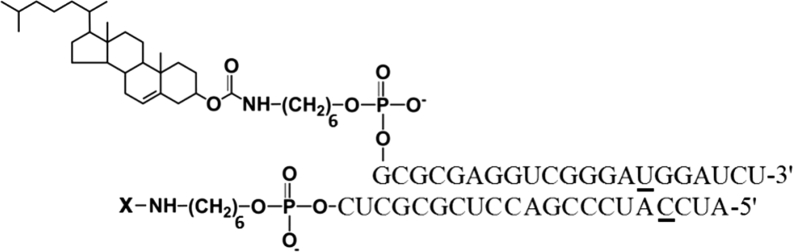Abstract
Chemical modifications are an effective way to improve the therapeutic properties of small interfering RNAs (siRNAs), making them more resistant to degradation in serum and ensuring their delivery to target cells and tissues. Here, we studied the carrier-free biodistribution and biological activity of a nuclease-resistant anti-MDR1 cholesterol-siRNA conjugate in healthy and tumor-bearing severe combined immune deficiency (SCID) mice. The attachment of cholesterol to siRNA provided its efficient accumulation in the liver and in tumors, and reduced its retention in the kidneys after intravenous and intraperitoneal injection. The major part of cholesterol-siRNA after intramuscular and subcutaneous injections remained in the injection place. Confocal microscopy data demonstrated that cholesterol-siRNA spread deep in the tissue and was present in the cytoplasm of almost all the liver and tumor cells. The reduction of P-glycoprotein level in human KB-8-5 xenograft overexpressing the MDR1 gene by 60% was observed at days 5–6 after injection. Then, its initial level recovered by the eighth day. The data showed that, regardless of the mode of administration (intravenous, intraperitoneal, or peritumoral), cholesterol-siMDR efficiently reduced the P-glycoprotein level in tumors. The designed anti-MDR1 conjugate has potential as an adjuvant therapeutic for the reversal of multiple drug resistance of cancer cells.
Keywords: cholesterol-containing siRNA, MDR1, 2’-O-methyl modification, tumor xenograft in SCID mice, multiple drug resistance, biodistribution of siRNA
Introduction
RNAi gene silencing technology has strong potential in biomedical applications for a broad range of diseases, from genetic disorders to cancer. Currently, challenges associated with nuclease stability and achieving efficient delivery to target cells and tissues significantly limit the biomedical applications of small interfering RNA (siRNA). Many of these restrictions could be resolved with the use of chemical modifications improving siRNA properties. It has been shown that the stability of siRNAs to nucleases present in blood serum and the cytoplasm of cells can be increased by using chemically modified analogs affecting the 2′ position of ribose, the 5′-terminal phosphate, and internucleoside phosphates.1, 2, 3, 4 The ability of cholesterol,5 α-tocopherol,6 aptamers,7 antibodies,8 and cell-penetration peptides9, 10 to alter the bioavailability and biodistribution of siRNA was demonstrated in several studies. Nevertheless, efficient gene silencing by conjugates could be achieved when they are used at high concentration, which is associated with retracting them from the bloodstream by renal filtration and trapping the drug that has entered the target cell in the endosomes.11 Combining different types of chemical modification in a given siRNA molecule could lead to the development of highly efficient modification patterns, tailored for specific applications.
Multidrug resistance of tumors presents a significant problem in the treatment of various tumor diseases.12, 13 The mechanisms of the emergence and development of such resistance include transportation of the drugs out of tumor cells reducing their effective concentration,14 changes in the efficiency of drug biotransformation,15, 16 accelerated repair of DNA damage in cells,17 apoptosis blocking,18 and alterations in the molecular targets.19 The most common cause of multidrug resistance in cancer chemotherapy is overexpression of the MDR1 gene that encodes the ATP-binding cassette (ABC) transporter family member P-glycoprotein.20 P-glycoprotein is a transmembrane ATP-dependent pump effluxing a wide spectrum of hydrophobic compounds from cells, including drugs commonly used in tumor therapy.21 A direct and universal approach to overcoming tumor drug resistance is to block the expression of genes encoding proteins causing the resistance with the help of siRNA, acting via the mechanism of RNAi.22
Previously, we proposed an experimental algorithm for the site-specific modification of siRNAs, based on mapping of their nuclease-sensitive sites in the presence of serum followed by incorporation of 2′-O-methyl analogs of ribonucleotides at the identified positions of cleavage.23 We demonstrated that protection of nuclease-sensitive sites considerably enhanced nuclease resistance of siRNA in the presence of serum and increased the duration of the gene-silencing effect. Based on RNA resistant to nucleases, we designed conjugates of anti-MDR1 siRNA with cholesterol attached through an optimized linker that are capable of penetrating efficiently into cells in a carrier-free mode, to silence the expression of P-glycoprotein and to restore the sensitivity of drug-resistant cancer cells to vinblastine.24
Within this study, we investigated carrier-free biodistribution and gene-silencing activity of cholesterol-containing conjugates of nuclease-resistant anti-MDR1 siRNA in healthy and tumor-bearing mice. Because of conjugation with cholesterol, these conjugates of nuclease-resistant siRNAs were able to accumulate mostly in the liver and in the tumors of mice, and to silence expression of the target MDR1 gene after intravenous, intraperitoneal, and peritumoral administration. The designed anti-MDR1 conjugates have great potential for the reversal of multiple drug resistance of cancer cells.
Results
In vivo, a number of additional factors constitute obstacles to the achievement of the desired biological effect of siRNA as compared with cell culture. These are degradation of siRNA by serum nucleases, binding of siRNA with serum protein, siRNA accumulation in non-target organs and tissues, excretion by the kidneys, low bioavailability, and inefficient silencing within the cells.11 To study the performance of anti-MDR1 siRNA and its conjugate with cholesterol in vivo, we chose siRNA targeted to the 411–431 nt region of human MDR1 mRNA (siMDR), shown in our previous study to have the highest bioactivity in cell culture.25 2′-O-methyl modifications were introduced into nuclease-sensitive sites according to the algorithm developed by us previously.23 These selective modifications prevent degradation of carrier-free siRNA in the bloodstream. The structure of the lipophilic conjugate containing cholesterol attached to the 5′ end of the sense strand of the molecule via optimized aminohexyl linker (Figure 1) was selected by analogy to our previous studies.24 This cholesterol-containing siRNA (Ch-siRNA) displayed optimal carrier-free uptake and gene-silencing activity in the cell culture experiments.
Figure 1.
Structure of Conjugates of siMDR and Cholesterol: Ch-siMDR
2′O-methyl (2′OMe) nucleotides are underlined. X, Cy5.5 or Cy7 conjugated via aminohexyl linker.
