Abstract
Exonic duplications account for 10%–15% of all mutations in Duchenne muscular dystrophy (DMD), a severe hereditary neuromuscular disorder. We report a CRISPR (clustered regularly interspaced short palindromic repeat)/Cas9-based strategy to correct the most frequent (exon 2) duplication in the DMD gene by targeted deletion, and tested the efficacy of such an approach in patient-derived myogenic cells. We demonstrate restoration of wild-type dystrophin expression at transcriptional and protein level in myotubes derived from genome-edited myoblasts in the absence of selection. Removal of the duplicated exon was achieved by the use of only one guide RNA (gRNA) directed against an intronic duplicated region, thereby increasing editing efficiency and reducing the risk of off-target effects. This study opens a novel therapeutic perspective for patients carrying disease-causing duplications.
Keywords: dystrophin, CRISPR/Cas9, duplication, lentivirus, gene editing
Graphical Abstract

Introduction
Duchenne muscular dystrophy (DMD; OMIM #310200) is an X-linked recessive muscle-wasting disease, characterized by progressive weakening of skeletal, respiratory, and cardiac muscle followed by necrosis and fibrosis. DMD affects ∼1:4,087 live male births and is associated with delayed motor milestones.1 DMD is caused by absent or reduced levels of the dystrophin protein, which plays an important role in the stabilization of sarcolemma. In the absence of dystrophin, muscle strength steadily declines in the first decade of life, accompanied by progressive degeneration and regeneration of muscle fibers and their replacement by fat and connective tissue.2 Lack of dystrophin affects as well the cardiac muscle and the diaphragm, resulting in respiratory and heart failure and premature death.
DMD occurs as a result of mutations in the DMD gene that lead to premature termination of translation. The most frequent mutations are exonic deletions and duplications that induce a frameshift in the protein-coding sequence.3 Skipping of in-frame mutated exons and deletions by antisense oligonucleotides (AONs) has long been considered a promising therapeutic strategy for DMD. This strategy aims at artificially inducing favorable exclusion of mutated/out-of-frame exons to restore the correct reading frame.4 The exciting prospect of an effective AON-based exon skipping therapy was raised by promising data from early clinical trials that were, however, not confirmed by double-blind, placebo-controlled phase IIb and III trials, which failed to meet efficacy endpoints.5 Independently from its efficacy, AON-based exon skipping can target only a limited number of mutations: skipping of exon 51, which could restore a correct reading frame in the largest proportion of DMD mutations, would be applicable to just ∼10% of DMD patients.6, 7 Moreover, this and similar treatments are not applicable to out-of-frame mutations and are of limited efficacy for exon duplications,8, 9 therefore requiring a multi-exon skipping approach.10
Among the reported DMD mutations, duplications represent a distinct group accounting for ∼10%–15% of those reported in the Leiden database,2 although their incidence may be higher,11 Duplications have been generally neglected by therapeutic approaches. Wein et al.12 recently reported a successful attempt at inducing out-of-frame skipping of exon 2, causing an alternative translation initiation in exon 6 and leading to expression of a functional N-truncated dystrophin. These results support a potential therapeutic approach for patients with mutations within the 5′ exons of DMD, but it would be ineffective for all the other duplications.
There is no definitive cure for DMD, and for a long time the only treatment option has been corticosteroid administration, a palliative management associated with many side effects. However, two drugs have recently obtained conditional approval in Europe (Ataluren, a read-through drug for premature stop mutations)13 and the United States (Eteplirsen, an antisense molecule to restore the open reading frame in patients with mutations correctable by skipping exon 51).14
Genome editing technology uses synthetic nucleases to introduce targeted modification at specific loci in the genome by exploiting the endogenous cellular DNA repair mechanisms. These engineered enzymes are commonly based on zinc finger nucleases (ZFNs),15 transcription activator-like effector nucleases (TALENs),16 meganucleases,17 and, lately, the RNA-guided clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system.18 These tools have been used to create new animal models19, 20, 21 and, recently, in vitro and in vivo therapeutic strategies for DMD22, 23, 24, 25 aimed at restoring disrupted reading frames by deleting instead of skipping mutated/out-of-frame exons. Compared to AON-mediated exon skipping, which involves repeated injection of the therapeutic molecules with toxicity and variable tissue uptake,26, 27 the exon deletion strategy would require only one administration of the gene-editing system to achieve full exclusion of the targeted exon. Restoration of the reading frame for some duplication by excising one copy of the duplicated exon could permit expression of a normal dystrophin transcript, as recently demonstrated for a multi-exonic duplication of 139 kb28 and allow synthesis of a normal dystrophin isoform as compared to the restoration of the reading frame around the more common deletions that can generate a less severe, Becker muscular dystrophy (BMD)-like phenotype.
We report a CRISPR/Cas9 strategy to remove the most frequent duplication in the DMD gene, applicable to all patients with exon 2 duplications. The deletion restores synthesis of wild-type dystrophin at the transcriptional and protein level, adding relevant information on the efficiency of the gene-editing approach and the correct localization of the resulting protein.
