Abstract
Muscle stem cells (MuSCs) hold great therapeutic potential for muscle genetic disorders, such as Duchenne muscular dystrophy (DMD). The CRISP/Cas9-based genome editing is a promising technology for correcting genetic alterations in mutant genes. In this study, we used fibrin-gel culture system to selectively expand MuSCs from crude skeletal muscle cells of mdx mice, a mouse model of DMD. By CRISP/Cas9-based genome editing, we corrected the dystrophin mutation in expanded MuSCs and restored the skeletal muscle dystrophin expression upon transplantation in mdx mice. Our studies established a reliable and feasible platform for gene correction in MuSCs by genome editing, thus greatly advancing tissue stem cell-based therapies for DMD and other muscle disorders.
Keywords: muscle stem cells, 3D fibrin gel, ex vivo expansion, CRISPR/Cas9, genome editing, Duchenne muscular dystrophy, DMD
Introduction
Duchenne muscular dystrophy (DMD) is one of the most common inherited muscular disorders worldwide. This genetic disorder is a recessive X-linked form of muscular dystrophy resulting from a mutation in the human X chromosome-localized dystrophin gene.1 The mutation disrupts production of the muscle fiber protein, leading to wasting of skeletal, respiratory, and cardiac muscles and advanced death. Early attempts in the treatment of muscular dystrophy have included intramuscular injection of lentivirus expressing a functional microdystrophin2 and targeted genetic correction of mutations in cultured myoblasts by single-stranded short-fragment homologous replacement (ssSFHR)3 or peptide nucleic acid single-stranded oligodeoxynucleotides (PNA-ssODNs).4 However, these approaches have not been effective due to insufficient expression of dystrophin or low gene correction editing efficiency.
The emerging genome-editing technologies, such as CRISPR/Cas9 and transcription activator-like effector nucleases (TALEN), have enabled correction of mutations using engineered nucleases in a wide variety of cell types and organisms. CRISPR/Cas9 genome editing is based on RNA-guided nuclease Cas9 and guided RNA with customizable specificities, which have shown rapid, easy, and efficient modification of endogenous genes in many types of cells.5, 6, 7 CRISPR/Cas9 technology brings incredible therapeutic benefit to genetic disorders.5 Indeed, CRISPR-induced deletion or CRISPR-mediated excision of mutated exon 23 of Dmd successfully restored a truncated Dystrophin protein expression in primary mdx or DMD myoblasts.8, 9 By using similar exon-skipping strategy, several independent groups recently reported that local or systemic delivery of adeno-associated virus (AAV) carrying CRISPR/Cas9 coupled with paired guide RNAs (gRNAs) flanking the mutated exon 23 of Dmd resulted in excision of intervening DNA and restored expression of a truncated version of the Dystrophin protein in myofibers, cardiomyocytes, and even muscle stem cells (MuSCs).10, 11, 12 The resulting improvement of muscle biochemistry and significant enhancement of muscle force in mdx mice are encouraging, which sheds light on a potential therapeutic benefit in humans. However, further studies demonstrating safety and assessing immune responses to the delivery vehicle and gene-editing system are in great demand before clinical applications.
Stem cell-based therapy is considered as one of the most promising methods for treating muscular dystrophies. Application of CRISPR/Cas9-based genome editing has thus far been widely explored in treating or preventing DMD by varying editing target from embryonic stem (ES) cells, induced pluripotent stem cells (iPSCs),13 to germline cells.14 However, the excitement of patient-specific iPSCs for the production of autologous cells for therapy has been tempered by the discovery of reprogramming-induced genomic mutations and the lack of efficient protocols for producing safe and transplantable muscle stem cells (MuSCs) in vitro. Moreover, germline cell-based gene therapies are not suitable for patients. Adult stem cells, compared to traditional embryonic stem cells (ESCs) and iPSCs, provide promising cell source in therapeutic utility for their properties of self-renewal, multi-potency, and defined cell lineages and functions. However, report on their effective application in regenerative and reparative therapeutics was rare except for human intestinal stem cells, which can be expanded in culture over long time periods as genetically and phenotypically stable epithelial organoids.15 In skeletal muscles, satellite cells are acknowledged as bona fide adult MuSCs being responsible for postnatal growth and regeneration of the muscle fiber.16 Transplantation-based studies have demonstrated the impressive potential of satellite cells in regenerating damaged or diseased muscle17, 18, 19, 20, 21 and thus highlighted the importance of such adult stem cells as therapeutic targets in treating inherited, acquired, as well as age-associated muscular disorders. However, the application of the CRISPR/Cas9 gene-editing technique in concert with satellite cell remains unseen in spite of the recognized importance and advantages of such adult stem cells.
Recently, we established a soft three-dimensional (3D) salmon fibrin gel culture system by the reaction of fibrinogen and thrombin, and showed that this system selectively expanded adult mouse MuSCs from bulk skeletal muscle preparations without the need for prior cell sorting.22 In this study, we performed selection and primary expansion of skeletal MuSCs in 3D soft fibrin gels, followed by customized genome editing through CRISPR/Cas9 technology, and secondary expansion to propagate the repaired MuSCs. Finally, the genetically repaired MuSCs were transplanted to restore dystrophin expression in mdx mice. The method provided a safer, easier, and more direct adult stem cell-based postnatal therapy for DMD patients, and further advanced tissue-specific stem cell-based therapies for clinical degenerative diseases.
Results
Selective Expansion of MuSCs from mdx Mice Using Soft 3D Fibrin Gel Culture System
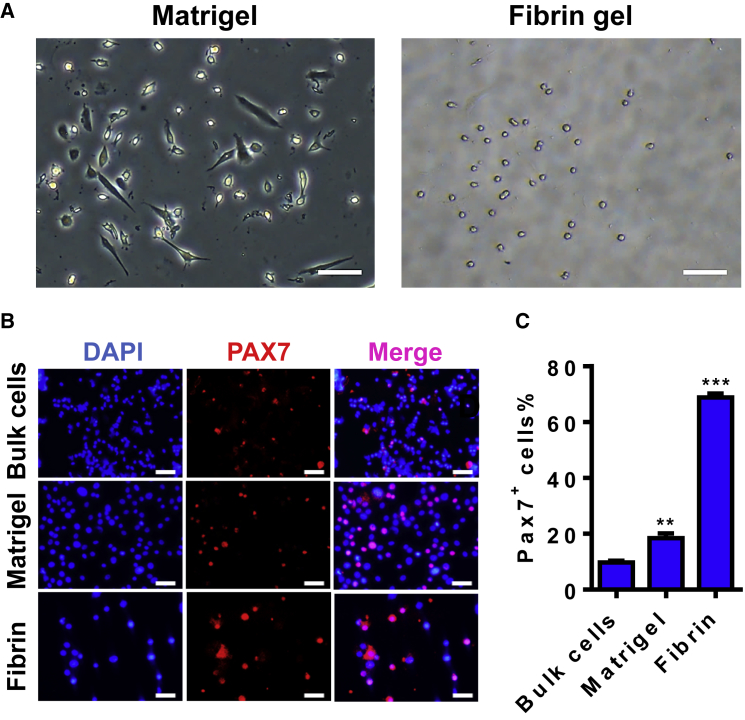
One of the roadblocks that hamper the application of adult MuSCs in regenerative therapeutics is their rare number in skeletal muscle tissue and rapid loss of stemness upon removal from the niche and culture ex vivo. Because of the limited source of patient specimen, a direct propagation of the highly enriched MuSCs from a small starting number of bulk skeletal muscle cells particularly benefits adult stem cell-based therapy. We have recently established a soft 3D salmon fibrin gel culture platform, which enables selective expansion of MuSCs in bulk skeletal muscle cells from wild-type mice. However, it has never been used for culturing MuSCs from mdx mice, which displays accelerated differentiation in primary cultures and in isolated myofibers under traditional culture condition.23 To expand mdx MuSCs, we seeded an equal number of bulk muscle cells on Matrigel or in soft 3D fibrin gel, respectively, and allowed the cells to propagate in complete growth medium for 7 days. Consistent with our previous observation for normal muscle cells, spindle-shaped cells exhibited an apparently superior growth over small round cells on Matrigel, whereas the vast majority of the expanded cells in soft 3D fibrin gel appeared round (Figure 1A). Immunofluorescence (IF) staining showed that less than 10% of cells in primary bulk skeletal muscle cells express PAX7 (Figures 1B and 1C). However, after 7 days of culture in soft 3D fibrin gel, greater than 70% of expanded cells were positive for PAX7 expression, which was over 3-fold higher than PAX7-expressing cells in Matrigel-expanded progeny (Figures 1B and 1C).