Ch-siRNA Biodistribution in Healthy Mice and the Effect of the Mode of Administration
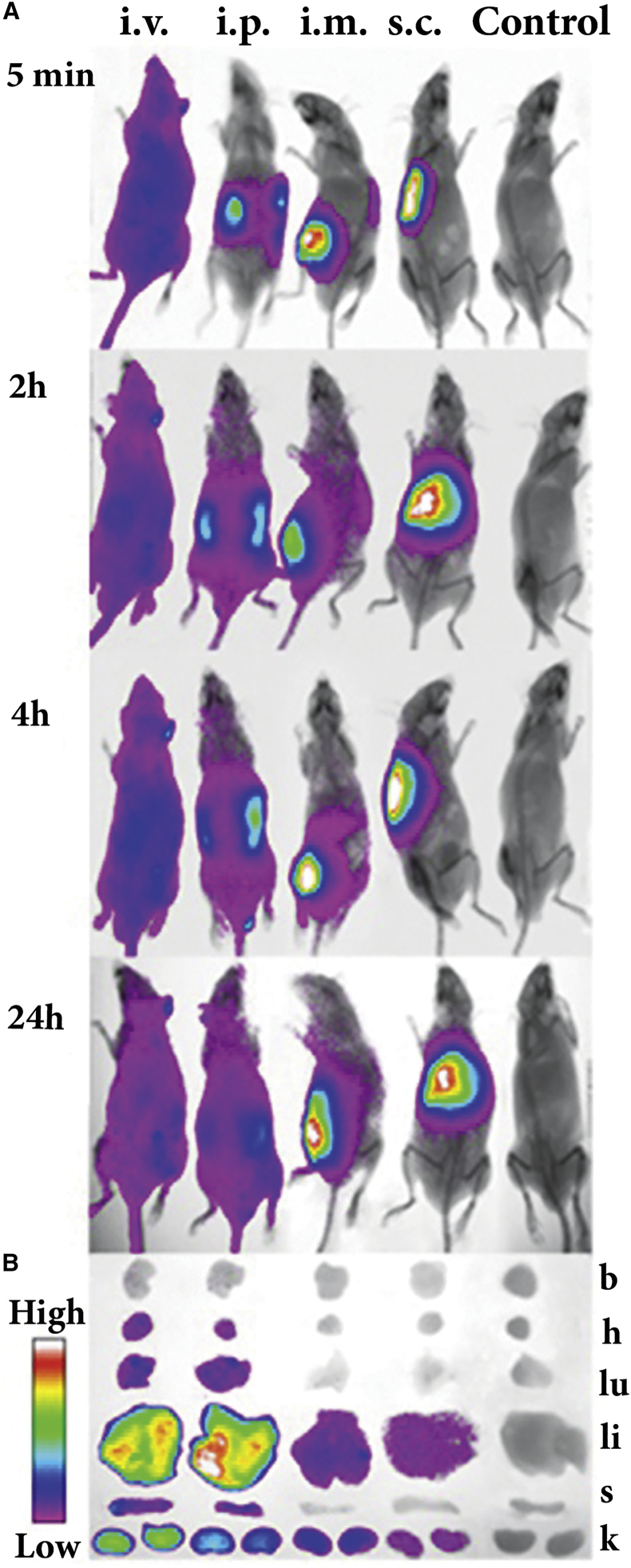
In the first stage, we investigated biodistribution of Ch-siRNA and the impact of the mode of its administration on the biodistribution pattern in healthy severe combined immune deficiency (SCID) mice. Equal amounts of Ch-siRNA bearing Cy7 at the 3′ end of the antisense strand were delivered into mice by intravenous (i.v.), intraperitoneal (i.p.), intramuscular (i.m.), and subcutaneous (s.c.) injections. The dosage of Ch-RNA-Cy7 (1.7 μg/g) was adjusted to get a high fluorescence signal. In vivo multispectral fluorescent imaging analysis was used to evaluate the dynamics of the biodistribution of Ch-siRNA in the mouse body. We showed (Figure 2A) that Ch-siRNA after i.v. injection rapidly spread throughout the mouse in the bloodstream, and 5 min after injection the fluorescent signal was detected from the whole body. With longer follow-up (Figure 2A), the distribution changed insignificantly, and a slight decrease in the total fluorescence of the body 24 hr post-injection (i.p.) could be observed. Shortly after i.p. injection, fluorescence was associated with the abdomen and later spread slowly throughout the body: 2–4 hr after i.p. injection, the lower part of the body displayed Ch-siRNA accumulation, and 24 hr after i.p. injection, the distribution pattern was similar to that of i.v.-injected mice. Ch-siRNA after i.m. and s.c. injection remained in the place of injection; the fluorescent zone increased slightly up to 2 hr after administration and then did not change significantly up to 24 hr (Figure 2A).
Figure 2.
Effect of Administration Mode on Biodistribution of Cy7-Labeled Ch-siRNA in Healthy SCID Mice
(A) In vivo fluorescence imaging of healthy SCID mice after i.v., i.p., i.m., and s.c. injection of Ch-siRNA at different time points (dorsal orientation view). Control, non-injected mice. (B) Images of dissected organs of SCID mice sacrificed 24 hr after injection. b, brain; h, heart; k, kidney; li, liver; lu, lungs; s, spleen.
Accumulation of Ch-siRNA in the internal organs was determined by measuring fluorescent signals from organs dissected 24 hr after administration. The data showed (Figure 2B) that 24 hr after i.v. and i.p. injections, the total amount of Ch-siRNA accumulated in the internal organs was similar: 466 relative fluorescence units (RFUs) after i.v. and 468 RFU after i.p. injection. The total amount of fluorescence accumulated in the internal organs was obtained by summing fluorescence from the brain, heart, lungs, liver, spleen, and kidneys, measured in RFU. The patterns of Ch-siRNA distribution between organs were similar: the major part of Ch-siRNA accumulated in the liver: 78% (i.v.) and 88% (i.p.), a moderate amount of Ch-siRNA accumulated in the kidneys: 18% (i.v.) and 9.3% (i.p.), and some Ch-siRNA was detected in the heart (0.1%–0.6%), lungs (1.5%–1.8%), and spleen (0.6%–1.6%) (Table 1).
Table 1.
Ch-siRNA-Cy7 Accumulation in the Internal Organs of Healthy SCID Mice 24 hr after Administration
| Total Fluorescence of the Organ (RFU) |
Total Florescence of the Organ Relative to the Total Fluorescence of All Organs (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| i.v. | i.p. | i.m. | s.c. | i.v. | i.p. | i.m. | s.c. | |
| Brain | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Heart | 2.7 ± 0.9 | 0.7 ± 0.4 | 0 | 0 | 0.6 ± 0.2 | 0.1 ± 0.1 | 0 | 0 |
| Lungs | 7.2 ± 2.8 | 8.6 ± 2.0 | 0 | 0 | 1.5 ± 0.6 | 1.8 ± 0.4 | 0 | 0 |
| Liver | 364 ± 45 | 412 ± 69 | 26 ± 5 | 8.1 ± 2.1 | 78 ± 10 | 88 ± 15 | 58 ± 12 | 80 ± 21 |
| Kidneys | 85 ± 18 | 44 ± 18 | 18 ± 4 | 2.0 ± 0.8 | 18 ± 4 | 9.3 ± 2.5 | 42 ± 9 | 20 ± 18 |
| Spleen | 7.7 ± 1.5 | 3.0 ± 1.5 | 0 | 0 | 1.6 ± 0.7 | 0.6 ± 0.3 | 0 | 0 |
| Total | 466 ± 70 | 468 ± 85 | 44 ± 9 | 10.1 ± 2.9 | 100 | 100 | 100 | 100 |
Mean ± SD (n = 3). RFU, relative fluorescence unit.
In order to evaluate the amount of intact Ch-siRNA in the internal organs of mice (liver and kidneys), we used stem-loop PCR. We showed that 24 hr after i.v. injection, intact antisense strand of Ch-siRNA is detected in liver (68.4 pmol) and kidneys (23.2 pmol). The data showed that the ratio of amounts of Ch-siRNA in liver and kidneys is similar to the ratio of data obtained by In-Vivo Imaging System (78% and 18%, respectively).
Accumulation of Ch-siRNA after i.m. and s.c. injections in the internal organs was very low; weak fluorescence signals were detected in the liver and kidneys of the animals, and total fluorescence of all investigated organs after i.m. and s.c. injection of Ch-siRNA was 10 and 46 times lower, respectively, than that after i.v. injection of the same amount of Ch-siRNA. Because i.v. administration of Ch-siRNA resulted in fast distribution and reliable accumulation in different organs, we selected this method of administration for the further experiments.
