the HEK293 cell line;17 however, the efficiency of longer ssODNs and ssODNs-mediated targeting in iPSCs has not been tested. Studies of dsDNA donors showed that longer homologous sequences can significantly improve the HDR rates when they are used with nucleases.18 Thus, four ssODNs were designed with different lengths (from 100- to 150-nt homologous sequences), all of which carried the same 4-base pair (bp) mismatch in the middle to correct for the deletion mutant (Figure 1A). Five days after transfection of iPSC-Δ41/42-EGFP, flow cytometry analysis was performed to determine the repair efficiency (Figure 1B). Cas9/gRNA2 combined with four ssODNs all achieved higher repair efficiencies than Cas9/gRNA1, which may be because Cas9/gRNA2 has a higher cleavage efficiency. Interestingly, longer homologous sequences did not improve the HDR efficiency, whereas the ssODNs with the 100-nt homologous sequence had the highest HDR efficiency in iPSCs with either Cas9/gRNA1 or Cas9/gRNA2 (Figure 1C). A possible reason may be that the ssODNs are used in an alternative genome repair process compared with dsDNA. Therefore, the combination of Cas9/gRNA2 and ssODN4 with a 100-nt homologous sequence was used in the subsequent genetic correction of the β-41/42 mutation in patient-specific iPSCs.
Figure 1.
Selection of the Optimal gRNAs and ssODN for Targeting the β-41/42 Deletion Mutant with a Dual Fluorescent Reporter
(A) Scheme of the RFP-GFP dual fluorescent reporter-based assay for assessing the gene correction efficiency of various gRNAs and ssODNs. The normal iPSCs were transfected with a dual fluorescent lentivirus vector harboring a 158-bp β-41/42 HBB genomic sequence that contains the 4-bp (TCTT) deletion resulting in disruption of EGFP. (B) Flow cytometry analysis of gene correction efficiency. After correction by Cas9/gRNA- and ssODNs-mediated HDR, the open reading frame of mCherry and EGFP could be restored and correctly translated. The relative efficiency of gene correction was measured by the ratio of double mCherry+ and EGFP+ iPSCs using flow cytometry. (C) Comparison of Cas9/gRNA- and ssODNs-mediated gene correction efficiency. Four ssODNs with different homologous sequences and two Cas9/gRNAs were co-transfected into iPSCs with the dual fluorescent reporter, and double mCherry+ and EGFP+ cells were counted using flow cytometry. Cells transfected without ssODNs were used as controls. Error bars indicate SEM from at least three independent experiments.
Seamless Correction of the HBB Deletion Mutant in Homozygous β-Thal iPSCs
To correct the deletion mutant of endogenous HBB, β-41/42 homozygous patient-specific fibroblast cells were reprogrammed into iPSCs that exhibited a TCTT deletion between the 41st and 42nd amino acids of the β-globin gene. The generation of β-thalassemia iPSCs was performed under virus-free, serum-free, and feeder-free conditions to enable further clinical treatment. CRISPR/Cas9 is highly efficient in genome editing and is widely used; however, wild-type (WT) spCas9 nucleases have significant off-target effects that cause unwanted mutations and have severe implications. High-fidelity Cas9 (SpCas9-HF1) nucleases with no detectable genome-wide off-target effects were recently reported.14 Thus, we used these newly developed SpCas-HF1 nucleases for our HBB gene repair in iPSCs. Because of the low efficiency of electroporation in iPSCs, we constructed a vector co-expressing gRNA and SpCas9-HF1 with EGFP linked by the 2A peptide to the N-terminal of Cas9 (Figure 2A). After electroporation with the Cas9/gRNA vector and ssODNs, the cells were recovered in normal human iPSCs (hiPSCs) medium with or without the small molecule L755507 in the first 24 hr. 48 hr post-electroporation, the EGFP-positive cells were harvested via FACS, re-plated at low density, and further grown for 1 week until the formation of distinct single colonies. The colonies were then picked up and expanded for further identification and characterization.
Figure 2.
