Abstract
Current acute myeloid leukemia (AML) disease models face severe limitations because most of them induce un-physiological gene expressions that do not represent conditions in AML patients and/or depend on external promoters for regulation of gene expression/repression. Furthermore, many AML models are based on reciprocal chromosomal translocations that only reflect the minority of AML patients, whereas more than 50% of patients have a normal karyotype. The majority of AML, however, is driven by somatic mutations. Thus, identification as well as a detailed molecular and functional characterization of the role of these driver mutations via improved AML models is required for better approaches toward novel targeted therapies. Using the IDH2 R140Q mutation as a model, we present a new effective methodology here using the RNA-guided clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system to reproduce or remove AML-associated mutations in or from human leukemic cells, respectively, via introduction of a DNA template at the endogenous gene locus via homologous recombination. Our technology represents a precise way for AML modeling to gain insights into AML development and progression and provides a basis for future therapeutic approaches.
Keywords: AML, CRISPR-Cas9, leukemia, genome editing
Introduction
Despite intense research into acute myeloid leukemia (AML) during past decades, the majority of AML patients still die from their disease. Thus, there is a high need for new AML therapies. AML is a heterogeneous disease harboring a multitude of genetic and epigenetic changes, and it is highly likely that the various AML subtypes require different targeted therapeutic approaches. Development of specific targeted therapies will require precise disease modeling, minimizing the amount of artificial effects in the leukemic cells. However, current AML disease models face severe limitations because most of them induce un-physiological gene expressions that do not represent conditions found in AML patients and/or depend on external promoters for regulation of gene expression/repression. Furthermore, many AML models are based on reciprocal chromosomal translocations that only reflect the minority of AML patients, whereas more than 50% of patients have a normal karyotype. The majority of AML, however, is driven by somatic mutations. Next-generation sequencing has revealed that even though AML usually harbors hundreds of mutated genes, only a limited number of these genes serve as so-called driver mutations that cause and/or maintain leukemia.1, 2 Thus, identification as well as a detailed molecular and functional characterization of the role of these driver mutations via improved AML models is required for better approaches toward novel targeted therapies.
During the last few years, the clustered regularly interspaced short palindromic repeats (CRISPR) and the associated Cas9 nucleases (CRISPR-Cas9) have revolutionized the options for targeted genome editing.3, 4, 5 These programmable RNA-guided endonucleases (RGENs) comprise two RNA elements, CRISPR RNA (cRNA) and its transactivating RNA (tracRNA), which can be fused together and used to induce a targeted double-strand break (DSB). Providing a corresponding DNA template, any specific gene sequence can be introduced via homologous recombination (HR).3 However, the efficiency of this system in primary hematopoietic stem and progenitor cells as well as AML cells, in which minimal transfection efficiency largely hinders effective genome editing, remains a major challenge.
Isocitrate dehydrogenases (IDHs) are digestive enzymes that catalyze the oxidative decarboxylation of isocitrate, producing alpha-ketoglutarate (α-ketoglutarate) and CO2. Mutations in IDH1/2 genes occur frequently in AML patients, and IDH2 R140Q has been shown to be the most frequent IDH mutation in AML.6 IDH mutants promote significant increases in repressive histone methylation marks, inducing a block in cell differentiation and aberrant self-renewal.7 Moreover, IDH2 R140Q has been identified as a key driver mutation in a transgenic mouse model, supporting its relevance as a therapeutic target for the treatment of human AML.8
Using the IDH2 R140Q mutation as a model, we present a new effective methodology here using the RNA-guided CRISPR-Cas9 system to reproduce or remove AML-associated mutations in or from human leukemic cells, respectively, which represents a precise way for AML modeling to gain insights into AML development and progression.