Results
Strategy for the Correction of Duplications in DMD and gRNA Rational Design
Our strategy to target duplications relies on the hypothesis that a single guide RNA (gRNA) designed over a tandem duplicated region will cut twice, leading to the deletion of the region between the two double-strand breaks (DSBs), thereby removing the duplication. This approach has been independently validated by the Cohn lab (University of Toronto) to remove a duplication of the exons 18–30 in the DMD gene.28
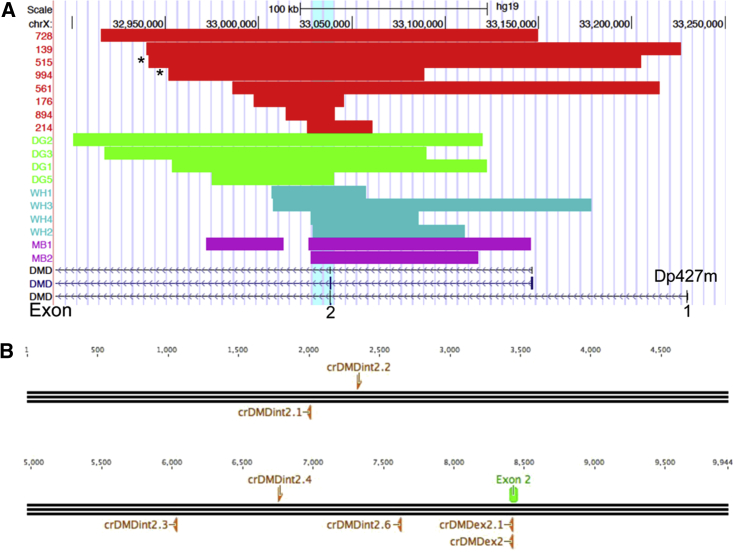
To identify editing targets able to correct the DMD duplication of the exon 2 (dup2) in all patients bearing this type of mutation, we collected DNA from eight patients and characterized the extension of the copy number variation with the DMD-CGH array.29 Data from ten more patients were collected from the literature29, 30, 31, 32 and aligned with the other duplications analyzed in order to identify a region shared among all the patients studied (Figure 1A). Interestingly, we identified duplicated regions spanning 25 to 286 kb with no common breakpoints.
Figure 1.
CGH Profile of DMD Exon 2 Duplications and CRISPR Design
(A) Breakpoint analysis of DMD exon 2 duplications from different cohorts (Cochin, red; Bovolenta et al.,29, 30 purple, DelGaudio et al.,31 green; White et al.,32 turquoise) allowed us to identify the minimal common duplicated region (light blue shade); (B) Diagram representing the ∼10 kb of the minimal common duplicated region and the position and orientation of the gRNAs (orange) designed with ZiFiT website in respect to the exon 2 of the DMD gene (green). The asterisks (*) indicate the mutation of the two patients tested in the present study.
A duplicated minimum common region (MCR) of about 10 kb was uploaded in ZiFiT and gRNAs of 18 nucleotides were selected based on the available guidelines and on the smallest number of potential off targets with a maximum of two mismatches. We also designed two gRNAs (one of which was 19 nt) targeting the coding sequence of the exon 2 in order to confirm the evidence that shifting the reading frame in the early codons should trigger the translational restart at exon 6 as previously demonstrated by an exon skipping approach (Figure 1B).12
gRNAs were named according to the following code: cr (crispr), DMD, int (intron), or ex (exon) 2 and numbered from 1 to 6 (the crDMD notation have been omitted in Figures 2E, 2F, and 3 to limit redundancy). Repeated regions and functional splicing sequences were excluded from the design. All gRNA sequences were cloned in the MLM3636 plasmid backbone (Table S1).
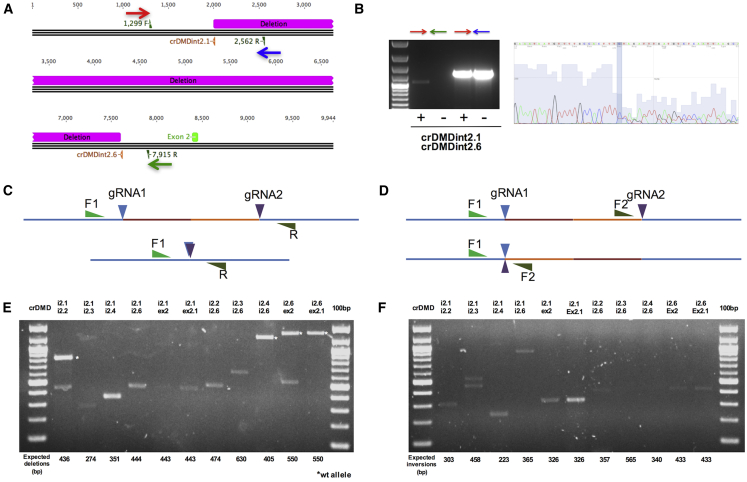
Figure 2.
Detection of Deletion and Inversions after Transfection of Two gRNAs in Myogenic Cells
(A) Schematic representation of the 5.6-kb deletion created by transfecting the crDMDint2.1 and int2.6 gRNAs in a control myogenic cell line. The primers used to amplify across the deletion breakpoint and the wild-type sequence are shown as colored arrows: red, forward common primer; blue, wild-type-specific reverse primer; green, deletion-specific reverse primer. (B) PCR amplification and sequencing of the wild-type and deleted products. The PCR amplification revealed a product corresponding to the deleted sequence of about 1.1 kb only in treated cells (left panel, first lane), whereas the amplification of the wild-type sequence was obtained in both treated and untreated cells with a reduced amplification evident in treated cells. Sequence analysis of the deleted product showed the presence of two products with the only difference of a thymine at the breakpoint causing a shifting starting from the shaded column (right panel). (C and D) Schemes representing the strategies used to amplify the deletions and inversions caused by two gRNAs. The same forward primer located upstream the first gRNA binding site (F1) was used in both approaches in combination with a reverse one downstream to the second gRNA binding site for deletions detection (C) and a second forward primer (F2) upstream the second gRNA binding site for inversions amplification (D). (E) PCR products resulting from the amplification of the deletion breakpoints. The combination of the gRNAs used is reported above the image, whereas the expected products size in base pairs is displayed below it. All the bands where excised and sequenced or directly sequenced (Figure S2). (F) PCR products resulting from the amplification of the inversions breakpoints. The combination of the gRNAs used is reported above the image, whereas the expected products size in base pairs is displayed below it. All the bands where excised and sequenced or directly sequenced (Figure S2). A PCR product resulting from the nonspecific amplification was found in the second lane (i2.1/i2.3, upper band). An unexpected product was found in the fourth lane upon treatment with i2.1 and i2.6; sequencing reported the presence of a complex rearrangement and coexistence of an inversion and deletion between the two gRNAs (Figure S2). T, treated; UT, untreated.
