Abstract
Lung cancer is the leading cause of cancer-related death worldwide. Although advanced drugs have benefitted patients, therapeutic success has largely been hampered because of rapid development of resistance. Here we report that PIWI-interacting RNA likes (piR-Ls), a novel type of functional sncRNAs, play key roles in chemoresistance to cisplatin (CDDP)-based chemotherapy in lung squamous cell carcinoma (LSCC). piR-L-138 was upregulated upon CDDP-based chemotherapy both in LSCC cells and in patient-derived xenograft (PDX) LSCC models. Further, targeting upregulated piR-L-138 led to increased apoptosis in CDDP-treated LSCC cells and LSCC xenograft mice treated with CDDP. In addition, piR-L-138 directly interacted with p60-MDM2 and inhibited CDDP-activated apoptosis in p53-mutated LSCC. We identified the upregulated piR-L-138 upon CDDP-based chemotherapy, confirmed the enhanced sensitivity of LSCC to agents by targeting the upregulated piR-L-138 both in vitro and in vivo, and revealed mechanisms underlying piR-L-138 in chemoresistance, bolstering a new emerging clinical modality where novel functional piR-Ls provide potential strategies to overcome chemoresistance for patients with LSCC.
Keywords: piRNA-likes, lung cancer, chemoresistance, sncRNA
Introduction
Lung cancer is the leading cause of cancer-related death worldwide. About 85% of lung cancers are non-small-cell lung cancers (NSCLCs), with lung adenocarcinoma (LADC) and lung squamous cell carcinoma (LSCC) as the major subtypes of NSCLC.1, 2 Although encouraging targeted therapies have dramatically afforded benefits to patients with LADC, few therapeutic targets have been validated for patients with LSCC.3, 4, 5 To date, the treatment options for patients with LSCC are limited, and platinum-based chemotherapy is still the gold standard for first-line treatment; however, some LSCCs are intrinsically resistant to chemotherapy. Virtually all patients, including initial responders, rapidly develop chemoresistance,6, 7, 8, 9 which, in the absence of alternative therapies, leads to an overall 5-year survival rate of less than 20%. Therefore, a better understanding of the molecular determinants of LSCC resistance to chemotherapy is a critically important step to overcome the clinical chemoresistance currently responsible for the dismal survival rates of patients with LSCC.
We recently reported, for the first time, that PIWI-interacting RNA likes (piR-Ls), a type of functional small non-coding RNAs (sncRNAs), played critical roles in NSCLC tumorigenesis and differentiation.10 We have profiled piR-Ls expression in normal human bronchial epithelial (HBE) and NSCLC cells, and we found that LSCC, LADC, and HBE cells could be clearly clustered together on the expression patters of all piR-Ls.10 We also found that piR-Ls were aberrantly differentially expressed in NSCLC and HBE cells as well as between LSCC and LADC subtypes. Using filtered log fold change = 1 and false discovery rate < 0.05 as criteria, we identified that there were seven piR-Ls that could distinguish LSCC from both LADC and HBE cells,10 indicating that these seven piR-Ls were LSCC-related piR-Ls. In this study, we observed the effects of LSCC-related piR-Ls in resistance caused by cisplatin (CDDP)-based chemotherapeutic agents commonly used for patients with LSCC. Characteristically, we identified that piR-L-138 was upregulated upon CDDP-based chemotherapy both in vitro and in vivo, and, further, we found that targeting the upregulated piR-L-138 resulted in enhanced sensitivity of LSCC to CDDP-based chemotherapy both in vitro and in vivo, indicating that piR-L-138 is a key contributor to CDDP-based chemoresistance and that targeting piR-L-138 could be a potential strategy to overcome chemoresistance for patients with LSCC.
Results
LSCC-Related piR-L-138 Is Upregulated upon CDDP-Based Chemotherapy Both In Vitro and In Vivo
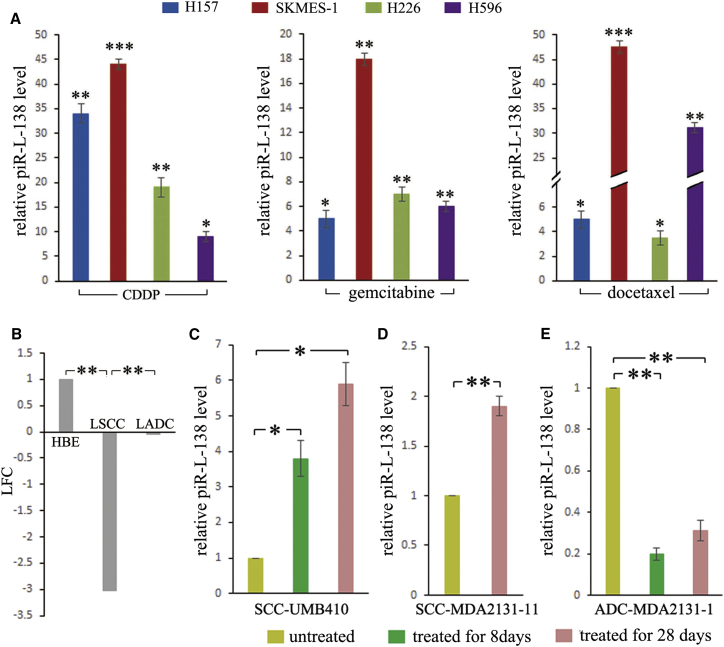
To determine whether these LSCC-related piR-Ls played roles in chemoresistance, we analyzed the relationships between their expression levels and the sensitivities to gold standard first-line platinum-based chemotherapy. Given that CDDP is the backbone of platinum-based chemotherapy, which is defined as a combination of a platinum compound (cisplatin or carboplatin) and one of the four chemotherapy agents (gemcitabine, docetaxel, paclitaxel, and vinorelbine), we evaluated expression levels of LSCC-related piR-Ls to these agents commonly used for patients with NSCLC. We observed that LSCC-related piR-Ls were upregulated upon exposure to agents (Figure S1). To further identify whether these piR-Ls were LSCC-related sncRNAs, we also observed both LSCC-related and non-LSCC-related piR-Ls in four LSCC as well as in four LADC cell lines (Figure S1), and piR-Ls levels were measured in cells treated with CDDP, gemcitabine, docetaxel, and their corresponding control solutions (PBS or DMSO) at a drug concentration required for 50% inhibition (IC50) for 24 hr (Table S1; Figure S2). Among piR-Ls, upon exposure to these chemotherapeutic agents, we observed a significant consistent and substantial increase of piR-L-138 levels in all four LSCC cells treated with CDDP, gemcitabine, and docetaxel (Figure 1A), indicating that piR-L-138 was upregulated upon CDDP-based chemotherapy in vitro. In contrast, neither piR-L-138 in all four LADCs nor any non-LSCC-related piR-Ls in all four LSCCs were observed to show an obviously consistent and substantial increase upon CDDP, gemcitabine, or docetaxel treatment (Figure S1), indicating that the upregulated piR-L-138 was LSCC-related, which was consistent with the data obtained using RNA sequencing (RNA-seq) (Figure 1B) as we reported previously.10 Together, these results suggest that LSCC-related piR-L-138 is upregulated upon CDDP-based chemotherapy in vitro.