Biodistribution of siRNA and Ch-siRNA in Tumor-Bearing Mice
Because i.v. and i.p. injections caused similar patterns of Ch-siRNA accumulation, in the next stage of the study we compared the biodistribution of Ch-siRNA, siRNA, and siRNA complexed with Lipofectamine 2000 (siRNA/Lipofectamine) in an SCID mice xenograft tumor model after i.v. administration through the tail vein. KB-8-5 human squamous carcinoma cells were chosen to induce tumors, because these cells overexpress MDR1 mRNA encoding P-glycoprotein responsible for the drug resistance phenotype. We proved the ability of non-lipophilic siMDR used in the current study to silence the expression of this gene and to restore the sensitivity of the cancer cells to vinblastine in experiments on KB-8-5 cells.26
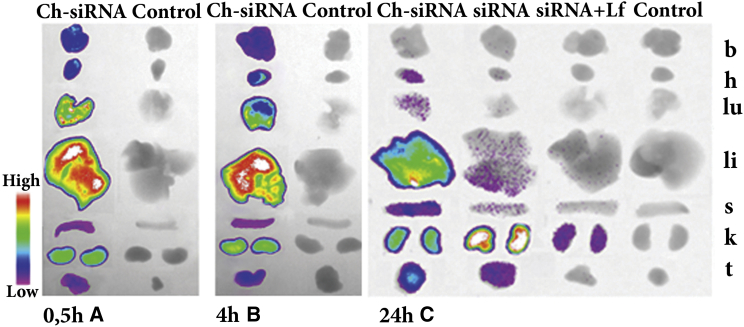
Tumors in mice were initiated as described in the Materials and Methods; when the tumor volume reached approximately 1 cm3, in each experiment three tumor-bearing mice were i.v. injected with 1.7 μg/g Cy5.5-labeled Ch-siRNA, siRNA, or siRNA/Lipofectamine; a non-injected tumor-bearing mouse was used as a control. All mice were imaged simultaneously at the indicated time points. The data showed that total accumulation of Ch-siRNA in internal organs was 2.4 times more efficient than accumulation of non-lipophilic siRNA with the same sequence, and almost 50 times more efficient than accumulation of siRNA/Lipofectamine (Figure 3; Table 2). It should be noted that the presence of a tumor in the body did not alter Ch-siRNA accumulation in the internal organs, and the biodistribution patterns were similar in healthy and tumor-bearing mice (Figures 2B and 3A), except that accumulation of Ch-siRNA was observed in the tumors. Data showed that Ch-siRNA was efficiently accumulated in the tumors, and despite their relatively small size, they accumulated about 6% of the Ch-siRNA (Table 2). The accumulation of non-lipophilic siRNA was substantially lower: the amount of accumulated siRNA (fluorescence signal) was 3.5 times less than in the case of Сh-siRNA, and only 4% of accumulated siRNA was in tumors. Because Ch-siRNA and siRNA contained the same antisense strand bearing a fluorescence label, their specific fluorescence was identical. The distribution pattern of siRNA also differed from the distribution of Ch-siRNA: the bulk of siRNA accumulated in the kidney (94%), 4% in the tumor, 1.6% in the liver, and less than 1% in the spleen (Table 2). When siRNA was administered in the complex with Lipofectamine, it accumulated mainly in the kidney (93%), and a small amount of the complex was detected in the liver (6.7%). No fluorescence signal was found in tumors after the injection of siRNA/Lipofectamine. Thus, conjugation of cholesterol to siRNA allowed an increase of siRNA accumulation in the tumor, both in absolute terms and in comparison with the total amount of siRNA accumulated in the internal organs.
Figure 3.
Accumulation of Cy5.5-Labeled Ch-siRNA, Non-modified siRNA, and siRNA/Lipofectamine Complex in the Organs of KB-8-5 Tumor-Bearing SCID Mice
Images of dissected organs and tumors of SCID mice sacrificed at 24 hr (A), 30 min (B), and 4 hr (C) after i.v. injection of corresponding siRNA. b, brain; h, heart; k, kidney; li, liver; lu, lungs; s, spleen.
Table 2.
Accumulation of Cy5.5-Labeled Ch-siRNA, Non-modified siRNA, and siRNA Complexed with Lipofectamine 2000 in the Organs of KB-8-5 Tumor-Bearing SCID Mice after i.v. Injection
| Total Fluorescence of the Organ (RFU) |
Total Florescence of the Organ Relative to the Total Fluorescence of All Organs (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ch-siRNA |
siRNA |
siRNA+Lf |
Ch-siRNA |
siRNA |
siRNA+Lf |
|||||
| 30 min | 4 hr | 24 hr | 24 hr | 24 hr | 30 min | 240 min | 24 hr | 24 hr | 24 hr | |
| Brain | 7 ± 1 | 4.3 ± 1.6 | 0 | 0 | 0 | 2.7 ± 0.5 | 2.1 ± 0.7 | 0 | 0 | 0 |
| Heart | 3.9 ± 1.2 | 2.8 ± 0.6 | 0.6 ± 0.2 | 0 | 0 | 1.4 ± 0.4 | 1.3 ± 0.3 | 0.2 ± 0.1 | 0 | 0 |
| Lungs | 42 ± 8 | 23 ± 6 | 0.4 ± 0.3 | 0 | 0 | 15 ± 3 | 11 ± 3 | 0.1 ± 0.1 | 0 | 0 |
| Liver | 189 ± 27 | 154 ± 20 | 235 ± 54 | 2.1 ± 2.1 | 0.40 ± 0.02 | 70 ± 10 | 73 ± 9 | 76 ± 17 | 1.6 ± 1.6 | 6.7 ± 0.4 |
| Kidneys | 26 ± 4 | 25 ± 4 | 45 ± 6 | 122 ± 19 | 5.6 ± 1.3 | 9.7 ± 1.5 | 12 ± 2 | 15 ± 2 | 94 ± 14 | 93 ± 21 |
| Spleen | 0.5 ± 0.3 | 0.1 ± 0.1 | 8.2 ± 2.6 | 0.2 ± 0.3 | 0 | 0.2 ± 0.1 | 0.04 ± 0.05 | 2.7 ± 0.9 | 0.2 ± 0.2 | 0 |
| Tumor | 1.4 ± 0.5 | 2 ± 0.7 | 19 ± 4 | 5.4 ± 1.8 | 0 | 0.5 ± 0.2 | 0.9 ± 0.3 | 6.1 ± 1.3 | 4.1 ± 1.4 | 0 |
| Total | 270 ± 43 | 211 ± 32 | 308 ± 67 | 130 ± 23 | 6 ± 1.3 | 100 | 100 | 100 | 100 | 100 |
Mean ± SD (n = 3). Lf, Lipofectamine 2000.
We measured the accumulation of Ch-siRNA in the organs at shorter time points after administration, 30 min and 4 hr, to evaluate the dynamics of accumulation (Figures 3B and 3C; Table 2). Time points were selected according to our previously obtained data on the accumulation of Ch-siRNA into KB-8-5 cells.24 It can be seen that maximal Ch-siRNA accumulation was observed in the liver as early as within 30 min after administration, and the further accumulation level barely changed in 24 hr. Accumulation in the kidneys, spleen, and tumor gradually increased with time, with the most significant increase in accumulation occurring in the tumor at 24 hr (Table 2). Fluorescence levels of the brain, heart, and lungs, on the contrary, were maximal at 30 min and gradually reduced to zero, indicating that Ch-siRNA was probably present in the blood that filled these organs at the initial time points.
Thus, the attachment of cholesterol to siRNA provided its efficient accumulation in the liver and in the tumor, and reduced its retention in the kidneys after i.v. injection. Weaker accumulation of free siRNA and lack of accumulation of its complexes with Lipofectamine did not allow assessment of the therapeutic effect of their use.
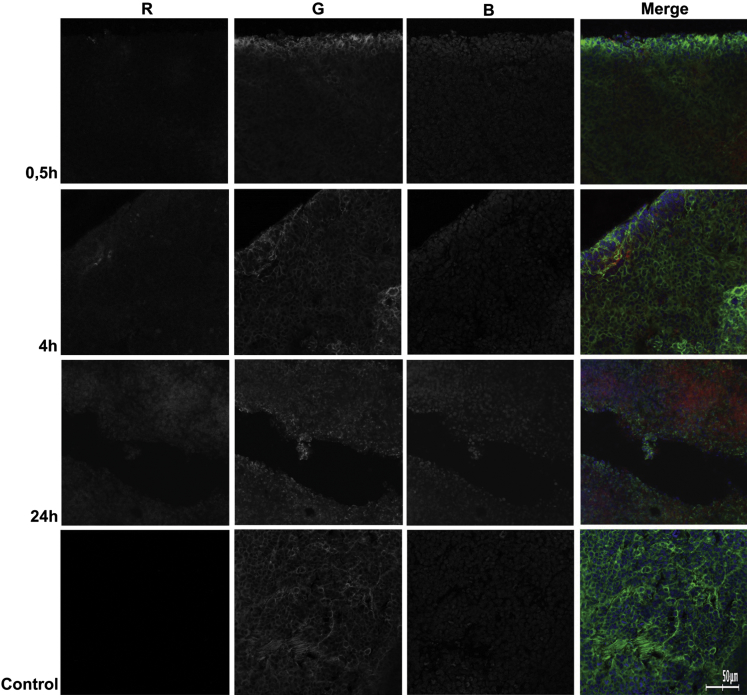
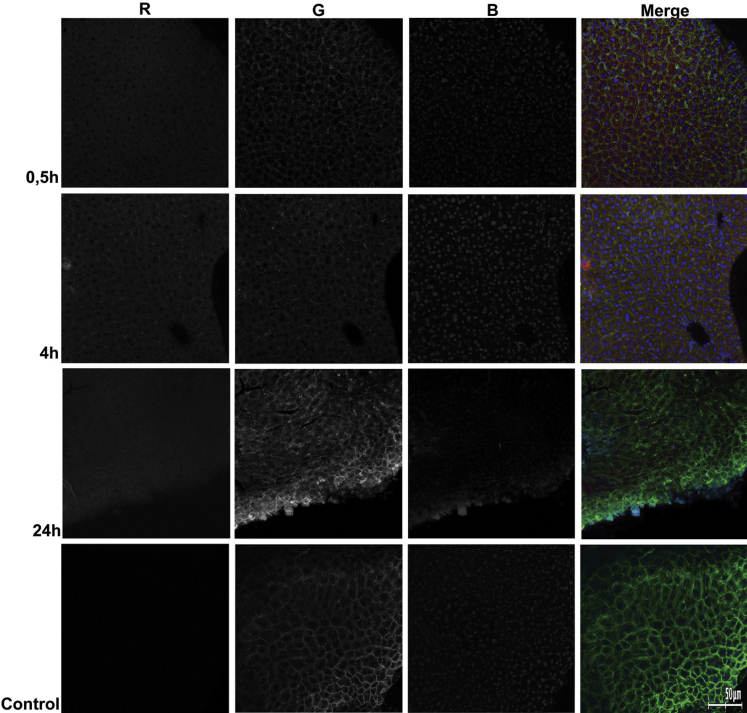
In order to assess how Ch-siRNA was distributed in the organs where its maximum accumulation was detected, we investigated cryosections of these organs by confocal microscopy. We selected 30 min, 4 hr, and 24 hr time points for cryosections of liver and tumor based on our in vivo imaging data (Table 2). The data showed that Ch-siRNA was distributed throughout the tumor volume, and the amount of Ch-siRNA in tumor tissue increased with increasing time after injection until 24 hr, which was consistent with the bioimaging data. Tumors presented as homogeneous round cells. At the 30 min and 4 hr time points, Ch-siRNA was unevenly distributed in the tumor volume. Within the cells, Ch-siRNA was accumulated in the cytoplasm, and Ch-siRNA was absent in the nuclei of cells or was detected in the nuclei in a smaller amount (Figure 4). Areas or even individual cells in the sections that contained higher concentrations of Ch-siRNA could be seen, but they were not visually distinguished from other areas or neighboring cells by morphology. At 24 hr after administration, siRNA distribution in tumor tissue became uniform.