Results
Introduction of IDH2 R140Q Mutation in Human Myeloid Leukemia Cells
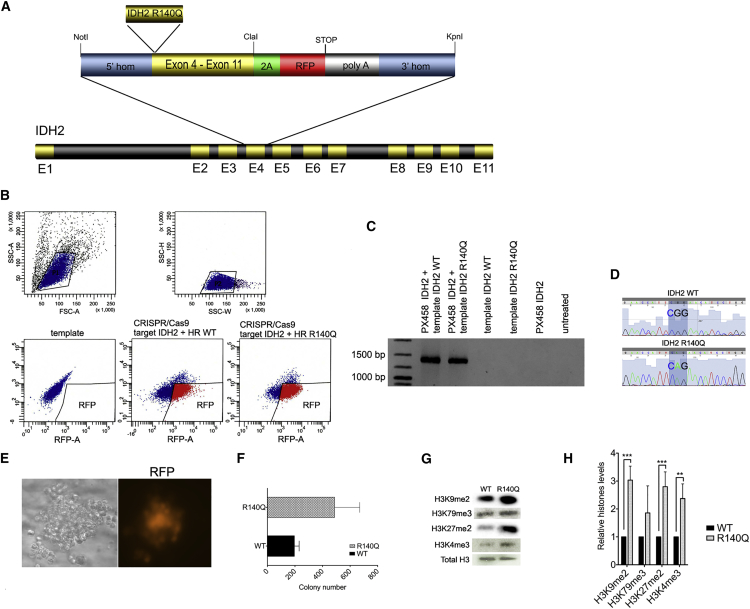
The human myeloid leukemia cell line K562 lacking the IDH2 R140Q mutation was chosen for this study. To target IDH2, we selected three small-guide (sg) RNAs (Table S1) according to published criteria mapping to different introns/exons of the gene. To test the efficiency of each sgRNA for induction of specific DSBs at its defined location, we co-transfected HEK293 cells with the sgRNA and Cas9-gene-expressing plasmids. To test efficiency for targeted gene disruption of each sgRNA at its defined location, we conducted surveyor nuclease digestion assay. DSB frequencies driven by each sgRNA ranged from 24% to 46% (Figure S1A). sgRNA showing the best ratio of DSB efficiency versus potential off-target disruptions was selected for further experiments (Figure S1B). The DNA template for targeted integration via HR after aimed DSB at the IDH2 locus was designed as displayed in Figure 1A and cloned into a pBS-SK(+) vector performing PCR-based site-directed mutagenesis. To avoid re-cutting after HR, silent point mutations were introduced into the IDH2 sequence at the sgRNA-binding site. The DNA template carried a red fluorescence protein (RFP) fused to the mutated IDH2 sequence via a 2A peptide (Figure 1A). Thus, only cells after successful genome editing with correct insertion of the DNA template at the IDH2 locus would show subsequent RFP expression.
Figure 1.
CRISPR-Cas9-Mediated Introduction of IDH2 R140Q Mutation in Human K562 Leukemia Cells
(A) Schematic of the DNA repair template for introduction of IDH2 R140Q mutation into human K562 cells via homologous recombination. (B) FACS sorting of K562 cells after transfection with the CRISPR-Cas9 plasmid and plasmid carrying the DNA template for HR; upper left: forward scatter (FSC); upper right: side scatter (SSC); lower left: K562 cells transfected with the plasmid carrying the DNA template only; lower middle: K562 cells transfected with the plasmid carrying the DNA template, including the wt IDH2 sequence plus CRIPSR-Cas9/sgRNA; lower right: K562 cells transfected with the plasmid carrying the DNA template, including the mutant IDH2 sequence plus CRIPSR-Cas9/sgRNA. (C) Targeted integration (TI) PCR confirming introduction of the DNA templates, including the IDH2 R140Q mutation or wt IDH2 sequence, respectively, at the IDH2 locus. (D) Sanger DNA sequences confirming accurate introduction of the IDH2 R140Q mutation (and IDH2 wt sequence) at the IDH2 gene locus. (E) Colony of human K562 cells plated into methylcellulose after FACS sorting for RFP expression; left: colony morphology in brightfield microscopy; right: RFP detection via fluorescence microscopy. (F) Histograph representing colony formation of K562 cells after successful genome editing with introduction of the IDH2 R140Q mutation and re-introduction of the IDH2 wt sequence, respectively, and after 7 days post-FACS sorting. Error bars indicate SD of three independent experiments. (G) Immunoblotting of purified histone extracts from K562 IDH2 wt and K562 IDH2 R140Q. (H) Bar chart representing quantification of specific histone levels relative to total histone 3. Error bars indicate SD of three independent experiments (±SD, **p < 0.01, ***p < 0.001).