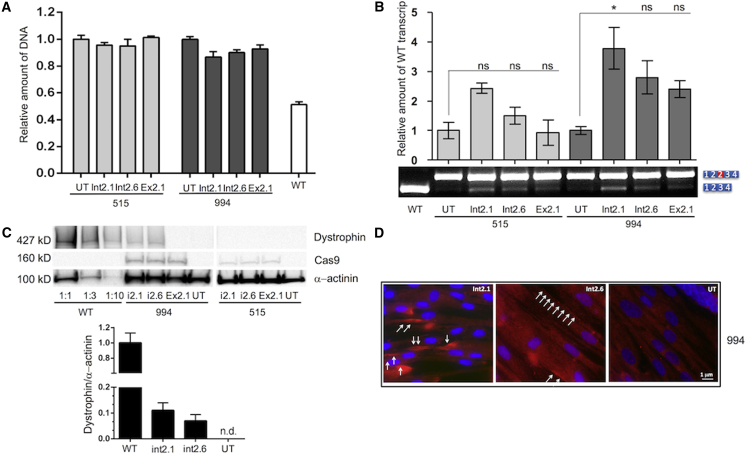
Figure 3.
DNA, RNA, and Protein Expression Analysis after Infection with LentiCRISPRv2
(A) Histograms reporting the genomic analysis by real-time PCR of the duplicated sequence. Transduced cells present reduction of the duplication in respect to untreated cells (UT) set to 1. Wild-type cells were analyzed as positive control. (B) RT-PCR was performed with primers designed on the exon 1 and 4. One microliter of the displayed PCR was loaded on a high-sensitivity DNA chip and run on the Bioanalyser 2100 (Agilent). Quantification of the peak obtained was performed calculating the ratio of the area of the normal transcript and the sum of the area of normal and duplicated transcript multiplied for 100 and normalized by setting the untreated (UT) as one. *p < 0.05 (Kruskal-Wallis test with post hoc Mann-Whitney test from three independent experiments). ns, not significant. (C) Western blot from treated and untreated myotubes of patient 994 and 515, with antibodies against dystrophin, Cas9 and α-actinin as loading normalizer. The amount of dystrophin was normalized to that of actinin by densitometric analysis. WT, wild-type. (D) Immunocytochemistry of dystrophin (red) and nuclei (blue) in myotubes from patient 994. Scale bar, 1 μm. All error bars represent the SD for three independent experiments.
Testing of gRNAs Activity by Plasmid Transfection in HEK293T and Myogenic Cell Lines
To validate the ability of gRNAs to induce DSBs, we transfected each gRNA and the Staphylococcus pyogenes’s Cas9 (spCas9, JDS246) in HEK293T cells and performed the T7E1 assay. Interestingly, the PCR system amplifying across the gRNA crDMDint2.1 and crDMDint2.2 regions incorporates a simple tandem repeat (TG) that creates a heteroduplex that is cleaved into two fragments when digested with the T7E1 enzyme (Figure S1).
To test the gRNAs in myogenic cells, we obtained primary myoblasts from two DMD patients with large duplications of 137 and 263 Kb, respectively, encompassing the exon 2 (Figure 1A; #994 and #515). These cells have been immortalized as previously described.33 Due to low transfection efficiency (<20%, data not shown) of myoblasts, we chose to test the presence of deletions caused by two gRNAs, as they can be easily discriminated from the wild-type sequences. Hence, we co-transfected Cas9 and different combinations of two gRNA in a wild-type myogenic cell line and tested the ability of the two gRNAs to delete a genomic region. We first tested the two gRNA crDMDint2.1 and crDMDint2.6 and detected a deletion of 5.6 kb resulting in about a 20% of reduction in the amplification of the wild-type region in treated cells when compared to untreated cells (Figure 2A). The PCR product corresponding to the deleted sequence was only detectable in treated cells. Sequencing of the latter revealed the coexistence of two products differing from each other for the insertion of one thymine at the breakpoint (Figure 2B).
As previously reported by Kraft et al.,34 the use of two synthetic gRNAs targeting two different positions of the same chromosome induces deletions, inversions and duplications through non-homologous end joining (NHEJ). Hence, we tested the occurrence of deletions and inversions by specifically designed PCR (Figures 2C and 2D) amplifying and sequencing the expected deletions and inversions of different range between the target sites of the tested gRNAs (Figures 2E and 2F). A representation of deletions and inversions obtained with two gRNAs and the sequence of the respective breakpoints is available in Figure S2.
Overall, five out of seven of our gRNAs resulted to have activity in HEK293T cells (Figure S1) and were able to introduce deletions (80%) and inversions (72%) when used in couples in a myogenic cell line (Figure 2).
Targeted Removal of a Large Duplication in Myogenic Cells by Lentiviral Delivery of CRISPR/Cas9 without Selection
In order to increase the efficacy and maximize our results in cell lines, we cloned the more efficient gRNAs in the LentiCRISPRv2 vector (Addgene Plasmid #52961).35 We did not select the infected cells by antibiotics resistance as the immortalization process takes advantage of two lentiviruses and one shares the puromycin resistance also present in the LentiCRISPRv2 vector. We analyzed vector copy number (VCN) 72 hr after transduction at a multiplicity of infection (MOI) of 20 (Table 1). #515 and #994 cells received an average of 0.6 and 2.7 copies of LentiCRISPRv2, respectively.
Table 1.
VCN Analysis
| Cells | LV.CRISPR | VCN (Total) | VCN (LV.CRISPR) |
|---|---|---|---|
| 515 | — | 3.4 | — |
| 994 | — | 3.6 | — |
| 515 | LV_int2.1 | 4.1 | 0.7 |
| 515 | LV_int2.6 | 3.8 | 0.4 |
| 515 | LV_ex2.1 | 4.2 | 0.8 |
| 994 | LV_int2.1 | 6.6 | 3 |
| 994 | LV_int2.6 | 6.2 | 2.6 |
| 994 | LV_ex2.1 | 6.2 | 2.6 |
LV, lentivirus.
To quantify the deletions of the duplicated region and indirectly the wild-type alleles created by the editing, we analyzed the genomic region targeted by our approach versus a region in intron 67 of the DMD gene by real-time PCR. We detected the highest rate of deletions in 994 cells treated with LV_int2.1 (about the 13%), whereas no deletions at all were revealed in 515 cells transduced with LV_Ex2.1 (Figure 3A).
