Abstract
Cell-penetrating peptides (CPPs) are delivery vectors widely used to aid the transport of biologically active cargoes to intracellular targets. These cargoes include small interfering RNAs (siRNA) that are not naturally internalized by cells. Elucidating the complexities behind the formation of CPP and cargo complexes is crucial for understanding the processes related to their delivery. In this study, we used modified analogs of the CPP transportan10 and investigated the binding properties of these CPPs to siRNA, the formation parameters of the CPP/siRNA complexes, and their stabiliy to enzymatic degradation. We conclude that the pH dependent change of the net charge of the CPP may very well be the key factor leading to the high delivery efficiency and the optimal binding strength between CPPs to siRNAs, while the hydrophobicity, secondary structure of the CPP, and the positions of the positive charges are responsible for the stability of the CPP/siRNA particles. Also, CPPs with distinct hydrophobic and hydrophilic regions may assemble into nanoparticles that could be described as core-shell formulations.
Keywords: cell-penetrating peptides, siRNA, nanoparticles, degradation, complexation
Graphical Abstract
Introduction
RNAi is the sequence-specific post-transcriptional silencing of a target gene. It was first discovered by Fire and Mello in 1998,1 and it is an important biological process occurring in cells with the function of regulating the endogenous RNA levels.2, 3, 4 The RNAi can be induced with specific RNAs, including short interfering RNAs (siRNAs). These can be spontaneously endocytosed by some cells,5 but have overall low cell membrane permeability because of the high number of anionic charges (∼40 negative charges per siRNA) and lack an efficient cellular uptake mechanism.6 Therefore the routine use of siRNA-based technology may rely on the development of a suitably modified siRNA or an efficient delivery method.
Because of the terminal overhangs, an unmodified siRNA is susceptible to nuclease degradation. Due to their small size, siRNA molecules may retain their initial structure and due to bigger persistence length (∼70 nm, compared to ∼50 nm for pDNA), render a stiffer molecule when compared to a plasmid DNA. Therefore, siRNA do not necessarily require the same process of condensation that leads to the collapse of pDNA into nanoparticles in the presence of cationic structures.4
Cell-penetrating peptides (CPPs) are amino acid sequences with an ability to enter live cells and facilitate the cellular uptake of diverse cargoes including plasmids (pDNA), RNA, or even proteins.7, 8 Various studies have addressed the internalization pathway, intracellular trafficking, physico-chemical parameters, and biological effect related to CPP/cargo complexes using pDNA as load.9, 10, 11, 12, 13, 14, 15 However, it is not possible to predict complex formation between siRNA and CPPs based only on these studies.9, 10 CPPs are able to form non-covalent complexes with nucleic acids due to positively charged groups in the CPP sequence and negative charges of the nucleic acid backbone.16 Formulation of non-covalent complexes is simple and enables varying the conditions of complex formation, ratios of siRNA per delivery vector, etc. and has several advantages over covalent conjugation strategy.17, 18 Understanding the parameters that control cargo-carrier interactions is imperative in order to develop new and efficient formulations.
CPPs used in this study (shown in Table 1) are further modifications of transportan10 (TP10) that have shown an increased efficacy in nucleic acid delivery compared to the parent peptide. TP10 is an analog of the amphipatic CPP Transportan, developed for the delivery of nucleic acids.19 PepFects (PF) and NickFects (NF) are TP10 analogs with an N-terminal fatty acid moiety20 and further modifications in the backbone of the CPP. PF3 is a direct analog of TP10 with only N-terminal fatty acid modification.21 PF3 was chosen to show the effect of fatty acid to the complex formation capability. PF6 is modified by covalent attachment of trifluoromethylquinoline-based moieties via a lysine tree in the Lys7 position.22 PF6 was chosen because it has been shown to be an efficient delivery reagent for pDNA, SCOs, and siRNA and has a pH sensitive moiety. It also has been used in vivo.22 PF14 is designed by replacing the lysines and isoleucines with ornithines and leucines, respectively, and adding an extra positive charge.23 Although PF14 C-terminal sequence is radically different from other CPPs used in this study, it does not change its net charge at biologically relevant pH. It has been used also in vivo.23 In NF51 and NF57, Lys7 is replaced with ornithine and subsequent synthesis is continued from the δ-NH2 group of ornithine instead of an ordinarily used α-NH2, creating a kink in the peptide7 and increase of percentage of α-helical structure, compared to PF3. NF57 and NF51 contain the same number of amino acids (including positively charged aa-s), but differ in their amphipathicity and positive charge distribution. In NF57, positive charges are located on the same side when projected as an α-helical wheel, increasing the amphipacy of the peptide. In the peptide sequences, lysines and ornithines are the aa-s with positively charged side groups that are able to interact with the negatively charged siRNA backbone. The net charge of each peptide in a pH range of 7.5–5.2 changes (Table 1, calculated using MarvinSketch 15.9.14) and therefore alters the interactions with negatively charged nucleic acid molecules.
Table 1.
CPPs Used in This Work, Their Calculated Charge, and the Molar Ratio that Contributes to Similar Charge Ratio for CPP/siRNA Complexes
NC, neutrality condition.
aCalculated with MarvinSketch, Chemaxon.
†MR that corresponds to CPP/siRNA CR 2.3–2.4.
Most of these CPPs use endosomal pathways when complexed with nucleic acids13, 14, 19, 23 and are mainly located in endosomal compartments after internalization. Therefore, the acidification processes inside endosomal compartments and also endosomal release are crucial factors to consider. The pH inside different cell organelles, such as late endosomes and lysosomes and cytosol differs, and the net charge of a CPP molecule changes throughout delivery from the extracellular to the intracellular organelles or cytosol. This change of net charge affects the interactions between a CPP and the cargo. Isothermal calorimetry (iTC) is an accurate method that has been used to study mainly protein-ligand interactions, but a growing number of studies harness iTC to study CPP-ligand interactions,24 CPP-siRNA interactions,25, 26 and CPP interactions with membranes.27 Furthermore, iTC enables to determine the binding constant of a CPP to cargo and calculate values of apparent dissociation constants. KD calculated from iTC measurements allows insight into the interactions between a CPP and a siRNA molecule and pH sensitive rearrangement of the nanoparticles during the different phases of the delivery.
We hypothesize that for efficient delivery, first an optimal binding between the CPP and the cargo, and thereafter further condensation into a nanoparticle by additional CPP are required. Optimal binding between siRNA and delivery vector is needed for complexation, but also for maintaining nucleic acid activity after delivery. Excess CPP is needed to protect the cargo from degradation during transport to the target site. For different types of nucleic acids, there is an optimal ratio between the number of delivery vectors and the cargo molecules.
In this study, the relationships between physico-chemical properties of the peptides, their influence on the CPP, and siRNA binding, the formation and stability of nanocomplexes were investigated by fluorescent dye intercalation assay, gel electrophoresis, enzymatic digestion experiments, and iTC. How these parameters are translated into biological activity of the delivered cargo molecule was investigated by using reporter gene downregulation after siRNA delivery with transport vectors. We draw correlations between the CPP-cargo binding parameters, complex stability, and formation and their relation to the properties of the CPP and propose a model for CPP/siRNA particle assembly.
Results
NickFects and PepFects Mediate Efficient Intracellular Delivery of siRNA and Promote luc2 Gene Downregulation without Inducing Permanent Membrane Damage
The biological effect (downregulation of a target gene) caused by the delivered siRNA cargo was assessed on U87 cells stably expressing luc2 gene. Delivery vectors PF/NFs (sequences shown in Table 1) were used to deliver siRNA into the cells and resulting gene downregulation was measured 24 hr post treatment.