Figure 1.
LSCC-Related piR-L-138 Was Upregulated upon CDDP-Based Chemotherapy Both In Vitro and In Vivo
(A) The expression levels of piR-L-138 upon CDDP, gemcitabine, and docetaxel treatment in four LSCC cell lines. The levels in cells treated with the corresponding control solutions (PBS or DMSO) were controls. (B) Log 2 fold change (LFC) of piR-L-138 from RNA-seq. (C–E) piR-L-138 was induced upon CDDP-based chemotherapy in LSCC tumors but not in LADC tumors in PDX models. All values are averages of three independent replicates, and the error bars reflect mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figures S1 and S2.
To determine whether the upregulated piR-L-138 upon CDDP-based chemotherapy was a cell culture artifact, we analyzed its expression levels in patient-derived xenograft (PDX) tumors developed from tissues of patients with LSCC and LADC, which were used in our previously published study.11 The PDX tumors were from untreated mice or mice treated with CDDP-based chemotherapy (CDDP plus gemcitabine) for 8 and 28 days, respectively. We observed that piR-L-138 levels were significantly increased in the tumors derived from LSCC tissues (patients UMB410 and MDA2131-11) at both time points after the mice had been treated with CDDP-based chemotherapy for 8 and 28 days (Figures 1C and 1D). On the contrary, piR-L-138 showed an obvious downregulation in tumors derived from LADC tissues (patient MDA2131-1) at both time points after the mice had been treated with CDDP-based chemotherapy for 8 and 28 days (Figure 1E), suggesting that piR-L-138 is induced upon CDDP-based chemotherapy in tumors developed from LSCC in vivo.
Next, we wanted to identify the roles of the upregulated piR-L-138 in chemoresistance of LSCC. Given that CDDP is the backbone agent of platinum-based chemotherapy for patients with NSCLC, we used CDDP as the representative agent to verify roles of piR-L-138 in chemoresistance.
Targeting piR-L-138 Enhanced CDDP-Induced Apoptosis in LSCC Cells
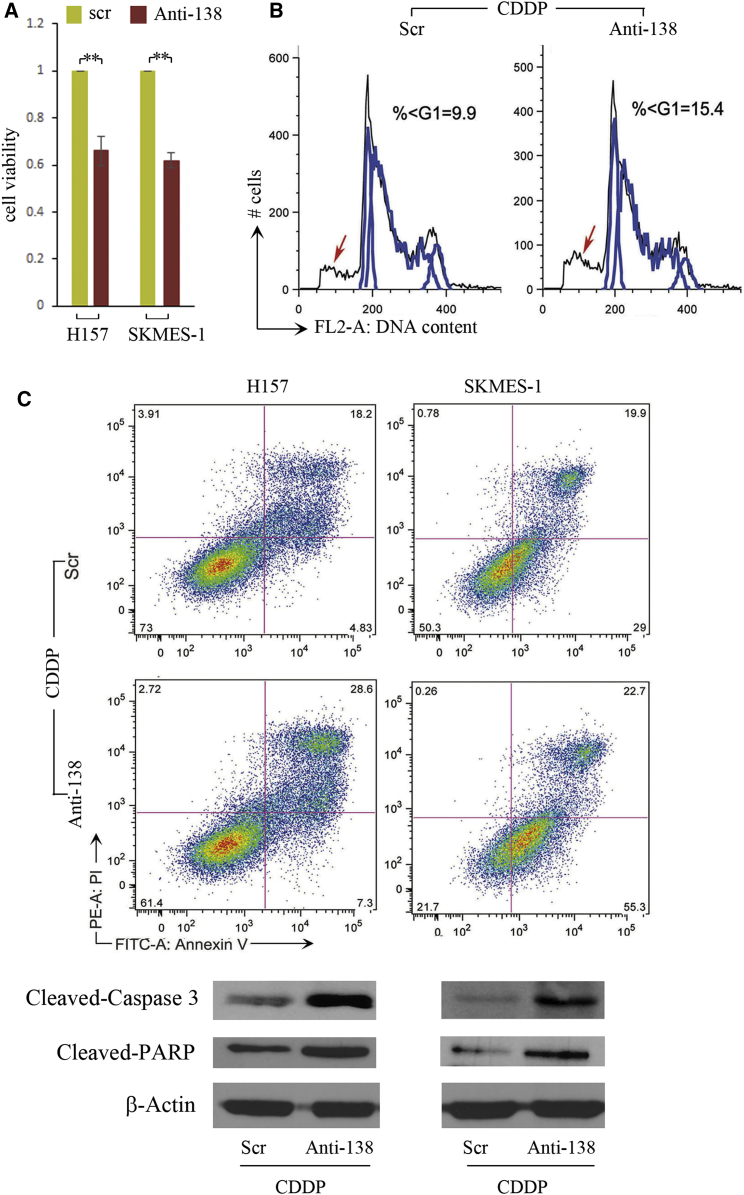
To determine whether the CDDP-induced piR-L-138 played functional roles in vitro, we used DNA oligonucleotides that were complementary to piR-L-138 as antisense (Anti-138) for blocking piR-L-138 because the cRNA oligonucleotides could trigger a short interfering RNA (siRNA)-like response.10, 12 Targeting piR-L-138 followed by CDDP treatment in two LSCC representative cell lines of H157 and SKMES-1, we found that blocking piR-L-138 resulted in decreased cell viability (Figure 2A), which was caused by an increased apoptotic cell population represented by a substantial increase of the sub-G1 fragment (Figure 2B; Figures S3A–S3C). In addition, H157 and SKMES-1 cells treated with Anti-138 showed a significantly increased percentage of Annexin V-positive cells, cleaved Capase-3, and cleaved poly ADP ribose polymerase (PARP) (Figure 2C; Figures S4A–S4C), further confirming that targeting piR-L-138 increases the sensitivity of LSCC cells to CDDP in vitro.