Seamless Gene Correction of the HBB Mutant in β-Thal iPSCs with spCas9-HF1/gRNA and ssODNs
(A) Schematic of the gene correction strategy for the human HBB CD41/42 mutation. β-Thal iPSCs were electroporated with the gRNA/spCas9-HF1-EGFP vector and ssODN templates and then recovered in mTeSR1 medium supplemented with or without L755507 for 24 hr. 48 hr post-transfection, the GFP-positive cells were collected, re-plated at low density, and later developed into distinct single colonies for further identification and characterization. (B) Diagram for the ssODN- and gRNA-mediated gene correction of the HBB mutation in iPSCs. (C) PCR analysis discovered the correctly rescued colonies derived from the sorted cells with or without L755507 treatment. As shown in (B), primers F and R flanked the ssODN donor and yielded a 695-bp fragment; primers RWT and RΔ were allele-specific, which distinguished the corrected and mutation DNA sequences, and yielded a 450-bp fragment. Red stars and green stars, respectively, indicate the biallelic and monoallelic gene-corrected colonies. (D) Summary of the detected colonies’ genotype. A total of 195 colonies were validated, 90 with L755507 treatment and 105 without L755507 treatment. (E) Comparison of the correction efficiency of the HBB mutation in β-thal iPSCs with and without L755507 treatment. (F) Comparison of the HDR and NHEJ efficiency of spCas9-HF1/gRNA-mediated gene targeting with and without L755507 treatment. (G) Sanger sequencing confirmed various genotypes from targeted colonies with PCR products of F and R primers.
To determine whether the ssODN-mediated HDR corrected the mutation, we designed a multiple PCR method with three primers (Figure 2B) in which primer F and primer R amplified the genomic DNA sequence outside the HDR arms and primer RWT and RΔ were allele-specific (to distinguish between the corrected and mutated DNA sequences). The correctly targeted clones would be positive for primer set 1 (F+ R/RWT) with two bands (695 bp and 450 bp) (Figure 2C). Primer set 2 (F+ R/RΔ) was applied to identify the biallelic or monoallelic correction of the deletion mutant. The PCR products of the F and R primers were used for sequencing to detect the genotype of an individual colony. Using this multiple PCR detection, we found a remarkable genetic correction efficiency (Figure 2D). Without small-molecule L755507 treatment, ssODN-mediated HDR can achieve 14.3% biallelic correction (WT/WT) and 49% monoallelic correction (WT/indel+WT/mut) (Figure 2E). With small-molecule L755507 treatment, the biallelic correction was significantly increased to 54%, and the monoallelic correction was 25.5%. Insertion or deletion (indel) mutations (indel/indel+indel/mut) caused by NHEJ repair were found in both the treated (12.2%) and untreated (26.6%) colonies with L755507 (Figure 2E). With the sequencing result (Figure 2G), we found that the small molecule L755507 can markedly enhance HDR frequency (Figure 2F) and repress NHEJ repair efficiency. These results indicated that the addition of L755507, when using Cas9/gRNA and ssODN, can achieve a high efficiency of precise iPSC editing, especially biallelic HDR-mediated gene editing.
Characterization of the Corrected β-Thalassemia iPSCs
To determine whether the gene targeting process affects the pluripotency of iPSC sub-clones, we randomly selected two corrected colonies that carried monoallelic (iPSC-C1WT/mut) and biallelic correction (iPSC-C2WT/WT), respectively, for further characterization. Both targeted colonies displayed typical iPSC morphology and retained normal karyotypes (Figure 3A). Immunofluorescence analysis revealed that the targeted clones retained uniform expression of typical pluripotency markers such as OCT4, SOX2, and SSEA-4. Additionally, the targeted iPSC clones were transplanted into severe combined immunodeficiency (SCID) mice, and teratoma formation was observed at 8 weeks to test their pluripotency in vivo. Histological examination revealed that the tumor generated by the CRISPR/Cas9-corrected iPS clones comprised cell types from all three germ layers. These results suggested that β-thalassemia patient-specific iPSCs retained pluripotency after gene repair by CRISPR/Cas9. Furthermore, to exclude cell line contamination, we performed short tandem repeat (STR) assays in untargeted and corrected iPSC-C1 and iPSC-C2 lines (Figure 3B). Within 15 STR loci, all three cell lines showed the same signal peaks, suggesting that the corrected colonies were derived from the untargeted cells with no contamination of other cells.
Figure 3.
Characterization of Gene-Corrected iPSC Clones
(A) The β-thal iPSCs and the two corrected iPSC clones (iPS-C1 and iPS-C2) expressed the pluripotent markers OCT4, SOX2, and SSEA4. All of the cells had normal karyotypes and could give rise to three germ layers (teratomas by H&E staining). Scale bars, 200 μm. (B) STR analysis confirmed that the corrected iPSC lines and the β-thal iPSCs were derived from the same parental cells.