Figure 1.
Soft 3D Fibrin Gel Selectively Expands Skeletal MuSCs from mdx-Derived Bulk Skeletal Muscle Cells
(A) Representative photographs of bulk cell progeny expanded on Matrigel or in 3D soft fibrin gel (n = 4 independent experiments). Bright-field images were taken on day 7 of culture. Scale bar, 100 μm. (B) Immunostaining for PAX7 in fresh bulk skeletal muscle cells and their progeny expanded on Matrigel or in soft 3D fibrin gel by day 7, with DAPI as nuclear counterstaining. Scale bar, 50 μm. (C) Quantification of PAX7+ cell subpopulations in fresh bulk skeletal muscle cells and their progeny expanded for 7 days on Matrigel or in soft 3D fibrin gel. Error bars represent means ± SEM of three independent experiments. **p < 0.01, ***p < 0.001 versus fresh bulk cells, Student’s t test.
Collectively, these data indicated that soft 3D fibrin gel selectively expands MuSCs from mdx mice-derived bulk muscle cells without a need for sorting pure MuSCs before culturing.
Design of CRISPR/Cas9 Components for Correcting the Dmd Mutation
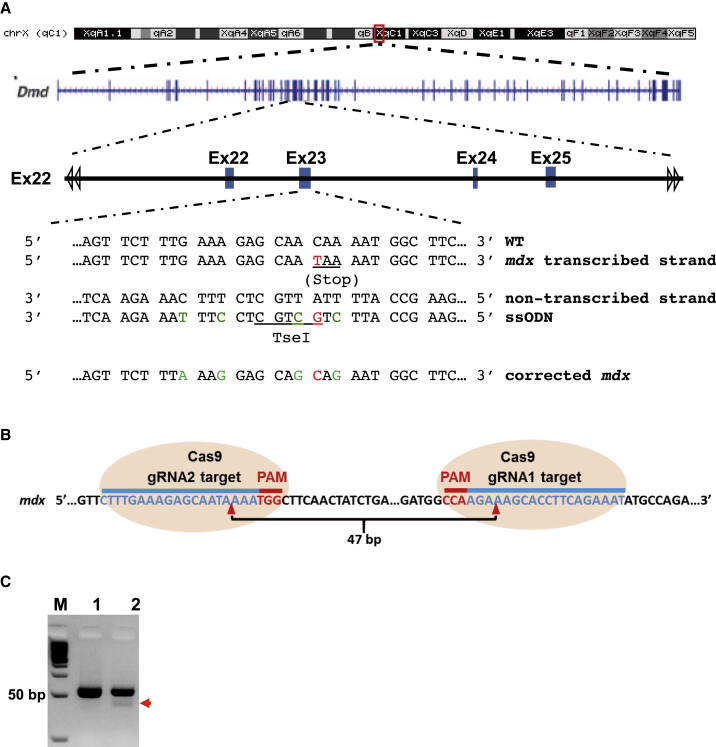
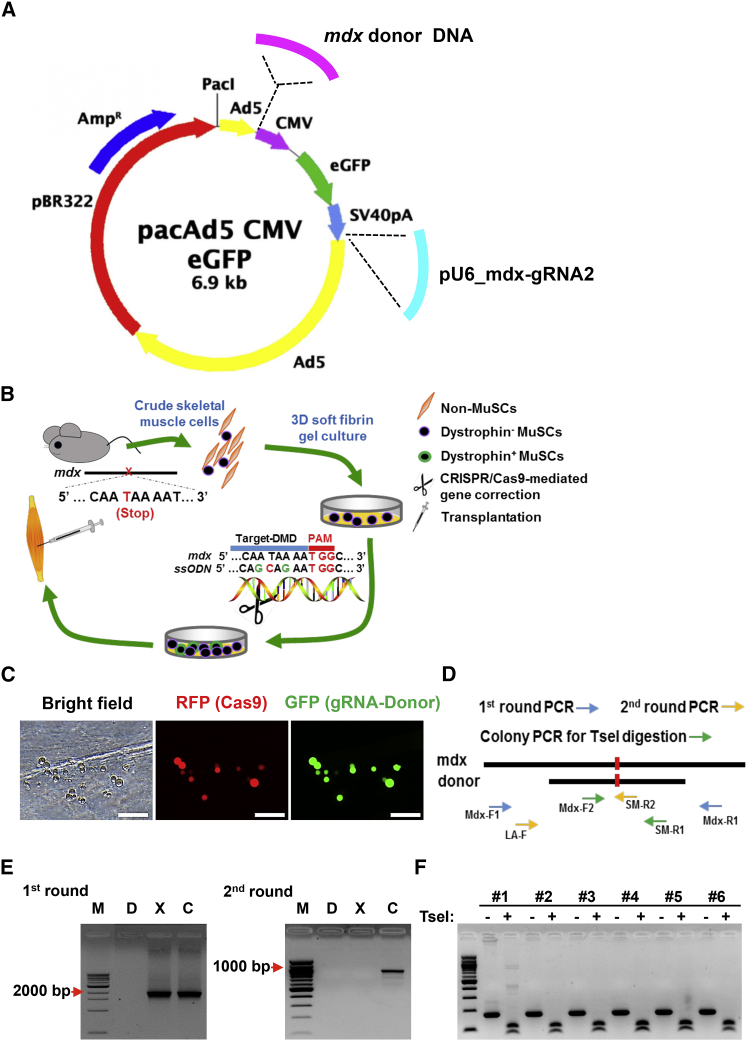
DMD is a lethal muscular degenerative disease caused by mutations in the dystrophin gene (Dmd) of X chromosome.1 The mdx mouse is a well-established model of DMD with one of several variants carrying a C-to-T nonsense transition in exon 23 of Dmd that causes a truncation of the expressed Dystrophin protein24, 25 (Figure 2A). Nuclease-induced double-stranded break (DSB) can be repaired by homology-directed repair (HDR) in the presence of exogenous homologous DNA donor templates.5 Single-stranded oligodeoxynucleotide (ssODN) serves as a powerful template directing HDR upon nuclease-mediated double-stranded DNA break in the genome.26, 27 Previous study demonstrated that ssODN length affects HDR efficiency, and an optimal HDR efficiency was achieved with an ssODN around 90 nt, whereas shorter or longer ssODNs decrease HDR efficiency.27 In addition, ssODN that hybridizes to the non-transcribe strand of genomic DNA was reported to be more effective in directing HDR.26 Based on these observations, we designed an 84-nt ssODN as HDR template, which targeted the non-transcribe strand of Dmd with 42 nt of homology sequence flanking each side of the mutated site (Figure 2A; Table S1). The ssODN was incorporated with four silent mutations, which together with the corrected mdx point mutation site added a TseI restriction enzyme site for facilitating genotyping (Figure 2A). To enhance the stability and lower the dosage of donor DNA, we added three phosphorothioate-modified terminal bases at on both ends of the ssODN28 (Table S1).
Figure 2.