Figure 2.
Targeting piR-L-138 Enhanced CDDP-Induced Apoptosis in LSCC Cells
(A–C) Blocking the upregulated piR-L-138 upon CDDP decreased cell viability (A), increased the sub-G1 population (B), and induced the Annexin V-positive percentage, cleaved Caspase-3, and cleaved PARP (C). All values are averages of three independent replicates, and the error bars reflect mean ± SEM. **p < 0.01. See also Figures S3 and S4.
Targeting piR-L-138 Enhanced CDDP-Induced Apoptosis In Vivo
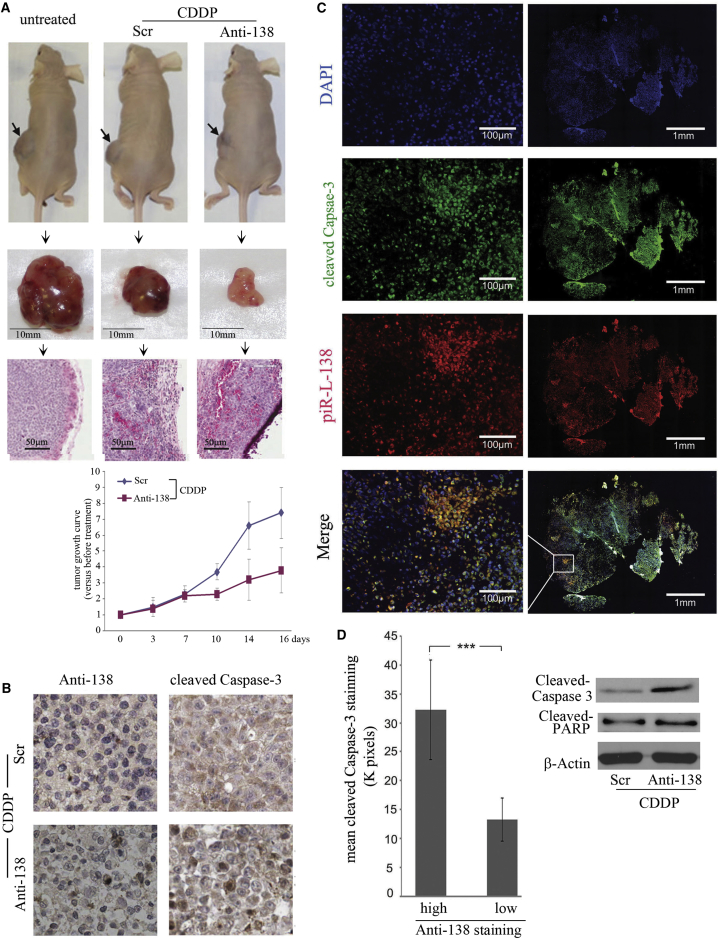
To further confirm the roles of CDDP-induced piR-L-138 in apoptosis, we used a tumor xenograft model to determine whether targeting piR-L-138 could enhance CDDP-induced cell apoptosis of tumors developed from H157 cells inoculated in Nu/Nu mice. H157 cells were inoculated into mice, tumors at least 5 mm in diameter were considered established, and all tumors were confirmed by pathology examination (Figure 3A, right). An intraperitoneal (i.p.) CDDP injection was given on day 1, followed by an intratumoral (i.t.) injection of Scr or Anti-138 oligonucleotides formulated with MaxSuppressor in vivo RNALancerII13 on days 3, 7, 10, 14, and 16, respectively. Compared with tumors formed in untreated mice and in mice treated with CDDP plus Scr, the tumors in mice treated with CDDP plus Anti-138 were significantly smaller (Figure 3A, left and center and growth curves), indicating that targeting piR-L-138 inhibited tumor growth. Further, compared with tumors treated with CDDP and Scr control, tumors treated with CDDP plus Anti-138 exhibited higher expression of cleaved Caspase 3 (Figure 3B), indicating that targeting piR-L-138 enhanced CDDP-induced cell apoptosis in tumors. In addition, we found that tumor areas that showed high concentrated Anti-138 also showed high levels of cleaved Caspase-3 and cleaved PARP (Figures 3C and 3D), further conforming that blocking CDDP-upregulated piR-L-138 accelerates CDDP-induced apoptosis of tumors derived from LSCC cells.