We then investigated the presence of inversion events by amplifying the region targeted by the gRNAs with only one primer close to the target region (Figure S3). We obtained no evidence of inversion event despite 45 cycles of PCR amplification (data not shown).
In order to better investigate dystrophin expression, we induced myoblast differentiation to myotubes. RNA analysis revealed an increased ratio between wild-type/duplicated transcripts ranging from 1.5 to about 4-fold in cells transduced by LentiCRISPRv2 (Figure 3B). Western blot analysis showed restoration of wild-type dystrophin in #994 cells transduced with LV_int2.1 (11%) and LV_int2.6 (7%) when compared to wild-type (Figure 3C), whereas #515 cells did not show any labeling; this is probably due to the low rate of transduction in #515 cells with VCN spanning from 0.4 to 0.8 while being around 3 in #994 cells.
Immunocytochemistry showed intense expression and correct localization of dystrophin in clustered myotubes in differentiated #994 cells infected with LV_int2.1 and LV_int2.6, even though these represented only a 5% of total cell population (Figure 3D).
We then investigated the on-target versus off-target activity of gRNA crDMDint2.1, which showed the highest activity in restoring dystrophin expression. We scored ∼73% on-target activity in 110 clones sequenced (Figure S4, upper table) and no off-target effect by T7E1 assay (Figure S4, bottom table).
Discussion
The recent development of the CRISPR/Cas9 system have brought great excitement in the research community and new perspectives for the treatment of several genetic diseases, and recently for DMD with convincing data in vitro and in vivo.36, 37, 38, 39, 40 For these purposes, the CRISPR/Cas9 system has been mainly exploited to remove out-of-frame or in-frame mutated exons and restore the dystrophin reading-frame by targeting the sequence to be removed with two gRNAs. This approach is applicable to out-of-frame deletions and point mutations optimistically accounting for the 85% of all mutations, restoring the expression of a smaller functional protein and converting the severe phenotype into a milder, Becker-like one, as aimed by exon skipping approaches.41
Duplications represent the third cause of DMD among all the dystrophin mutations. The attempt to treat duplications has been recently reported, in a multi-exonic duplication in the DMD gene.28 Here, we described a strategy to correct the most frequent duplication in the DMD gene that could be extended to all other, less frequent duplications, leading to the synthesis of full-length, wild-type dystrophin. CGH analysis allowed us to identify a minimal region that is duplicated and shared among all the patients studied, against which we screened gRNAs with the aim of finding a deletion strategy potentially addressing all those patients (Figure 1). We used lentiviral vectors to bring our gRNAs and the spCas9 in patient-derived myoblasts, in order to maximize the efficiency of delivery and have both elements in a single vector. We were able to quantify the efficiency of the Cas9-induced deletions directly in treated cells without selection, and assay the results at both transcriptional and protein expression levels by cytochemical analysis of myotubes differentiated in vitro from treated myoblasts. The sequence analysis of inversions and deletions breakpoints indicated that rearrangements caused by two gRNAs are mostly neat and show the mutational spectrum introduced by NHEJ only in few cases (Figure S2). Interestingly, we did not detect inversions when dup2 patient’s myoblasts were treated, backing up the concept that deletions occur more frequently than inversions in our system, as described elsewhere.34
Treatment of immortalized myoblasts derived from two dup2 patients resulted in the deletion of the duplication at the genomic level (Figure 3A) and in an increased expression of wild-type dystrophin transcripts (Figure 3B). This expression was followed by a detectable and correctly localized dystrophin only in the #994 cell line (Figures 3C and 3D), which carries a smaller duplication of 137 kb, and not in the #515 cell line, which carries a larger, 263-kb duplication, probably due to the limited transduction efficiency in this cell line. Our results are concordant with those reported by Wojtal et al.28 in terms of restoration of protein expression in cells bearing deletions of comparable size (139 and 137 kb). We extended the significance of those findings by reporting the correct localization of dystrophin at the sarcolemma in myotubes derived from #994 cells transduced with two out of three lentiviral vectors, despite the small number of myoblasts positive for DMD expression (Figure 3D).
Even if we did not detect off-target activity for the best gRNA in our hands (Figure S4, bottom table), the recent improvements in the CRISPR design,42 specificity of the spCas943 and repertoire of Cas9 enzymes44, 45 let foresee a safer application of the technology, which opens perspectives for the clinical investigation.
In fact, by delivering Cas9 and gRNAs with adeno-associated virus (AAV) vectors it will be possible to specifically drive the correction of duplications to the muscles in vivo.37 It has been demonstrated that the AAV viral genomes are lost in the regenerating dystrophic skeletal muscle.46 This could mimic a transient expression of the CRISPR/Cas9 system, which reduces the risk of immune response and off-target activity while still producing stable genome modifications in satellite cells that could be able to repair or restore the muscles.37
Taken together, these data indicate that the removal of a duplication event by Cas9-mediated genome editing is feasible with only one gRNA directed against a duplicated intronic region (which is duplicated in a relevant number of DMD patients), increasing editing efficiency and reducing the risk of off-target effects. These findings provide proof of concept for a gene-editing approach in DMD patients carrying duplications and lay out the foundation for further research into the application such an approach in other inherited disorders caused by gene duplications.
Materials and Methods
Patients
We have studied eight DMD patients known to have duplications of the exon 2 by multiple ligase probe amplification (MLPA) analysis. DNA from these patients was obtained thanks to the collaboration with Dr. France Leturcq through the AFM-Telethon Biobank.
Myoblasts from two of these patients (ID #515 and #994), available through the Biobank, were immortalized thanks to the collaboration with the Institut de Myologie as previously described.33
Informed consent was obtained from all patients to conduct the study, which was in accordance with the Helsinki Declaration. All the experimental procedures were approved by Genethon.
CGH Analysis
According to manufacturer’s instructions, DNA samples were processed and hybridized to the DMD-CGH array as previously reported.29 The array was analyzed with the Agilent scanner and the Feature Extraction software (v.12.0). A graphical overview and analysis of the data were obtained using the Agilent Genomic Workbench (v.7.0). For identifying duplications and deletions, we used the standard setup of the ADM-2 statistical analysis provided by Genomic Workbench software (Agilent).