Off-Target and Exome Sequencing Analysis of Corrected iPSCs
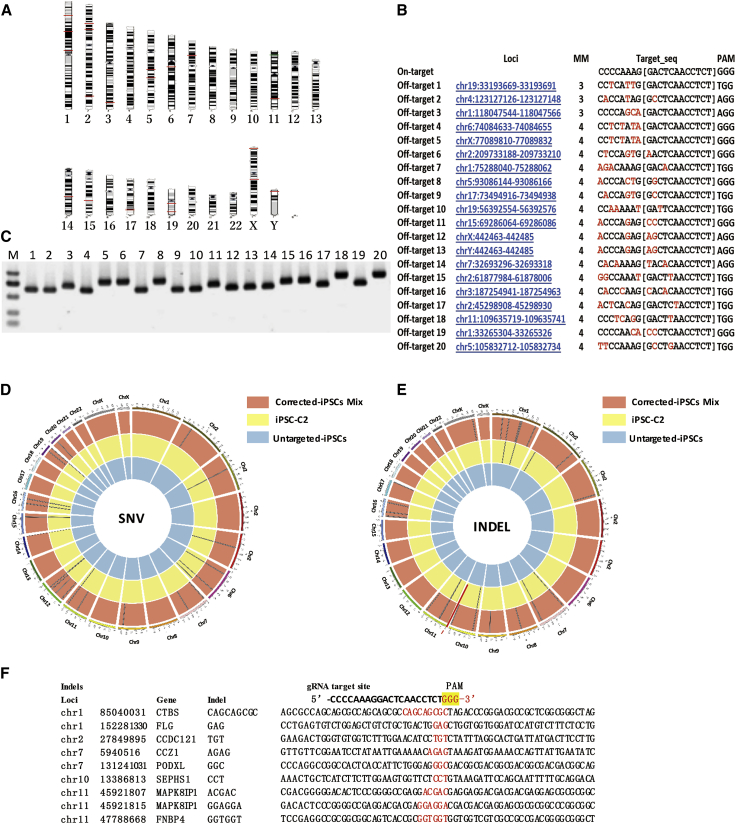
CRISPR/Cas9 has highly improved gene targeting efficiency while it has some propensity to cause off-target mutagenesis, which is crucial for regenerative therapy using gene-corrected iPSCs. Although we used the latest high-fidelity SpCas9-HF1 to target patient-specific iPSCs, the off-target effects remained to be addressed. We first predicted the potential off-target sites using the online software CCTop and found top 20 off-target sites within less than four mismatches (Figures 4A and 4B). T7E1 assay and Sanger sequencing showed no off-target mutagenesis in these sites (Figure 4C). Because of the poor sensitivity of the T7E1 assay and Sanger sequencing to discover rare off-target events, high-throughput sequencing was applied to detect the whole-exome sequence of the iPSCs prior to and after gene targeting. Genomic DNA from iPSC-C2 lines, the correctly targeted single colony, was detected with whole-exon sequencing. Additionally, the rest of the corrected colonies were mixed and then detected in the same way. Compared with the uncorrected iPSCs, we detected 12 single-nucleotide variants (SNVs) (Figure 4D) and 9 indels (Figure 4E) in the corrected iPSC-C2. For the rest of the corrected colonies, 21 SNVs and 35 indels were detected. The variations in the gene-corrected clones were then compared with the uncorrected iPSCs to enable the generation of a list of potential variations that may arise during the gene editing process (Figure 4F; Table S1). One important consideration was how many of these detected SNVs were the results of off-target mutagenesis by the CRISPR/Cas9 endonuclease. All sequences of the SNVs and indel sites were compared with the gRNA target sequence, and none were within a potential off-target region, which is consistent with previous analyses studying predicted off-target sites, although some off-target mutagenesis in the non-coding regions of chromosomal may be undetected. The absence of recurring mutations and the fact that none of the mutations reside in any putative off-target sites according to the bioinformatics predictions strongly suggest that these mutations were randomly accumulated during regular cell expansion and are not direct results of off-target activities by Cas9.
Figure 4.