Figure 3.
Targeting piR-L-138 Enhanced CDDP-Mediated Apoptosis in a Xenograft Tumor Model
(A) Representative animals, tumors, and H&E-stained sections of animals with different treatment and growth curves of tumors treated with Scr and Anti-138. (B) The presence of Anti-138 and cleaved Caspase-3 in tumor sections with different treatment. (C) Immunofluorescence-stained cleaved Caspase-3 (red) and Anti-138 (green) in tumor sections of animals treated with CDDP plus Anti-138 (left, 10× magnification of the images at the right). (D) Intensities of cleaved Caspase-3 signals in tumor regions with high (>20,000 pixels) and low (≤20,000 pixels) Anti-138 signals (20 regions for each category). The levels of cleaved Caspase-3 and cleaved PARP of tumors with different treatments were determined by western blot analysis. All values are averages of four independent tumor areas, and error bars represent mean ± SEM. ***p < 0.001.
piR-L-138 Directly Interacts with p60-MDM2
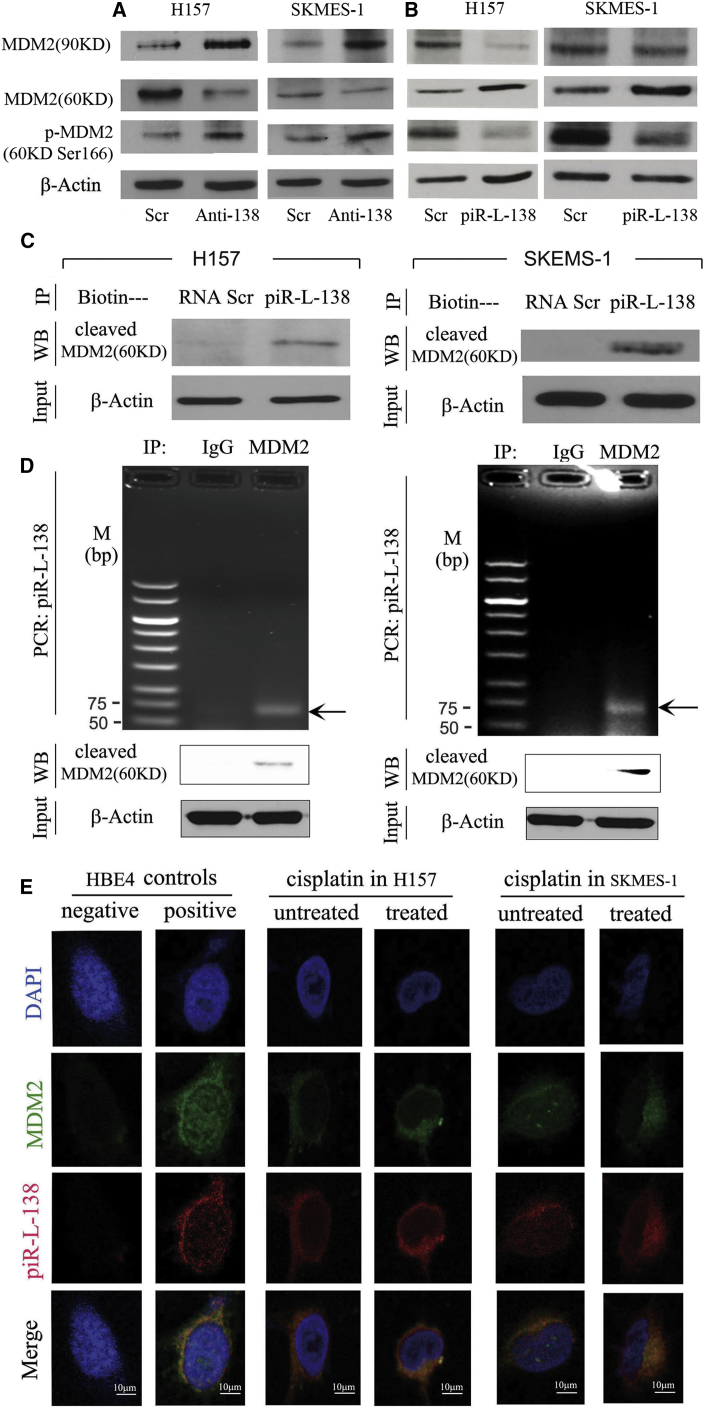
Finally, we wanted to decipher the potential mechanisms underlying CDDP-induced piR-L-138 in chemoresistance. Given that CDDP-induced piR-L-138 inhibits apoptosis and that these two LSCC-representative cell lines harbor p53 mutations,14 we hypothesized that piR-L-138 could be involved in regulating functions of mouse double minute 2 homolog (MDM2) because MDM2 and its three isoforms (90, 75, and 60 kDa) have been implicated in p53-independent apoptosis as well as in chemoresistance.15, 16, 17 To test our hypothesis, we measured the expression levels of total MDM2 and its cleaved isoforms after blocking and ectopic expression of piR-L-138, respectively. As shown in Figure 4A, blocking piR-L-138 increased full-length MDM2 and decreased the 60-kDa cleaved isoform (referred to hereafter as p60-MDM2) upon CDDP treatment, and, in contrast, ectopic expression of piR-L-138 decreased total MDM2 and increased p60-MDM2 upon CDDP treatment (Figure 4B), indicating that the role of piR-L-138 in apoptosis inhibition was related to p60 MDM2. Strikingly, however, we observed that reduced p60-MDM2 was accompanied with an increase in its serine-166 phosphorylation and that increased p60-MDM2 was accompanied by a decrease in its serine-166 phosphorylation (Figures 4A and 4B). Based on the positive relationship between piR-L-138 and p60-MDM2, a negative correlation with serine-166-p60-MDM2, and our experiences with this type of sncRNAs,11 we hypothesized that piR-L-138 could directly bind to p60-MDM2 to play roles in apoptosis.