The array platform and data-related information have been submitted to the online data repository Gene Expression Omnibus (GEO, NCBI): GSE83658.
A graphical representation of the duplications was produced by creating a custom track and displaying it in UCSC Genome Browser GRCh37/hg19 assembly (http://genome.ucsc.edu). Ten more characterized duplications of the exon 2 from DMD patients were collected from the literature27, 28, 29, 30 and represented with a custom track (Figure 1A).
gRNA Design and Off-Target Prediction
The minimal common duplicated region shared among all the patients was uploaded in ZiFiT (http://zifit.partners.org/ZiFiT/) and used as a template to design six truncated gRNAs47 of 18 bp and one (crDMDex2) of 19 bp (Figure 1B). Oligos used to clone the gRNAs are shown in the Table S1. According to the available guidelines48 on CRISPR design we selected and cloned into the MLM3636 backbone (Addgene number 43860) those with less than ten off-target regions with two mismatches as predicted by ZiFiT (Figure S4).
Cell Cultures
To preliminarily screen the activity of the candidate gRNAs, we transfected HEK293T cells with the Cas9 plasmid JDS246 (Addgene number 43861) and with each gRNA, using Lipofectamine2000 (Thermo Fisher Scientific). DNA extraction (Blood & Cell Culture DNA Mini Kit, QIAGEN) was performed 72 hr after transfection for PCR amplification and T7E1 assay.
Immortalized DMD myoblasts were maintained in Skeletal Muscle Growth Medium (Promocell) and differentiated into myotubes for at least 7 days in Skeletal Muscle Differentiation Medium (Promocell) at 37°C with 5% CO2 incubation. Transfection of spCas9 and DMD gRNAs was performed with JetPEI (Polyplus) following manufacturer’s instructions. After 72 hr, DNA was extracted to perform PCR amplification.
T7EI Assay
PCR amplicons for targeted and off-target genomic regions were obtained by amplification with Platinum Taq DNA Polymerase High Fidelity (Thermo Scientific). The PCRs were carried out under the following conditions: preheating 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 64°C for 30 s, and 68°C for 1 min 20 s.
The PCR products were resolved on 1.5% agarose gel and then purified with Qiaquick PCR Purification Kit (QIAGEN). For T7E1 cleavage assay purified PCR products were denatured and annealed in NEBuffer 2 (NEB) using a thermocycler. Hybridized PCR products were digested with T7 endonuclease 1 (NEB, M0302L) for 15 min at 37°C and subjected to 2% agarose gel electrophoresis. Percentage of DNA modification was calculated using the formula %DNA modification = 100*(1-(1-fraction cleaved)1/2) reported on the NEB website (https://www.neb.com/protocols/2014/08/11/determining-genome-targeting-efficiency-using-t7-endonuclease-i). All PCR primer sequences are listed in Table S2.
Detection of Deletions and Inversions
The gDNA samples obtained from cells transfected with two gRNAs were amplified with Platinum Taq DNA Polymerase High Fidelity (Thermo Scientific). We prepared two specific PCRs with a common forward primer upstream the first gRNA binding site and a second forward primer localized between the two gRNA binding sites to detect inversions or a reverse primer downstream the second gRNA binding site to detect inversions (Figures 2C and 2D). The PCRs were conducted in order to detect genomic deletions and inversions under these conditions: preheating 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 60°C for 30 s, and 68°C for 40 s. Inversions were amplified with 40 cycles.
Inversions in cells with duplication of the exon 2 were amplified with only one forward primer upstream of the binding site of the gRNA used. If inversions occur, the primer would bind to the wild-type sequence in forward orientation and the inverted one in reverse orientation (Figure S3). PCR conditions were preheating 95°C for 5 min, followed by 45 cycles of 95°C for 30 s, 60°C for 30 s, and 68°C for 40 s.
Amplicons were resolved through 1.5% agarose gel electrophoresis. All PCR primer sequences are listed in Table S3.
Lentivirus Production, Titration, and Transduction
In order to increase the efficacy and maximize our results in cell lines, we cloned selected gRNAs in the LentiCRISPRv2 vector (Addgene Plasmid #52961).35
Research Grade Lentiviral Vector Production
Third-generation lentiviral vectors were produced by calcium phosphate transient transfection of 293T cells of the selected transfer vector, the packaging plasmid pKLg/p.RRE, pK.REV, the vesicular stomatitis virus glycoprotein G (VSV-G) envelope plasmid pK.G as previously described.49
Lentiviral Vector Titration
Three hundred thousand (3 × 105) HCT116 cells were transduced with serial vector dilutions in the presence of polybrene (6 μg/mL). Genomic DNA (gDNA) was extracted 72 hr after transduction. gDNA is extracted by using MagNaPure 96 system (Roche Diagnostics, Promega) according to manufacturer’s instructions. Vector copies per diploid genome (VCN) were quantified by quantitative PCR (qPCR) starting from 100 ng of template gDNA using primers (HIV sense: 5′-TACTGACGCTCTCGCACC-3′; HIV antisense: 5′-TCTCGACGCAGGACTCG-3′) and a probe FAM 5′- ATCTCTCTCCTTCTAGCCTC-3′) against the packaging signal region of LVs. Endogenous DNA amount was quantified by a primers/probe set against the human albumin gene (ALB sense: 5′ GCTGTCATCTCTTGTGGGCTGT-3′; ALB antisense: 5′-ACTCATGGGAGCTGCTGGTTC-3′; ALB probe: VIC 5′-CCTGTCATGCCCACACAAATCTCTCC-3′). Copies per genome were calculated by the formula = [ng LV/ng endogenous DNA] × [no. of LV integrations in the standard curve]. The standard curve was generated by using a plasmid containing the appropriate sequences in cis from the vectors and ALB gene. Vector particles were measured by HIV-1 Gag p24 antigen immunocapture assay (PerkinElmer) according to manufacturer’s instructions. Vector infectivity is calculated as the ratio between titer and particles.50
Titers obtained ranged from 5.8E8 transducing units (TU)/mL to 1.1E9 TU/mL. LV_int2.1, LV_int2.6 and LV_ex2.1 were used at MOI 20 together with 8 μg/mL of polybrene in exon 2 duplicated myoblasts cell lines.