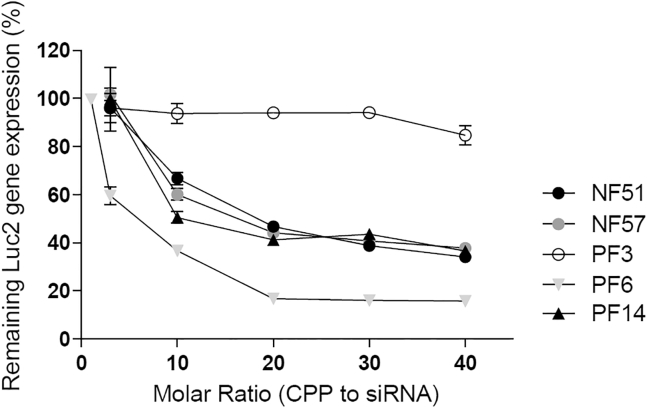
Various siRNA concentrations (100, 50, and 25 nM) were tested to optimize the conditions at which the differences between peptides are most pronounced (Figures S1A–S1C) and the 25 nM siRNA final concentration was chosen (Figure S1E). For downregulation, the PF6/siRNA complexes already at molar ratio (MR)20 were most effective, resulting in a 80% downregulation compared to 65% downregulation with NF51/siRNA, NF57/siRNA, or PF14/siRNA complexes. Treatment with PF3/siRNA complexes did not result in efficient downregulation of luc2 gene (Figure 1A) at given conditions. Molar ratio takes into account only how many peptide molecules are per siRNA molecule, but as net charge of each peptide is different, we compared the results also at the same range of charge ratio. Charge ratio expresses how many positive charges from the peptide are per negative charge from the nucleic acid. To our surprise at a similar charge ratio (CR, ratio of positive charges to negative charges 2.3–2.4, corresponding molar ratio is shown in Table 1), all complexes except PF3/siRNA had a similar effect on gene knockdown (Figure S1D). This indicates that an optimal number of positive charges per siRNA molecule may be needed.
Figure 1.
Downregulation of luc2 Gene
Experiments carried out in 10% FBS containing media and complexes formed at CPP/siRNA range MR1 to 40 with siRNA final concentration 25 nM. The Luc2 gene activity was measured 24 hr post treatment with complexes. The results are expressed as SEM with mean and error of three separate experiments.
In addition to transfection efficiency, the toxicity of the peptides in serum free conditions was tested. It has been shown, that addition of serum (10% FBS) into the media reduces toxicity of the CPP/cargo complexes. Only low toxicity was detected with complexes or free peptide compared to 25 nM siRNA. Lactate dehydrogenase leakage (LDH) indicates membrane disruption, and when detecting increased LDH levels from media, we may assume at least some disruption of the cell membrane. Out of tested peptides, the complexes formed with PF6 and free peptide at a higher concentration led to LDH leakage, indicating toxic effects on the cell membrane (Figure S2A). Neutral red assay is an easy method to detect lyso-toxicity in the cells. Neutral red accumulates in acidic organelles, such as lysosomes, and when lysosome is damaged or pH buffered, the dye also leaks from the vesicle or is not accumulated. In this assay PF6, PF14, and NF51 and their CPP/siRNA complexes at higher concentrations showed toxic and/or buffering effects (Figure S2B).
Effect of pH on the Packing of siRNA into a CPP/siRNA Complex
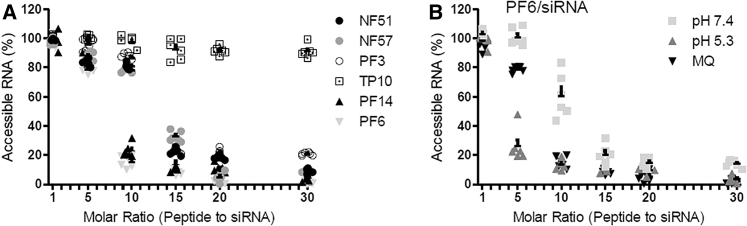
The packing of the siRNA into a CPP/siRNA complex and complex formation was assessed by PicoGreen (PG) intercalation assay and gel electrophoresis. The dye intercalates into accessible nucleic acid molecules and indicates the amount of the siRNA that is not incorporated into the complex or is not shielded by the CPP in the complex, whereas gel electrophoresis is able to indicate the fraction of the siRNA that is free. The increase of CPP concentration leads to the decrease of free siRNA in the solution. Only at higher MRs most of the peptides were able to fully shield the siRNA form the dye. Exceptionally with PF6, the complexes were fully condensed already at low MRs (Figures 2 and S3). In order to validate the packing ability of the CPP and also detect differences induced by a pH change, complexes were formed in HEPES buffer (pH 7.4), acetate buffer (pH 5.26), and MQ (pH 5.3–6.3). The formation of the complexes at different pH was assessed both on gel and with PG assay. In the case of PF6, the MR that is critical for complex formation in MQ was MR10, in HEPES (pH 7.4) buffer MR15, and in acetate buffer (pH 5.25) MR6 (Figures 2 and S3).
Figure 2.
Complex Formation Assessed with PicoGreen Intercalation Assay
(A) Complexes were formed in MQ, incubated at RT for 1 hr, and PicoGreen was added. After 15 min of incubation, fluorescence was measured. The results are normalized to the free siRNA in the same concentration as used in the complexes and RNA free sample. The results are shown as a mean of six separate experiments with error. (B) The formation of PF6/siRNA complexes in HEPES buffer (pH 7.4) or acetate buffer (pH 5.26) is compared to complex formation in MQ.
We hypothesize that there are two distinct processes in packing of the siRNA into a particle with CPPs. First binding and packing, that forms the particle and second shielding, which requires excess CPP in order to fully protect the siRNA. The packing can be detected with gel electrophoresis indicated by the lack of free siRNA band, and shielding can be measured by adding nucleic acid binding dye into the solution. When comparing the amount of CPP that is needed to pack or shield the siRNA (Table S1), it does not differ significantly when measured in the biologically relevant pH range. We could assume that the net charge of the peptide and, more importantly, the pH dependency of the net charge allow for a more stable and dynamic particle. For NF51, NF57, PF14, and PF3, the pH change did not significantly influence the needed MR (MR15 or MR10) to form condensed CPP/siRNA complexes (Figure S4; Table S1). Only with PF6, the complex formation is strongly influenced by pH change.
In cell experiments, higher than MR15 complexes give sufficient downregulation mediated by delivered siRNA. This could mean that a higher CPP concentration is needed for successful delivery in addition to packing of siRNA molecule. The complex formation was also confirmed by DLS. Representative DLS results for NF51/siRNA (net charge not significantly affected by pH) and particles of PF6/siRNA (net charge changes with pH) (Table S2) show that both types of CPPs are able to form a complex with a size of 98–180 nm.