Using the CRISPR-Cas9 system and DNA template described above, we induced specific DNA double-strand breaks in the IDH2 gene of K562 myeloid leukemia cells and introduced the IDH2 R140Q point mutation via HR, inserting the specific DNA template at the determined IDH2 gene locus. After specific insertion, the endogenous IDH2 gene promoter regulated the integrated IDH2 R140Q mutation as well as RFP expression. Thus, successfully targeted cells were selected via fluorescence-activated cell sorting (FACS) of the RFP-positive polyclonal cell population (Figure 1B). Editing efficiency varied between 3% and 10% (n = 7). As a control, K562 cells were transfected using the same procedure but reintroducing the wild-type (wt) IDH2 sequence. Correct insertion of the mutation and the wt IDH2 sequence in the control cells, respectively, was confirmed by performing targeted-integration PCR (Figure 1C) as well as genomic DNA sequencing (Figure 1D). K562 cells maintained their proliferation and colony formation capacity in methylcellulose after successful genome editing. IDH2 protein levels remained unchanged in the modified cells (Figure S1C). Moreover, the genome-engineered K562 cells maintained a strong RFP expression (Figure 1E).
Proliferation and Epigenetic Changes upon Induction of IDH2 R140Q Mutation in Human Myeloid Leukemia Cells
We next addressed whether the IDH2 R140Q mutation driven by CRISPR-Cas9-mediated genome engineering was able to induce functional changes characteristic for this mutation. Strikingly, K562 cells carrying the IDH2 R140Q mutation showed a significantly enhanced in vitro colony forming capacity as compared with controls carrying wt IDH2 introduced analogously (Figure 1F). Moreover, K562 cells carrying the IDH2 R140Q point mutation after successful CRISPR-Cas9-directed genome editing showed a significant increase in H3K9me2, H3K27me2, and H3K4me3 (Figures 1G and 1H). Thus, mutated K562 cells displayed typical histone hypermethylation expected upon IDH2 R140Q mutation.7
Thus, our results present a new strategy for the generation of leukemia-related gene mutations at defined positions and under the control of the endogenous gene promoter. Moreover, our methodology allows accurate selection of modified cells and provides a promising new methodology for AML disease modeling, largely minimizing the limitations of current AML models.
CRISPR-Cas9-Mediated Removal of IDH2 R140Q Mutation from Primary AML Patient Blasts
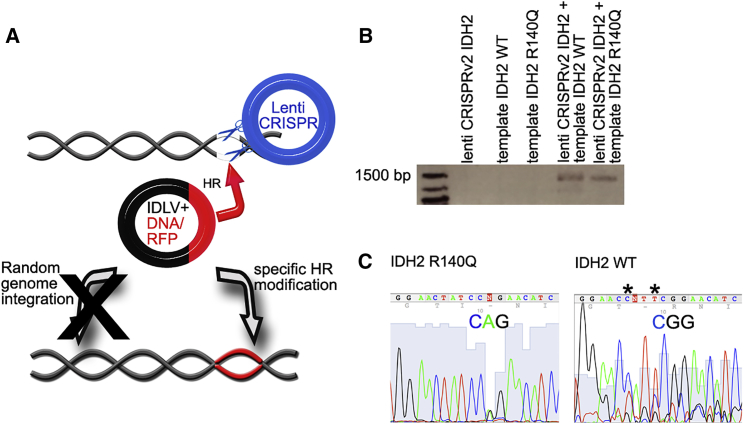
Recently, it has been demonstrated that the CRISPR-Cas9 system can also be used for genome editing for functional repair of gene mutations in intestinal stem cells.9 However, a key challenge in leukemia research is still genome engineering of primary hematopoietic stem and progenitor cells as well as patient-derived primary leukemia cells. Nucleofection/transfection efficiencies are particularly low in these cells, and the viability and shape of the cells after the corresponding procedure tend to be dismal. Thus, we used lentiviral transduction to increase our genome engineering efficiency in primary AML cells. Via sequencing of genomic DNA, we identified an IDH2 R140Q mutation in myeloid blasts of a 66-year-old patient with AML subtype FAB M4. Tempting to remove this mutation from the patient cells by replacing the mutated IDH2 sequence with the corresponding IDH2 wt sequence, primary blasts were lentivirally transduced with Lenti CRISPRv2 carrying an sgRNA targeting IDH2 R140Q at the same locus as in the experiments for Figure 1. The sgRNA sequence was modified according to the sequence of the IDH2 point mutation as compared to experiments shown in Figure 1. Moreover, AML cells were co-transduced with a lentiviral plasmid carrying a DNA template with the IDH2 wt sequence and with the same design as depicted in Figure 1A. As a control, primary AML blasts from the same patient were transfected using the same procedure but reintroducing the IDH2 R140Q mutation. Because one of the major hurdles for targeted genome engineering using lentiviral transduction approaches is lentiviral integration into the genome, our DNA template was cloned into an integrase-deficient lentivirus (IDLV) (Figure 2A). Experimental procedures were carried out analogously to the approaches, as explained for Figure 1. Again, correct insertion of the wt IDH2 sequence and in the control cells, respectively, was confirmed by performing targeted integration PCR (Figure 2B) as well as genomic DNA sequencing (Figure 2C).