Quantification of Wild-Type Alleles Created after Removal of the Exon 2 Duplication by One gRNA
Quantification of the efficacy of the gRNAs to remove the duplicated sequence at the DNA level was performed by Real-Time PCR using SsoFast EvaGreen Supermix (Bio-Rad) for 40 cycles and primers designed on the minimal common duplicated region (forward CCAAACCCATAGTCGTTCAAAT, reverse AGATGCTGCAAAGGAAGGAA) and on intron 67 of the DMD gene as control (forward GCATTTGGAAAGATTTGCTGA, reverse TGCTGGTGCAGACTTCTCTC). Real-Time PCR was performed in triplicate in 96-well plates using 50 ng genomic DNA from wild-type, untreated, and lentiviral transduced cells. The calculation of the fold changes was based on the 2−ΔΔCT method (Applied Biosystems User Bulletin #2) and results were expressed in relation to the untreated cells set to 1.
RNA Analysis
Total RNA was isolated from myotubes three days after differentiation using the RNeasy Kit (QIAGEN) and reverse-transcribed by means of a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to the manufacturer’s instructions.
Before cDNA synthesis, RNA was treated with DNase I (Roche) and checked for residual DNA contamination by a 55-cycle PCR.
RT-PCR of the exon 2 duplication was performed with Platinum Taq DNA Polymerase High Fidelity (Thermo Scientific) and oligos 21F and 448R (designed complementary to exon 1 and 4 of the DMD muscle isoform and taken from http://dmd.nl/) and resolved on 2% agarose gel. One microliter of the RT-PCR was loaded on high-sensitivity DNA chips (Agilent) for the quantification of the duplication excision, which was performed calculating the ratio of the area of the normal transcript and the sum of the area of normal and duplicated transcripts multiplied for 100 and normalized by setting to one the untreated (UT) samples. Experiments were performed in triplicate, mean results and SDs are shown. Significance was evaluated by Kruskal-Wallis test with post hoc Mann-Whitney test for p values less than 0.05 considered significant.
Western Blot and Immunofluorescence Analysis
Immortalized myoblasts seeded in 30-cm Petri dishes and differentiated for 10 days were washed with cold PBS and lysed in radioimmunoprecipitation assay (RIPA) buffer in presence of Complete Mini protease inhibitor mixture (Roche) for 30 min on ice. Cell lysates were cleared and quantified.
To check the expression of dystrophin, 30 μg of protein from myotubes was mixed with 4 × NuPage LDS buffer (Invitrogen) supplemented with 50 mM DTT, heated for 2 min at 85°C, loaded on Novex 3%–8% Tris-Acetate Midi Gel (Invitrogen), and electrophoresed at 150 V with Tris-Acetate SDS running buffer (Invitrogen) for 70 min at room temperature. Proteins were then transferred at 45 V overnight at 4°C onto polyvinylidene fluoride (PVDF) membranes as described above using transfer buffer with no methanol.
Membranes were blocked in Tris-buffered saline-tween (TBS-T) (20 mM Tris, 500 mM NaCl [pH = 7.5], 0.1% Tween 20) supplemented with 3% (Dystrophin and Cas9) or 5% (α-actinin) non-fat dry milk (Bio-Rad) for 1 hr at room temperature.
Membranes were incubated for 1 hr at room temperature with rabbit polyclonal anti-Dystrophin (ab15266, Abcam), anti-CRISPR-Cas9 (ab191468, Abcam), and mouse monoclonal α-actinin (Santa Cruz Biotechnology) antibodies, washed three times for 10 min in TBS-T, incubated with immunopure goat anti-rabbit or rabbit anti-mouse immunoglobulin Gs (IgGs) (Dako) conjugated to horseradish peroxidase (HRP, 1:2,000 in TBS-T 3 or 5% milk) for 1 hr at room temperature, and revealed with Pierce ECL Western Blotting Substrate (Pierce Biotechnology). Chemiluminescence was acquired by the ChemiDoc MP System (Bio-Rad) and quantified using ImageJ v.1.37 (NIH image). Immortalized control myoblasts differentiated into myotubes in vitro were used as positive control.
In vitro differentiated immortalized wild-type, #515 and #994 myoblasts were washed with PBS (Sigma-Aldrich), fixed with 4% paraformaldehyde (Sigma-Aldrich) at room temperature for 10 min, and permeabilized with ethanol 75% (Sigma-Aldrich) at room temperature for 1 min. As a blocking solution, 20% fetal bovine serum (Sigma-Aldrich) was used at room temperature for 30 min to reduce secondary antibody background signal. The cells were subsequently incubated overnight at 4°C with the anti-dystrophin (NCL-DYS2 Novocastra) primary antibody. After incubation, cells were washed with PBS and then incubated with the 594-fluorochrome conjugated secondary antibodies (Thermo Fisher) together with DAPI for nucleic acid staining (Ibidi) for 1 hr at room temperature in PBS containing 0.2% Triton X-100. After three successive washings with PBS, dishes were mounted using fluorescent mounting medium (Ibidi) and examined by fluorescence microscopy (DMI6000B, Leica).
Author Contributions
Conceptualization, M.B.; Methodology, M.B.; Investigation, A.L., S.D., A.M., A.I., and E.B.; Writing – Original Draft, M.B. and A.L.; Writing – Review & Editing, V.M., F.B., F.M., and M.B.; Funding Acquisition, F.M. and M.B.; Resources, S.M., K.M., and V.M.; Supervision, F.M. and M.B.
Conflicts of Interest
We declare no competing financial interests.
