Abstract
The CRISPR-Cas (clustered regularly interspaced short palindromic repeats-associated proteins) technology enables rapid and precise genome editing at any desired genomic position in almost all cells and organisms. In this study, we analyzed the impact of different repair templates on the frequency of homology-directed repair (HDR) and non-homologous end joining (NHEJ). We used a stable HEK293 cell line expressing the traffic light reporter (TLR-3) system to quantify HDR and NHEJ events following transfection with Cas9, eight different guide RNAs, and a 1,000 bp donor template generated either as circular plasmid, as linearized plasmid with long 3′ or 5′ backbone overhang, or as PCR product. The sequence to be corrected was either centrally located (RS55), with a shorter 5′ homologous region (RS37), or with a shorter 3′ homologous region (RS73). Guide RNAs targeting the transcriptionally active strand (T5, T7) showed significantly higher NHEJ frequencies compared with guide RNAs targeting the transcriptionally inactive strand. HDR activity was highest when using the linearized plasmid with the short 5′ backbone overhang and the RS37 design. The results demonstrate the importance of the design of the guide RNA and template DNA on the frequency of DNA repair events and, ultimately, on the outcome of treatment approaches using HDR.
Keywords: HDR, NHEJ, CRISPR-Cas9, DNA donor template
Introduction
The CRISPR-Cas (clustered regularly interspaced short palindromic repeats-associated proteins) systems are prokaryotic immune systems, which provide adaptive immunity against invading phages and foreign nucleic acids.1, 2 In the past 3 years, this system was used in a wide range of cells and organisms, and has shown a high efficiency for creating insertions and deletions via error-prone non-homologous end joining (NHEJ) or high-fidelity homology-directed repair (HDR) using a template DNA,3, 4, 5, 6, 7, 8 although the efficiency of the latter is substantially lower.9, 10 HDR-based precise genome engineering has become a powerful tool for the development of gene therapeutic approaches, but several limitations are still present including the quest for optimal target sites near the mutant DNA sequence and the optimal frequency at which HDR takes place.
Maximum activity of the Cas9 protein to induce double-strand breaks (DSBs) at the target site is a prerequisite for efficient genome editing. However, this activity is dependent on the sequences surrounding the target site and other factors, necessitating to search for the optimal guide RNA sequence prior to any steps optimizing gene repair activities.11 Different approaches have been developed to enhance HDR, including the manipulation of the cell cycle and the regulation of expression of key repair pathway proteins.12 However, these invasive manipulations may be undesirable for therapeutic applications because they can alter the cellular response to DNA damage at other non-target sites in the genome and lead to tumor formation. In contrast, designing optimal DNA donor templates can increase HDR frequencies and at the same time leave cell cycle regulation untouched. In mammalian cells, a plasmid donor with at least 1–2 kb of total homology is usually used for creating large sequence changes in the presence of target cleavage.13, 14 For small sequence changes, single-stranded DNA (ssDNA) sequences are usually more efficient than plasmid donors. Recently, enhanced HDR rates have been reported by using optimized asymmetric ssDNA donor templates for conversion of a blue fluorescent protein (BFP) reporter gene into a GFP reporter gene via mutation of 3 nt within the BFP reading frame.15 It was shown that optimizing a donor template at the 5′ and 3′ homology regions flanking the DSB site could boost the frequency of HDR in the absence of chemical and genetic intervention.
Here, we have induced the site-specific DSBs by employing the CRISPR-Cas9 system and used the modified traffic light reporter (TLR-3) system to quantitatively determine the DNA repair outcomes in the standard cell line HEK293.16 To understand the influence of DNA donor templates on the results of repair pathway choices, double-stranded DNA plasmid, linearized plasmids, as well as PCR products were used, and the length of the 5′ and 3′ homology arms were altered. Furthermore, plasmid donors were linearized at different sites in the backbone for the generation of differing 5′ and 3′ overhangs to investigate whether this also influences HDR activity.