Figure 2.
CRISPR-Cas9-Mediated Conversion of IDH2 R140Q Mutation from Primary AML Patient Blasts
(A) Schematic diagram illustrating lentiviral infection of human primary AML blasts with the lenti-CRISPR-Cas9 plasmid carrying sgRNA targeting the IDH2 gene as well as the IDLV plasmid carrying the DNA template for subsequent HR. (B) TI PCR confirming correct insertion of the DNA template at the IDH2 locus via HR. (C) Sanger DNA sequences of the genome modified AML patient samples after removal of the IDH2 R140Q mutation (right) and IDH2 wt sequence of the original patient sample as a control (left), respectively. Asterisks indicate the positions of the silent point mutations introduced to avoid CRISPR-Cas9-system-mediated re-cutting after successful genome editing.
Thus, lentiviral CRISPR-Cas9 systems can be used successfully for genome editing of primary AML patient samples. Introduction of specific DNA templates via IDLV enables engineering of AML mutations under endogenous gene promoters and avoids random integration into the genome of the cells.
Discussion
The majority of AML is characterized by a detailed census of mutated genes.10, 11 Although next-generation sequencing has revealed hundreds of mutated genes in AML patients, identification as well as a detailed molecular and functional characterization of the few driver mutations among them is likely to pave the avenue toward novel targeted therapies. Today, one of the biggest challenges for AML studies is precise disease modeling, which avoids artificial phenomena that do not accurately reflect genetic conditions found in AML patients. Recently, the CRISPR-Cas9 system has been used successfully for combined disruption of up to five genes in single murine hematopoietic cells, at the same time leading to clonal outgrowth and myeloid malignancy.12 This technique is able to elucidate the effect of cooperating genetic alterations and reflect the complexity of human cancer in an elegant approach. However, gene disruption still does not precisely recapitulate the most frequent genetic event found in AML: somatic gene mutations. Moreover, combined gene disruption will not enable investigation of the direct effects of somatic mutations on genetic and epigenetic events in leukemia cells nor confirmation of single mutations as driver mutations critical for disease induction and maintenance.
Genome engineering via the RNA-guided CRISPR-Cas9 system provides a novel methodology, allowing induction of genomic modifications under the endogenous gene promoters.13 However, primary hematopoietic stem and leukemia cells remain challenging for transfection/transduction strategies, and reported efficiencies, including the CRIPSR-Cas9 technology, are low.
Our data present a new strategy for efficient generation or removal of leukemia-related gene mutations at defined positions and under maintained control of the endogenous gene promoter in leukemia cell lines as well as in primary AML blasts using the IDH2 R140Q point mutation as a model. Our approach is able to recapitulate genetic and epigenetic as well as functional changes known for IDH2 mutations.
To our knowledge, this is the first report describing CRISPR-Cas9-mediated introduction of an AML-associated driver mutation into human leukemia cells under the endogenous gene promoter as well as an accurately corrected acquired gene mutation in primary AML patient cells restoring the wt status. Optimizations of this approach will provide a promising new methodology for AML disease modeling and the development of novel targeted therapies.
Materials and Methods
Cell Lines and Culture Conditions
K562 cells were purchased from ATCC (CCL-243 American Type Culture Collection [ATCC]). Cells were cultured using Iscove’s modified Dulbecco’s medium (IMDM; Thermo Scientific) supplemented with 1% penicillin/streptomycin (Life Technologies) and 10% fetal calf serum (FCS) (Biochrom).