Effect of pH on the CPP and siRNA Dissociation Constants
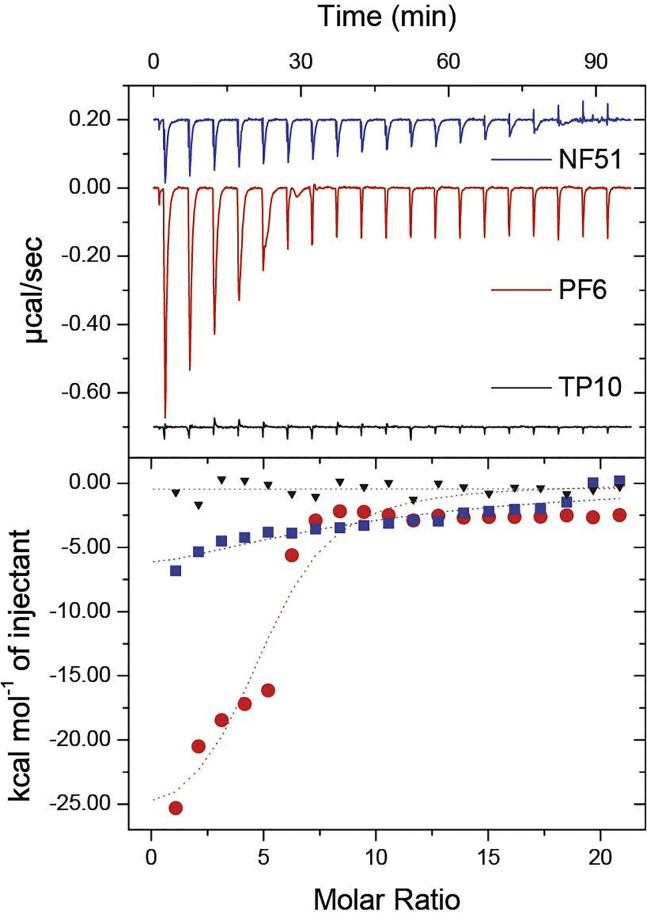
CPP and siRNA binding and CPP/siRNA complex dissociation can be expressed by constants. iTC enables measurement of K (M−1) out of which apparent KD (M) can be calculated. Due to the higher concentrations iTC requires, increased concentrations than used in standard complex formation (200 μM CPP and 2–5 μM siRNA) were used. Still, stepwise addition of CPP enables to cover all the MRs that were used in other experiments (MR1–40). The experiment was done at different pHs. At pH 7.4, the binding of CPP to a siRNA molecule required higher CPP concentrations than at other pH values. The strongest difference between binding at pH 7.4 and pH 5.26 was observed for PF6, indicating a pH dependent correlation between the charge of the CPP and its binding efficiency, as PF6 also exhibits the biggest change in charges per CPP from pH 7.5 to pH 5.5. The KD values were similar for NF51, NF57, and PF14 (Table 2). TP10 was not able to form stable complexes at given conditions as indicated by apparent KD values highly above 1 μM. The binding between PF6 and siRNA clearly differs from NF51 and TP10 (Figure 3). As indicated also by apparent KD values, the binding is similar for NF51, NF57, PF14, and PF3.
Table 2.
KD Values at Different pH Conditions Calculated from iTC Measurement Results, Where CPP Is Titrated into siRNA
| CPP | KD (nM) |
|||
|---|---|---|---|---|
| pH 7.4 | pH 6.26 | pH 5.26 | MQa | |
| NF51 | >1 μM | 510 ± 10 | 150 ± 10 | 160 ± 40 |
| NF57 | >1 μM | 615 ± 20 | 300 ± 15 | 418 ± 28 |
| PF3 | 490 ± 20 | 320 ± 17 | 800 ± 100 | 190 ± 44 |
| TP10 | >1 μM | >1 μM | >1 μM | >1 μM |
| PF6 | 90 ± 20 | 58 ± 15 | 6.31 ± 0.36 | 25 ± 5 |
| PF14 | 404 ± 50 | 280 ± 40 | 150 ± 40 | 220 ± 22 |
Results are shown as average of three separate experiments with deviation.
pH 5.6–6.6.
Figure 3.
Comparison of Binding between CPPs NF51 or PF6 with siRNA in MQ Water by iTC
NF51, PF6, and TP10 were chosen on as representatives of CPPs tested. PF6 has stearyl, pH sensitive moiety, charge +10. NF51 has kinked structure, stearyl, and charge +4. TP10 has charge +4.
Role of CPP in the Stability of Formed Complexes
In order to characterize the stability of the complexes to enzymatic degradation, Proteinase K was used. Proteinase K is a protease able to degrade the CPP component from the complex and therefore decrease the amount of the CPP in the complex or solution. The results are expressed as the first major release of siRNA (more than 40%) detectable with PG, at the same charge ratio for all the CPPs (results at the same MR shown in Figure S5). PF6 formed the most stable complexes with the siRNA compared to other tested CPPs. Even though at MR10 there was 2–3 times less PF6 present in the complexes, the stability to proteinase was 10–20 times higher than with other tested CPPs. Surprisingly, PF14, that mediated high biological effect from the delivered siRNA, was susceptible to enzymatic degradation and both PF14 and PF3 complexes with siRNA showed lower stability to degradation compared to NF51, NF57, and PF6. The NF51/siRNA and NF57/siRNA complexes were almost equally resistant to degradation, indicating that a small difference in the peptide sequence did not alter the stability (Figure 4A; Table 3).
Figure 4.
Shielding of CPP/siRNA Complexes from Enzymatic Degradation and Resistance of Complexes to Heparin Displacement
Resistance to degradation was measured for each peptide at the same charge ratio 2.3–2.4 (corresponding to MRs: PF6–MR10; NF51 and NF57–MR25; PF3–MR25; and PF14–MR20). To pre-formed complexes, PicoGreen dye was added and accessible siRNA was measured. (A) Proteinase K added to the complexes and release of siRNA was measured over a time period of 10 hr. (B) Heparin sodium salt in range of concentrations was added and complexes incubated in heparin solution for 1 hr at 37°C. Thereafter, PG was added and free siRNA measured. The results are normalized to free siRNA at the same concentration.
Table 3.
Stability of Complexes Detected with Fluorescence Dye
| CPPa | Stability and Resistanceb |
|
|---|---|---|
| Proteinasec (hr) | Heparin (mg/mL) | |
| NF51 | 1.3 ± 0.3 | 1.2 ± 0.05 |
| NF57 | 1.5 ± 0.3 | 1.9 ± 0.08 |
| PF3 | 0.6 ± 0.3 | 1 ± 0.02 |
| TP10 | ND | ND |
| PF6 | 12 ± 0.6 | 2.9 ± 0.05 |
| PF14 | 0.5 ± 0.3 | <1 |
CPP in CPP/siRNA complex.
Proteinase K treatment results are expressed at a similar CR 2.3–2.4. The heparin displacement assay is expressed at MR30. The results are calculated as the point when 50% of siRNA is released from the complexes from best fitting curve.
Proteinase K treated CPP/siRNA complexes. The time of first release of siRNA, where at least 40% of the siRNA can be detected (h).
The stability of CPP/siRNA complexes to heparin sodium salt displacement was assessed by adding different concentrations of heparin solution to pre-formed complex solution. Heparin is negatively charged and functions as a competitive binding partner for CPPs instead of siRNA. After PG addition and fluorescence measurement for each peptide, heparin concentration resulting in 50% of siRNA release was calculated. (Figure 4B; Table 3) Highest heparin resistance was expressed by PF6/siRNA complexes, which required almost twice as much heparin in order to release siRNA. TP10 and PF14 released siRNA at a heparin concentration below 1 mg/mL. NF57 exhibited a higher resistance to heparin displacement, compared to NF51 (Table 3).
Discussion
Delivery of bioactive nucleic acid cargoes holds a great potential in biotechnology, gene therapy, and modern synthetic biology.8 Synthetic siRNA constitutes a popular tool, but caused by the low cell membrane permeability of extracellular siRNA, a high number of anionic charges, and lack of efficient cellular uptake mechanism, necessitate the use of a delivery vector to efficiently deliver siRNA. There have been several studies describing the cellular entry and different physicochemical parameters of CPP/pDNA complexes,7, 8, 28 but finding the characteristics essential for an efficient delivery vector for the siRNA remains cumbersome.