Acknowledgments
France Leturcq (Laboratoire de biochimie et génétique, moléculaire, Hôpital Cochin et Institut de Myologie, Groupe hospitalier La Pitié Salpétrière, APHP, France) and AFM-Telethon Biobank are gratefully acknowledged for providing patients samples and cells. Plasmids MLM3636 (Addgene plasmid # 43860), JDS246 (#43861), and LentiCRISPRv2 (#52961) were a gift from Keith Joung and Feng Zhang. This work was supported by grants from the European Research Council (ERC-2010-AdG, GT-SKIN), the association “Amici di Edy,” AFM-Téléthon (Trampoline grant no. 17247), and Horinzon 2020 Marie Sklodowska-Curie Individual Fellowship Action (Project DMD2CURE #703093) to M.B. Federica Brugnoli (Section of Anatomy and Histology, Department of Morphology, Surgery and Experimental Medicine, University of Ferrara, Ferrara, Italy) is also acknowledged for the technical support.
Footnotes
Supplemental Information includes four figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.02.004.
Supplemental Information
References
- 1.Mendell J.R., Shilling C., Leslie N.D., Flanigan K.M., al-Dahhak R., Gastier-Foster J., Kneile K., Dunn D.M., Duval B., Aoyagi A. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann. Neurol. 2012;71:304–313. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 2.Kinali M., Arechavala-Gomeza V., Cirak S., Glover A., Guglieri M., Feng L., Hollingsworth K.G., Hunt D., Jungbluth H., Roper H.P. Muscle histology vs MRI in Duchenne muscular dystrophy. Neurology. 2011;76:346–353. doi: 10.1212/WNL.0b013e318208811f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aartsma-Rus A., Van Deutekom J.C., Fokkema I.F., Van Ommen G.J., Den Dunnen J.T. Entries in the Leiden Duchenne muscular dystrophy mutation database: An overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 4.Wilton S.D., Veedu R.N., Fletcher S. The emperor’s new dystrophin: Finding sense in the noise. Trends Mol. Med. 2015;21:417–426. doi: 10.1016/j.molmed.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Lu Q.L., Cirak S., Partridge T. What can we learn from clinical trials of exon skipping for DMD? Mol. Ther. Nucleic Acids. 2014;3:e152. doi: 10.1038/mtna.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki Y., Nakamura A., Yokota T., Saito T., Okazawa H., Nagata T., Takeda S. In-frame dystrophin following exon 51-skipping improves muscle pathology and function in the exon 52-deficient mdx mouse. Mol. Ther. 2010;18:1995–2005. doi: 10.1038/mt.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arechavala-Gomeza V., Graham I.R., Popplewell L.J., Adams A.M., Aartsma-Rus A., Kinali M., Morgan J.E., van Deutekom J.C., Wilton S.D., Dickson G., Muntoni F. Comparative analysis of antisense oligonucleotide sequences for targeted skipping of exon 51 during dystrophin pre-mRNA splicing in human muscle. Hum. Gene Ther. 2007;18:798–810. doi: 10.1089/hum.2006.061. [DOI] [PubMed] [Google Scholar]
- 8.Aartsma-Rus A., Janson A.A., van Ommen G.J., van Deutekom J.C. Antisense-induced exon skipping for duplications in Duchenne muscular dystrophy. BMC Med. Genet. 2007;8:43. doi: 10.1186/1471-2350-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greer K.L., Lochmüller H., Flanigan K., Fletcher S., Wilton S.D. Targeted exon skipping to correct exon duplications in the dystrophin gene. Mol. Ther. Nucleic Acids. 2014;3:e155. doi: 10.1038/mtna.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Vliet L., de Winter C.L., van Deutekom J.C., van Ommen G.J., Aartsma-Rus A. Assessment of the feasibility of exon 45-55 multiexon skipping for Duchenne muscular dystrophy. BMC Med. Genet. 2008;9:105. doi: 10.1186/1471-2350-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor P.J., Maroulis S., Mullan G.L., Pedersen R.L., Baumli A., Elakis G., Piras S., Walsh C., Prósper-Gutiérrez B., De La Puente-Alonso F. Measurement of the clinical utility of a combined mutation detection protocol in carriers of Duchenne and Becker muscular dystrophy. J. Med. Genet. 2007;44:368–372. doi: 10.1136/jmg.2006.047464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wein N., Vulin A., Falzarano M.S., Szigyarto C.A., Maiti B., Findlay A., Heller K.N., Uhlén M., Bakthavachalu B., Messina S. Translation from a DMD exon 5 IRES results in a functional dystrophin isoform that attenuates dystrophinopathy in humans and mice. Nat. Med. 2014;20:992–1000. doi: 10.1038/nm.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan N.J. Ataluren: First global approval. Drugs. 2014;74:1709–1714. doi: 10.1007/s40265-014-0287-4. [DOI] [PubMed] [Google Scholar]
- 14.Traynor K. Eteplirsen approved for Duchenne muscular dystrophy. Am. J. Health Syst. Pharm. 2016;73:1719. doi: 10.2146/news160063. [DOI] [PubMed] [Google Scholar]
- 15.Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joung J.K., Sander J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva G., Poirot L., Galetto R., Smith J., Montoya G., Duchateau P., Pâques F. Meganucleases and other tools for targeted genome engineering: Perspectives and challenges for gene therapy. Curr. Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura K., Fujii W., Tsuboi M., Tanihata J., Teramoto N., Takeuchi S., Naito K., Yamanouchi K., Nishihara M. Generation of muscular dystrophy model rats with a CRISPR/Cas system. Sci. Rep. 2014;4:5635. doi: 10.1038/srep05635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Zheng Y., Kang Y., Yang W., Niu Y., Guo X., Tu Z., Si C., Wang H., Xing R. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum. Mol. Genet. 2015;24:3764–3774. doi: 10.1093/hmg/ddv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larcher T., Lafoux A., Tesson L., Remy S., Thepenier V., François V., Le Guiner C., Goubin H., Dutilleul M., Guigand L. Characterization of dystrophin deficient rats: A new model for Duchenne muscular dystrophy. PLoS ONE. 2014;9:e110371. doi: 10.1371/journal.pone.0110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ousterout D.G., Perez-Pinera P., Thakore P.I., Kabadi A.M., Brown M.T., Qin X., Fedrigo O., Mouly V., Tremblay J.P., Gersbach C.A. Reading frame correction by targeted genome editing restores dystrophin expression in cells from Duchenne muscular dystrophy patients. Mol. Ther. 2013;21:1718–1726. doi: 10.1038/mt.