Primary AML blasts were cultured as previously described.14 In brief, blasts were thawed in IMDM +20% FCS + 100 μg/mL DNase I at 37°C and maintained in IMDM containing 15% BIT 9500 serum substitute (Stemcell Technologies), 100 ng/mL stem cell factor (Preprotech), 50 ng/mL FMS-like tyrosine kinase 3 ligand (FLT3L; Preprotech), 20 ng/mL interleukin (IL)-3 (Preprotech), 20 ng/mL granulocyte-colony stimulation factor (Preprotech), 50 ng/mL thrombopoietin (Preprotech), 10 μg/mL low-density lipoprotein (LDL) (Stem Cell Technologies), 0.1 mM β-mercaptoethanol (Thermo Scientific), 1% penicillin/streptomycin (Life Technologies), 0.5% ciprofloxacin, 500 nM stemregenin 1 (Focus Bioscience), 1 μM UM729 (kindly provided by Caroline Pabst), and 10 μL/mL Glutamax (Life Technologies) at 37°C, 5% CO2. The study was reviewed and approved by the ethics committee of the physician’s chamber of Westfalen-Lippe and the medical faculty of the University of Muenster (2007-524-f-S and 2007-390-f-S) before the study began.
CRISPR Vectors, sgRNA, and DNA Template Cloning
sgRNAs were designed using the website http://crispr.mit.edu/. Sequences of complementary oligos for each sgRNA are listed in Table S1. For transfection of K562 cells, sgRNAs were cloned into the pX458 vector (Addgene) expressing Cas9, as described by Ran et al.3 For lentiviral transductions, sgRNAs were cloned into lentiCRISPR v2 (Addgene).15 The DNA template was designed as depicted in Figure 1A and cloned into pBS-SK(+) for transfection of K562 cells and into pLVdLacZGFPiNwpre IDLV for lentiviral transductions. The helper plasmids used were pMDL.gpD64A and pMDG. Both lentiviral plasmids as well as the helper plasmids were a kind gift from Tony Cathomen and Tatjana Cornu.16 To avoid continued CRISPR-Cas9-mediated DSB after successful genome editing, two silent point mutations were introduced in the spacer area of the cDNA. All constructs were generated either by direct cloning or PCR-based site-directed mutagenesis and verified by Sanger DNA sequencing. Primer sequences for cloning of the DNA template are available upon request.
Nucleofection
K562 cells were nucleofected using the Amaxa Cell Line Nucleofector Kit V (Lonza) following the instructions of the supplier. Nucleofection was carried out using the Nucleofector Device I program T-16.
FACS
K562 cells harboring the correctly inserted and located homology-directed repair template indicated by tomato expression were isolated via FACS (BD FACS Aria II). Sorting was carried out 4 to 5 days post nucleofection.
Colony Assays
Human K562 cells were plated after FACS in methylcellulose MethoCult H4034 (Stem Cell Technologies). Colonies were scored after 7 days.
Lentivirus Production and Lentiviral Transduction
Lentivirus production and lentiviral transduction were performed on K562 cells and human primary blasts as previously described.17 In brief, K562 spinoculation was carried out by centrifugation at 800 × g in the presence of 5 mg/mL polybrene (Sigma-Aldrich) at 32°C for 2 hr.
For transduction of primary AML blasts on the day of transduction, 2 × 105 primary AML patient cells were pelleted and resuspended in 500 μL of CRISPR-Cas9 lentivirus containing IDH2-targeting sgRNA and 500 μL of lentivirus carrying the DNA template for HR. Afterward, cytokines were added at the same concentration as described above. Cells were spinoculated at 32°C (90 min, 800 × g). After centrifugation, supernatant was removed and cells were resuspended in culture medium. The same procedure was repeated 24 hr later.
Antibodies
We used the following specific antibodies in this study: polyclonal rabbit anti-human anti-histone H3 tri methyl K79 (Abcam), anti-histone H3 di methyl K27 (Abcam), anti-histone H4 mono methyl K20 (Abcam), anti-histone H3 di methyl K9 (Abcam), and anti-histone H3 (Abcam).
Histone Purification and Western Blot
Cells were grown and collected under the specific experimental conditions required. The cells were washed in PBS and lysed in Triton extraction buffer (TEB) (PBS containing 0.5% Triton X-100 [v/v], 2 mM phenylmethylsulfonyl fluoride [PMSF], and 0.02% [w/v] NaN3) at a cell density of 106 cells per mL. Lysed cells were incubated on ice for 10 min with gentle stirring and centrifuged at 6,500 × g for 10 min at 4°C to spin down the nuclei. Afterward, nuclei were washed in half the volume of TEB and centrifuged as before, and the pellet was re-suspended in 0.2 N HCl at a density of 4 × 106 nuclei per mL and incubated overnight at 4°C. The following day, samples were centrifuged, and supernatant containing the histone protein was used to determine the protein content using the Bradford assay. The histone proteins were resolved by SDS-PAGE. Membranes were probed with the described antibodies.