A non-covalent complex formation strategy that is mainly based on electrostatic interactions between the cationic CPPs and an anionic nucleic acid molecule allows a versatile nanocomplex formulation using a simple mixing technique. However, many siRNA delivery systems that incorporate their payload by electrostatic interaction are prone to instability and premature release of the nucleic acid. A high density of positive charges allows complex formation at a lower vector concentration and may help to overcome this problem. Still, an optimal balance between the stability of a particle and the release of the siRNA is important.
In addition to positive charges in the peptide sequence, hydrophobic interactions contribute to particle formation.29, 30 Addition of fatty acid increases delivery efficiency considerably,21, 31, 32, 33, 34 and amphipathy may have a considerable impact on CPP efficiency.35 For example, PF3 was able to form complexes at MR15, while TP10 did not form stable complexes at tested MRs, although it was reported to form complexes at MR50.36 Only the charge may not be enough to form stable particles;36 therefore, an additional force (e.g., hydrophobic interactions) may be needed. PF3, NF51, and NF57 differ from TP10 mainly by the presence of the fatty acid and modifications in the peptide sequence. Addition of fatty acids enables to increase the hydrophobicity of a CPP and creates distinct hydrophilic and hydrophobic regions in the sequence, which may contribute to the formation of particles. Although PF3 formed complexes with siRNA, only low levels of gene knockdown were achieved compared to other NFs and PFs. This could likely be due to less effective endosomal release and premature complex dissociation before target site. It has been shown that PF3/oligonucleotide complexes are not endocytosed actively, cannot efficiently destabilize endosomes, and remain sedentary after vesicle rupture.34 Other tested CPPs mediate high gene downregulation at MR30, and treatment with CPP/siRNA resulted in 65% downregulation for NF51, NF57, and PF14 and 80% downregulation for PF6 (Figure 1). In addition, the advantage of PF6 is that it requires lower MR to achieve similar biological effect (Figure 1).
During the delivery and intracellular trafficking of siRNA, the CPP/siRNA complexes face changing pH conditions. The pH drops from physiological pH of 7.5 to an acidic pH of 5 in the cell compartments, lysosomes. The change in the pH also affects the net charge of the peptides (Table 1), depending on their sequence and modifications. In a simplified system, it may be possible to characterize the complex formation depending only on the pH change. The net charges of CPPs NF51, NF57, and PF3 are similar (∼+4), and these peptides have also similar complex formation capabilities, needing approximately 15 molecules per siRNA. PF14 has a higher net charge than aforementioned CPPs, and it does not change at biologically relevant pH range, maintaining a net charge of +5. Due to the higher charge, only a theoretical ten peptides are needed per siRNA in the case of PF14 (Figure S3; Table S1). The net charge of the PF6 changes drastically from +7.7 to +11.2 when pH decreases from 7.5–5.5. Caused by this change, 15 peptide molecules per siRNA are needed at pH 7.4 and only six CPPs at pH 5.26 to form stable complexes (Figure 2; Table 1). Similar tendencies were observed for the apparent KD values. The tested CPPs, except for PF6, had apparent KD values in biologically relevant pHs from 100 nM to 2 μM. In PF6, there is an additional pH sensitive moiety. The apparent KD value for PF6 changed from 90 nM at pH 7.4 to 6 nM at pH 5.26 (Table 2). Some other studies have measured KD values for CPP siRNA binding using various methods. For example, CPP CADY with +5 charge and no fatty acid modification has estimated siRNA binding to CPP KD at 15 nmol/L concentrations in MQ.37, 38 Another study reported a KD value of 224 nM in MQ measured by iTC for a peptide with charge 5.0–5.5 in water.25 For a C6 peptide with seven arginines in its sequence, a K value of 9.23 × 106 M−1 was reported in water, resulting in a KD value of ∼108 nM.26 Apparent KD values between 100 nM to >1 μM were reported for cationic peptides and DNA.36
The efficient intracellular uptake and protection of siRNA from degradation are important limiting factors in their delivery.10 Free siRNA may be rapidly degraded by nucleases when not protected in a complex.39 The resistance of the complexes to the degradation and avoiding premature dissociation of siRNA from the complex are fundamental for CPP-mediated cargo delivery. This is compromised by the cleavage of the CPP by proteases before the complex reaches the targeted cells. Enzyme Proteinase K was used to test stability of the complexes to degradation, and heparin displacement was used to show the resistance of the complex to competitive binding. Complexes with PF6 were most stable to heparin displacement and enzymatic degradation, compared to other tested CPPs. Stability to heparin may be the consequence of 2–3 times higher net charge compared to other tested CPPs, resulting in stronger interactions between PF6 and siRNA (Table 3). In another experimental setup, it was shown that the CQ modification in the PF6 might protect the CPP from the degradation by the proteinase (as indicated by the coincubation of CPP with the enzyme and lack of a considerable increase of detectable peptide fragments in UPLC after 2 hr incubation at 37°C) (data not shown). Surprisingly, PF14 was less stable to enzyme treatment than NFs (Table 3), although PF14 has more positive charges (Table 1). The higher stability of the NF/siRNA complexes could be contributed to efficient packing that is increased with the kinked structure caused by the synthesis from the ornithine's side chain. In PF6, the pH sensitive moieties on a lysine tree may also cause the conformational changes of peptide similar to the kinked structure in NF51 and NF57, as positive charges repulse at low pH and if they are not neutralized by the negatively charged cargo. Furthermore, the difference in stability to degradation may be caused by the particle properties. Transmission electron microscopy analysis showed that PF6/siRNA particles were tightly packed and almost perfectly spherical as are particles with NFs, but PF14 formed oval shaped particles with a larger surface area exposed to enzymes.40 The stability of PFs to heparin displacement and serum has been compared to CPPs R9 and Tat,41 and the results indicated that an optimal stability to heparin displacement and a high stability to serum could be crucial for higher delivery efficiency.
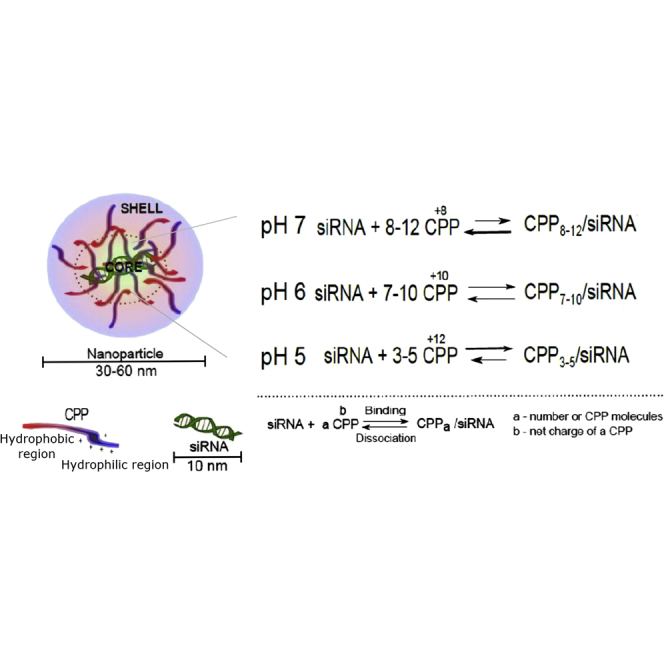
Endosomotropic modification, such as chloroquine moiety has been shown to be ∼90% protonated at physiological pH and at low pH it is fully protonated. This protonation is donating to the net charge increase of PF6 at the lower pH. In the complex formation hydrogen bonding, electrostatic and lipophilic interactions are considered dominant and strong binding enables the siRNA to accompany CPP into the cell and through membranes and endosomes.42 This leads to a pH dependent increase of the net charge in PF6 and enables the endosomal release and the dissociation of a CPP/siRNA complex only after entering the cell. iTC graph values indicate that only 3 to 5 CPPs are needed at low pH 5.26, whereas 7–10 CPPs are needed in MQ at pH 5.6–6.6 and 8–12 CPPs at pH 7.4.