Figure 4.
piR-L-138 Interacted with p60MDM2
(A and B) Levels of total, cleaved p60-MDM2, and p-cleaved p60-MDM2 in cells with blocking (A) and ectopic expression (B) of piR-L-138, respectively. (C) Pull-down experiment using immobilized Scr RNA or piR-L-138 followed by western blot to detect cleaved p60-MDM2 in CDDP-treated cells. (D) Immunoprecipitation experiment using either immunoglobulin G (IgG) or an anti-p60-MDM2 antibody followed by RT-PCR to detect piR-L-138 in CDDP-treated SCC cells. (E) Co-localization of piR-L-138 and p60-MDM2 analyzed in HBE4 cells and H157 and SKMES-1 cells left untreated or treated with CDDP, as indicated by FISH analysis.
To determine whether piR-L-138 directly interact with p60-MDM2, biotin-conjugated piR-L-138 RNA oligonucleotides were used as bait to pull down its potential binding proteins in CDDP-treated H157 and SKEMS-1 cells. We found that p60-MDM2 was readily detectable in the pull-down products using bio-piR-L-138 but not in the products of bio-Scr RNA control (Figure 4C), indicating that piR-L-138 bound to p60-MDM2. To confirm the interaction of piR-L-138 and p60-MDM2, we performed an immunoprecipitation (IP) assay using an anti-p60-MDM2 antibody followed by RT-PCR using primers specific for piR-L-138. Although piR-L-138 was not detectable in the IP products of a control IgG antibody, piR-L-138 was easily detected in the products using the anti-p60-MDM2 antibody (Figure 4D).
To further validate the interaction between piR-L-138 and p60-MDM2 at the individual cell level, we performed a fluorescence in situ hybridization (FISH) assay using a digoxin (DIG)-labeled DNA probe complementary to piR-L-138 and anti-p60-MDM2 antibody. We first tested the probes in HBE4 cells, which express a high level of piR-L-138, to ensure the sensitivity and specificity of the piR-L-138 probe and p60-MDM2 antibody (Figures 1B and 4E), and we then measured their distribution upon CDDP treatment. We found that both piR-L-138 and p60-MDM2 were predominantly co-localized at the perinuclear area when unchallenged and showed an enhanced and polarized perinuclear pattern upon CDDP treatment in both H157 and SKMES-1 cells (Figure 4E). Together, these results suggest that piR-L-138 directly interacts with p60-MDM2 in response to CDDP treatment in p53-mutated LSCC H157 and SKMES-1 cells.
Discussion
PIWI-interacting RNAs (piRNAs) have been identified to play roles in transposon silencing, heterochromatin modification, and germ cell maintenance.18, 19 Because of our limited knowledge regarding their biogenesis, most studies focused on their functions in the renewal of germline cells and development.20, 21, 22, 23 Their potential roles in somatic tissues, particularly in pathogenesis such as tumorigenesis, although poorly studied, have recently been recognized.10, 18, 19, 24
In our recently published study, we demonstrated the presence of a type of functional sncRNA class that has certain similarity with piRNAs but is expressed in somatic lung bronchial epithelial cells, and we termed them piR-Ls.10 Importantly, piR-Ls not only showed differentially expressed patterns among normal HBE, LSCC, and LADC cells but also played key roles in fundamental biological activities of HBE, LSCC and LADC cells by directly binding to their targeted phosphorylated proteins,10 suggesting a biological role of piRNA-Ls in lung tumorigenesis and, possibly, as deterring factors of cellular behaviors for individual tumors. Because little has been studied regarding this type of sncRNAs, in this study, we explored the potential effect of piR-Ls in chemoresistance to the gold standard first-line platinum-based chemotherapy for treating patient with NSCLC.
Based on the expression levels of a piR-L that could distinguish a certain type of lung cancer from all other subtypes, we optimized a panel of LSCC-related and LADC-related piR-Ls, respectively. Our initial intention was to identify an association between these LSCC-/LADC-related piR-Ls and sensitivity to the chemotherapeutic agents commonly used for patients with NSCLC. We first focused on LSCC because few therapeutic targets are validated for patients with LSCC.5 We observed some relationships between piR-Ls and the sensitivities to agents; characteristically, piR-L-138 was found to be consistently and substantially upregulated after CDDP-based chemotherapy treatment in LSCC cell lines. The results that piR-L-138 was significantly downregulated in LSCC, could distinguish LSCC from both HBE and LADC cells, and was consistently upregulated by CDDP-based agents suggested that piR-L-138 might play a critical role in CDDP-based chemotherapy in LSCC. It is important to note that the upregulation of piR-L-138 was not only observed in LSCC cell lines but also in the PDX LSCC models treated with a CDDP-based regimen, indicating that the effect also occurs in LSCC tissues in vivo.
Our results clearly showed that the upregulated piR-L-138 was critical in protecting LSCC cells from CDDP-induced apoptosis and that inhibiting piR-L-138 enhanced CDDP-mediated apoptosis, suggesting targeting piR-L-138 as a strategy to boost CDDP efficacy for patients with LSCC. However, a major challenge of using a nucleotide-based treatment strategy is the delivery. Our study provides a proof-of-concept result for the potential use of Anti-138 to enhance CDDP-mediated apoptosis of LSCC cells in vivo because we were using local injection with a sub-optimal dose, which produced only modest anti-tumor activity. We were, therefore, focusing on determining whether Anti-138 increased apoptosis at the sites of Anti-138 injection by analyzing tumor tissue sections for cleaved Caspase-3 and cleaved PARP, which are indicators of apoptosis. The concept was supported by the close relationship between higher levels of cleaved Caspase-3 and cleaved PARP and higher concentrated Anti-138 (Figure 3).