Genomic DNA Extraction
Genomic DNA extraction was performed with the DNeasy Blood & Tissue Kit (QIAGEN) using the manufacturer’s instructions.
Targeted Integration PCR and DNA Sequencing
Targeted integration PCR was performed in an Eppendorf nexus gradient cycler using standard PCR protocols. The primers used for targeted integration PCR were the following: forward primer 5′GACCGGAGCATGTGGAGT 3′ and reverse primer 5′ CCTCAATCGTCTTCCCATCA 3′. PCR products were verified by Sanger DNA sequencing.
Statistical Analysis
A two-tailed Student’s test was used to determine the statistical significance for all bar charts. p values of less than 0.05 were considered as indicating statistically significant differences.
Author Contributions
O.B. carried out the molecular biology, cell culture, and flow cytometry experiments and analysis. V.A., L.A., and C.S. performed and analyzed the biochemistry experiments. J-H.M., M.-F.A., and W.E.B. designed the experiments. M.-F.A and J.-H.M. supervised the experimental work and analysis. M.-F.A. and J.-H.M. wrote the manuscript, with critical input from W.E.B. J.-H.M. designed the overall study. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare that they have no competing interests.
Acknowledgments
We would like to thank T. Cathomen and T. Cornu for providing us with the pLVdLacZGFPiNwpre IDLV plasmid as well as the pMDL.gpD64A and pMDG helper plasmids. We would like to thank D. Schwammbach for help with the experimental work. W.E.B.’s laboratory was supported by Deutsche Forschungsgemeinschaft (grant DFG EXC 1003).
Footnotes
Supplemental Information includes one figure and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2016.12.012.
Contributor Information
Maria-Francisca Arteaga, Email: marifrancis.arteaga@uni-muenster.de.
Jan-Henrik Mikesch, Email: jan-henrik.mikesch@ukmuenster.de.
Supplemental Information
References
- 1.Papaemmanuil E., Gerstung M., Bullinger L., Gaidzik V.I., Paschka P., Roberts N.D., Potter N.E., Heuser M., Thol F., Bolli N. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metzeler K.H., Herold T., Rothenberg-Thurley M., Amler S., Sauerland M.C., Görlich D., Schneider S., Konstandin N.P., Dufour A., Bräundl K., AMLCG Study Group Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–698. doi: 10.1182/blood-2016-01-693879. [DOI] [PubMed] [Google Scholar]
- 3.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 6.Paschka P., Schlenk R.F., Gaidzik V.I., Habdank M., Krönke J., Bullinger L., Späth D., Kayser S., Zucknick M., Götze K. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J. Clin. Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 7.Lu C., Ward P.S., Kapoor G.S., Rohle D., Turcan S., Abdel-Wahab O., Edwards C.R., Khanin R., Figueroa M.E., Melnick A. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kats L.M., Reschke M., Taulli R., Pozdnyakova O., Burgess K., Bhargava P., Straley K., Karnik R., Meissner A., Small D. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. 2014;14:329–341. doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwank G., Koo B.-K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welch J.S., Ley T.J., Link D.C., Miller C.A., Larson D.E., Koboldt D.C., Wartman L.D., Lamprecht T.L., Liu F., Xia J. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heckl D., Kowalczyk M.S., Yudovich D., Belizaire R., Puram R.V., McConkey M.E., Thielke A., Aster J.C., Regev A., Ebert B.L. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dow L.E. Modeling disease in vivo with CRISPR/Cas9. Trends Mol. Med. 2015;21:609–621. doi: 10.1016/j.molmed.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pabst C., Krosl J., Fares I., Boucher G., Ruel R., Marinier A., Lemieux S., Hébert J., Sauvageau G. Identification of small molecules that support human leukemia stem cell activity ex vivo. Nat. Methods. 2014;11:436–442. doi: 10.1038/nmeth.2847. [DOI] [PubMed] [Google Scholar]
- 15.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornu T.I., Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol. Ther. 2007;15:2107–2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- 17.Arteaga M.F., Mikesch J.-H., Qiu J., Christensen J., Helin K., Kogan S.C., Dong S., So C.W. The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia. Cancer Cell. 2013;23:376–389. doi: 10.1016/j.ccr.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.