Figure 4.
Accumulation of Ch-siRNA in a KB-8-5 Tumor Xenograft in SCID Mice
Localization of Ch-siMDR was analyzed by confocal fluorescence microscopy at 20× magnification. Three-channel (RGB) pictures were obtained using Cy5.5 (R), attached to Ch-siRNA; actin filaments were stained by TRITC-phalloidin (G), and DNA was stained with DAPI (B).
Total fluorescent signal in the liver cross sections was more intense than in the sections of the tumor and, unlike the tumor, was virtually the same at the time points examined, which also agrees well with the biodistribution data (Table 2; Figure 5). Ch-siRNA was uniformly distributed throughout the tissue of the liver within 30 min after administration. It should be noted that, at this time, an increased amount of Ch-siRNA was observed in the intercellular space than after later observations: at 4 and 24 hr, Ch-siRNA disappeared from the intercellular space and was located mostly in the cytoplasm of the cells. Just as in the tumor, liver cell nuclei were depleted with Ch-siRNA.
Figure 5.
Accumulation of Ch-siRNA in the Liver of KB-8-5 Tumor-Bearing SCID Mice
Localization of Ch-siMDR was analyzed by confocal fluorescence microscopy at 20× magnification. Three-channel (RGB) pictures were obtained using Cy5.5 (R), attached to Ch-siRNA; actin filaments were stained by TRITC-phalloidin (G), and DNA was stained with DAPI (B).
Silencing of P-glycoprotein Expression in the Tumor by Cholesterol-Containing siRNA
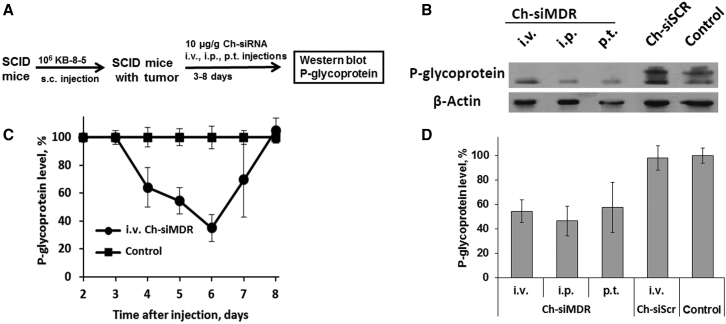
The gene-silencing effect of Ch-siRNA targeting P-glycoprotein (Ch-siMDR) was evaluated in drug-resistant KB-8-5 human xenograft tumors in SCID mice using western blot analysis (Figure 6). Tumors in mice were initiated as described above. When the tumor volume reached approximately 0.3 cm3, three mice per time point per group were i.v. injected with 10 μg/g Ch-siMDR; a non-injected tumor-bearing mouse was used as a control. Because study of the efficacy of treatment with cytostatics, together with the siRNA, was not the aim of this work, we used a single injection of siRNA. The kinetics of the silencing effect of Ch-siMDR on the levels of P-glycoprotein in the tumor were monitored for days 3–8 post i.v. injection (Figure 6A). The starting time point (3 days) was selected taking into account the half-life of P-glycoprotein: 48–72 hr.27 A reduction of P-glycoprotein level relative to controls was observed on the fourth day after siRNA administration, where it had decreased by 40%; at days 5–6, the level continued to decrease, reaching a minimum level (40% of control) on day 6. Then, a gradual increase in P-glycoprotein level recovered to its initial level by the eighth day was observed (Figure 6A). An example of a western blot image representing P-glycoprotein levels in tumor lysates obtained at day 6 after Ch-siRNA i.v. injection is shown in Figure 6B.
Figure 6.
Silencing of P-glycoprotein Expression in KB-8-5 Human Xenograft Tumors in SCID Mice by Ch-siMDR
(A) Experimental scheme. (B) Example of western blot analysis of P-glycoprotein levels in tumor lysates obtained at day 5 after i.v., i.p., and p.t. injection of Ch-siRNA. (C) Kinetics of P-glycoprotein suppression in tumors of SCID mice after i.v. injection of Ch-siMDR. (D) Levels of P-glycoprotein in tumors of SCID mice 5 days after i.v., i.p., and p.t. injection of Ch-siRNA. Human β-actin protein was used as an internal standard. Data were normalized to the ratio of the P-glycoprotein/β-actin levels in the tumors of untreated mice (control). Mean values (±SD) from three independent experiments are shown.
The effect of the route of administration of Ch-siMDR on its biological activity was studied using the same tumor model in SCID mice. For these purposes, Ch-siMDR or Ch-siScr was administered i.v., i.p., or p.t.; then, 5 days after injection, the mice were sacrificed and P-glycoprotein levels in tumors were detected by western blot (Figure 6D). The data showed that, regardless of the mode of administration (i.v., i.p., or peritumorally [p.t.]), Ch-siMDR efficiently reduced the P-glycoprotein level by more than 50% compared with the control. The observed differences in P-glycoprotein silencing levels between i.v., i.p., and p.t. modes of administration were not statistically significant. These data prove that for Ch-siRNA administration, the most comfortable and least invasive method can be chosen.