We identified a mechanism underlying chemoresistance caused by CDDP-based chemotherapy for patients with LSCC, where the functional piR-L-138 is upregulated by CDDP-based agents and interacts with p60-MDM2 to inhibit apoptosis, and targeting its upregulation increases LSCC sensitivity to chemotherapy agents, indicating that piR-L-138 is a key contributor to CDDP-based chemoresistance and that targeting piR-L-138 could be a strategy to overcome chemoresistance for patients with LSCC. In our previous study, we have shown that piR-L-163 could directly bind and regulate phosphorylated protein activities.10 In this study, we explored the possibility that piR-L-138 directly interacts with the p60-MDM2 fragment upon CDDP treatment in LSCC cells. The direct interaction between piR-L-138 and p60-MDM2 was demonstrated by immunoprecipitation, piR-L-138-specific pull-down, and immunofluorescence co-localization assays. Although p60-MDM2 was reported to be induced in p53-wild-type,25, 26, 27, 28 we identified that the upregulated piR-L-138, upon CDDP-based chemotherapy, inhibited CDDP-induced apoptosis by interacting with the p60-MDM2 in P53-mutated LSCC cells. Strikingly and interestingly, we found that there was a negative correlation between p60-MDM2 and its serine-166 phosphorylation (Figure 4A), indicating the possibility that the interaction of piR-L-138 and p60-MDM2 could inhibit its phosphorylation to enhance apoptosis. Also, there could be a totally unknown mechanism underlying p60-MDM2 and serine-166-p60-MDM2 in regulating apoptosis by chemotherapy in p53-mutated LSCC cells. In addition, further studies are necessary to determine how piR-L-138 affects p60-MDM2 phosphorylation.
Materials and Methods
Cell Culture and Oligonucleotides
For detailed information about all cell lines used here, please refer to our previous publication.10 All cell lines were recently genotyped for their authentication and routinely treated to prevent mycoplasma growth. All DNA or RNA oligonucleotides were synthesized by Integrated DNA Technologies.
Cell Transfection
Cell transfection was performed as described previously.10, 29, 30 In brief, transfection experiments were performed by using Opti-MEM medium and Lipofectamine 3000 (Invitrogen) for DNA oligonucleotides or Lipofectamine RNAiMAX (Invitrogen) for RNA oligonucleotides according to the manufacturer’s instructions.
sncRNA Extraction, RT-PCR, and Real-Time PCR
sncRNA extraction, RT-PCR, and real-time PCR were performed as described previously.10 In brief, we used adaptors for sncRNA ligation, genome-specific primer (GSP) for reverse transcription, and specific primers for target piR-Ls amplification. An adaptor (/5′rapp/5′-CTGTAGGCACCATCAAT-3′/3′ddc/) with both 5′ and 3′ modification, GSP (5′-CAAGCAGAAGACGGCATACGAATTGATGGTGCCTACAG-3′), a common reverse primer (5′- CAAGCAGAAGACGGCATACGA-3′), and a piR-L-138-specific forward primer (5′- ACTTTAGCTCTAGAATTACTCTGAGAC-3′) were used. The amplification conditions were as follows: denaturation at 95°C for 30 s (5 min for the first cycle), annealing at 60°C for 20 s, and extension at 72°C for 20 s in 25 cycles for RT-PCR and in 40 cycles for real-time PCR.
Chemotherapeutic Drug Preparation
A stock solution of cisplatin (Sigma) was dissolved in PBS to a concentration of 1.7 mmol/L (0.5 mg/mL), a stock solution of docetaxel (Enzo Life Sciences) was dissolved in DMSO to a concentration of 10 μmol/L (8 μg/mL), and a stock solution of gemcitabine (Gimzar, Lilly) was dissolved in PBS to a concentration of 10 mmol/L (3 mg/mL). All working solutions were freshly prepared in cell culture medium to the needed concentrations.
Cell Viability and IC50 Determination
Cell viability and IC50 were determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Life Technologies) assay as performed previously,10, 29, 30 and IC50 was defined as the drug concentration that was required for 50% inhibition.
Use of Anti-138 for Blocking piR-L-138 Expression
To avoid triggering uncertain siRNA effects,10, 12 we used complementary DNA oligonucleotides as antagonists targeting piR-L-138. The sequence was 5′-AGGTCTCAGAGTAATTCTAGAGCTAAAGT-3′, and the corresponding scrambled sequence was 5′-GATACCAGGGACATACGCTTGATCCTAGC-3′. Transfection experiments were performed by using Opti-MEM medium and Lipofectamine 3000 (Invitrogen), and the final concentration was 100 nM.
Cell Cycle and Apoptosis Analysis
Cell cycle and apoptosis analyses were performed as described previously.10, 29, 30 In brief, cell cycle distribution and fluorescein isothiocyanate (FITC) Annexin V/PI (BD Pharmingen) for the apoptosis assay were analyzed by flow cytometry (Becton Dickinson), and DNA content and percentage of apoptosis cells were analyzed on a FACScan flow cytometer in combination with BD lysis software (Becton Dickinson).