Whole-Exome Sequencing of the Parental and Gene-Corrected iPSCs
(A and B) The top 20 potential off-target sites predicted using the online software CCTop, the distribution on the chromosomes (A), and detailed information (B). (C) T7E1 assays assessed mutagenesis at the predicted off-target sites. (D) Single-nucleotide variations in iPSCs via exome sequencing. The black bars represent individual SNVs. Compared with the untargeted β-thal iPSCs, iPSC-C2 holds 12 SNVs, and the remaining corrected colonies hold 21 SNVs. (E) Idles in the gene-corrected iPSCs via exome sequencing. The red arrow bar indicates the HBB locus. Compared with the untargeted β-thal iPSCs, iPSC-C2 holds 9 indels, and the remaining corrected colonies hold 35 indels. (F) Idle sequence detected in corrected iPSC-C2 via exome sequencing. Compared with the gRNA target site, no homologous sequences were found in the indel locus.
Transcription of HBB Restoration after Gene Correction
To determine whether gene correction of patient-specific iPSCs could restore the normal expression of full-length β-globin, we employed the OP9 co-culture system to induce hematopoietic differentiation of β-thalassemia iPSCs prior to and after gene correction. The morphologies of the CRISPR/Cas9-corrected iPSCs and the parental lines changed rapidly upon differentiation in OP9 co-culture (Figure 5A). The expression of hematopoietic differentiation-related genes, including CD31, CD34, CD45, GATA1, and HBG, was examined by RT-PCR (Figure 5B). FACS analysis showed that 12.2% of the CRISPR/Cas9-corrected iPSC-C1 and 15.7% of iPSC-C2 differentiated into CD34+CD31− hematopoietic progenitor cells (HPCs) compared with 10.5% of the parental cell lines and 12.3% of the wild-type iPSCs (Figure 5C). Using conventional RT-PCR to amplify the HBB cDNA from the iPSCs-derived erythroblasts, we sequenced the cDNA and confirmed that the expression of the HBB cDNA was successfully restored after gene correction (Figures 5D and 5E).
Figure 5.
Erythroblast Differentiation of Corrected iPSCs
(A) Images of the differentiated iPSCs on days 2, 6, 12, and 21. Scale bars, 100 μm. (B) RT-PCR analysis of the pluripotent gene OCT4 and hematopoietic genes, including CD31, CD34, CD45, GATA1, and HBG, in differentiated iPSC-C2 cells on day 21. Glyceraldehyde-phosphate dehydrogenase (GAPDH) was used as an internal control. (C) Flow cytometric analysis of β -thal iPSCs, corrected iPSC-C1, and iPSC-C2 using the surface markers CD34 and CD31 after differentiation for 14 days. (D) Conventional RT-PCR amplifies HBB cDNA in erythroblasts derived from indicated cells. cDNA from normal blood was used as a positive control, and GAPDH was used as an internal control. (E) The Sanger sequencing result confirmed that the HBB gene was successfully corrected in erythroblasts derived from iPSC-C2.
Discussion
MGDs caused by single gene mutations have serious effects on human health.19 β-Thalassemia is one of the most common MGD-associated diseases and represents more than 200 types of mutants in HBB gene-induced blood disorders.20 iPSCs techniques, combined with genetic correction, provide insights into potential treatments for MGDs using patient-derived stem cells. In this study, we developed the most efficient and safe approach for the seamless correction of the deletion mutant (β41-42 (-TCTT)) in the HBB gene. Using a dual fluorescent reporter, we optimized the most efficient combination of gRNA and ssODNs to correct the deletion mutant (β41-42 (-TCTT)). A FACS approach was applied to select the targeted cells without drug selection, and, with a small molecule, L755507, the gene correction efficiency was remarkably improved to 79.5%; notably, biallelic correction was 54%. Off-targeting analysis and whole-exon sequencing showed that the strategy we developed here can safely and seamlessly target patient-derived iPSCs, which is crucial for clinical applications. We further characterized the gene-corrected colonies and showed that corrected iPSCs retained pluripotency, multiple differentiation ability, and normal karyotypes. Most importantly, upon differentiating these iPSCs into HPCs and then erythroblasts, we confirmed the restoration of the normal expression of full-length mRNA of β-globin in the corrected cells.
A traditional method to correct a mutated gene utilizes a homologous targeting vector combined with gene targeting tools such as ZFNs, TALENs, and Cas9 nucleases, which require drug resistance genes to select positive clones. Additionally, the drug selection markers need to be removed from the correctly targeted clones to restore the transcription unit of the targeted gene, which is typically performed using recombinase (Cre or Flp) or transposase (PiggyBac or Sleeping Beauty); nevertheless, this leaves a residual footprint in the genome, which may affect functional elements and is undesirable in clinical application. The successful use of ssODNs as donors combined with customized nucleases to target endogenous genes initiated the concept of seamlessly correcting the mutant gene without any footprint; nevertheless, ssODN-mediated gene editing is inefficient for iPSCs. Moreover, unlike the dsDNA donor, the optimal ssODN length was 100 nt for HDR. A longer homologous sequence did not improve repair efficiency, which may indicate that 50-nt homologous recombination arms are sufficiently long for HDR to repair the CRISPR/Cas9-mediated DSB with ssODN as a donor.