Design of gRNAs and ssODN for CRISPR/Cas9-Mediated Gene Editing
(A) Design of ssODN as HDR DNA template. ssODN, which contains 41 bp of homology arms flanking each side of the mutation site and targets the non-transcribed strand of Dmd gene. ssODN was phosphorothioate-modified at both ends (denoted with * [refer to Table S1]) and incorporated four silent mutations (green), which prevent binding of gRNA2/Cas9 and add a TseI restriction enzyme site for genotyping and verification of HDR-mediated gene correction. (B) Schematic of gRNAs targeting sequences of Dmd (blue) and PAM sequence (red). Red arrowhead indicates the cleavage site by CRISPR/Cas9. gRNA1 and gRNA2 were the two gRNAs specific for Dmd. (C) PCR validation of CRISPR/Cas9-mediated DNA cleavage of genomic DNA target. Mouse muscle-derived fibroblasts were transfected with CRISPR/Cas9, gRNA1, and gRNA2 by Lipofectamine 3000. CRISPR/Cas9 cut at each of the two gRNA-targeted sequences in Dmd, which resulted in 47-bp shorter PCR products when amplified with two primers of Mdx-F2 and Mdx-R2 (see Table S1), which locate in 5′ end of gRNA2 site and 3′ ends of gRNA1 site, respectively.
A previously reported gRNA (mdx-gRNA2) targeting Dmd exon 2314 was adapted to direct Cas9 cutting near the mdx point mutation site (Figure 2B). To confirm the successful cleavage of Dmd genomic DNA via Cas9 and mdx-gRNA2, we first designed another gRNA (mdx-gRNA1) targeting 47 bp upstream of the 5′ end of gRNA2 (Figure 2B). Next, we transfected mouse muscle-derived fibroblasts with Cas9 and the two gRNAs (mdx-sgRNA1 and 2) using Lipofectamine 3000. Three days later, we extracted genomic DNA from the transfected cells and performed PCR with two primers locating at the 5′ end of sgRNA2 site and the 3′ ends of sgRNA1 site, respectively. We detected smaller DNA band with an expected size only in the cells transfected with both of the gRNAs and Cas9, indicating that mdx-gRNA1- and 2-guided Cas9 nuclease successfully cleaved the target sites in Dmd (Figure 2C).
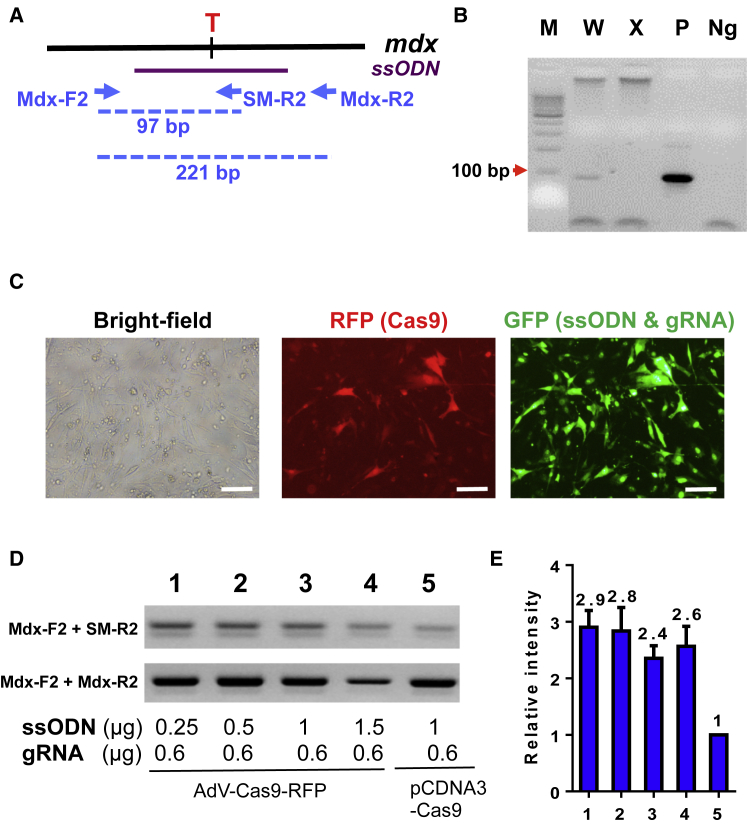
Before assessing the ssODN-directed HDR of Dmd in MuSCs, we designed a pair of primers for genotyping by allele-specific PCR (Figure 3A). Under the optimal condition, the two primers (Mdx-F2 and Mdx-R2) amplified a 97-bp PCR product only from the ssODN containing the silent mutations (Figure 3B). To improve HDR efficiency, we continued to optimize the ratio of ssODN to gRNA. We transfected muscle-derived fibroblasts with 0.6 μg of gRNA2 and an increasing dosage of ssODN using Lipofectamine 3000. Six hours after transfection, we infected the transfected cells with Cas9-expressing adenovirus (AdR-Cas9) (Figure 3C). Four days after infection, the cells were harvested for genomic extraction, followed by allele-specific PCR with two pairs of primers (Mdx-F2 + SM-R2 and Mdx-F2 + Mdx-R2) (Figure 3A). HDR-directed correction of Dmd mutation was further confirmed by TseI endonuclease digestion and DNA sequencing (Figure S1). Our result showed that lower dosage of ssODN actually directed higher HDR efficiency at the expected site of Dmd (Figures 3D and 3E). It appeared that delivery of Cas9 nuclease by adenovirus had a significantly higher HDR efficiency than that of co-transfection of Cas9-harboring plasmids with ssODN and gRNA (Figures 3D and 3E). To calculate the efficiency of the ssODN-mediated HDR, we amplified DNA fragments with both corrected and uncorrected Dmd mutation site by PCR with Mdx-F2 and Mdx-R2 primers and cloned them into TOPO-TA cloning vector, and transformed Escherichia coli. Single bacterial colonies were screened for gene correction by allele-specific PCR using Mdx-F2 and SM-R2. It turned out that about 1% of total cells underwent ssODN-mediated HDR correction of the Dmd mutation.
Figure 3.
Assessment of ssOND-Mediated Dmd Gene Correction via HDR in Muscle Fibroblasts
(A) Scheme of allele-specific PCR-based genotyping. A pair of common forward (Mdx-F2) and reverse (Mdx-R2) primers, which located out of ssODN template sequence in Dmd gene, was designed to amplify a 221-bp product. Another reverse primer (SM-R2) ended with the corrected mutation site was used to amplify a 97-bp product from gene template after HDR-mediated Dmd correction. (B) Allele-specific PCR for genotyping of corrected Dmd gene. W, wild-type genomic DNA template; X, genomic DNA template from mdx mice; P, synthesized DNA template containing corrected mutation site and silent mutations; Ng, PCR negative control containing no cDNA template. (C–E) Optimization of CRISPR/Cas9-mediated Dmd correction by HDR in muscle-derived fibroblasts. Representative images showing delivery of gRNA, ssODN, and pmax-GFP by Lipofectamine 3000 and Cas9 by adenovirus infection (designated as “AdV-Cas9-RFP”) or transfection of pCDNA3 plasmids containing Cas9 (designated as pCDNA3-Cas9) using Lipofectamine 3000. Scale bar, 50 μm. RFP was used to track infecting efficiency of adenovirus expressing Cas9, whereas GFP was used to reflect transfection efficiency of ssODN and gRNA (C). Allele-specific PCR was performed to assess HDR-mediated Dmd correction in muscle-derived fibroblasts (D). Semi-quantification of the allele-specific PCR results in (D) was analyzed using ImageJ (E).