Discussion
Multidrug resistance is an attractive target for siRNAs, because temporal silencing of the gene responsible for the emergence of resistance, which can be achieved using synthetic siRNA, could overcome tumor resistance to therapy and provide successful treatment.22 Inhibition of MDR1 gene expression in cultures of tumor cells of different origin allows their killing in the presence of previously tolerated concentrations of cytotoxic drugs.16, 28 At the same time, this gene is expressed normally only in a limited number of tissues, and its temporal suppression has no significant negative effect on the entire body. The main problem to be solved in the transition of this effect from cell culture to the level of the organism is to ensure delivery of siRNA to the tumor in amounts sufficient to inhibit expression of the target gene. For siRNA delivery at the body level, different approaches are used: siRNA delivery in complexes with cationic lipids or cationic polymers,29 use of different types of nanoparticle30, 31 and conjugation of siRNA with transport molecules such as cholesterol,5 folate,32, 33 transferrin,34 peptides,35, 36, 37 or aptamers.38, 39
Application of different siRNA complexes with lipids and polymers faces with the need to address different requirements for effective formation of the complex, its stability in serum, and effective release from the endosome, which necessitates construction of multilevel structures.40 The issue of toxicity is also very important.41 Phagocytic uptake by the mononuclear phagocyte system in the bloodstream is a major challenge when using nanoparticles for siRNA delivery, which forces the use of chemical shields (for example, polyethylene glycol [PEG]) to prevent recognition.42 Application of siRNA conjugates with a different transport molecule covalently attached to the siRNA eliminates the need for complexation with liposomes and polymers with a high density of positive charge, which are usually toxic.43 Progress in improving the stability of the siRNA to nuclease cleavage using chemical modification makes redundant such a function of complexes as a protection against the action of ribonucleases.44 The most successful examples of such conjugates have been obtained using molecules with affinity for the target organ, e.g., antibodies, aptamers, and receptor ligands.44 Published data show that, in some cases, attaching addressing groups to siRNA provides effective accumulation in target cells without biological activity,45 and the way from empirical screening to a rational choice of such groups is not obvious. In the present study, we used siRNA conjugated with cholesterol, which does not have a specific target among the organs and tissues. The cholesterol transport system is active in all cells of the body, but the transportation of its conjugates with oligonucleotides is carried out differently.46, 47 Conjugation of cholesterol to siRNA has been used in a number of studies;5, 9, 47, 48, 49, 50, 51, 52, 53 however, linker optimization and sustainability of nuclease action were necessary to ensure the effective delivery of such conjugates in cells in a carrier-free mode and to maintain their biological activity.23, 24
After i.v. injection, siRNA is subjected to renal clearance, and increasing its molecular weight by attachment of ligands, incorporation into larger particles, or binding to plasma proteins efficiently saves siRNA from elimination.54 Our results showed that the attachment of cholesterol to siRNA altered its distribution between internal organs: accumulation in the kidneys was significantly reduced, whereas total accumulation and accumulation in the liver were enhanced (Figure 3; Table 2). We can assume that this was accompanied by an increase in the circulation time of siRNA in the bloodstream, which was indirectly confirmed by the rapid accumulation of the fluorescent signal in organs with a good blood filling—brain, heart, and lungs—and then its gradual disappearance. These data are in good agreement with the results of Wolfrum et al.,47 who showed that Ch-siRNA in mouse bloodstream was present in the unbound fraction (free) and in fractions that contained either high-density lipoprotein (HDL) particles or plasma proteins of ∼60 kDa. On the contrary, unconjugated siRNA was detected only in the unbound fractions. It should be noted that the kidney filtration limit for nucleic acids was found to be 20–40 kDa.54 Slowing excretion by the kidneys allows improved siRNA accumulation in the tumor because of the enhanced permeability and retention (EPR) effect, and in other organs through the endothelium vasculature depending on capillary pore size.55 Our data showed that Ch-siRNA distribution in mouse internal organs does not depend on the presence or absence of a tumor (Figures 2 and 3) and apparently depends only on the conjugate nature, concentration of siRNA, and its circulation time in the bloodstream.
The speed of action of conventional medicines administered by i.p. injection approaches that of the i.v. mode of injection. Our data showed that the same dependence was observed for siRNA: 24 hr after injection, the siRNA distribution pattern and efficiency of accumulation in internal organs differed slightly (Figure 2; Table 1).
Despite the fact that i.v. administration is most often used for therapeutic nucleic acids, local methods for siRNA administration are actively studied in order to achieve a local effect, to prevent systemic toxicity, and to reduce the required dose of the drug.56 For example, siRNA administered via local routes, such as intravitreal injection, subconjunctival injection, or topical instillation, has been found to be effective in the treatment of ocular diseases.57, 58 One of the most advanced therapeutic siRNA conjugates, equipped with three N-acetylgalactosamine molecules developed by Alnylam for treatment of transthyretin-mediated amyloidosis, is intended for subcutaneous administration.44 Our data showed that, after subcutaneous or intramuscular administration, Ch-siRNA remained for a relatively long time at the injection site, did not extend in a substantial amount to the bloodstream, and, therefore, did not accumulate in the internal organs (Figure 2; Table 2).
Efficient siRNA accumulation in the organ alone is not sufficient for effective gene silencing; its dissemination to all parts of the target tissue and its penetration from capillaries and the intercellular space inside the cells is important for successful silencing. Correlations of lipophilicity to delivery efficiency in cells and biological effects are rather complicated. In the study of Wolfrum et al.,47 it was shown that siRNA conjugated with more lipophilic molecules than cholesterol such as docosanyl (C22) and stearoyl (C18), which show stronger binding to HDLs, better accumulated in the cells, and more effectively inhibited target gene expression in the liver. Other authors52 have used siRNA conjugates containing varying numbers of cholesterol residues (0–3) and showed that the conjugate with one cholesterol more effectively inhibited target gene expression in the liver than the conjugate with two cholesterols, and only pre-formation of the complex with albumin reversed the situation. Previously, we studied the carrier-free cellular uptake of nuclease-resistant siRNA equipped with lipophilic residues (cholesterol, lithocholic acid, oleyl alcohol, and lithocholic acid oleylamide) attached to the 5′ end of the sense strand via oligomethylene linkers of various length in different tumor cell lines, and demonstrated that cholesterol-conjugated siRNAs with linkers containing from 6 to 10 carbon atoms demonstrated optimal uptake and gene-silencing properties: shortening of the linker reduced the efficiency of cellular uptake of siRNA conjugates, whereas lengthening of the linker facilitated uptake but retarded the gene-silencing effect and decreased silencing efficiency.24 To conduct the current study, we initially chose siRNA that had previously shown a good ability to penetrate into KB-8-5 cells in culture without the aid of a carrier and to exit from the endosomes into the cytoplasm of cells.24 In this study, we demonstrated that Ch-siRNA was able to spread deep into the tissue and to be present in almost all cells of the liver (Figure 4) and the tumor (Figure 5). Initially, after 30 min to 4 hr, siRNA accumulation in the intercellular space and a higher concentration near the surface of the tumor or capillaries could be seen; then, after 24 hr, distribution became uniform and siRNA was located mainly in the cytoplasm of the cell.
Most of the known ways of siRNA internalization into cells involve the participation of receptor-mediated or adsorption endocytosis, as well as macropinocytosis. In all cases, siRNA within the cell is isolated from the cytoplasm by the endosomal membrane.40 Endosomal escape is a limiting step in achieving effective gene silencing.59 It has been suggested that endosomal escape should occur before late endosomes fuse with lysosomes, which contain biopolymer-degrading enzymes.60 The lack of a dotted pattern of siRNA distribution in tissue cross sections testifies in favor of the theory that Ch-siRNA is not trapped in the endosomes or lysosomes, but is distributed in the cytoplasm, which is important for achieving biological effect (Figures 4 and 5). Cytoplasmic localization of siRNA was also confirmed by the biological activity data: after a single i.v. injection of siRNA, the level of P-glycoprotein in the tumor was reduced by 40%–60% 4–7 days after injection, with a maximum decrease at day 6 (Figure 6). Such efficiency and duration of P-glycoprotein reduction are therapeutically significant, because clinically observed overexpression levels of the MDR1 gene that lead to tumor drug resistance are 1.7- to 2.3-fold higher than levels in drug-sensitive tumors.61 The administration mode (i.v., i.p., or p.t.) had little or no impact on the efficiency of gene silencing, providing a wide selection to determine the best treatment strategy in each case (Figure 6). In cases of known tumor localization, peritumoral administration of Ch-siRNA could reduce the load on the body and provide more targeted action, whereas i.v. or i.p. administration could be used in cases when the presence of metastasis could be expected.
In conclusion, the attachment of cholesterol to siRNA provided its efficient accumulation in the liver and reduced its retention in the kidneys, both in healthy and tumor-bearing mice after i.v. and i.p. administration. Cholesterol-containing selectively 2′-O-methyl-modified siRNA after i.v., i.p., or p.t. injection was able to overcome all barriers from the injection site to the cytoplasm of tumor cells, accumulated there in sufficient amounts, and suppressed expression of the target gene. Cholesterol-containing nuclease-resistant siRNAs provide a promising tool for the development of anticancer RNAi-based therapeutics for overcoming multidrug resistance of tumors.