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ousterout D.G., Kabadi A.M., Thakore P.I., Perez-Pinera P., Brown M.T., Majoros W.H., Reddy T.E., Gersbach C.A. Correction of dystrophin expression in cells from Duchenne muscular dystrophy patients through genomic excision of exon 51 by zinc finger nucleases. Mol. Ther. 2015;23:523–532. doi: 10.1038/mt.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H.L., Fujimoto N., Sasakawa N., Shirai S., Ohkame T., Sakuma T., Tanaka M., Amano N., Watanabe A., Sakurai H. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4:143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long C., McAnally J.R., Shelton J.M., Mireault A.A., Bassel-Duby R., Olson E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goyenvalle A., Griffith G., Babbs A., El Andaloussi S., Ezzat K., Avril A., Dugovic B., Chaussenot R., Ferry A., Voit T. Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat. Med. 2015;21:270–275. doi: 10.1038/nm.3765. [DOI] [PubMed] [Google Scholar]
- 27.Vila M.C., Klimek M.B., Novak J.S., Rayavarapu S., Uaesoontrachoon K., Boehler J.F., Fiorillo A.A., Hogarth M.W., Zhang A., Shaughnessy C. Elusive sources of variability of dystrophin rescue by exon skipping. Skelet. Muscle. 2015;5:44. doi: 10.1186/s13395-015-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wojtal D., Kemaladewi D.U., Malam Z., Abdullah S., Wong T.W., Hyatt E., Baghestani Z., Pereira S., Stavropoulos J., Mouly V. Spell checking nature: Versatility of CRISPR/Cas9 for developing treatments for inherited disorders. Am. J. Hum. Genet. 2016;98:90–101. doi: 10.1016/j.ajhg.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bovolenta M., Neri M., Fini S., Fabris M., Trabanelli C., Venturoli A., Martoni E., Bassi E., Spitali P., Brioschi S. A novel custom high density-comparative genomic hybridization array detects common rearrangements as well as deep intronic mutations in dystrophinopathies. BMC Genomics. 2008;9:572. doi: 10.1186/1471-2164-9-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bovolenta M., Scotton C., Falzarano M.S., Gualandi F., Ferlini A. Rapid, comprehensive analysis of the dystrophin transcript by a custom micro-fluidic exome array. Hum. Mutat. 2012;33:572–581. doi: 10.1002/humu.22017. [DOI] [PubMed] [Google Scholar]
- 31.del Gaudio D., Yang Y., Boggs B.A., Schmitt E.S., Lee J.A., Sahoo T., Pham H.T., Wiszniewska J., Chinault A.C., Beaudet A.L., Eng C.M. Molecular diagnosis of Duchenne/Becker muscular dystrophy: Enhanced detection of dystrophin gene rearrangements by oligonucleotide array-comparative genomic hybridization. Hum. Mutat. 2008;29:1100–1107. doi: 10.1002/humu.20841. [DOI] [PubMed] [Google Scholar]
- 32.White S.J., Aartsma-Rus A., Flanigan K.M., Weiss R.B., Kneppers A.L., Lalic T., Janson A.A., Ginjaar H.B., Breuning M.H., den Dunnen J.T. Duplications in the DMD gene. Hum. Mutat. 2006;27:938–945. doi: 10.1002/humu.20367. [DOI] [PubMed] [Google Scholar]
- 33.Mamchaoui K., Trollet C., Bigot A., Negroni E., Chaouch S., Wolff A., Kandalla P.K., Marie S., Di Santo J., St Guily J.L. Immortalized pathological human myoblasts: Towards a universal tool for the study of neuromuscular disorders. Skelet. Muscle. 2011;1:34. doi: 10.1186/2044-5040-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraft K., Geuer S., Will A.J., Chan W.L., Paliou C., Borschiwer M., Harabula I., Wittler L., Franke M., Ibrahim D.M. Deletions, inversions, duplications: Engineering of structural variants using CRISPR/Cas in mice. Cell Rep. 2015;10:833–839. doi: 10.1016/j.celrep.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ousterout D.G., Kabadi A.M., Thakore P.I., Majoros W.H., Reddy T.E., Gersbach C.A. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabebordbar M., Zhu K., Cheng J.K., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L., Park K.H., Zhao L., Xu J., El Refaey M., Gao Y., Zhu H., Ma J., Han R. CRISPR-mediated Genome Editing Restores Dystrophin Expression and Function in mdx Mice. Mol. Ther. 2016;24:564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu Q.L., Mann C.J., Lou F., Bou-Gharios G., Morris G.E., Xue S.A., Fletcher S., Partridge T.A., Wilton S.D. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat. Med. 2003;9:1009–1014. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- 42.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Hir M., Goyenvalle A., Peccate C., Précigout G., Davies K.E., Voit T., Garcia L., Lorain S. AAV genome loss from dystrophic mouse muscles during AAV-U7 snRNA-mediated exon-skipping therapy. Mol. Ther. 2013;21:1551–1558. doi: 10.1038/mt.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Y., Reyon D., Joung J.K. Targeted genome editing in human cells using CRISPR/Cas nucleases and truncated guide RNAs. Methods Enzymol. 2014;546:21–45. doi: 10.1016/B978-0-12-801185-0.00002-7. [DOI] [PubMed] [Google Scholar]
- 48.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cantore A., Ranzani M., Bartholomae C.C., Volpin M., Valle P.D., Sanvito F., Sergi L.S., Gallina P., Benedicenti F., Bellinger D. Liver-directed lentiviral gene therapy in a dog model of hemophilia B. Sci. Transl. Med. 2015;7:277ra28. doi: 10.1126/scitranslmed.aaa1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zanta-Boussif M.A., Charrier S., Brice-Ouzet A., Martin S., Opolon P., Thrasher A.J., Hope T.J., Galy A. Validation of a mutated PRE sequence allowing high and sustained transgene expression while abrogating WHV-X protein synthesis: Application to the gene therapy of WAS. Gene Ther. 2009;16:605–619. doi: 10.1038/gt.2009.3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.