To date, the transfection efficiency achieved has been unsatisfactory, even with electroporation (only achieving 30%);21 this limits the gene editing efficiency in iPSCs. The transient or consistent expression of marker genes combined with drug selection was often used to enrich the transfected cells; however, this process may affect cell vitality and pluripotency.22 Here we generated a Cas9/gRNA and EGFP co-expression vector that can be used to highly enrich the transfected cells via FACS. The EGFP marker can also be applied to negatively select the targeted colonies without foreign gene integration. Several small-molecule compounds have recently been identified to enhance CRISPR/Cas9-mediated gene targeting efficiency,23 some of which have a significant effect on HDR-mediated gene editing. Among these, L755507 has been shown to greatly enhance ssODN-mediated gene targeting efficiency in iPSCs. Consistent with a previous study, we found that L755507 can substantially improve ssODN-mediated HDR efficiency and that it depresses NHEJ-mediated gene editing. This result indicates that L755507 can enhance the activity of enzymes employed in ssODN-mediated HDR. By combining FACS and L755507, we here achieved a remarkable efficiency for correcting deletion mutants in patient-specific iPSCs through ssODN-mediated HDR.
One remaining and unavoidable concern in the use of a CRISPR/Cas9 system is off-target cleavage, which may have unpredictable deleterious phenotypic consequences. SpCas9-HF1 was reported to display undetectable off-target effects compared with the wild-type Cas9 and may thus provide an alternative choice for precise gene correction.14 To minimize the clinical concerns of using these gene-corrected cells, we first assessed off-targeting effects and compared the results with the untargeted cells; no off-target mutagenesis was found in the gene-corrected cells. We cannot completely rule out other genomic changes without whole-genome sequencing; however, we performed whole-exome sequencing to assess the genomic variations during the CRISPR/Cas9-mediated gene correction process. Only 12 SNVs and 9 indels were detected in the corrected iPSC-C2, and there were a total of 21 SNVs and 35 indels detected in the rest of the corrected colonies. Compared with the gRAN target sequence, none of those sites were within a potential off-target region. Our result is consistent with recent studies reporting that the genome editing tools did not appear to generate more intolerable variations at the single-nucleotide level, such as SNVs or indels.8 We also confirmed functionality restoration in the transcription of the HBB gene via RT-PCR and sequencing in erythroid cells differentiated from corrected iPSCs compared with those obtained from parental iPSCs.
In summary, we describe optimized approaches for achieving the most efficient and seamless biallelic correction of the HBB mutant in homologous β-thal iPSCs via ssODN and CRISPR/Cas9-mediated HDR combined with the small molecule L755507. Deep sequencing revealed a minimal mutational load and no off-target mutagenesis in the corrected iPSCs, and the expression of the HBB gene was also restored in erythrocytes differentiated in vitro. Our gene targeting strategy is a significant step toward the genetic correction of MGDs associated with disease-causing mutants in patient-specific iPSCs and for future clinical stem cell therapies.
Experimental Procedures
Animals and Ethics Statement
The patients in this study provided written informed consent for donating fibroblasts for stem cell generation. The experiments regarding animal research were approved by the Institutional Review Board at The Third Affiliated Hospital of Guangzhou Medical University. The experiments using human cells and mice were approved by the ethics committee of The Third Affiliated Hospital of Guangzhou Medical University, and all animal care and experiments were carried out in accordance with the institutional ethical guidelines for animal experiments.