Materials and Methods
Synthesis of siRNAs and Conjugates
The sense and antisense strands of siRNA were synthesized on an automatic ASM-800 synthesizer using solid-phase phosphoramidite synthesis protocols10, 62 optimized for the instrument, with a 10 min coupling step for 2′-O-TBDMS-protected phosphoramidites and a 6 min coupling step for 2′-O-methylated phosphoramidites. A C6 CPG 3′-PT-amino-modifier (Glen Research) was used for synthesis of the 3′-aminohexyl-containing antisense strand. The following siRNAs were used in the present study (2′-О-methyl-modified С and U nucleotides are designated as Cm and Um): siMDR, homologous to region 411–431 nt of mRNA of the human MDR1 gene (sense strand 5′-GCGCGAGGUCGGGAUmGGAUCU-3′; antisense strand 5′-AUCCmAUCCCGACCUCGCGCUC-3′), and siScr, with no significant homology to any known mouse, rat, or human mRNA sequences (sense strand 5′-GCUUGAAGUCUUUmAAUUmAAGG-3′; antisense strand 5′-UUmAAUUmAAAGACUUCmAAGCGG-3′).
A combination of H-phosphonate and phosphoramidite methods was applied to synthesize 5′-cholesterol conjugates of siRNAs as described by us previously.24 Cyanine5.5 (Cy5.5) or Cyanine7 (Cy7) was attached to the 3′ end of the antisense strand of siRNA equipped with a 3′-aminohexyl linker according to the manufacturer’s protocol using Cy5.5 or Cy7 N-hydroxysuccinimide esters (Biotech Industry) in 0.1 M Tris buffer (pH 8.4). Isolation of the oligoribonucleotides and their conjugates from reaction mixtures was accomplished by denaturing polyacrylamide gel (dPAAG). The purified oligoribonucleotides were characterized by electrophoretic mobility in 12% dPAAG and by MALDI-TOF-MS and liquid chromatography-electrospray ionization mass spectrometry (LC-ESI-MS). siRNAs were obtained by annealing 300 μM sense and antisense strands in buffer containing 30 mM HEPES-potassium hydroxide (KOH) (pH 7.4), 100 mM sodium acetate, and 2 mM magnesium acetate, and were stored at −20°C.
Mice and Tumor Model
All animal procedures were carried out in strict accordance with the recommendations for proper use and care of laboratory animals (ECC Directive 86/609/EEC). The protocol was approved by the Inter-Institute Bioethics Commission of the Siberian Branch of the Russian Academy of Sciences (SB RAS) (22.11 from 30.05.2014). The experiments were conducted in the Center for Genetic Resources of Laboratory Animals at the Institute of Cytology and Genetics, SB RAS (RFMEFI61914X0005 and RFMEFI62114Х0010). Eight- to ten-week-old female SCID (SHO-PrkdcscidHrhr) mice with an average weight of 20–22 g from the same center were used.
Multiple drug-resistant human cell line KB-8-5 growing in the presence of 300 nM vinblastine was generously provided by Prof. M. Gottesman (NIH). The cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin at 37°C in a humidified atmosphere containing 5% CO2/95% air. Tumors were initiated in mice by inoculating 106 KB-8-5 cells in 200 μL 0.9% saline solution subcutaneously into the right side of mice. Once tumors had reached a palpable volume of at least 50 mm3, mice were randomly assigned to experimental or control groups.
In Vivo Biodistribution Studies
In vivo real-time fluorescence imaging analysis was used to evaluate the distribution of Cy7- or Cy5.5-labeled (for subsequent microscopy) siRNAs in healthy and tumor-bearing mice. An In-Vivo MS FX PRO Imaging System (Carestream) was used to obtain X-ray and, concurrently, near-infrared fluorescence (NIRF) images (Cy5.5: excitation 620 nm, emission 700 nm; Cy7: excitation 760 nm, emission 830 nm). Tumors in mice were initiated as described above and were allowed to grow to approximately 1 cm3 volume. In each experiment, four tumor-bearing or healthy mice were injected intravenously (i.v.), intraperitoneally (i.p.), intramuscularly (i.m.), subcutaneously (s.c.), or peritumorally (p.t.) with 1.7 μg/g Cy7 or Cy5.5-labeled siRNA, Ch-siRNA, or siRNA/Lipofectamine 2000 complex in 100 μL Opti-MEM medium (Invitrogen), and the fifth mouse was left intact as a control. Animals were anesthetized with Avertin (150 mg/kg, i.p.) and placed on a heated tray (37°C). The fluorescence (10 s exposure) and X-ray (15 s exposure) scans were performed at various time points (15 min, 1, 2, 4, 6, and 24 hr) post-injection; five mice were scanned simultaneously on the tray. At the end of the experiment, mice were sacrificed and brain, lungs, heart, liver, spleen, kidneys, and tumor were collected. Each organ was rinsed with PBS and the fluorescence intensity was detected. Data were obtained from at least three independent sets of experiments with identical experimental setup.
Two parameters reflecting the efficiency of siRNA accumulation in the organs were used for comparison: organ fluorescence intensity, measured in relative fluorescent units (RFU), and the percentage of organ fluorescence intensity relative to the total fluorescent intensity of all organs.
Quantitative Stem-Loop Real-Time PCR Analysis
Healthy mice were intravenously injected with 1.7 μg/g Ch-siMDR. After 24 hr mice were sacrificed and siRNA was extracted from organs using Triton X-100 according to Landesman et al.63 siRNA-specific stem-loop RT-qPCR assays were designed according to the instructions of Czimmerer et al.64 using UPL-probe based stem-loop quantitative PCR assay design software (freely available online at http://genomics.dote.hu:8080/mirnadesigntool). Synthesis of cDNA and stem-loop PCR was carried out using SuperScript III Reverse Transcriptase (Thermo); qPCR mix contained BioMaster qPCR SYBR Blue (Biosan).
Confocal Microscopy
After imaging, all tumors and organs were immediately frozen with Tissue-Tek O.C.T. (Sakura) in liquid nitrogen. The container with samples was kept at −70°C until processing. Sections 7 μm thick were cut in a Microm HM 505N cryostat (Microm) at −21°C. The cryosections were stained with DAPI and phalloidin-TRITC according to the standard protocol, and mounted in ProLong Gold Antifade Mountant (Life Technologies). Finally, the cryosections were observed using an LSM 780 confocal fluorescent microscope (Carl Zeiss) at 20× magnification using band-pass (BP) 420–480 nm, BP 505–530 nm, and long-pass (LP) 560 nm optical filters.
Western Blotting
The level of P-glycoprotein in KB-8-5 tumors in SCID mice was evaluated by western blotting. Tumors in mice were initiated as described above and were allowed to grow to approximately 0.3 cm3. Mice with KB-8-5 tumors (three mice per time point per group) were injected i.v., i.p., or p.t. with 10 μg/g siMDR, Ch-siMDR, or Ch-siScr. At different time points (3–8 days), mice were sacrificed and tumors were excised and cut into 100- to 200-mg sections. The sections were weighed and homogenized in radioimmunoprecipitation assay (RIPA) buffer freshly supplemented with 1 mM phenylmethanesulfonyl fluoride (Thermo Scientific) using 300 μL buffer per 100 mg tumor tissue. The samples were stirred for 30 min at 4°C, then cleared by centrifugation at 10,000 × g for 10 min (4°C). Supernatants were diluted by two volumes of sample buffer (Sigma-Aldrich), and 10 μL of each sample was loaded onto a 10% SDS-polyacrylamide gel and then separated at 60 mA for 1 hr. The proteins were transferred from PAAG to polyvinylidene fluoride (PVDF) membrane (Millipore) using SemiPhor (Hoefer); then the membrane was blocked overnight in 1% non-fat dried milk in PBS. The membranes were incubated with monoclonal anti-P-glycoprotein and anti-β-actin antibodies (Sigma-Aldrich) at 1:800 and 1:5,000 dilutions, respectively, for 1 hr. After the membranes were washed in PBS with 0.1% Tween 20, they were subsequently incubated for 1 hr with secondary rabbit anti-mouse antibodies conjugated with peroxidase (Abcam). Visualization was performed using a Western Blotting Chemiluminescent Reagent Kit (Abcam) and X-ray film (Carestream). Human β-actin protein was used as an internal control. Data were analyzed using GelPro 4.0. software (Media Cybernetics).
Statistical Analysis
Variables are expressed as mean ± SD. Mean values were considered to be significantly different when p < 0.05 by use of Student’s t test or one-way ANOVA.