Western Blot and Antibodies
Primary antibodies used were as follows: anti-cleaved PARP (1:1,000, clone Asp214, catalog no. 9541s, Cell Signaling Technology), anti-MDM2 (1:200, clone N-20, lot L099, catalog no. 813, Cell Signaling Technology), anti-P21 (1:200, clone H-164, catalog no. 756, Cell Signaling Technology), anti-phospho-FoxO3a (1:1,000, clone Ser253, catalog no. 9444, Cell Signaling Technology), anti-β-actin (1:1,000, catalog no. A228, Sigma-Aldrich), human/mouse cleaved Caspase-3 (Asp175) antibody (1:1,000, catalog no. MAB835, R&D Systems), and cleaved PARP (1:1,000, catalog no. 9546, Cell Signaling Technology). Secondary antibodies included goat anti-mouse (1:5,000, catalog no. 31160, Pierce) and goat anti-rabbit (1:5,000, catalog no. 31460, Pierce).10, 11, 29, 30 Protein extraction from cells was performed as we did before, and proteins were extracted from formalin-fixed, paraffin-embedded (FFPE) tissues using the Qproteome FFPE tissue kit (catalog no. 37623, QIAGEN) according to the instructions.
Pull-Down Analysis and IP
We followed the steps for sncRNAs pull-down analysis and IP as we did previously,10 and the specific sequences were as follows:
piR-L-138 /5′Biosg/AGGTCTCAGAGTAATTCTAGAGCTAAAGT-3′
Control /5′Biosg/GAUACCAAGGACAUACGCUUAUGCAUGCUA-3′.
Immunohistochemistry
Immunohistochemistry (IHC) was performed as we described previously.10, 11 Tumor samples were fixed in 10% neutral formalin and embedded in paraffin, and sections were prepared less than 5 mm thick, deparaffinized in Xylene, and rehydrated. For H&E staining, slides were processed according to our standard protocols.
FISH and Immunofluorescence Staining
Cell chamber slides (Fisher Scientific) were fixed with 4% paraformaldehyde (PFA) for half an hour at room temperature (RT), and fissure slides were deparaffinized and rehydrated using standard protocols.10, 11 Slides were incubated with 40 μg/mL protein kinase K (Roche) at 37°C for 5 min, followed by a termination reaction in 0.2% Glycine (Affymetrix USB) for 1 min before processed to pre-hybridization buffer (50% Formamide, 1× saline-sodium citrate (SCC), 0.5 μg/mL yeast RNA, and 1× Denhardt solution) at 40°C for 2 hr. Probes for piR-L-138 and Anti-138 were as follows:
piR-L-138: /5′DigN/AGGTCTCAGAGTAATTCTAGAGCTAAAGT-3′
Anti-138: /5′DigN/ACTTTAGCTCTAGAATTACTCTGAG-3′.
We added hybridization buffer (Enzo) with DIG-labeled probes and incubated this overnight at 37°C. Then we incubated the slides with the primary antibodies MDM2 (1:500) or cleaved Caspase3 (1:1,000) at 4°C overnight after washing the slides in 2× SCC, 1× SCC, and 0.2× SCC for 10 min, followed by blocking in phosphate-buffered saline Tween 20 (PBST) with 5% goat serum (Vector Laboratories) at RT for 2 hr. Then we incubated the slides in 2.5% goat serum with DyLight 594 anti-DIG (1:500, catalog no. DI-7594, Vector Labs) or FITC anti-rabbit (1:750, catalog no. FI-1000, Vector Labs) for 2 hr in the dark at RT. We sealed the slides using mounting medium with DAPI for fluorescence (Vector Labs) and prepared them for scanning. For images, we used Zeiss LSM 700 for scanning the slides with the HBE4, H157 and SKMES-1 cell lines, Tumor tissues slides were scanned by a Cytation 3 cell imaging multi-mode reader (BioTek Instruments) and analyzed BioTek’s new Gen5 software. The high and low staining was distinguished by pixels of 20,000 as the cutoff, and 20 paired lesions were calculated.
Animal Models
Tumor tissues used to determine piR-L-138 expression levels were from patient-derived NSCLC xenograft models we already prepared in our previous study.11 For xenograft mouse models, 5 × 106 H157 cells in 100 μL culture medium were inoculated into the lower back and anterior chest of 6- to 8-week-old female athymic nude (Nu/Nu) mice, which are defective for development of thymic epithelium and lack cell-mediated immunity, according to the protocols approved by Institutional Animal Care and Use Committees (IACUC no.120901). Tumors that reached at least 5 mm in diameter were considered established. The tumor sizes were measured twice a week, and tumor volumes were calculated using the formula (π/6) × length (L) × width (W) × height (H) of tumors, where L and W represent the longest and shortest tumor dimensions, respectively. Each established tumor model was confirmed by pathology examination. An i.p. CDDP (1 mg/kg body weight/week) injection was given on day 1, followed by an i.t. injection of a Scramble or Anti-138 DNA oligo formulated with MaxSuppressor in vivo RNALancerII (BIOO Scientific) at 3 μg/20 μL on days 4, 7, 11, and 14, respectively. Six mice were used for each group, and the treatments lasted 16 days.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism 6 (GraphPad). Significance of the difference between groups was calculated by Student’s unpaired t test as we did before10, 11, 29, 30 and by two-way ANOVA (Tukey’s and Bonferroni’s multiple comparisons test). Welch’s corrections were used when variances between groups were unequal. The results are reported as mean ± SEM, and p < 0.05 is considered statistically significant.
Author Contributions
Y.M. and L.M. provided conceptual framework. Y.M. designed experiments. Y.M., Y.W., T.G., M.Z.M., D.C., and J.Z. performed the experiments and statistical analysis. Y.Z. and W.L. contributed to data interpretation and provided intellectual input. Y.M., L.M., and Y.W. wrote the manuscript. All authors reviewed, participated in revision, and approved the final version of the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
We thank Drs. Abraham Schneider and Tao Mao for their assistance and discussions.
Footnotes
Supplemental Information includes four figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.01.003.