β-Thal iPSC Generation and Characterization
A skin biopsy was obtained from a β-thal patient with a β-41/42 homozygous 4-bp (TCTT) deletion in exon 2, which is the most common mutation in the Chinese population. Human fibroblasts were cultured in fibroblast medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (HyClone), 1 mM glutamine (Gibco), l% non-essential amino acids (NEAA) (Gibco), and 100 IU/ml penicillin/streptomycin (Gibco). To generate non-integrating iPSCs, fibroblast cells were transduced with the CytoTune-iPS 2.0 Sendai reprogramming kit (Life Technologies, A16517). The transduced cells were then plated onto Matrigel-coated culture dishes according to the manufacturer’s instructions. The β-thal iPSCs used in this study were incubated at 38.5°C for 4 days to completely remove the Sendai viruses and, thus, were free of the reprogramming factors. All of the iPSCs were cultured on Matrigel-coated tissue culture dishes (ES-qualified, BD Biosciences) with mTeSR1 (STEMCELL Technologies) at 37°C and 5% CO2 in a 100% humidified atmosphere incubator. The culture medium was refreshed daily until the cells were ready to passage or harvest. The cells were passaged every 3–4 days using Accutase (STEMCELL Technologies).
Dual Fluorescence Assay for Gene Correction Efficiency
The dual fluorescent report vector pEF1α-mCherry-2A-Δ41/42-EGFP was constructed using an EGFP expression lentivirus vector, pWPSLd. The red fluorescent mCherry cassette was linked to the green fluorescent EGFP cassette with a T2A sequence and a 158-bp β-41/42 HBB genomic sequence. Because of the 4-bp TCTT deletion in the dual fluorescent reporter vector, the EGFP cassette was out of frame and could not be correctly translated; thus, only the red fluorescent protein could be expressed. The dual fluorescent report vector was first packaged in a lentivirus via co-transfection with the packing plasmids in HEK293T cells to optimize the gRNA and ssODN donors for the seamless correction of β-41/42. The iPSCs were infected with the dual fluorescent reporter lentivirus. Seventy days after infection, the red fluorescent cells were sorted using FACS (termed iPSC-Δ41/42-EGFP) and then used to test the efficiency of ssODN-mediated HDR with Cas9/gRNA.
Guide RNAs were designed using a web tool (http://crispr.mit.edu) and followed the rule 5′-GN (18–20) NGG-3′. The oligonucleotides were purchased from Invitrogen and inserted into the pX458 vector according to the protocol from Dr. Zhang’s laboratory. Briefly, the pX458 vector expressing Cas9 and the single guide RNA (sgRNA) were linearized with BbsI digestion and gel-purified. A pair of oligos for each targeting site was phosphorylated, annealed, and ligated to the linearized pX458 vector.
The ssODNs were purchased from GENEWIZ and were complementary to the HBB genomic sequence. Approximately 1 × 106 iPSC-Δ41/42-EGFP cells were co-electroporated with 2 μg of ssODN and 8 μg of the Cas9/gRNA plasmid. Five days after transfection, mCherryEGFP-positive cells were observed using a fluorescence microscope and then analyzed using FACS.
Gene Targeting of β-Thal iPSCs by ssODNs and spCas9-HF1
The spCas9-HF1 expression vector was constructed using the wild-type spCas9 expression vector pX458. Briefly, the coding sequence of spCas9-HF1 (N497A, R661A, Q695A, and Q926A) was generated via standard PCR mutagenesis of wild-type spCas9 as reported previously. Following this, the spCas9 was replaced by spCas9-HF1 in the pX458 vector.
For gene targeting, 1 × 106 iPSCs were electroporated with the 8-μg spCas9/HF1vector containing 2 μg ssODN4 template using the Neon transfection system (Thermo Fisher). The cells were then recovered in mTeSR1 medium supplemented with 10 μM Rho-associated kinase (ROCK) inhibitor Y-27632 (10 μM, Sigma) with or without L755507 for 24 hr after electroporation. 48 hr post-transfection, the GFP-positive cells were collected via FACS, plated in 6-well plates, and cultured for 10 days.
Genomic DNA was extracted using the TIANamp genomic DNA kit (Tiangen) for PCR analysis. Approximately 100–200 ng of the genomic DNA templates and LA Taq (Takara) were used in all PCRs. PCR screening primers were as follows. The forward primer (F: 5′ ACGGCTGTCATCACTTAGACCT3′) was located 430 bp upstream of the mutant site, and the reverse primer (R: 5′ TCCCCTTCCTATGACATGAACT3′) was located 243 bp downstream of the mutant site. Primers RWT (5′ TCCCCAAAGGACTCAAAGAACC 3′) and RΔ (5′ AGATCCCCAAAGGACTCAACC3′) were allele-specific and located on the mutant site. Then the PCR products (F/R) were sequenced to confirm the corrected clones.
Immunofluorescence Staining
Immunofluorescence (IF) staining was performed using primary antibodies (all at 1:200 dilutions) to detect OCT4 (Abcam), SOX2 (Abcam), and SSEA-4 (Abcam). The nuclei were stained with DAPI at a final concentration of 0.01 mg/mL for 10 min.