Author Contributions
I.V.C., M.A.Z., V.V.V., and E.L.C. conceived and designed the experiments. I.V.C., D.V.G., and M.I.M. performed the experiments. I.V.C., D.V.G., M.I.M., A.G.V., and E.L.C. analyzed the data. M.A.Z., V.V.V., and E.L.C. contributed reagents, materials, and analysis tools. I.V.C., M.A.Z., and E.L.C. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
The authors acknowledge Mrs. Albina V. Vladimirova (Institute of Chemical Biology and Fundamental Medicine SB RAS) for cell maintenance and Dr. Valery P. Nikolin (Institute of Cytology and Genetics SB RAS) for his excellent assistance with intravenous injections of mice. This work was supported by Russian Scientific Foundation grant 14-14-00697.
References
- 1.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Watts J.K., Deleavey G.F., Damha M.J. Chemically modified siRNA: tools and applications. Drug Discov. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Behlke M.A. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 4.Chernolovskaya E.L., Zenkova M.A. Chemical modification of siRNA. Curr. Opin. Mol. Ther. 2010;12:158–167. [PubMed] [Google Scholar]
- 5.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 6.Nishina K., Unno T., Uno Y., Kubodera T., Kanouchi T., Mizusawa H., Yokota T. Efficient in vivo delivery of siRNA to the liver by conjugation of alpha-tocopherol. Mol. Ther. 2008;16:734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- 7.McNamara J.O., 2nd, Andrechek E.R., Wang Y., Viles K.D., Rempel R.E., Gilboa E., Sullenger B.A., Giangrande P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 8.Xia C.F., Boado R.J., Pardridge W.M. Antibody-mediated targeting of siRNA via the human insulin receptor using avidin-biotin technology. Mol. Pharm. 2009;6:747–751. doi: 10.1021/mp800194y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moschos S.A., Jones S.W., Perry M.M., Williams A.E., Erjefalt J.S., Turner J.J., Barnes P.J., Sproat B.S., Gait M.J., Lindsay M.A. Lung delivery studies using siRNA conjugated to TAT(48-60) and penetratin reveal peptide induced reduction in gene expression and induction of innate immunity. Bioconjug. Chem. 2007;18:1450–1459. doi: 10.1021/bc070077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakase I., Tanaka G., Futaki S. Cell-penetrating peptides (CPPs) as a vector for the delivery of siRNAs into cells. Mol. Biosyst. 2013;9:855–861. doi: 10.1039/c2mb25467k. [DOI] [PubMed] [Google Scholar]
- 11.Wang J., Mi P., Lin G., Wáng Y.X., Liu G., Chen X. Imaging-guided delivery of RNAi for anticancer treatment. Adv. Drug Deliv. Rev. 2016;104:44–60. doi: 10.1016/j.addr.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luqmani Y.A. Mechanisms of drug resistance in cancer chemotherapy. Med. Princ. Pract. 2005;14(Suppl 1):35–48. doi: 10.1159/000086183. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman M.M. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman M.M., Hrycyna C.A., Schoenlein P.V., Germann U.A., Pastan I. Genetic analysis of the multidrug transporter. Annu. Rev. Genet. 1995;29:607–649. doi: 10.1146/annurev.ge.29.120195.003135. [DOI] [PubMed] [Google Scholar]
- 15.Gamcsik M.P., Dubay G.R., Cox B.R. Increased rate of glutathione synthesis from cystine in drug-resistant MCF-7 cells. Biochem. Pharmacol. 2002;63:843–851. doi: 10.1016/s0006-2952(01)00931-5. [DOI] [PubMed] [Google Scholar]
- 16.Deng H.B., Parekh H.K., Chow K.C., Simpkins H. Increased expression of dihydrodiol dehydrogenase induces resistance to cisplatin in human ovarian carcinoma cells. J. Biol. Chem. 2002;277:15035–15043. doi: 10.1074/jbc.M112028200. [DOI] [PubMed] [Google Scholar]
- 17.Synold T.W., Dussault I., Forman B.M. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat. Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 18.Lowe S.W., Ruley H.E., Jacks T., Housman D.E. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 19.Gorre M.E., Mohammed M., Ellwood K., Hsu N., Paquette R., Rao P.N., Sawyers C.L. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 20.Sharom F.J. Complex interplay between the P-glycoprotein multidrug efflux pump and the membrane: its role in modulating protein function. Front. Oncol. 2014;4:41. doi: 10.3389/fonc.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szakács G., Paterson J.K., Ludwig J.A., Booth-Genthe C., Gottesman M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 22.Essex S., Navarro G., Sabhachandani P., Chordia A., Trivedi M., Movassaghian S., Torchilin V.P. Phospholipid-modified PEI-based nanocarriers for in vivo siRNA therapeutics against multidrug-resistant tumors. Gene Ther. 2015;22:257–266. doi: 10.1038/gt.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volkov A.A., Kruglova N.S., Meschaninova M.I., Venyaminova A.G., Zenkova M.A., Vlassov V.V., Chernolovskaya E.L. Selective protection of nuclease-sensitive sites in siRNA prolongs silencing effect. Oligonucleotides. 2009;19:191–202. doi: 10.1089/oli.2008.0162. [DOI] [PubMed] [Google Scholar]
- 24.Petrova N.S., Chernikov I.V., Meschaninova M.I., Dovydenko I.S., Venyaminova A.G., Zenkova M.A., Vlassov V.V., Chernolovskaya E.L. Carrier-free cellular uptake and the gene-silencing activity of the lipophilic siRNAs is strongly affected by the length of the linker between siRNA and lipophilic group. Nucleic Acids Res. 2012;40:2330–2344. doi: 10.1093/nar/gkr1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrova N.S., Meschaninova M.I., Venyaminova A.G., Zenkova M.A., Vlassov V.V., Chernolovskaya E.L. Silencing activity of 2′-O-methyl modified anti-MDR1 siRNAs with mismatches in the central part of the duplexes. FEBS Lett. 2011;585:2352–2356. doi: 10.1016/j.febslet.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Logashenko E.B., Vladimirova A.V., Repkova M.N., Venyaminova A.G., Chernolovskaya E.L., Vlassov V.V. Silencing of MDR 1 gene in cancer cells by siRNA. Nucleosides Nucleotides Nucleic Acids. 2004;23:861–866. doi: 10.1081/NCN-200026032. [DOI] [PubMed] [Google Scholar]
- 27.Richert N.D., Aldwin L., Nitecki D., Gottesman M.M., Pastan I. Stability and covalent modification of P-glycoprotein in multidrug-resistant KB cells. Biochemistry. 1988;27:7607–7613. doi: 10.1021/bi00420a006. [DOI] [PubMed] [Google Scholar]
- 28.Yhee J.Y., Song S., Lee S.J., Park S.G., Kim K.S., Kim M.G., Son S., Koo H., Kwon I.C., Jeong J.H. Cancer-targeted MDR-1 siRNA delivery using self-cross-linked glycol chitosan nanoparticles to overcome drug resistance. J. Control Release. 2015;198:1–9. doi: 10.1016/j.jconrel.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Serramía M.J., Álvarez S., Fuentes-Paniagua E., Clemente M.I., Sánchez-Nieves J., Gomez R., de la Mata J., Muñoz-Fernández M.Á. In vivo delivery of siRNA to the brain by carbosilane dendrimer. J. Control. Release. 2015;200:60–70. doi: 10.1016/j.jconrel.2014.12.042. [DOI] [PubMed] [Google Scholar]
- 30.Perepelyuk M., Thangavel C., Liu Y., Den R.B., Lu B., Snook A.E., Shoyele S.A. Biodistribution and pharmacokinetics study of siRNA-loaded anti-NTSR1-mAb-functionalized novel hybrid nanoparticles in a metastatic orthotopic murine lung cancer model. Mol. Ther. Nucleic Acids. 2016;5:e282. doi: 10.1038/mtna.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J., Park J., Pei Y., Xu J., Yeo Y. Pharmacokinetics and biodistribution of recently-developed siRNA nanomedicines. Adv. Drug Deliv. Rev. 2016;104:93–109. doi: 10.1016/j.addr.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dohmen C., Fröhlich T., Lächelt U., Röhl I., Vornlocher H.P., Hadwiger P., Wagner E. Defined folate-PEG-siRNA conjugates for receptor-specific gene silencing. Mol. Ther. Nucleic Acids. 2012;1:e7. doi: 10.1038/mtna.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas M., Kularatne S.A., Qi L., Kleindl P., Leamon C.P., Hansen M.J., Low P.S. Ligand-targeted delivery of small interfering RNAs to malignant cells and tissues. Ann. N Y Acad. Sci. 2009;1175:32–39. doi: 10.1111/j.1749-6632.2009.04977.x. [DOI] [PubMed] [Google Scholar]