Supplemental Information
References
- 1.Herbst R.S., Heymach J.V., Lippman S.M. Lung cancer. N. Engl. J. Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender E. Epidemiology: The dominant malignancy. Nature. 2014;513:S2–S3. doi: 10.1038/513S2a. [DOI] [PubMed] [Google Scholar]
- 3.Paez J.G., Jänne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Kwak E.L., Bang Y.J., Camidge D.R., Shaw A.T., Solomon B., Maki R.G., Ou S.H., Dezube B.J., Jänne P.A., Costa D.B. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pao W. New approaches to targeted therapy in lung cancer. Proc. Am. Thorac. Soc. 2012;9:72–73. doi: 10.1513/pats.201112-054MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottesman M.M., Lavi O., Hall M.D., Gillet J.P. Toward a Better Understanding of the Complexity of Cancer Drug Resistance. Annu. Rev. Pharmacol. Toxicol. 2016;56:85–102. doi: 10.1146/annurev-pharmtox-010715-103111. [DOI] [PubMed] [Google Scholar]
- 7.Dempke W., Voigt W., Grothey A., Hill B.T., Schmoll H.J. Cisplatin resistance and oncogenes--a review. Anticancer Drugs. 2000;11:225–236. doi: 10.1097/00001813-200004000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer. 2011;71:3–10. doi: 10.1016/j.lungcan.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 9.Derman B.A., Mileham K.F., Bonomi P.D., Batus M., Fidler M.J. Treatment of advanced squamous cell carcinoma of the lung: a review. Transl. Lung Cancer Res. 2015;4:524–532. doi: 10.3978/j.issn.2218-6751.2015.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mei Y., Wang Y., Kumari P., Shetty A.C., Clark D., Gable T., MacKerell A.D., Ma M.Z., Weber D.J., Yang A.J. A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells. Nat. Commun. 2015;6:7316. doi: 10.1038/ncomms8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J., Ma M.Z., Ren H., Liu Z., Edelman M.J., Pan H., Mao L. Anti-HDGF targets cancer and cancer stromal stem cells resistant to chemotherapy. Clin. Cancer Res. 2013;19:3567–3576. doi: 10.1158/1078-0432.CCR-12-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 13.Wiggins J.F., Ruffino L., Kelnar K., Omotola M., Patrawala L., Brown D., Bader A.G. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thierry, S. (2007). Handbook of p53 Mutation in Cell Lines. pp. 97–101. https://www.researchgate.net/profile/Thierry_Soussi/publication/238766403_Handbook_of_p53_mutation_in_cell_lines/links/53e8e6f90cf2fb1b9b642b9e/Handbook-of-p53-mutation-in-cell-lines.pdf.
- 15.Zhang Z., Li M., Wang H., Agrawal S., Zhang R. Antisense therapy targeting MDM2 oncogene in prostate cancer: Effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc. Natl. Acad. Sci. USA. 2003;100:11636–11641. doi: 10.1073/pnas.1934692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z., Wang H., Li M., Agrawal S., Chen X., Zhang R. MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J. Biol. Chem. 2004;279:16000–16006. doi: 10.1074/jbc.M312264200. [DOI] [PubMed] [Google Scholar]
- 17.Bouska A., Eischen C.M. Mdm2 affects genome stability independent of p53. Cancer Res. 2009;69:1697–1701. doi: 10.1158/0008-5472.CAN-08-3732. [DOI] [PubMed] [Google Scholar]
- 18.Mei Y., Clark D., Mao L. Novel dimensions of piRNAs in cancer. Cancer Lett. 2013;336:46–52. doi: 10.1016/j.canlet.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross R.J., Weiner M.M., Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353–359. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 21.Lee H.C., Gu W., Shirayama M., Youngman E., Conte D., Jr., Mello C.C. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashe A., Sapetschnig A., Weick E.M., Mitchell J., Bagijn M.P., Cording A.C., Doebley A.L., Goldstein L.D., Lehrbach N.J., Le Pen J. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirayama M., Seth M., Lee H.C., Gu W., Ishidate T., Conte D., Jr., Mello C.C. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Z., Hu H.Y., Jiang X., Maierhofer V., Neb E., He L., Hu Y., Hu H., Li N., Chen W., Khaitovich P. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 2011;39:6596–6607. doi: 10.1093/nar/gkr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L., Marechal V., Moreau J., Levine A.J., Chen J. Proteolytic cleavage of the mdm2 oncoprotein during apoptosis. J. Biol. Chem. 1997;272:22966–22973. doi: 10.1074/jbc.272.36.22966. [DOI] [PubMed] [Google Scholar]
- 26.Pochampally R., Fodera B., Chen L., Shao W., Levine E.A., Chen J. A 60 kd MDM2 isoform is produced by caspase cleavage in non-apoptotic tumor cells. Oncogene. 1998;17:2629–2636. doi: 10.1038/sj.onc.1202206. [DOI] [PubMed] [Google Scholar]
- 27.Araki T., Shinoda S., Schindler C.K., Quan-Lan J., Meller R., Taki W., Simon R.P., Henshall D.C. Expression, interaction, and proteolysis of death-associated protein kinase and p53 within vulnerable and resistant hippocampal subfields following seizures. Hippocampus. 2004;14:326–336. doi: 10.1002/hipo.10184. [DOI] [PubMed] [Google Scholar]
- 28.Oliver T.G., Meylan E., Chang G.P., Xue W., Burke J.R., Humpton T.J., Hubbard D., Bhutkar A., Jacks T. Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol. Cell. 2011;43:57–71. doi: 10.1016/j.molcel.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei Y.P., Liao J.P., Shen J., Yu L., Liu B.L., Liu L., Li R.Y., Ji L., Dorsey S.G., Jiang Z.R. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31:2794–2804. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mei Y., Zhang P., Zuo H., Clark D., Xia R., Li J., Liu Z., Mao L. Ebp1 activates podoplanin expression and contributes to oral tumorigenesis. Oncogene. 2014;33:3839–3850. doi: 10.1038/onc.2013.354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.