Results
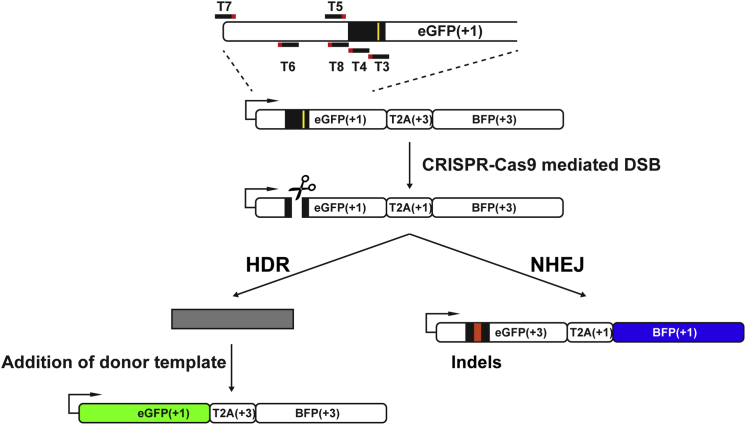
The TLR system, which has been used frequently to monitor DNA repair activities, allows rapid observation of repair pathway choices in cells based on fluorescence microscopy and fluorescence-activated cell sorting (FACS).16 Our modified TLR system (TLR3) comprises a bicistronic expression system of a non-functional green fluorescent protein (GFP), followed by a self-cleaving T2A peptide and a second BFP in a reading frame shifted by 2 bp (Figure 1). The GFP cDNA sequence contains an insertion comprising an ISce-I site and a stop codon, which disrupts the normal gene function. Upon repair of the DSB induced upstream of the stop codon, different fluorescent signals will appear depending on whether NHEJ or, in the presence of a DNA template, HDR takes place. Mutagenic NHEJ causes insertion and/or deletions, thus shifting the downstream BFP sequence in frame in about one-third of all cases, resulting in a blue fluorescent signal, whereas HDR restores the GFP with the help of the DNA donor template, resulting in a green fluorescent signal (Figure 1). Because the NHEJ rates cannot be easily corrected for the 1/3 ratio of events, the focus of the data analysis lies on the comparison of different guide RNAs and the different template compositions, rather than on the absolute values. This approach has also been used by others in the field.9
Figure 1.
Design of CRISPR-Cas9 Guide RNAs Targeting TLR3 Sequence
Targets (T3–T8) around the stop codon and I-SceI site (yellow line) have been designed for the specific cleavage within the GFP sequence in the TLR3 system. Depending on the addition of the donor templates, DSBs can be repaired through either NHEJ (BFP) or HDR (GFP). T, target.
Activity of Guide RNAs to Induce NHEJ Differs When Targeting the Transcriptionally Active or Inactive Strand of the TLR3 System
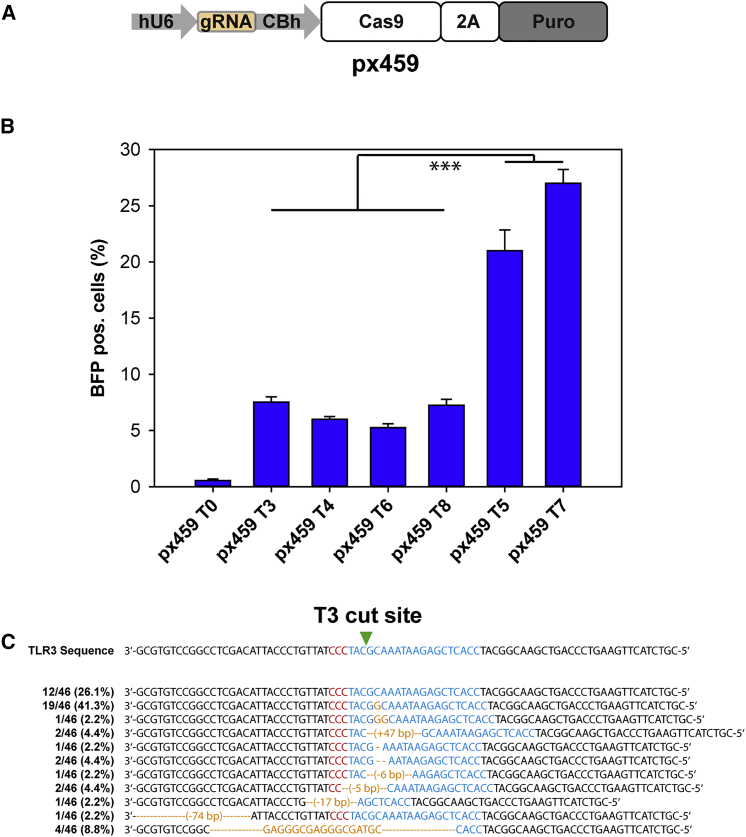
Six different target sites (Table 1; Figure 1) were identified upstream or downstream of the stop codon within the GFP sequence or within the ISce-I site containing the initiating 5′-G and the 3′-PAM (NGG), and the respective guide RNA sequences were cloned into the px459 expression vector that produces the guide RNA and the Cas9 protein simultaneously (Figure 2A). A stable HEK293 cell line was created using the linearized pcDNA3.1(−)-TLR3 expression vector, in which the TLR3 system is under the control of a CMV promoter. To examine the efficiency of