There may be a differing neutrality condition for CPP/siRNA complexes at different pHs as the net charge of the peptide is different at pH 5.5 or pH 7.5. With PF6, the difference is more pronounced, because the net charge of the peptide changes drastically, compared to other tested CPPs. PF6's higher net charge yields in stronger binding at pH 5.5. This binding could enable some of the peptide to be released from the complex and mediate endosomal escape. The pH of the cytosol is higher than in endosomes. After CPP/siRNA is released from the acidic endosomes/lysosomes, the binding properties of the CPP also change. At a less acidic environment, the complex may be destabilized, as more peptides may be needed. Self-assembly of CPPs and nucleic acids have been reported for many CPPs reviewed by Pujals et al.11, 43 For example, for chitosan-polyethylene glycol-TAT-siRNA nanoparticles, an increase in the pH led to a gradual decrease in the total positive surface charge of the nanoparticles, decreasing the electrostatic repulsion, leading to the particle aggregation at pH 7.0, but still remaining biologically active. Several studies indicate that CPP-nucleic acid nanoparticles consist of several layers where a portion of CPPs is bound to nucleic acids and another portion is responsible for cell membrane interactions and essentially acting as naked CPPs. For example, CADY-siRNA nanocomplexes molecular dynamic calculations revealed that an average of 14 CADYs were required to condense one siRNA. However, 40 CADYs were detected per one siRNA, which corresponds to two layers of CADYs per siRNA.38 For PF/NF complexes, MR30 was optimal to achieve higher gene downregulation from siRNA. Gel electrophoresis and PG assay experiments indicated that 10–15 CPPs per siRNA are needed for complex formation, and an enzymatic degradation assay indicated that more CPP was needed to protect siRNA from degradation and premature dissociation of complexes.
To cover siRNA in PF6/siRNA complexes, 6–15 CPPs were needed, depending on the pH. To avoid enzymatic degradation, a lot of 40–50 CPPs per siRNA molecule were sufficient (Figure S4) for most tested CPPs. Similar tendency was reported for the CADY peptide, where the peptides formed a stable complex despite the electrostatic repulsions. Thermodynamic calculations proved that the peptide cage is stabilized by hydrophobic CADY-CADY contacts due to CADY polymorphism.38 The same could apply of PF/NFs, as they are also quite hydrophobic. A similar layer theory was proposed by Québatte et al.,44 where the authors speculated that a portion of riDOM, their developed CPP, is not involved in DNA binding and will instead bind to a sulfated GAG, thereby starting an endocytotic translocation process. Nanoparticles formed with CPPs have been reported to use scavenger receptors for internalization and also interact with heparin sulfate proteoglycans,13, 14, 15, 24, 45 indicating the presence of CPP able to interact with cell membrane structures.
According to the obtained data, we propose a model for CPP/siRNA complexes. We hypothesize that at physiological pH, part of the CPP molecules are bound to siRNA, while excess CPPs form self-assemblies around this core. This kind of self-assembly is especially pronounced when CPPs come into contact with the cell membrane, as reported in Bürck et al.46 The extra layer of CPPs may be destabilizing the cellular membrane and enable the complexes to enter cells. Once in acidic endosomes, these CPP self-assemblies can be gradually released and contribute to endosomal escape. This event could be triggered by changes in the CPP ionization properties, as the peptide becomes more positively charged at acidic pH. The charge repulsion between CPPs starts to increase and the self-assemblies start collapsing, separating the core of the complex, consisting of an optimal number of CPPs bound to siRNA. Once the CPP/siRNA complex is released from the endosomes, the siRNA could be recognized by its more favored binding partner, the multi-subunit RNA-induced silencing complex,47 and the peptides are concurrently degraded.48, 49 Whereas this siRNA release process might seem quite inefficient, it has been reported that in the case of lipid nanoparticle mediated siRNA delivery, only 1%–2% siRNA needs to escape from endosomes into the cytosol, and only during a limited window of time when the nanoparticles reside in a specific compartment sharing early and late endosomal characteristics, to induce efficient gene knockdown.50
Although in this study PF6 was the most stable and efficient for CPP/siRNA complex formation, alternatives for this CPP would be desired, because PF6, due to its chloroquine moieties, exhibits toxicity, which may limit its use in vivo. In order to design CPPs with similarly high or higher efficiency than PF6, understanding the processes behind complex formation and stability are needed. The dissociation constants of each CPP give valuable background for understanding the binding properties of the CPP to a siRNA and complex characteristics at different pHs. The idea of conditional modulating of binding CPPs to siRNA may provide the basis for maintaining CPP/siRNA stability and release of siRNA at its target site.51 This study shows that a pH sensitive change of net charge is highly desirable, and, in addition, an optimal number of charges per peptide and distinctive hydrophobic and hydrophilic regions in the CPP may be needed to have an efficient and stable CPP for siRNA delivery. We also propose a core-shell model for CPP/siRNA complexes that could be applied for amphipathic CPPs in complex with siRNAs.
Conclusions
Taken together, this study demonstrates that data from iTC measurements provide a basis for determining the properties needed for an efficient siRNA carrier. High charge density, hydrophobicity, amfipaticity, and pH sensitivity are all desired traits for the amphipathic CPPs. In addition, a complex needs to be stable against enzymatic degradation, and CPP should be able to pack siRNA into a complex and shield it from degrading enzymes. Out of all tested CPPs, PF6 remained superior for siRNA delivery and stability; although the pH sensitive moiety, chloroquine, may lead to some lyso-toxicity. Therefore, a CPP with similar characteristics and without the chloroquine may be advantageous.
We propose a model describing how CPPs interact with siRNA to form nanoparticles at different pHs, where enough molecules bind to each siRNA molecule to neutralize negative charges, an additional shell of CPPs is needed to form a stable complex and extra CPPs to protect and cover the siRNA. The binding affinity between the CPP and a siRNA define the complex stability to degradation and siRNA release at target site are key factors for efficient siRNA delivery and should be considered when designing CPP based delivery vehicles for siRNA.
Materials and Methods
Peptide Synthesis
Peptides were synthesized on an automated peptide synthesizer (Applied Biosystems) using fluorenylmethyloxycarbonyl (Fmoc) solid-phase peptide synthesis strategy with Rink-amide methylbenzylhydrylamine resin (0.41 mmol/g loading) to obtain C-terminally amidated peptides. The stearic acid was coupled manually to the N terminus of the peptide overnight, at room temperature with 5 eq. stearic acid. For the synthesis of NF51 and NF57 Boc-L-Orn(Fmoc)-OH (Iris Biotech) was used. Reaction was carried out using HOBT/HBTU as coupling reagents in DMF with DIEA as an activator base. For the synthesis of PF6, a chloroquine analog trifluoromethylquinoline (QN) was coupled to the lysine tree in the side chain of TP10 as described in Andaloussi et al.22
Cleavage was performed with trifluoroacetic acid (TFA), 2.5% triisopropylsilane, and 2.5% water for 2 hr at room temperature. Peptides were purified by reversed-phase high-performance liquid chromatography on C4 column (Phenomenex Jupiter C4, 5 μm, 300A, 250 × 10 mm) using a gradient of acetonitrile/water containing 0.1% TFA. The molecular weight of the peptides was analyzed by matrix-assisted laser desorption-ionization/time of flight mass spectrometry (The Voyager-DE PRO Biospectrometry System). Concentration of the peptides was determined based on dilutions of accurately weighed substances.