Karyotype Analysis
For the chromosome analysis, iPS cells were incubated in culture medium with 0.25 mg/mL colcemid (Gibco, Invitrogen) for 4 hr, harvested, and incubated in 0.4% sodium citrate and 0.4% chloratum Kaliumat (1:1, v/v) at 37°C for 5 min. The cells were then fixed three times in a methanol:acetic acid solution (3:1, v/v). Subsequently, after Giemsa staining, at least 20 cells were examined in each group for the chromosome analysis.
Teratoma Formation
The iPSCs from a confluent 10-cm plate were harvested by digestion with 2 mg/ml dispase, resuspended in Matrigel, and injected into the inguinal grooves of 6-week-old male SCID mice. Eight weeks later, the resulting tumors were removed, fixed for 4–8 hr in 4% paraformaldehyde, and embedded in paraffin. After staining with H&E, the sections were examined using a light microscope for the presence of derivatives from the three germ layers.
DNA Fingerprinting Analysis
For the DNA fingerprinting analysis, genomic DNA was extracted from two corrected iPSC-C1, iPSC-C2, and the patient’s iPSCs. The extracted DNA was amplified for 15 different genetic loci using the Promega PowerPlex 16 system kit (Promega). Capillary electrophoresis was performed on an automated ABI 3100 genetic analyzer (Applied Biosystems).
Exome Sequencing
We used SeqCap EZ Exome 64M (Roche NimbleGen) and a TruSeq DNA sample preparation kit (Illumina) to capture the exome and establish the exome sequencing library according to the manufacturers’ instruction manuals. All sequencing was performed on an Illumina HiSeq2000 sequencer with a paired end 2× 150-nt multiplex.
Hematopoietic Differentiation
iPSCs were harvested via treatment with 2 mg/ml dispase (Invitrogen) and co-cultured with OP9 stromal cells at an approximate density of 5 × 106 cells/10-cm dish in 20 mL of MEM Alpha (Gibco) supplemented with 10% FBS (HyClone), 100 mM monothioglycerol (MTG, Sigma), and 100 μM vitamin C. The co-cultures of OP9 cells and pluripotent cells were incubated for 8 days; half of the medium was replaced on days 4 and 6. Differentiated hiPSCs were harvested on day 9. The CD34+/CD31− cells were sorted using a direct CD34+ progenitor cell isolation kit (Miltenyi Biotec).
Statistical Analysis
All statistical analyses were performed using SPSS 19.0 software to detect significant differences in measured variables among groups. A value of p < 0.05 was considered to indicate a statistically significant difference.
Author Contributions
Y.F., L.B., and X.L. designed the study. Y.L., Y.Y., X.K., B.L., Q.Y., G.G., and B.S. performed the experiments. Y.C. and X.S. contributed reagents/materials. Y.F., L.B., and X.L. analyzed the data and wrote the paper. Y.F. provided administrative support and final approval of the manuscript.
Conflicts of Interest
The authors have declared that no competing interests exist.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (81370766, 81401254, 81570101, 81671121, and 31601187), Guangdong Province Higher Education Funding (Yq2013135), the Guangdong Province Science and Technology Project (2014TQ01R683, 2014A02011029, 2016A020216023, and 2015A030310119), and the Bureau of Science and Technology of Guangzhou Municipality (201505011111498).Y.F. was supported by State 863 Project 2015AA020307. L.B. was supported by NYU startup funding.
Footnotes
Supplemental Information includes two tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2016.11.010.
Contributor Information
Xiaoping Li, Email: li_xiaoping@gibh.ac.cn.
Lei Bu, Email: lei.bu@med.nyu.edu.
Yong Fan, Email: yongfan011@gzhmu.edu.cn.