these six targets, we transfected the respective px459 vectors into the HEK293-TLR3 stable cell line and counted fluorescing cells by FACS (Figure 2B). By measuring the BFP-positive cells, we demonstrate NHEJ rates of up to 27% using T7 and T5, representing approximately four times the rates observed for the targets T3, T4, T6, and T8 (about 5%–7%; ANOVA, p < 0.005). Interestingly, the latter four target sites with lower NHEJ activity are all located on the transcriptionally inactive minus strand of the DNA, whereas the two sites with significantly higher activity are located on the transcriptionally active plus strand of the DNA, indicating that DSB induction and/or NHEJ rates are dependent on the location of the target site with regard to the transcriptionally active DNA strand.
Table 1.
Guide RNA Sequences Targeted TLR3 System
| Name | Sequences (GN19NGG) |
|---|---|
| T3 | 5′-GGTGAGCTCTTATTTGCGTAGGG-3′ |
| T4 | 5′-GGGATAACAGGGTAATGTCGAGG-3′ |
| T5 | 5′-GGCCACAAGTTCAGCGTGTCCGG-3′ |
| T6 | 5′-GCCGTCCAGCTCGACCAGGATGG-3′ |
| T7 | 5′-GAGCGCCACCATGGTGAGCAAGG-3′ |
| T8 | 5′-GGCCGGACACGCTGAACTTGTGG-3′ |
Figure 2.
Activity of the CRISPR-Cas9 Targets in the TLR3 System
(A) Guide RNAs are cloned into the px459 vector under the hU6 promoter. (B) px459 Cas9 targets were expressed in the HEK-TLR3 cell line. Seventy-two hours after transfection, flow cytometric analysis displayed about 27% of BFP+ (NHEJ repair) cells by px-459-T7. The graphs represent triplicate data from three independent experiments. Statistical analysis: ANOVA, ***p < 0.001. (C) Analysis of sequence modifications after T3 cleavage. Sequence changes were analyzed using Sanger sequencing via TOPO Cloning. Sequence in red: PAM site; sequence in blue: guide RNA target sequence; sequence in yellow: changes to the wild-type sequence. T0, px459 without guide RNA; T3, target 3.
To analyze the sequence changes in the TLR3 system after NHEJ-mediated DSB repair, we isolated genomic DNA from cells transfected with the guide RNA T3, and the sequence around the target site was sequenced and compared (Figure 2C). Twelve of 46 analyzed clones (26.1%) were unchanged. The majority of changes (41.3%) observed were insertions of 1 nt (G) right after the cut site, whereas insertions of 2 nt or deletions of 1 or 2 nt were less often the case. Sporadically, larger deletions or insertions were observed, showing the importance of analyzing sequence changes in genome-editing approaches based on NHEJ.