CRISPR/Cas9-Mediated Dmd Correction in Fibrin Gel-Expanded MuSCs from mdx Mice
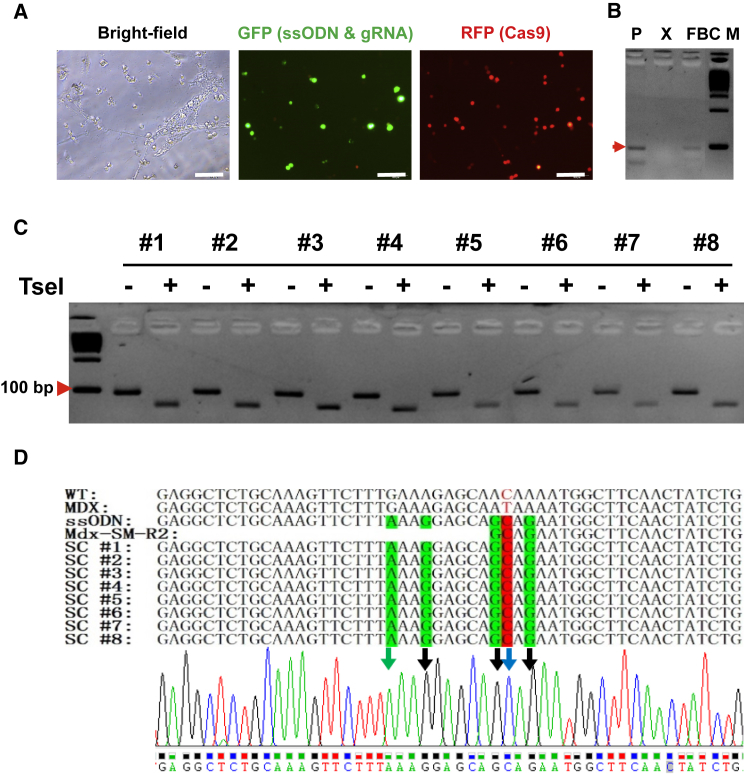
Following the above established ssODN-mediated HDR and allele-specific PCR conditions, we continued to correct Dmd mutation in fibrin gel-expanded MuSCs from mdx mice. First, bulk skeletal muscle cells were isolated from hindlimb of mdx mice and directly cultured in soft 3D fibrin gel. After morphologically round MuSCs became the dominant population in culture, cells were transfected with ssODN and gRNA2 by Lipofectamine 3000, followed by infection with AdR-Cas9, a Cas9-expressing adenovirus (Figure 4A). Allele-specific PCR detected a target band in genomic DNA from harvested MuSCs, indicating ssODN-mediated HDR in fibrin-expanded MuSCs from mdx mice (Figure 4B). To confirm CRISPR/Cas9-mediated Dmd correction, we subcloned the allele-specific PCR product in pCR4-TOPO vector. Individual clones were used as templates for PCR using Mdx-F2 and SM-R2 primers (Figure 3A). PCR products were directly subjected to digestion using TseI restriction endonuclease. As shown in Figure 4C, a smaller band was detected in all the TseI-digested PCR products, indicating a successful correction of Dmd mutation using designed ssODN as template. We further confirmed the Dmd correction by DNA sequencing for those randomly picked-up clones (Figure 4D).
Figure 4.
Correction of the Dmd Mutation Using ssODN Donor with CRISPR/Cas9 System in Fibrin-Expanded MuSCs
(A) Representative images showing delivery of CRISPR/Cas9 components into fibrin-expanded cells. Bulk skeletal muscle cells were cultured in soft 3D fibrin gel. When round-shaped MuSCs became the dominant cell type in soft 3D fibrin gel, cells were transfected with a cocktail of gRNA, ssODN, and pmax-GFP by Lipofectamine 3000. Six hours after transfection, cells were then infected with adenovirus AdV-Cas9-RFP, which co-expresses RFP and CRISPR/Cas9. GFP was used to reflect transfection efficiency of ssODN and gRNA. Scale bar, 100 μm. (B) Genotyping PCR using Mdx-F2 and SM-R2 indicating HDR-mediated Dmd correction in the fibrin-expanded MuSCs (n = 3). P, positive control using synthesized donor DNA fragment; X, genomic DNA from uncorrected mdx muscle cells; FBC, genomic DNA from corrected fibrin-expanded bulk muscle cells from mdx mice; M, 100-bp DNA marker. (C) TseI digestion confirming HDR-mediated Dmd correction in fibrin gel-expanded MuSCs. Allele-specific PCR products amplified by Mdx-F1 and SM-R2 from genomic DNA of expanded MuSC were sub-cloned into TOPO cloning vector, followed by colony-PCR with the same pair of allele-specific primers. The 97-bp PCR products from individual colonies were directly digested by TseI, which was incorporated by ssODN-directed HDR, and resulted in two fragments of 73 and 24 bp, respectively. Only the 73-bp fragment was visible by electrophoresis in 3% agarose gel. (D) Confirmation of HDR-mediated Dmd correction in fibrin-expanded MuSCs by DNA sequencing. TOPO clones referred to in (C) were sequenced. Silent mutations were indicated with green letters. Point mutations were highlighted in red.
CRISPR/Cas9-Mediated Dmd Correction in Fibrin Gel-Expanded MuSCs Restores Dystrophin Expression in mdx Mice upon Transplantation
We had established a procedure for ssODN-mediated HDR in ex vivo expanded MuSCs. However, it is very challenging to achieve high transfection efficiency in MuSCs. To solve this problem, we used adenoviral vector for delivering both gRNA expression cassette and a 1,314-bp donor DNA with 591- and 722-bp homology arms flanking the mutation site (AdG-gRNA-Donor) (Figure 5A). We co-infected fibrin-expanded MuSCs with AdR-Cas9 and AdG-gRNA-Donor (Figure 5B) and achieved very high infection efficiency (Figure 5C). We designed a nested PCR strategy to examine the HDR-mediated Dmd correction in MuSCs (Figure 5D). To avoid the false-positive PCR result caused by donor DNA, we first amplified DNA fragments (2,188 bp) containing both corrected and uncorrected Dmd mutation site using a pair of primers (Mdx-F1 and Mdx-R1) that resided outside of the donor DNA sequence. Next, we purified the DNA fragment amplified by the first PCR and used it in the second-round allele-specific PCR (LA-F and SM-R2). By this strategy, we confirmed that adenoviral delivery of designed CRISPR/Cas9 components successfully corrected the Dmd mutation in fibrin-expanded MuSCs from mdx mice (Figure 5E). To calculate the efficiency of the donor-mediated HDR repair by adenoviral system, we amplified DNA fragments with corrected and uncorrected Dmd mutation site by PCR with Mdx-F1 and Mdx-R1 primers and cloned them into TOPO-TA cloning vector, and transformed E. coli. Single bacterial colonies were screened for gene correction by allele-specific PCR. The positive clones were further amplified by PCR with Mdx-F2 and SM-R1 primers (Figure 5D), and the PCR products were digested using TseI to confirm the corrections (Figure 5F). By such an approach, we found that approximately 6% of infected MuSCs underwent donor-mediated HDR correction of the Dmd mutation.
Figure 5.
Correction of Dmd Gene in Soft 3D Fibrin-Expanded MuSCs Using Adenoviral Vector Delivery of RNA-Guided CRISPR/Cas9 and Donor DNA
(A) Diagram of adenoviral vectors expressing Cas9 (AdV-Cas9) and harboring gRNA expression cassette and Dmd-specific donor template. The mdx-gRNA2 shown in Figure 2B was used. The donor template was a 1,314-bp DNA fragment with 591- and 722-bp homology arms flanking the mutation site. The silent mutations constituting TseI restriction enzyme site in ssODN were also incorporated in this longer donor template. (B) Scheme of adult MuSC-based gene therapy for DMD in mdx mice. Bulk skeletal muscle cells were isolated from mdx mice and then cultured in soft 3D fibrin gel. When morphologically round MuSCs became evident (3∼4 days), CRISPR/Cas9 and donor DNA complexes were delivered to initiate targeted genome editing for correcting Dmd mutations. Cells were allowed to expand in fibrin gel for 3 more days to propagate Dmd-corrected MuSCs. Expanded MuSCs were then transplanted in mdx mice. (C) Representative images showing adenoviral delivery of CRISPR/Cas9 components into fibrin-expanded cells. Bulk skeletal muscle cells were cultured in soft 3D fibrin gel. When round MuSCs became the dominant cell type in soft 3D fibrin gel, cells were coinfected with adenoviruses expressing CRISPR/Cas9 (red) and carrying gRNA and donor DNA (green). RFP was used to track infection efficiency of adenovirus expressing Cas9, whereas GFP was used to reflect transfection efficiency of donor DNA and gRNA. Scale bar, 100 μm. (D) Schematic diagram for assessing correction of Dmd mutation by adenoviral delivery of CRISPR/Cas9 complexes. To avoid interference of donor DNA as template in PCR, genome DNA was amplified first by a pair of primers (Mdx-F1 and Mdx-R1) residing outside of donor DNA sequence. The resultant PCR products containing both original and mdx-corrected fragments were then purified and subcloned into TOPO-TA cloning vector. After transformation, single bacterial colonies were picked up for allele-specific PCR using primers of LA-F and SM-R2 to screen for the corrected clones. To confirm allele-specific PCR results, colony PCR was further performed using primers of Mdx-F2 and SM-R1, and the resultant product was subjected to TseI digestion. TseI site was designed and introduced in the donor DNA. (E) Genotyping result of HDR-mediated Dmd correction in the fibrin-expanded MuSCs (n = 3 independent experiments). M, 100-bp DNA marker; D, donor DNA plasmids; X, genomic DNA from uncorrected mdx muscle cells; C, genomic DNA from corrected fibrin-expanded mdx muscle cells. Mdx-F1 and Mdx-R1 were used for the first round of PCR, and LA-F and SM-R2 were used for the second round of PCR. (F) TseI digestion confirming HDR-mediated Dmd correction in fibrin gel-expanded MuSCs. DNA fragments (lane C, left panel of D) from first round of PCR were sub-cloned into TOPO-TA cloning vector, followed by colony-PCR with primers of Mdx-F2 and SM-R1. PCR products from individual colonies were directly digested by TseI.