Cell Culture
U87 MG-luc2 human glioma cells, stably expressing luc2 and U87 cells, were grown on 0.1% gelatine (Naxo) treated plates/dishes at 37°C, 5% CO2 in DMEM with glutamax, and supplemented with 0.1 mM non-essential amino acids, 1.0 mM sodium pyruvate, 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin (Invitrogen).
Dynamic Light Scattering
Hydrodynamic mean diameter of CPPs and the particles resulting from the condensation of the siRNA with the CPPs were determined by DLS studies using a Zetasizer Nano ZS apparatus (Malvern Instruments). Particle formulation followed the same protocol as for siRNA transfection, using peptide at molar ratio 30 with 1 μM siRNA concentration. DLS studies were conducted at 25°C in MQ (pH 5.3–6.3) and in buffers. All DLS results are based on three to four measurements from two independent samples. All data were converted to “relative by intensity” plots from where the mean hydrodynamic diameter was derived.
Formation of Peptide-siRNA Complexes and siRNA Delivery
Cells were seeded (1–6 × 104) 24 hr prior the experiment on 48-well plates. For luc2, gene knockdown evaluation cell media was replaced with fresh serum containing free media. For complex formation, peptide (100 μM stock solutions) was mixed with luc2 siRNA (5′-GGACGAGGACGAGCACUUCUU, 3′-UUCCUGCUCCUGCUCGUGAAG, Life Technologies) in RNase free water (MQ) (pH 5.3–6.3) or buffers in 1/10 of final treatment volume, using molar ratios 1 to 40 (CPP over siRNA). After mixing and 1 hr incubation at room temperature, complexes were added to the cells, grown to 60% confluence, in 225 μL serum containing medium yielding a 100, 50, or 25 nM final siRNA concentration. After 4 hr, media was removed and fresh complete medium (with 10% FBS) was added per well and cells were incubated for additional 20 hr. Luc2 gene downregulation was measured with Promega Luciferase Assay System (Promega), in combination with GLOMAX 96 Microplate Luminometer obtaining LUs (light unit). The LU values were normalized to the protein content of each sample (Bio-Rad).
Lyso-toxicity and Membrane Activity of the CPP and CPP/siRNA Complexes in Serum-free Conditions
To reveal the lyso-toxicity or membrane activity of tested CPPs, the harshest conditions were used; e.g., only reduced serum media (Transfectagro, Corning) compatible with used kits. Whereas the presence of serum decreases the transfection efficiency, it also drastically decreases toxic effects that may be present due to CPP or cargo. To have detectable effects, addition of serum was excluded from these experiments.
To detect lyso-toxicity, a neutral red based In Vitro Toxicology Assay Kit (Sigma-Aldrich) was used. Neutral red is a dye that accumulates in acidic organelles. When these organelles are damaged (for example by chloroquine), the dye is released and not detected afterward from the cell lysate. For this, 105 U87 cells were seeded on a black 96-well plate with transparent bottom and grown for 24 hr in 250 μL serum containing media. The next day, media was removed and 10 μL of complexes or peptide solution in 90 μL of reduced serum media was transferred to each well. Untreated cells, cells treated with 10 μM CQ, and cell free controls were added. Thereafter, neutral red dye was added to each well and incubated for 6 hr at 37°C, 5% CO2. After incubation media was removed, cells washed with 1× PBS, and detection solution was added. After 10 min, incubation absorbance was measured at a wavelength of 540 nm and results normalized to controls.
LDH based CytoTox-ONE Homogeneous Membrane Integrity Assay (Promega) was used to detect membrane activity. Again, only reduced serum media was used. For this, 105 U87 cells were seeded to black 96-well plate with transparent bottom and grown for 24 hr in 250 μL serum containing media. The next day, media was removed and 10 μL of complexes or peptide solution in 90 μL of media was transferred to each well. Untreated cells, cells treated with 0.1% Triton X-100, and cell free controls were added. After incubation for 6 hr at 37°C, 5% CO2 LDH activity was measured according to manufacturer’s protocol. The results were normalized to controls.
Isothermal Titration Calorimetry
iTC was performed on a MicroCal ITC200 Microcalorimeter with Origin 5.0 software. The experiments were performed at 25°C in MQ (pH 5.3–6.3) in acetate buffer (5 mM, pH 5.26) and in HEPES (20 mM, pH 7.4). Luc2 siRNA (250 μL of 2 μM or 5 μM) was placed in the sample cell while water or the corresponding buffer (250 μL) was placed into the reference cell. The CPPs (200 μM) were then injected into the siRNA solution (250 μL) using a micro syringe at volumes of 1.4–2 μL after every 300 s for 24 times with low feedback for noise reduction. This covers the range of MRs used in other experiments. Blank titrations were also performed. For that, CPPs were titrated into buffer and the heats of reaction from this blank titration were subtracted from the experimental heats. Prior to analyzing the data, the first injection was discarded (initial injection is used to remove any air bubble from the end of the syringe and reduce any diffusion of nucleotide into the syringe during the equilibration period prior to initializing the titration). Data were analyzed using a single-site independent binding model where all the binding sites are considered energetically equal and binding of subsequent CPPs is not affected by already occupied binding sites.
Peptide Modeling
All peptide sequences were modeled and the charge calculations were performed using MarvinSketch 15.9.14, ChemAxon.
Fluorescent Dye Intercalation Assay
Complexation of siRNA and CPPs was assessed by Quant-iT PG assay in MQ and buffers. Complexes were prepared as described previously, following addition of diluted PG (Thermo Fisher Scientific). For detection, complexes (1/5 of final volume), MQ water or heparin (3/5), and PG (1/5) working dilution were added and incubated for 5 min and fluorescence was measured by fluorimeter (Synergy Mx, BioTek). Complex formation was confirmed by gel electrophoresis. Briefly, pre-formed complexes were mixed with RNA loading buffer and transferred to 1.8% agarose gel in 0.5 × TAE buffer (Bio-Rad).
To assess protection of siRNA from enzymatic degradation to pre-formed complexes, PG dilution was added. It was incubated for 10 min and fluorescence was measured. Proteinase K (Thermo Scientific) was added coincubated with complexes and fluorescence was measured over a 10 hr period. Proteinase K final concentration in working solution was chosen according to manufacturer’s recommendations.
Author Contributions
L.P. provided substantial contributions to the conception or design of the work, interpreted data for the work, revising it critically for important intellectual content, and drafted the work. P.A. provided substantial contributions to the conception or design of the work, interpreted data for the work, revising it critically for important intellectual content, and provided final approval of the version to be published. K.L. made contributions to the conception or design of the work and the acquisition, analysis, or interpretation of data for the work, revising it critically for important intellectual content. T.T. contributed to the the acquisition, analysis, or interpretation of data for the work, revising it critically for important intellectual content, and provided final approval of the version to be published. Ü.L. provided substantial contributions to the conception or design of the work, drafting the work or revising it critically for important intellectual content, and provided final approval of the version to be published.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by IUT20-26 (Ü.L.) and IUT2-22 (T.T.) from the Estonian Ministry of Education and Research, the Estonian Science Foundation (ETF9438) (Ü.L.), the DoRa Program of The European Social Fund (K.L.), the EU through the European Regional Development Fund through the Center of Excellence in Chemical Biology and project Tumor-Tech (3.2.1001.11-0008), Estonia (Ü.L., T.T., and L.P.), and the Swedish Research Council, VR-NT (Ü.L.). Special thanks to Vasili Hauryiliuk for technical support on iTC.