HDR Frequency Depends on the Characteristics of the DNA Template
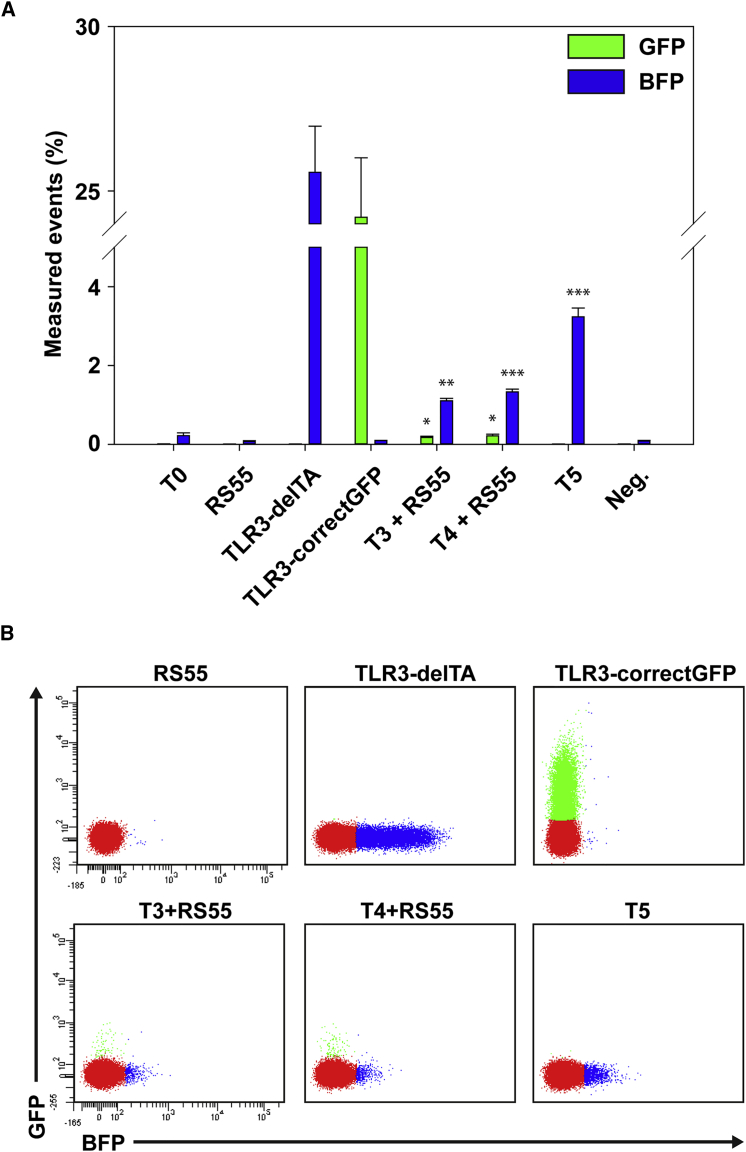
In order to test HDR frequency in our TLR3 system, we co-transfected the stable cell line with the px459 plasmid and the RS55 template plasmid, the latter containing approximately 1,000 bp of the natural GFP sequence without start region and the sequence to be repaired (i.e., the original GFP sequence that had been replaced by the I-SceI site together with the stop codon) centrally (Figure 3A). The application of guide RNAs T3 and T4 resulted in significant HDR signal, whereas the application of RNA guide T5 did not show any GFP-positive signal, even though the NHEJ activity was higher, indicating the presence of DSBs at the target site (Figure 4A). This is due to the fact that the guide RNA targets a site within the natural GFP sequence (which is also present in the template DNA), representing a reliable control that GFP signaling in our experiments is indeed originating from the HDR activity at the target site.
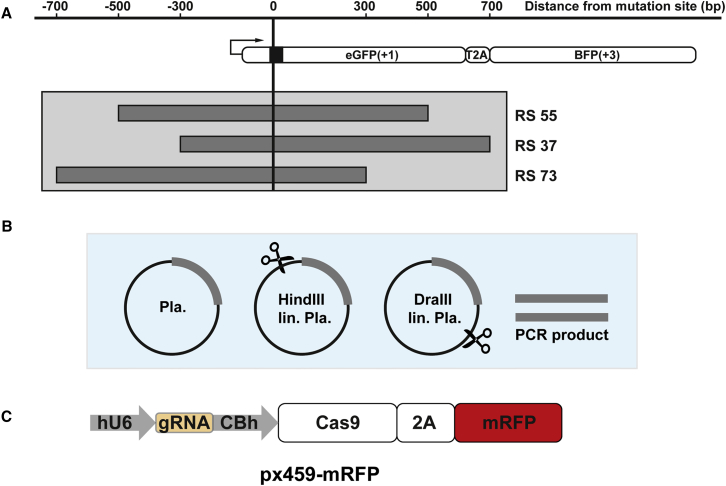
Figure 3.