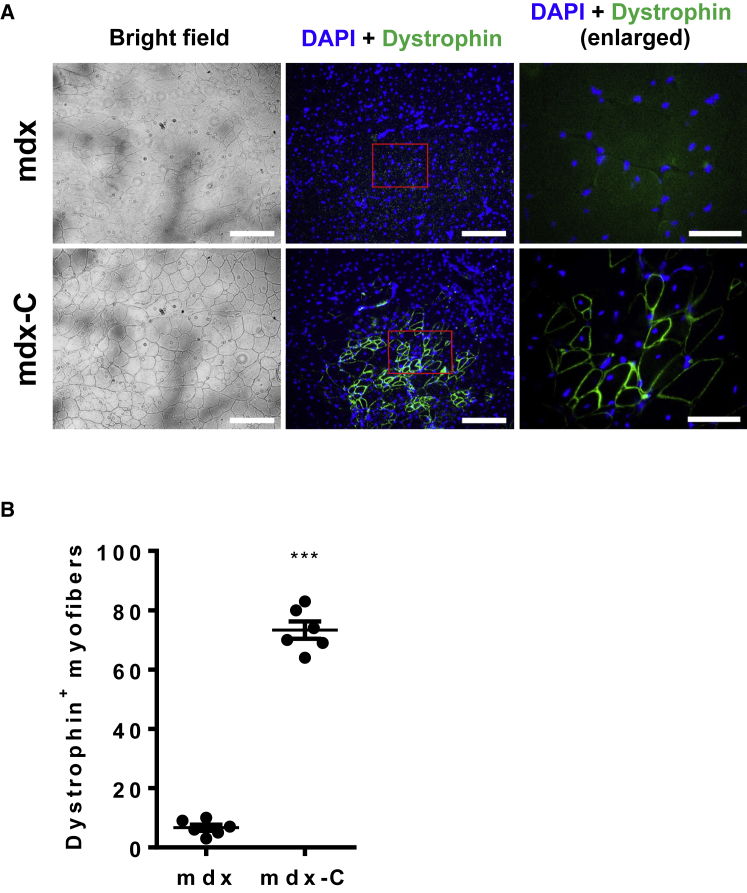
To determine whether the Dmd mutation-corrected MuSCs can restore dystrophin in vivo, we expanded gene-corrected MuSCs in fibrin gel for 3 more days, and then transplanted them into hindlimb of mdx mice (Figure 5B). One month after transplantation, we performed IF analysis and found that gene-corrected MuSCs successfully restored the expression of dystrophin in skeletal muscles of mdx recipients (Figures 6A and 6B). Together, our results demonstrated that CRISPR/Cas9-mediated genome editing of ex vivo expanded MuSCs holds great potential for the treatment of DMD and other genetic muscle disorders.
Figure 6.
Restoration of Dystrophin Expression in mdx Mice upon Transplantation of Corrected MuSCs
(A) Representative dystrophin immunostaining (scale bar, 500 μm) and magnification of boxed area (scale bar, 50 μm) of TA muscles from control (mdx) and mutation-corrected MuSCs (mdx-C)-transplanted mdx mice. Muscle was dissected from recipient mice 4 weeks after transplantation with gene-corrected MuSCs and subjected to dystrophin immunostaining. A partial restoration of dystrophin expression in transplanted TA muscles of mdx mice was evident from three independent experiments. (B) Quantification of dystrophin-expressing myofibers in TA muscles of recipient mdx mice (n = 6 mice) shown in (A). Error bars represent means ± SEM. Three independent experiments were performed. ***p < 0.001, Student’s t test.
Discussion
DMD is a lethal muscular degenerative disease caused by mutations in dystrophin gene of X chromosome.1 Lack of the Dystrophin protein in muscle cells causes them to be fragile and easily damaged. Before heart and respiratory muscles are affected by the early teens, symptoms of skeletal muscle weakness begin first in the arms, legs, and trunk as early as age 3. To restore the Dystrophin expression in skeletal and cardiac muscle is essential for relieving the muscle wasting and weakness as well as extending life span of the patients. In the current study, based on our recently established novel soft 3D salmon fibrin gel culture system,22 which enables selectively expansion of MuSCs with highly reserved regenerative competency from a small amount of bulk skeletal muscle cells, we corrected disease-driving mutation directly in adult skeletal MuSCs from mdx mice using CRISPR/Cas9-based genome editing, followed by transplantation to restore Dystrophin protein expression in vivo in mdx mice.
Stem cell-based therapy provides an effective approach for many inherited diseases. Patient-derived iPSCs have been favorably considered as a therapeutic target for treating DMD.29, 30, 31 However, these cells are not yet the perfect alternative. Generation of iPSCs relies on ectopic expression of Oct4, Sox2, Klf4, and c-Myc genes in somatic cells, whereas this set of genes is associated with the development of multiple tumors.32, 33 In addition, the efficiency of directed differentiation of iPSCs to myogenic lineage cells is a key step for therapies and needs to be further improved. Traditional protocol based on the embryoid body formation method got a fairly low yield of heterogeneous myogenic population.34 Recently, some new stepwise monolayer cell differentiation strategies have been developed to differentiate ESCs35 or iPSCs36 to myogenic progenitors in high efficiency. However, the engraftment efficiency, functionality, and safety of these induced myogenic progenitor cells remain to be determined by in vivo studies.
In contrast, adult skeletal MuSCs have been acting as a potent cell source in therapeutic utility for their properties of self-renewal, multi-potency, and more importantly defined identities and functions. More importantly, it was recently revealed that the cause for muscle wasting in DMD was more than commonly acknowledged myofiber fragility, but also included intrinsically impaired MuSC functions.37 Thus, strategies targeting MuSCs will provide more pertinent, promising, and essential therapeutic approaches for DMD disease. Despite the acknowledgment of such renewable source of cells in muscle tissues, direct application of MuSCs in genetically degenerative muscle diseases was rare. The application of adult MuSCs is hampered by the emerging roadblocks of rare native MuSC and severely reduced regenerative competency upon culture, which also accounts for the failure in targeting primary myoblasts gene editing for DMD.38 Salmon fibrins are well established for their nontoxicity and low immunogenicity.39, 40 Our recent work demonstrated that soft 3D fibrin gel culture system maintains MuSC properties during expansion in comparison to Matrigel. Meanwhile, because of the scarcity of MuSCs in adult muscle tissue and limited patient tissue, acquiring a sufficient starting number of pure MuSCs by fluorescence-activated cell sorting (FACS) for ex vivo expansion is impractical for clinical therapy. Selective expansion of regenerable MuSCs from a small number of bulk skeletal muscle cells is especially valuable for solving the problem in comparison to other established MuSC ex vivo culture systems.41, 42 Because of extended culture time with this ex vivo culture system, it is now feasible to correct disease-driving mutations directly in MuSCs using emerging genome editing. Combination of MuSC ex vivo expansion and CRISPR/Cas9-based genome editing greatly advances MuSC-based gene therapies for treatment of genetic muscle disorders.