Footnotes
Supplemental Information includes five figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.02.003.
Supplemental Information
References
- 1.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Martinez J., Patkaniowska A., Urlaub H., Lührmann R., Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 3.Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 4.Cardoso A.L., Simões S., de Almeida L.P., Pelisek J., Culmsee C., Wagner E., Pedroso de Lima M.C. siRNA delivery by a transferrin-associated lipid-based vector: a non-viral strategy to mediate gene silencing. J. Gene Med. 2007;9:170–183. doi: 10.1002/jgm.1006. [DOI] [PubMed] [Google Scholar]
- 5.Detzer A., Overhoff M., Mescalchin A., Rompf M., Sczakiel G. Phosphorothioate-stimulated cellular uptake of siRNA: a cell culture model for mechanistic studies. Curr. Pharm. Des. 2008;14:3666–3673. doi: 10.2174/138161208786898770. [DOI] [PubMed] [Google Scholar]
- 6.Ren Y., Hauert S., Lo J.H., Bhatia S.N. Identification and characterization of receptor-specific peptides for siRNA delivery. ACS Nano. 2012;6:8620–8631. doi: 10.1021/nn301975s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arukuusk P., Pärnaste L., Oskolkov N., Copolovici D.M., Margus H., Padari K., Möll K., Maslovskaja J., Tegova R., Kivi G. New generation of efficient peptide-based vectors, NickFects, for the delivery of nucleic acids. Biochim. Biophys. Acta. 2013;1828:1365–1373. doi: 10.1016/j.bbamem.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Kurrikof K., Gestin M., Langel Ü. Recent in vivo advances in cell-penetrating peptide-assisted drug delivery. Expert Opin. Drug Deliv. 2015;13:373–387. doi: 10.1517/17425247.2016.1125879. [DOI] [PubMed] [Google Scholar]
- 9.Williford J.M., Wu J., Ren Y., Archang M.M., Leong K.W., Mao H.Q. Recent advances in nanoparticle-mediated siRNA delivery. Annu. Rev. Biomed. Eng. 2014;16:347–370. doi: 10.1146/annurev-bioeng-071813-105119. [DOI] [PubMed] [Google Scholar]
- 10.Scholz C., Wagner E. Therapeutic plasmid DNA versus siRNA delivery: common and different tasks for synthetic carriers. J. Control. Release. 2012;161:554–565. doi: 10.1016/j.jconrel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Pujals S., Giralt E. Proline-rich, amphipathic cell-penetrating peptides. Adv. Drug Deliv. Rev. 2008;60:473–484. doi: 10.1016/j.addr.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Bartz R., Fan H., Zhang J., Innocent N., Cherrin C., Beck S.C., Pei Y., Momose A., Jadhav V., Tellers D.M. Effective siRNA delivery and target mRNA degradation using an amphipathic peptide to facilitate pH-dependent endosomal escape. Biochem. J. 2011;435:475–487. doi: 10.1042/BJ20101021. [DOI] [PubMed] [Google Scholar]
- 13.Arukuusk P., Pärnaste L., Margus H., Eriksson N.K., Vasconcelos L., Padari K., Pooga M., Langel Ü. Differential endosomal pathways for radically modified peptide vectors. Bioconjug. Chem. 2013;24:1721–1732. doi: 10.1021/bc4002757. [DOI] [PubMed] [Google Scholar]
- 14.Ezzat K., Helmfors H., Tudoran O., Juks C., Lindberg S., Padari K., El-Andaloussi S., Pooga M., Langel Ü. Scavenger receptor-mediated uptake of cell-penetrating peptide nanocomplexes with oligonucleotides. FASEB J. 2012;26:1172–1180. doi: 10.1096/fj.11-191536. [DOI] [PubMed] [Google Scholar]
- 15.Lindberg S., Muñoz-Alarcón A., Helmfors H., Mosqueira D., Gyllborg D., Tudoran O., Langel Ü. PepFect15, a novel endosomolytic cell-penetrating peptide for oligonucleotide delivery via scavenger receptors. Int. J. Pharm. 2013;441:242–247. doi: 10.1016/j.ijpharm.2012.11.037. [DOI] [PubMed] [Google Scholar]
- 16.Deshayes S., Morris M., Heitz F., Divita G. Delivery of proteins and nucleic acids using a non-covalent peptide-based strategy. Adv. Drug Deliv. Rev. 2008;60:537–547. doi: 10.1016/j.addr.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Moschos S.A., Jones S.W., Perry M.M., Williams A.E., Erjefalt J.S., Turner J.J., Barnes P.J., Sproat B.S., Gait M.J., Lindsay M.A. Lung delivery studies using siRNA conjugated to TAT(48-60) and penetratin reveal peptide induced reduction in gene expression and induction of innate immunity. Bioconjug. Chem. 2007;18:1450–1459. doi: 10.1021/bc070077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Figueiredo I.R., Freire J.M., Flores L., Veiga A.S., Castanho M.A. Cell-penetrating peptides: A tool for effective delivery in gene-targeted therapies. IUBMB Life. 2014;66:182–194. doi: 10.1002/iub.1257. [DOI] [PubMed] [Google Scholar]
- 19.Lindgren M., Gallet X., Soomets U., Hällbrink M., Bråkenhielm E., Pooga M., Brasseur R., Langel Ü. Translocation properties of novel cell penetrating transportan and penetratin analogues. Bioconjug. Chem. 2000;11:619–626. doi: 10.1021/bc990156s. [DOI] [PubMed] [Google Scholar]
- 20.Mäe M., El Andaloussi S., Lundin P., Oskolkov N., Johansson H.J., Guterstam P., Langel Ü. A stearylated CPP for delivery of splice correcting oligonucleotides using a non-covalent co-incubation strategy. J. Control. Release. 2009;134:221–227. doi: 10.1016/j.jconrel.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 21.Lehto T., Simonson O.E., Mäger I., Ezzat K., Sork H., Copolovici D.M., Viola J.R., Zaghloul E.M., Lundin P., Moreno P.M. A peptide-based vector for efficient gene transfer in vitro and in vivo. Mol. Ther. 2011;19:1457–1467. doi: 10.1038/mt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andaloussi S.E., Lehto T., Mäger I., Rosenthal-Aizman K., Oprea I.I., Simonson O.E., Sork H., Ezzat K., Copolovici D.M., Kurrikoff K. Design of a peptide-based vector, PepFect6, for efficient delivery of siRNA in cell culture and systemically in vivo. Nucleic Acids Res. 2011;39:3972–3987. doi: 10.1093/nar/gkq1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ezzat K., Andaloussi S.E., Zaghloul E.M., Lehto T., Lindberg S., Moreno P.M., Viola J.R., Magdy T., Abdo R., Guterstam P. PepFect 14, a novel cell-penetrating peptide for oligonucleotide delivery in solution and as solid formulation. Nucleic Acids Res. 2011;39:5284–5298. doi: 10.1093/nar/gkr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziegler A. Thermodynamic studies and binding mechanisms of cell-penetrating peptides with lipids and glycosaminoglycans. Adv. Drug Deliv. Rev. 2008;60:580–597. doi: 10.1016/j.addr.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Chen B., Xu W., Pan R., Chen P. Design and characterization of a new peptide vector for short interfering RNA delivery. J. Nanobiotechnology. 2015;13:39. doi: 10.1186/s12951-015-0098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jafari M., Xu W., Naahidi S., Chen B., Chen P. A new amphipathic, amino-acid-pairing (AAP) peptide as siRNA delivery carrier: physicochemical characterization and in vitro uptake. J. Phys. Chem. B. 2012;116:13183–13191. doi: 10.1021/jp3072553. [DOI] [PubMed] [Google Scholar]