Generation of Different Donor Templates and px459-mRFP Target
(A) One kilobase of double-stranded DNA (dsDNA) donor templates was generated with varying homology sequence overlaps on the 5′ and 3′ side of the mutation site. (B) Donor templates were generated as plasmid, linearized plasmid with 5′ or 3′ backbone overhang, or PCR product. (C) mRFP gene sequence was cloned into the px459 expression vector instead of the puromycin resistance gene.
Figure 4.
FACS Analysis of HDR and NHEJ Events Using the Circular Plasmid RS55
(A) The TLR3 plasmid was cotransfected with nucleases and donor template for the quantification of both HDR and NHEJ events. Artificial NHEJ (TLR3-delTA) and HDR (TLR3-correctGFP) control for the TLR systems were generated through deletion of 2 bp nucleotides (thymin and adenine) and correction of mutant sequence, respectively. (B) FACS data of the respective samples and controls. Statistical analysis: paired t test, NHEJ values compared with T0 plasmid transfection, HDR values compared with RS55 plasmid alone. *p < 0.001; **p < 0.0001; ***p < 0.00005.
In order to study the effect of different kinds of DNA donor templates, we generated, in addition to the RS55 construct, DNA templates of 1,000 bp with a shorter 5′ homologous region (RS37) or a shorter 3′ homologous region (RS73) (Figure 3A). The different lengths of the homologous regions are 300 and 700 bp and vice versa. Four different kinds of DNA were employed in the experiments: uncut plasmid, linearized plasmid with 5′ or 3′ backbone overhang, or PCR product (Figure 3B). Because we could use only T3 or T4 in subsequent experiments and HDR levels for these two sites were rather low, we exchanged within the px459 plasmid the puromycin gene by a red fluorescent protein (mRFP) gene (px459-mRFP) and sorted prior to the DNA repair activity-based signaling (blue or green signals) for red fluorescent cells, thus enriching the population of successfully transfected cells with our plasmids of interest (Figure 3C).
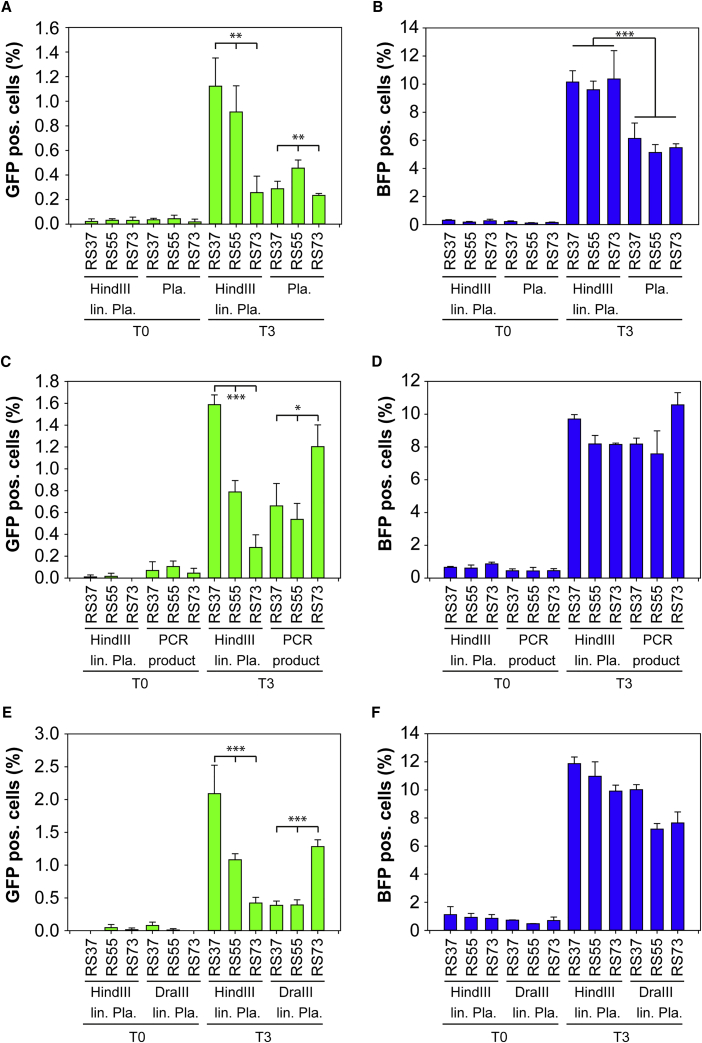
Upon co-transfection of the px459-mRFP vector and the different template versions, FACS-based quantification of NHEJ and HDR was performed and different template versions compared (Figure 5). Most importantly, the use of different donor templates had an effect on the repair outcomes for both HDR and NHEJ, with rates ranging from 0.2% to 2.1% and 5% to 12%, respectively (Figure 5).