In current study, about 80 dystrophin-positive myofibers were observed in the tibialis anterior (TA) muscle after transplantation, which takes up about 8.7% total myofibers in transplanted muscles. Hoffman et al.43 previously characterized dystrophin in muscle-biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy, and demonstrated that most patients with typically severe Duchenne’s dystrophy had dystrophin less than 3% of normal levels. Their test on two biopsy specimens from female patients with a phenotype intermediate between those of Duchenne’s and Becker’s dystrophy indicated that a content of dystrophin of less than 5% correlated with severe symptoms, whereas the other, with a content of over 20% of normal levels, was a mild disease patient who suffered only proximal weakness at the age of 5 years and was still ambulatory at age 12.43 Accordingly, 8.7% of dystrophin-positive myofibers in the TA muscle after transplantation in our study should have reached a clinically significant percentage of dystrophin-positive myofibers in a muscle. Nevertheless, it could be further improved to reach a percentage of dystrophin-positive myofibers to more than 20% of normal level.
To achieve the goal, the regenerative capacity of expanded MuSCs should be further enhanced. Adult stem cells inevitably undergo replicative stress during ex vivo expansion,44, 45, 46 which potentially causes stem cell senescence and limits their therapeutic efficacy.47, 48 Inhibition of p38MAPK by SB202190 could restore the regenerative capacity of MuSCs from aged mice.49 Strikingly, inhibition of p38 not only reversibly prevented differentiation, enabling expansion of human MuSCs in culture, propagated MuSCs exhibited even greater engrafting frequency than freshly isolated satellite cells.50 Therefore, it is tempting to test whether combinations of myogenic growth factors and small molecules can significantly enhance regenerative capability of fibrin-expanded MuSCs to further advance the intervention of effective stem cell-based muscular disease therapy.
In addition, efficiency of HDR-directed gene editing needs further improvement so as to significantly increase the percentage of effective cells. It has been reported that small molecules such as Scr7, RS-1, and L755507 enhanced precise genome editing by inhibiting non-homologous end joining or promoting HDR.51, 52, 53, 54, 55 Future work on optimizing dose and time, as well as combinations for applying these small molecules during CRISPR/Cas9-mediated gene correction in fibrin gel, is therefore highly demanded.
However, it has to be realized that DMD is a chronic and progressive disorder affecting all muscles in the body. Unfortunately, delivery of myogenic stem cells to the site of muscle lesions via systemic circulation like intravenous injection was previously shown to result in only a small portion entering the muscle microvasculature and migrating into dystrophic muscles.56 Instead, intramuscular injections have been repeatedly proved to ensure a good uptake of satellite cell-derived myoblasts in skeletal muscles.57 Obviously, our approach helps to preserve autonomy and life quality in DMD patients by slowing down or stopping muscle degeneration and increasing force in wasted muscles, but not for a respiratory muscle such as the diaphragm. Efficient systemic delivery of adult stem cells to the whole-body muscles will undoubtedly advance stem cell-based therapy for muscular disease.
Materials and Methods
Mice
C57BL/6 and C57BL/10ScSn-Dmdmdx/J mice were obtained from The Jackson Laboratory. All experiments were performed with 4- to 8-week-old mice and were in compliance with the institutional guidelines of University of Illinois at Chicago.
Preparation of Mouse Skeletal Muscle Cells
Hindlimb TA and gastrocnemius muscles or gastrocnemius muscle were dissected, cleaned by removal of non-muscle tissues including tendons, fat, and vessels, followed by cutting into small fragments and being subjected to collagenase (100 U/mL; catalog no. 171001015; Life Technologies) and dispase (2 U/mL; catalog no. 17105041; Life Technologies) digestions in DMEM at 37°C water bath for 1 hr. After enzymatic digestion, muscle mass was re-suspended in DMEM with 10% horse serum and further mechanically triturated through vigorous and consecutive passing through a 10-mL glass pipette and subsequently 9” cotton-plugged glass Pasteur pipette. The resultant cell suspension was passed through sequential 70- and 40-μm strainer (BD Biosciences) to generate bulk skeletal muscle single-cell suspensions.
Skeletal Muscle Cell Culture
Mouse skeletal muscle cells were cultured using DMEM/F10 (1:1) with 20% FBS, 2.5 ng/mL basic fibroblast growth factor (bFGF) (PROSPEC), and 1% penicillin/streptomycin. For coating 24-well plate with Matrigel, proper volume of stock Matrigel (BD Biosciences) was diluted into 1 mg/mL working solution by ice-cold DMEM in a pre-chilled 50-mL conical tube on ice. Diluted Matrigel solution (150 μL) was then added into 24-well plate through a chilled 1-mL glass pipette. Coated plates were left on ice for 7 min followed by incubation in tissue culture incubator for at least 1 hr after removal of working Matrigel solution in each well. For preparing 3D fibrin gel culture of muscle cells, salmon fibrinogen (Reagent Proteins) was diluted into gradient concentrations. For one well of 24-well plate, 125 μL of fibrinogen and 125 μL of cell solution were mixed together. A total volume of 250 μL of cell/fibrinogen mixture was transferred into each well of 24-well plate and mixed well with pre-added 5 μL of salmon thrombin (Reagent Proteins) (0.1 U/μL). The plate was then moved into 37°C cell culture incubator for no less than 30 min to allow sufficient gel-forming reaction. Afterward, 1 mL of growth medium was added and replenished every other day.
Histology and Immunohistochemistry of Muscle Cryosections
Fresh TA muscles were embedded in Tissue-Tek O.C.T. compound (Fisher), frozen in liquid nitrogen-cooled isopentane, and stored at −80°C until analysis. Frozen muscles were cross-sectioned (10 μm) using a Leica CM1850 cryostat. For immunohistochemistry study, air-dried muscle sections were fixed with 4% paraformaldehyde (PFA) and permeabilized in 0.2% Triton X-100. Tissue sections were then blocked in PBS with 5% goat serum, 2% BSA, and 1% Tween 20 for 1 hr, followed by incubation with primary antibodies overnight at 4°C. Rat anti-laminin (clone A5; Pierce; 1:100 dilution) and mouse anti-dystrophin (Sigma; 1:100 dilution) antibodies were used to denote myofiber boundaries and indicate restoration of Dystrophin protein, respectively.
IF Staining
Cells were cytospun onto the slides and fixed with 4% PFA. After permeabilization by 0.2% Triton X-100, cells were then blocked in PBS with 1% BSA for 1 hr at room temperature, followed by staining with primary antibody overnight at 4°C. Cells were washed three times with PBS and incubated with secondary antibody for 1 hr at room temperature. Nuclei were labeled with DAPI. Primary antibodies used were mouse anti-PAX7 (1:10; DSHB).
Transplantation
Experimental mdx mice were anesthetized with isoflurane (Merial). TA muscles were injected with 30 μL of cardiotoxin (Naja mossambica mossambica, 10 μM; Sigma) 1 day before transplantation. The next day, 2 × 105 fibrin-expanded skeletal muscle cells were resuspended in 10 μL of PBS and injected into pre-injured TA muscle. Transplanted muscles were harvested 4 weeks after transplant and analyzed by cryosectioning and microscopy.
gRNA and Donor Template DNA
Complementary oligonucleotides for gRNA were designed according to the published sequences14 and ordered from IDT (Integrated DNA Technologies). Two complementary oligonucleotides were annealed and cloned into Esp3I site of phU6-gRNA, an gRNA expression vector constructed in our laboratory. An 84-nt ssODN targeting the non-transcribed strand was used as HDR template and synthesized from IDT with phosphorothioate modification at both ends.