- 34.Cardoso A.L., Simões S., de Almeida L.P., Pelisek J., Culmsee C., Wagner E., Pedroso de Lima M.C. siRNA delivery by a transferrin-associated lipid-based vector: a non-viral strategy to mediate gene silencing. J. Gene Med. 2007;9:170–183. doi: 10.1002/jgm.1006. [DOI] [PubMed] [Google Scholar]
- 35.Huang Y., Wang X., Huang W., Cheng Q., Zheng S., Guo S., Cao H., Liang X.J., Du Q., Liang Z. Systemic administration of siRNA via cRGD-containing peptide. Sci. Rep. 2015;5:12458. doi: 10.1038/srep12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X., Wang W., Samarsky D., Liu L., Xu Q., Zhang W., Zhu G., Wu P., Zuo X., Deng H. Tumor-targeted in vivo gene silencing via systemic delivery of cRGD-conjugated siRNA. Nucleic Acids Res. 2014;42:11805–11817. doi: 10.1093/nar/gku831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arthanari Y., Pluen A., Rajendran R., Aojula H., Demonacos C. Delivery of therapeutic shRNA and siRNA by Tat fusion peptide targeting BCR-ABL fusion gene in chronic myeloid leukemia cells. J. Control. Release. 2010;145:272–280. doi: 10.1016/j.jconrel.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 38.Chu T.C., Twu K.Y., Ellington A.D., Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai W.Y., Wang W.Y., Chang Y.C., Chang C.J., Yang P.C., Peck K. Synergistic inhibition of lung cancer cell invasion, tumor growth and angiogenesis using aptamer-siRNA chimeras. Biomaterials. 2014;35:2905–2914. doi: 10.1016/j.biomaterials.2013.12.054. [DOI] [PubMed] [Google Scholar]
- 40.Hirsch M., Helm M. Live cell imaging of duplex siRNA intracellular trafficking. Nucleic Acids Res. 2015;43:4650–4660. doi: 10.1093/nar/gkv307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shukla R.S., Jain A., Zhao Z., Cheng K. Intracellular trafficking and exocytosis of a multi-component siRNA nanocomplex. Nanomedicine (Lond.) 2016;12:1323–1334. doi: 10.1016/j.nano.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moghimi S.M., Hunter A.C., Murray J.C. Long-circulating and target-specific nanoparticles: theory to practice. Pharmacol. Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 43.Raouane M., Desmaële D., Urbinati G., Massaad-Massade L., Couvreur P. Lipid conjugated oligonucleotides: a useful strategy for delivery. Bioconjug. Chem. 2012;23:1091–1104. doi: 10.1021/bc200422w. [DOI] [PubMed] [Google Scholar]
- 44.Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 45.Cuellar T.L., Barnes D., Nelson C., Tanguay J., Yu S.F., Wen X., Scales S.J., Gesch J., Davis D., van Brabant Smith A. Systematic evaluation of antibody-mediated siRNA delivery using an industrial platform of THIOMAB-siRNA conjugates. Nucleic Acids Res. 2015;43:1189–1203. doi: 10.1093/nar/gku1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunzell J.D., Davidson M., Furberg C.D., Goldberg R.B., Howard B.V., Stein J.H., Witztum J.L., American Diabetes Association. American College of Cardiology Foundation. Lipoprotein management in patients with cardiometabolic risk: consensus statement from the American Diabetes Association and the American College of Cardiology Foundation. Diabetes Care. 2008;31:811–822. doi: 10.2337/dc08-9018. [DOI] [PubMed] [Google Scholar]
- 47.Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K., Rajeev K.G., Nakayama T., Charrise K., Ndungo E.M. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 48.Lorenz C., Hadwiger P., John M., Vornlocher H.P., Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg. Med. Chem. Lett. 2004;14:4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 49.Kuwahara H., Nishina K., Yoshida K., Nishina T., Yamamoto M., Saito Y., Piao W., Yoshida M., Mizusawa H., Yokota T. Efficient in vivo delivery of siRNA into brain capillary endothelial cells along with endogenous lipoprotein. Mol. Ther. 2011;19:2213–2221. doi: 10.1038/mt.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding Y., Wang W., Feng M., Wang Y., Zhou J., Ding X., Zhou X., Liu C., Wang R., Zhang Q. A biomimetic nanovector-mediated targeted cholesterol-conjugated siRNA delivery for tumor gene therapy. Biomaterials. 2012;33:8893–8905. doi: 10.1016/j.biomaterials.2012.08.057. [DOI] [PubMed] [Google Scholar]
- 51.Oe Y., Christie R.J., Naito M., Low S.A., Fukushima S., Toh K., Miura Y., Matsumoto Y., Nishiyama N., Miyata K., Kataoka K. Actively-targeted polyion complex micelles stabilized by cholesterol and disulfide cross-linking for systemic delivery of siRNA to solid tumors. Biomaterials. 2014;35:7887–7895. doi: 10.1016/j.biomaterials.2014.05.041. [DOI] [PubMed] [Google Scholar]
- 52.Bienk K., Hvam M.L., Pakula M.M., Dagnaes-Hansen F., Wengel J., Malle B.M., Kragh-Hansen U., Cameron J., Bukrinski J.T., Howard K.A. An albumin-mediated cholesterol design-based strategy for tuning siRNA pharmacokinetics and gene silencing. J. Control. Release. 2016;232:143–151. doi: 10.1016/j.jconrel.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 53.Gilleron J., Paramasivam P., Zeigerer A., Querbes W., Marsico G., Andree C., Seifert S., Amaya P., Stöter M., Koteliansky V. Identification of siRNA delivery enhancers by a chemical library screen. Nucleic Acids Res. 2015;43:7984–8001. doi: 10.1093/nar/gkv762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexis F., Pridgen E., Molnar L.K., Farokhzad O.C. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolkestein M., de Blois E., Koelewijn S.J., Eggermont A.M., Grosveld F., de Jong M., Koning G.A. Investigation of factors determining the enhanced permeability and retention effect in subcutaneous xenografts. J. Nucl. Med. 2016;57:601–607. doi: 10.2967/jnumed.115.166173. [DOI] [PubMed] [Google Scholar]
- 56.Vicentini F.T., Borgheti-Cardoso L.N., Depieri L.V., de Macedo Mano D., Abelha T.F., Petrilli R., Bentley M.V. Delivery systems and local administration routes for therapeutic siRNA. Pharm. Res. 2013;30:915–931. doi: 10.1007/s11095-013-0971-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guzman-Aranguez A., Loma P., Pintor J. Small-interfering RNAs (siRNAs) as a promising tool for ocular therapy. Br. J. Pharmacol. 2013;170:730–747. doi: 10.1111/bph.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thakur A., Fitzpatrick S., Zaman A., Kugathasan K., Muirhead B., Hortelano G., Sheardown H. Strategies for ocular siRNA delivery: potential and limitations of non-viral nanocarriers. J. Biol. Eng. 2012;6:7. doi: 10.1186/1754-1611-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 60.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 61.Zenkov A.N., Scvortsova N.V., Chernolovskaya E.L., Pospelova T.I., Vlassov V.V. Expression of the MDR1 and MRP genes in patients with lymphoma with primary bone marrow involvement. Nucleosides Nucleotides Nucleic Acids. 2004;23:843–847. doi: 10.1081/NCN-200026029. [DOI] [PubMed] [Google Scholar]
- 62.Bellon L. Oligoribonucleotides with 2′-O-(tert-butyldimethylsilyl) groups. Curr. Protoc. Nucleic Acid Chem. 2001;Chapter 3 doi: 10.1002/0471142700.nc0306s01. Unit 3.6. [DOI] [PubMed] [Google Scholar]
- 63.Landesman Y., Svrzikapa N., Cognetta A., 3rd, Zhang X., Bettencourt B.R., Kuchimanchi S., Dufault K., Shaikh S., Gioia M., Akinc A. In vivo quantification of formulated and chemically modified small interfering RNA by heating-in-Triton quantitative reverse transcription polymerase chain reaction (HIT qRT-PCR) Silence. 2010;1:16. doi: 10.1186/1758-907X-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Czimmerer Z., Hulvely J., Simandi Z., Varallyay E., Havelda Z., Szabo E., Varga A., Dezso B., Balogh M., Horvath A. A versatile method to design stem-loop primer-based quantitative PCR assays for detecting small regulatory RNA molecules. PLoS ONE. 2013;8:e55168. doi: 10.1371/journal.pone.0055168. [DOI] [PMC free article] [PubMed] [Google Scholar]