Figure 5.
Optimizing HDR Events Using Different Donor Templates
Cas9-mRFP targets and donor templates were coexpressed in the HEK-TLR3 cell line. (A–F) Seventy-two hours after transfection, flow cytometric analysis of mRFP+ gated cells displayed GFP+ (A, C, and E, HDR repair) and BFP+ (B, D, and F, NHEJ repair) cells. The graphs represent triplicate data from three independent experiments. lin., linearized; Pla., plasmid; RS, repair substrate; T0, px459-mRFP without guide RNA. Statistical analysis: ANOVA, *p < 0.05; **p < 0.005; ***p < 0.001.
The linearized plasmid with short 5′ backbone overhang (HindIII lin. Pla.) was compared with the other three template versions in three different experiments, proving the reproducibility of the results (Figures 5A, 5C, and 5E). Although the absolute values varied depending on the transfection efficiency in each experiment, the overall outcome of the RS37-based template resulted in highest levels of HDR, followed by RS55, and RS73, revealing the lowest HDR activity, was constantly reproducible and significant (ANOVA, p < 0.001). In contrast, uncut plasmid DNA template RS55 showed the highest HDR activity (Figure 5A), and for PCR product and the linearized plasmid with the long 5′ backbone overhang (DraIII lin. Pla.) the template version RS73 revealed highest HDR activity (Figures 5C and 5E). Notably, linearized plasmid with short 5′ backbone overhang and short 5′ homology arm resulted in the highest HDR rates observed compared with any other templates (Figures 5A, 5C, and 5E).
When looking at the NHEJ activity measured in each experiment, different template overhangs (RS37, RS55, RS73) did not seem to alter the NHEJ activity when added as circular plasmid, linearized plasmid with short 5′ backbone overhang, or as PCR product (Figures 5B and 5D), whereas small differences could be observed when adding the linearized plasmid with the long 5′ backbone overhang (Figure 5F). Here, the presence of RS55 and RS73 resulted in lower NHEJ rates. More importantly, the presence of circular plasmid template generally reduced significantly the NHEJ activity when compared with the linearized plasmid template (Figure 5B), indicating that there are interactions between the template and the NHEJ repair machinery.
Discussion
We report herein that the choice of the guide RNA target site has an impact on the activity of DSB repair as measured by NHEJ activity, and that target sites on the transcriptionally active strand result in higher DSB repair activity compared with sites on the inactive DNA strand. More importantly, we show that the choice of the template DNA has tremendous effects on the outcome of HDR rates, revealing important information for the subsequent preparation of genome-editing approaches to treat disease-causing mutations. It is acknowledged that the observations are currently restricted to one cell line, HEK293, and differences among cell lines and tissues exist with regard to DNA repair activities. However, we consider the results of this proof-of-concept work still very useful for the scientific community, because the HEK293 cell line is widely used for the development of genome-editing approaches,17, 18 as well as the study of DNA repair mechanisms in general.11
The position of the RNA guide target sequence being on the transcriptionally active or inactive strand has already been analyzed in a HDR setting using single-stranded oligonucleotides (ssOGNs) of less than 100 bp, and the authors observed increased DSB repair activity when the site was positioned on the active strand.11 Similar to the results of nicking, we observed that NHEJ rates on the transcriptionally active strand are four times more effective than the targets on the inactive strand. Whether this observation is due to a mechanistically relevant aspect related to the accessibility of the nuclease to the transcriptionally active strand remains elusive, and further work needs to be done in order to shed more light on this aspect.