Generation of Recombinant Adenoviruses
Recombinant adenovirus expressing human CRISPR/Cas9 was generated by using the AdEasy technology as described.58, 59 Specifically, the hCas9 expression cassette was excised from Addgene’s pX330 vector,60 subcloned into the NotI/XbaI sites of adenoviral shuttle vector pAdTrack, and subsequently used to generate and amplify recombinant adenovirus in 293T cells.61 The resulting adenovirus was designated as AdV-Cas9-RFP, which also co-expresses Cas9 and RFP. PhU6-sgRNA fragment was excised from homemade sgRNA expression vector and, together with a double-stranded donor DNA fragment with 591- and 722-bp homology arms flanking the mutation site of Dmd, was subcloned into pacAd5 CMV eGFP shuttle vector. Linearized shuttle vector and pacAd5 9.2-100 backbone vector were then co-transfected in 293T cells to generate and amplify recombinant adenovirus (AdG-gRNA-Donor).
Genome Editing with ssODN Template and CRISPR/Cas9
For delivery of DNA into MuSCs cultured in soft 3D fibrin gel, gRNA, ssODN, and pmax-GFP were transfected with Lipofectamine 3000 (Life Technologies) as instructed and then followed by infection with CRISPR/Cas9-expressing adenovirus.
Genomic DNA Extraction and PCR
Cells were added with 50 μL of tail lysis buffer with proteinase K and 0.3% β-mercaptoethanol and digested at 55°C overnight. After 30 min at 99°C to inactivate the proteinase K, crude genomic DNA samples were kept at 4°C for short-term storage. PCR was carried out in a 10-μL volume containing 5 μL of 2xGoTaq Green Master Mix (Promega), and 0.3 μM of each forward and reverse primer and 1 μL of genomic DNA lysate. MF and MR2 primers were used for amplify Cas9-corrected genomic DNA product, and PCR conditions was set as 95°C for 2 min; 32 times (95°C for 20 s, 65°C for 20 s, 72°C for 10 s); 72°C for 5 min; followed by holding at 4°C. MF and MR primers were used to quantify the total input of genomic DNA, with PCR condition similar to the allele-specific PCR except for a lower annealing temperature of 55°C and a longer extension term of 30 s. PCR products were analyzed by 3% agarose gel electrophoresis and purified using Zymoclean Gel DNA Recovery Kit (Zymo Research). Purified PCR products were subcloned into pCR4-TOPO vector (Life Technologies) according to the manufacturer’s instructions. Individual clones were picked for direct sequencing or restriction enzyme digestion.
Analysis of Indels in PCR Products Using Restriction Fragment Length Polymorphism
To confirm allele-specific PCR products, individual clones containing pCR4-TOPO vector were used as templates for PCR using MF and MR2. PCR products were directly digested by TseI (New England BioLabs) for 3 hr at 65°C and analyzed by 3% agarose gel electrophoresis. Only PCR products amplified from HDR-mediated genomic editing DNA could be digested by TseI.
Data Analysis
All the experimental data are presented as mean ± SEM. Significant differences between means for single comparisons were determined by unpaired two-tailed Student’s t test.
Author Contributions
P.Z. and W.-S.W. conceived the project and wrote the manuscript. P.Z. and F.W. performed the most of the experiments. J.M. contributed to the writing of the manuscript. H.Z. and T.-C.H generated adenoviruses-expressing Cas9.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
We would like to thank members of the W.-S.W. Laboratory for their suggestions. We also thank the Research Resources Center Flow Cytometry Service at UIC for cell analysis and sorting. This work was supported by NIA/NIH grant R01AG040182.
Footnotes
Supplemental Information includes one figure and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.02.007.
Supplemental Information
References
- 1.Koenig M., Hoffman E.P., Bertelson C.J., Monaco A.P., Feener C., Kunkel L.M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 2.Kimura E., Li S., Gregorevic P., Fall B.M., Chamberlain J.S. Dystrophin delivery to muscles of mdx mice using lentiviral vectors leads to myogenic progenitor targeting and stable gene expression. Mol. Ther. 2010;18:206–213. doi: 10.1038/mt.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapsa R., Quigley A., Lynch G.S., Steeper K., Kornberg A.J., Gregorevic P., Austin L., Byrne E. In vivo and in vitro correction of the mdx dystrophin gene nonsense mutation by short-fragment homologous replacement. Hum. Gene Ther. 2001;12:629–642. doi: 10.1089/104303401300057324. [DOI] [PubMed] [Google Scholar]
- 4.Nik-Ahd F., Bertoni C. Ex vivo gene editing of the dystrophin gene in muscle stem cells mediated by peptide nucleic acid single stranded oligodeoxynucleotides induces stable expression of dystrophin in a mouse model for Duchenne muscular dystrophy. Stem Cells. 2014;32:1817–1830. doi: 10.1002/stem.1668. [DOI] [PubMed] [Google Scholar]
- 5.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 7.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyombe-Engembe J.P., Ouellet D.L., Barbeau X., Rousseau J., Chapdelaine P., Lagüe P., Tremblay J.P. Efficient restoration of the dystrophin gene reading frame and protein structure in DMD myoblasts using the CinDel method. Mol. Ther. Nucleic Acids. 2016;5:e283. doi: 10.1038/mtna.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L., Park K.H., Zhao L., Xu J., El Refaey M., Gao Y., Zhu H., Ma J., Han R. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol. Ther. 2016;24:564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabebordbar M., Zhu K., Cheng J.K., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H.L., Fujimoto N., Sasakawa N., Shirai S., Ohkame T., Sakuma T., Tanaka M., Amano N., Watanabe A., Sakurai H. Precise correction of the dystrophin gene in Duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4:143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long C., McAnally J.R., Shelton J.M., Mireault A.A., Bassel-Duby R., Olson E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwank G., Koo B.K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Messina G., Cossu G. The origin of embryonic and fetal myoblasts: a role of Pax3 and Pax7. Genes Dev. 2009;23:902–905. doi: 10.1101/gad.1797009. [DOI] [PubMed] [Google Scholar]
- 17.Cerletti M., Jurga S., Witczak C.A., Hirshman M.F., Shadrach J.L., Goodyear L.J., Wagers A.J. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montarras D., Morgan J., Collins C., Relaix F., Zaffran S., Cumano A., Partridge T., Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 19.Sacco A., Doyonnas R., Kraft P., Vitorovic S., Blau H.M. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherwood R.I., Christensen J.L., Conboy I.M., Conboy M.J., Rando T.A., Weissman I.L., Wagers A.J. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell. 2004;119:543–554. doi: 10.1016/j.cell.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka K.K., Hall J.K., Troy A.A., Cornelison D.D., Majka S.M., Olwin B.B. Syndecan-4-expressing muscle progenitor cells in the SP engraft as satellite cells during muscle regeneration. Cell Stem Cell. 2009;4:217–225. doi: 10.1016/j.stem.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu P., Zhou Y., Wu F., Hong Y., Wang X., Shekhawat G., Mosenson J., Wu W.S. Selective expansion of skeletal muscle stem cells from bulk muscle cells in soft three-dimensional fibrin gel. Stem Cells Transl Med. 2017 doi: 10.1002/sctm.16-0427. Published online February 28, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yablonka-Reuveni Z., Anderson J.E. Satellite cells from dystrophic (mdx) mice display accelerated differentiation in primary cultures and in isolated myofibers. Dev. Dyn. 2006;235:203–212. doi: 10.1002/dvdy.20602. [DOI] [PubMed] [Google Scholar]
- 24.Ryder-Cook A.S., Sicinski P., Thomas K., Davies K.E., Worton R.G., Barnard E.A., Darlison M.G., Barnard P.J. Localization of the mdx mutation within the mouse dystrophin gene. EMBO J. 1988;7:3017–3021. doi: 10.1002/j.1460-2075.1988.tb03165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Im W.B., Phelps S.F., Copen E.H., Adams E.G., Slightom J.L., Chamberlain J.S. Differential expression of dystrophin isoforms in strains of mdx mice with different mutations. Hum. Mol. Genet. 1996;5:1149–1153. doi: 10.1093/hmg/5.8.1149. [DOI] [PubMed] [Google Scholar]