- 27.Voievoda N., Schulthess T., Bechinger B., Seelig J. Thermodynamic and biophysical analysis of the membrane-association of a histidine-rich peptide with efficient antimicrobial and transfection activities. J. Phys. Chem. B. 2015;119:9678–9687. doi: 10.1021/acs.jpcb.5b04543. [DOI] [PubMed] [Google Scholar]
- 28.Copolovici D.M., Langel K., Eriste E., Langel Ü. Cell-penetrating peptides: design, synthesis, and applications. ACS Nano. 2014;8:1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
- 29.Soomets U., Lindgren M., Gallet X., Hällbrink M., Elmquist A., Balaspiri L., Zorko M., Pooga M., Brasseur R., Langel U. Deletion analogues of transportan. Biochim. Biophys. Acta. 2000;1467:165–176. doi: 10.1016/s0005-2736(00)00216-9. [DOI] [PubMed] [Google Scholar]
- 30.Pooga M., Hällbrink M., Zorko M., Langel Ü. Cell penetration by transportan. FASEB J. 1998;12:67–77. doi: 10.1096/fasebj.12.1.67. [DOI] [PubMed] [Google Scholar]
- 31.Lehto T., Abes R., Oskolkov N., Suhorutsenko J., Copolovici D.M., Mäger I., Viola J.R., Simonson O.E., Ezzat K., Guterstam P., Samir El Andaloussi Delivery of nucleic acids with a stearylated (RxR)4 peptide using a non-covalent co-incubation strategy. J. Control. Release. 2010;141:42–51. doi: 10.1016/j.jconrel.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura Y., Kogure K., Futaki S., Harashima H. Octaarginine-modified multifunctional envelope-type nano device for siRNA. J. Control. Release. 2007;119:360–367. doi: 10.1016/j.jconrel.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Kim H.J., Ishii A., Miyata K., Lee Y., Wu S., Oba M., Nishiyama N., Kataoka K. Introduction of stearoyl moieties into a biocompatible cationic polyaspartamide derivative, PAsp(DET), with endosomal escaping function for enhanced siRNA-mediated gene knockdown. J. Control. Release. 2010;145:141–148. doi: 10.1016/j.jconrel.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 34.Oskolkov N., Arukuusk P., Copolovici D.M., Lindberg S., Margus H., Padari K., Pooga M., Langel Ü. NickFects, phosphorylated derivatives of transportan 10 for cellular delivery of oligonucleotides. Int. J. Pept. Res. Ther. 2011;17:147–157. [Google Scholar]
- 35.Freimann K., Arukuusk P., Kurrikoff K., Vasconcelos L.D., Veiman K.L., Uusna J., Margus H., Garcia-Sosa A.T., Pooga M., Langel Ü. Optimization of in vivo DNA delivery with NickFect peptide vectors. J. Control. Release. 2016;241:135–143. doi: 10.1016/j.jconrel.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 36.Xiong K., Blainey P.C. Molecular sled sequences are common in mammalian proteins. Nucleic Acids Res. 2016;44:2266–2273. doi: 10.1093/nar/gkw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crombez L., Aldrian-Herrada G., Konate K., Nguyen Q.N., McMaster G.K., Brasseur R., Heitz F., Divita G. A new potent secondary amphipathic cell-penetrating peptide for siRNA delivery into mammalian cells. Mol. Ther. 2009;17:95–103. doi: 10.1038/mt.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crowet J.M., Lins L., Deshayes S., Divita G., Morris M., Brasseur R., Thomas A. Modeling of non-covalent complexes of the cell-penetrating peptide CADY and its siRNA cargo. Biochim. Biophys. Acta. 2013;1828:499–509. doi: 10.1016/j.bbamem.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Lin P.J., Tam Y.Y., Hafez I., Sandhu A., Chen S., Ciufolini M.A., Nabi I.R., Cullis P.R. Influence of cationic lipid composition on uptake and intracellular processing of lipid nanoparticle formulations of siRNA. Nanomedicine (Lond.) 2013;9:233–246. doi: 10.1016/j.nano.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Margus H., Arukuusk P., Langel Ü., Pooga M. Characteristics of cell-penetrating peptide/nucleic acid nanoparticles. Mol. Pharm. 2015;13:172–179. doi: 10.1021/acs.molpharmaceut.5b00598. [DOI] [PubMed] [Google Scholar]
- 41.van Asbeck A.H., Beyerle A., McNeill H., Bovee-Geurts P.H., Lindberg S., Verdurmen W.P., Hällbrink M., Langel Ü., Heidenreich O., Brock R. Molecular parameters of siRNA--cell penetrating peptide nanocomplexes for efficient cellular delivery. ACS Nano. 2013;7:3797–3807. doi: 10.1021/nn305754c. [DOI] [PubMed] [Google Scholar]
- 42.García-Sosa A.T., Tulp I., Langel K., Langel Ü. Peptide-ligand binding modeling of siRNA with cell-penetrating peptides. Biomed. Res. Int. 2014;2014:257040. doi: 10.1155/2014/257040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pujals S., Fernández-Carneado J., López-Iglesias C., Kogan M.J., Giralt E. Mechanistic aspects of CPP-mediated intracellular drug delivery: relevance of CPP self-assembly. Biochim. Biophys. Acta. 2006;1758:264–279. doi: 10.1016/j.bbamem.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Québatte G., Kitas E., Seelig J. riDOM, a cell-penetrating peptide. Interaction with DNA and heparan sulfate. J. Phys. Chem. B. 2013;117:10807–10817. doi: 10.1021/jp404979y. [DOI] [PubMed] [Google Scholar]
- 45.Walrant A., Bechara C., Alves I.D., Sagan S. Molecular partners for interaction and cell internalization of cell-penetrating peptides: how identical are they? Nanomedicine (Lond.) 2012;7:133–143. doi: 10.2217/nnm.11.165. [DOI] [PubMed] [Google Scholar]
- 46.Bürck J., Roth S., Wadhwani P., Afonin S., Kanithasen N., Strandberg E., Ulrich A.S. Conformation and membrane orientation of amphiphilic helical peptides by oriented circular dichroism. Biophys. J. 2008;95:3872–3881. doi: 10.1529/biophysj.108.136085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dominska M., Dykxhoorn D.M. Breaking down the barriers: siRNA delivery and endosome escape. J. Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 48.Han L., Tang C., Yin C. Effect of binding affinity for siRNA on the in vivo antitumor efficacy of polyplexes. Biomaterials. 2013;34:5317–5327. doi: 10.1016/j.biomaterials.2013.03.060. [DOI] [PubMed] [Google Scholar]
- 49.Palm C., Netzereab S., Hällbrink M. Quantitatively determined uptake of cell-penetrating peptides in non-mammalian cells with an evaluation of degradation and antimicrobial effects. Peptides. 2006;27:1710–1716. doi: 10.1016/j.peptides.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 50.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 51.Simeoni F., Morris M.C., Heitz F., Divita G. Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003;31:2717–2724. doi: 10.1093/nar/gkg385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.