The presence of a varying number of sequence changes at the target site has been shown in many studies and further corroborates the importance of analyzing these changes, most importantly in a therapeutic setting. Indel formation is always critical in the development of therapeutic approaches, because it can well be the cause of unwanted effects in therapeutic settings,18 or the desired change in order to destroy pathology DNA sequence.19 However, the absence of controlling the DNA changes renders NHEJ as a therapeutic approach problematic. Consequently, in our setting, the quantification of NHEJ events was just to have a measurement tool to study guide RNA activity.
The efficiency of HDR is determined by many factors, of which the donor template is considered to be among the most important ones.15 Linearized or double-stranded DNA plasmid sequences, as well as ssDNA oligonucleotides, are often used as a donor template for large or small sequence changes in the presence of target cleavage.15, 20 The length of the homology arm also plays an important role in increasing HDR rate because the efficiency of recombination increases as the length of homology arms increases.21 Here, we chose 1 kb total homology sequence for the recombination either without CMV promoter sequence or without intact GFP sequence. We have generated the templates as double-stranded DNA, PCR product, and linearized plasmid at two different backbone sites.
Our observation that linearized plasmid is more effective than plasmid donor suggests that circular DNAs can be broken randomly at undesired sites in the homology arm. In contrast with results from our study in HEK cells, it was observed that plasmid donor is more effective than linearized donor in Drosophila embryos probably because of the degradation of the linear DNA by exonucleases or conversion of long concatemers.20 The fate of the plasmid donor and linearized donor could also be determined by delivery system because the duration of the donor in the cytoplasm increases the possibility of degradation rates.
The issue of linearized plasmid versus PCR product is also an interesting one. Upon generation of the plasmid DNA, post-translational modification of the bacteria could be a limiting factor for HDR. Another possible reason for the higher HDR efficiency of the linearized plasmid is the protection of the backbone sequence from degradation of the template sequence, which is not the case for the PCR product.
Furthermore, we have linearized the plasmid at the 5′ and 3′ end for the generation of the backbone overhang at different sites. Surprisingly, the cut side of the plasmid has generated different HDR and NHEJ rates. It is possible that the overhangs resulting from HindIII and DraIII have different affinity to the DNA ends generated from the Cas9 protein, so that the random integration of the templates into the TLR3 cut site could be different, which in turn influences the NHEJ efficiency. Our observation that linearized RS37 showed the best HDR rate suggests that 3′ end degradation takes places at the break, whereas the overhang of the backbone protects the degradation of the 3′ end. Our data show the importance of choosing the right DNA template molecule as one important aspect in the development of future gene therapeutic approaches.
Materials and Methods
Generation of CRISPR-Cas9 Targets and Cas9-mRFP Constructs
The Cas9 expression vector, pSpCas9(BB)-2A-Puro (PX459) V2.0, was purchased from Addgene (plasmid 62988). The Cas9 target was cloned into the px459 expression vector using In-Fusion Cloning Kit (Clontech), according to the manufacturer’s instructions. The puromycin resistance gene was exchanged with mRFP gene using In-Fusion Cloning Kit. The plasmid containing mRFR gene was purchased from Addgene (plasmid 13032). Plasmids were prepared by using the QIAGEN Maxi plasmid kit (QIAGEN).
Analysis of Sequence Modification after T3 Cleavage
T3-mRFP was transfected into the HEK-TLR3 stable cell line, and the mRFP-positive cells were sorted by FACS. The genomic DNA was isolated from the cells, and an 821 bp DNA sequence was amplified using PCR, in which the T3 recognition site is located centrally. The amplicons were subcloned into TOPO TA (Life Technologies) plasmid vector, and individual colonies were sequenced by Sanger sequencing using PCR forward primer. The sequences were analyzed using Vector NTI (Life Technologies).