- 26.Strouse B., Bialk P., Niamat R.A., Rivera-Torres N., Kmiec E.B. Combinatorial gene editing in mammalian cells using ssODNs and TALENs. Sci. Rep. 2014;4:3791. doi: 10.1038/srep03791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Guell M., Byrne S., Yang J.L., De Los Angeles A., Mali P., Aach J., Kim-Kiselak C., Briggs A.W., Rios X. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonner M., Strouse B., Applegate M., Livingston P., Kmiec E.B. DNA damage response pathway and replication fork stress during oligonucleotide directed gene editing. Mol. Ther. Nucleic Acids. 2012;1:e18. doi: 10.1038/mtna.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazuki Y., Hiratsuka M., Takiguchi M., Osaki M., Kajitani N., Hoshiya H., Hiramatsu K., Yoshino T., Kazuki K., Ishihara C. Complete genetic correction of ips cells from Duchenne muscular dystrophy. Mol. Ther. 2010;18:386–393. doi: 10.1038/mt.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filareto A., Parker S., Darabi R., Borges L., Iacovino M., Schaaf T., Mayerhofer T., Chamberlain J.S., Ervasti J.M., McIvor R.S. An ex vivo gene therapy approach to treat muscular dystrophy using inducible pluripotent stem cells. Nat. Commun. 2013;4:1549. doi: 10.1038/ncomms2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young C.S., Hicks M.R., Ermolova N.V., Nakano H., Jan M., Younesi S., Karumbayaram S., Kumagai-Cresse C., Wang D., Zack J.A. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell. 2016;18:533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 33.Ben-Porath I., Thomson M.W., Carey V.J., Ge R., Bell G.W., Regev A., Weinberg R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Awaya T., Kato T., Mizuno Y., Chang H., Niwa A., Umeda K., Nakahata T., Heike T. Selective development of myogenic mesenchymal cells from human embryonic and induced pluripotent stem cells. PLoS One. 2012;7:e51638. doi: 10.1371/journal.pone.0051638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shelton M., Metz J., Liu J., Carpenedo R.L., Demers S.P., Stanford W.L., Skerjanc I.S. Derivation and expansion of PAX7-positive muscle progenitors from human and mouse embryonic stem cells. Stem Cell Reports. 2014;3:516–529. doi: 10.1016/j.stemcr.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chal J., Oginuma M., Al Tanoury Z., Gobert B., Sumara O., Hick A., Bousson F., Zidouni Y., Mursch C., Moncuquet P. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 2015;33:962–969. doi: 10.1038/nbt.3297. [DOI] [PubMed] [Google Scholar]
- 37.Dumont N.A., Wang Y.X., von Maltzahn J., Pasut A., Bentzinger C.F., Brun C.E., Rudnicki M.A. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 2015;21:1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ousterout D.G., Kabadi A.M., Thakore P.I., Majoros W.H., Reddy T.E., Gersbach C.A. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uibo R., Laidmäe I., Sawyer E.S., Flanagan L.A., Georges P.C., Winer J.P., Janmey P.A. Soft materials to treat central nervous system injuries: evaluation of the suitability of non-mammalian fibrin gels. Biochim. Biophys. Acta. 2009;1793:924–930. doi: 10.1016/j.bbamcr.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ju Y.E., Janmey P.A., McCormick M.E., Sawyer E.S., Flanagan L.A. Enhanced neurite growth from mammalian neurons in three-dimensional salmon fibrin gels. Biomaterials. 2007;28:2097–2108. doi: 10.1016/j.biomaterials.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert P.M., Havenstrite K.L., Magnusson K.E., Sacco A., Leonardi N.A., Kraft P., Nguyen N.K., Thrun S., Lutolf M.P., Blau H.M. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu X., Xiao J., Wei Y., Li S., Liu Y., Yin J., Sun K., Sun H., Wang H., Zhang Z. Combination of inflammation-related cytokines promotes long-term muscle stem cell expansion. Cell Res. 2015;25:1082–1083. doi: 10.1038/cr.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffman E.P., Fischbeck K.H., Brown R.H., Johnson M., Medori R., Loike J.D., Harris J.B., Waterston R., Brooke M., Specht L. Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy. N. Engl. J. Med. 1988;318:1363–1368. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- 44.Piacibello W., Gammaitoni L., Pignochino Y. Proliferative senescence in hematopoietic stem cells during ex-vivo expansion. Folia Histochem. Cytobiol. 2005;43:197–202. [PubMed] [Google Scholar]
- 45.Wagner W., Horn P., Castoldi M., Diehlmann A., Bork S., Saffrich R., Benes V., Blake J., Pfister S., Eckstein V., Ho A.D. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One. 2008;3:e2213. doi: 10.1371/journal.pone.0002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner W., Bork S., Horn P., Krunic D., Walenda T., Diehlmann A., Benes V., Blake J., Huber F.X., Eckstein V. Aging and replicative senescence have related effects on human stem and progenitor cells. PLoS One. 2009;4:e5846. doi: 10.1371/journal.pone.0005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chambers S.M., Shaw C.A., Gatza C., Fisk C.J., Donehower L.A., Goodell M.A. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schallmoser K., Bartmann C., Rohde E., Bork S., Guelly C., Obenauf A.C., Reinisch A., Horn P., Ho A.D., Strunk D., Wagner W. Replicative senescence-associated gene expression changes in mesenchymal stromal cells are similar under different culture conditions. Haematologica. 2010;95:867–874. doi: 10.3324/haematol.2009.011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cosgrove B.D., Gilbert P.M., Porpiglia E., Mourkioti F., Lee S.P., Corbel S.Y., Llewellyn M.E., Delp S.L., Blau H.M. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med. 2014;20:255–264. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charville G.W., Cheung T.H., Yoo B., Santos P.J., Lee G.K., Shrager J.B., Rando T.A. Ex vivo expansion and in vivo self-renewal of human muscle stem cells. Stem Cell Reports. 2015;5:621–632. doi: 10.1016/j.stemcr.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 53.Pinder J., Salsman J., Dellaire G. Nuclear domain “knock-in” screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015;43:9379–9392. doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song J., Yang D., Xu J., Zhu T., Chen Y.E., Zhang J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016;7:10548. doi: 10.1038/ncomms10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu C., Liu Y., Ma T., Liu K., Xu S., Zhang Y., Liu H., La Russa M., Xie M., Ding S., Qi L.S. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J., Sanberg P.R., Li Y., Wang L., Lu M., Willing A.E., Sanchez-Ramos J., Chopp M. Intravenous administration of human umbilical cord blood reduces behavioral deficits after stroke in rats. Stroke. 2001;32:2682–2688. doi: 10.1161/hs1101.098367. [DOI] [PubMed] [Google Scholar]
- 57.Skuk D., Roy B., Goulet M., Chapdelaine P., Bouchard J.P., Roy R., Dugré F.J., Lachance J.G., Deschênes L., Hélène S. Dystrophin expression in myofibers of Duchenne muscular dystrophy patients following intramuscular injections of normal myogenic cells. Mol. Ther. 2004;9:475–482. doi: 10.1016/j.ymthe.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 58.He T.C., Zhou S., da Costa L.T., Yu J., Kinzler K.W., Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo J., Deng Z.L., Luo X., Tang N., Song W.X., Chen J., Sharff K.A., Luu H.H., Haydon R.C., Kinzler K.W. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 60.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu N., Zhang H., Deng F., Li R., Zhang W., Chen X., Wen S., Wang N., Zhang J., Yin L. Overexpression of Ad5 precursor terminal protein accelerates recombinant adenovirus packaging and amplification in HEK-293 packaging cells. Gene Ther. 2014;21:629–637. doi: 10.1038/gt.2014.40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.