Supplemental Information
References
- 1.Rund D., Rachmilewitz E. β-thalassemia. N. Engl. J. Med. 2005;353:1135–1146. doi: 10.1056/NEJMra050436. [DOI] [PubMed] [Google Scholar]
- 2.Lucarelli G., Gaziev J. Advances in the allogeneic transplantation for thalassemia. Blood Rev. 2008;22:53–63. doi: 10.1016/j.blre.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Bernardo M.E., Zecca M., Piras E., Vacca A., Giorgiani G., Cugno C., Caocci G., Comoli P., Mastronuzzi A., Merli P. Treosulfan-based conditioning regimen for allogeneic haematopoietic stem cell transplantation in patients with thalassaemia major. Br. J. Haematol. 2008;143:548–551. doi: 10.1111/j.1365-2141.2008.07385.x. [DOI] [PubMed] [Google Scholar]
- 4.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woods N.-B., Bottero V., Schmidt M., von Kalle C., Verma I.M. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440 doi: 10.1038/4401123a. 1123–1123. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Zheng C.G., Jiang Y., Zhang J., Chen J., Yao C., Zhao Q., Liu S., Chen K., Du J. Genetic correction of β-thalassemia patient-specific iPS cells and its use in improving hemoglobin production in irradiated SCID mice. Cell Res. 2012;22:637–648. doi: 10.1038/cr.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma N., Shan Y., Liao B., Kong G., Wang C., Huang K., Zhang H., Cai X., Chen S., Pei D. Factor-induced Reprogramming and Zinc Finger Nuclease-aided Gene Targeting Cause Different Genome Instability in β-Thalassemia Induced Pluripotent Stem Cells (iPSCs) J. Biol. Chem. 2015;290:12079–12089. doi: 10.1074/jbc.M114.624999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma N., Liao B., Zhang H., Wang L., Shan Y., Xue Y., Huang K., Chen S., Zhou X., Chen Y. Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free β-thalassemia induced pluripotent stem cells. J. Biol. Chem. 2013;288:34671–34679. doi: 10.1074/jbc.M113.496174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie F., Ye L., Chang J.C., Beyer A.I., Wang J., Muench M.O., Kan Y.W. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24:1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu P., Tong Y., Liu X.Z., Wang T.T., Cheng L., Wang B.Y., Lv X., Huang Y., Liu D.P. Both TALENs and CRISPR/Cas9 directly target the HBB IVS2-654 (C > T) mutation in β-thalassemia-derived iPSCs. Sci. Rep. 2015;5:12065. doi: 10.1038/srep12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L., Guell M., Byrne S., Yang J.L., De Los Angeles A., Mali P., Aach J., Kim-Kiselak C., Briggs A.W., Rios X. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu C., Liu Y., Ma T., Liu K., Xu S., Zhang Y., Liu H., La Russa M., Xie M., Ding S., Qi L.S. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshimi K., Kunihiro Y., Kaneko T., Nagahora H., Voigt B., Mashimo T. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat. Commun. 2016;7:10431. doi: 10.1038/ncomms10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Wang K., Wu H., Jin Q., Ruan D., Ouyang Z., Zhao B., Liu Z., Zhao Y., Zhang Q. Genetically humanized pigs exclusively expressing human insulin are generated through custom endonuclease-mediated seamless engineering. J. Mol. Cell Biol. 2016;8:174–177. doi: 10.1093/jmcb/mjw008. [DOI] [PubMed] [Google Scholar]
- 18.Orlando S.J., Santiago Y., DeKelver R.C., Freyvert Y., Boydston E.A., Moehle E.A., Choi V.M., Gopalan S.M., Lou J.F., Li J. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin Y., Gao W.Q. Concise Review: Patient-Derived Stem Cell Research for Monogenic Disorders. Stem Cells. 2016;34:44–54. doi: 10.1002/stem.2112. [DOI] [PubMed] [Google Scholar]
- 20.Finotti A., Breda L., Lederer C.W., Bianchi N., Zuccato C., Kleanthous M., Rivella S., Gambari R. Recent trends in the gene therapy of β-thalassemia. J. Blood Med. 2015;6:69–85. doi: 10.2147/JBM.S46256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore J.C., Atze K., Yeung P.L., Toro-Ramos A.J., Camarillo C., Thompson K., Ricupero C.L., Brenneman M.A., Cohen R.I., Hart R.P. Efficient, high-throughput transfection of human embryonic stem cells. Stem Cell Res. Ther. 2010;1:23. doi: 10.1186/scrt23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Z., Verma N., González F., Shi Z.-D., Huangfu D. ACRISPR/Cas-mediated selection-free knockin strategy in human embryonic stem cells. Stem Cell Reports. 2015;4:1103–1111. doi: 10.1016/j.stemcr.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song J., Yang D., Xu J., Zhu T., Chen Y.E., Zhang J. RS-1 enhances CRISPR/Cas9-and TALEN-mediated knock-in efficiency. Nat. Commun. 2016;28:10548. doi: 10.1038/ncomms10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.