Design and Generation of Donor Templates
For the generation of the donor templates, the mutant GFP gene of the TLR3 system was corrected with wide-type GFP gene sequence by using In-Fusion Cloning Kit (Clontech), according to the manufacturer’s instructions. PCR donor templates were amplified using Phusion polymerase (NEB), and the PCR products were further cloned into the TOPO TA Cloning Kit (Life Technologies) for the generation of the plasmid donor templates. Linearized plasmid donor templates were generated by digesting the plasmids using HindIII and DraIII endonuclease (NEB). The PCR products and linearized plasmid DNA were purified using NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel). Plasmids were prepared by using the QIAGEN Maxi plasmid kit (QIAGEN).
Cell Culture and Transfection
A stable cell line of HEK293 expressing TLR3 was generated following transfection of a pcDNA3.1(−)-TLR3 vector (V795-20; Life Technologies) and selected using neomycin (Geneticin; Life Technologies). The cell line was maintained in DMEM supplemented with 10% FBS, 2 mM l-glutamine, and 50 U/mL penicillin-streptomycin (Life Technologies), and cultured at 37°C with 5% CO2 incubation. Transfection of CRISPR-Cas constructs and donor templates was performed using Lipofectamine LTX (Life Technologies), with 500 ng of plasmid and 500 ng of donor templates per well of a 24-well plate, according to the manufacturer’s instructions. Fluorescence images were visualized 3 days post-transfection with a Keyence microscope (Keyence) to control the transfection efficacy.
FACS Analysis
Cells were collected 3 days after transfection by trypsinization (Accutase; PAN-Biotech), then washed and resuspended with PBS. FACS analysis of mRFP/BFP/GFP-positive events was performed using a FACSCanto II flow cytometer (BD Biosciences).
Author Contributions
F.S. performed the experiments and wrote the paper. K.S. designed the study and edited the paper.
Acknowledgments
We thank the Institute for Clinical Immunology and Transfusion Medicine Giessen for support with the FACS analysis. We thank Claudia Lopez, Tobias Wimmer, Jessica Grebe, Annabella Janise, and Bettina Gill for technical assistance and support. We also thank Mert Yanik and Brigitte Müller for critical reading of the manuscript.
References
- 1.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R., van Pijkeren J.P. Exploiting CRISPR-Cas immune systems for genome editing in bacteria. Curr. Opin. Biotechnol. 2016;37:61–68. doi: 10.1016/j.copbio.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., Xiong J.W., Xi J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiCarlo J.E., Chavez A., Dietz S.L., Esvelt K.M., Church G.M. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 2015;33:1250–1255. doi: 10.1038/nbt.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedland A.E., Tzur Y.B., Esvelt K.M., Colaiácovo M.P., Church G.M., Calarco J.A. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 8.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 10.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis L., Maizels N. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc. Natl. Acad. Sci. USA. 2014;111:E924–E932. doi: 10.1073/pnas.1400236111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srivastava M., Raghavan S.C. DNA double-strand break repair inhibitors as cancer therapeutics. Chem. Biol. 2015;22:17–29. doi: 10.1016/j.chembiol.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Dickinson D.J., Ward J.D., Reiner D.J., Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 16.Certo M.T., Ryu B.Y., Annis J.E., Garibov M., Jarjour J., Rawlings D.J., Scharenberg A.M. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods. 2011;8:671–676. doi: 10.1038/nmeth.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J., Hatanaka F., Yamamoto M., Araoka T., Li Z. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G., Zhao N., Berkhout B., Das A.T. CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mol. Ther. 2016;24:522–526. doi: 10.1038/mt.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeder M.L., Mepani R., Gloskowski S.W., Skor M.N., Collins M.A., Gotta G.M., Marco E., Barrera L.A., Jayaram H., Bumcrot D. Therapeutic correction of an LCA-causing splice defect in the CEP290 gene by CRISPR/Cas-mediated genome editing. Mol. Ther. 2016;23(Suppl 1):S273–S274. [Google Scholar]
- 20.Carroll D., Beumer K.J. Genome engineering with TALENs and ZFNs: repair pathways and donor design. Methods. 2014;69:137–141. doi: 10.1016/j.ymeth.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li K., Wang G., Andersen T., Zhou P., Pu W.T. Optimization of genome engineering approaches with the CRISPR/Cas9 system. PLoS ONE. 2014;9:e105779. doi: 10.1371/journal.pone.0105779. [DOI] [PMC free article] [PubMed] [Google Scholar]