Abstract
Doxorubicin is a chemotherapeutic drug typically administered systemically which frequently leads to cardiac and hepatic toxicities. Local delivery to a tumor has a chance to mitigate some of these toxicities and can further be mitigated by including a means of tumor-specific drug release. Our laboratory has explored the use of molecular interactions to control the rate of drug release beyond that capable of diffusion alone. To this system, we added an additional affinity group (adamantane) to doxorubicin through a pH-sensitive hydrazone bond. The result was a modified doxorubicin which had an even higher affinity to our drug delivery polymer, and virtually no release in normal conditions, but showed accelerated release of drug in tumor-like low pH. Further, we show that adamantane-modified doxorubicin (adamantane-doxorubicin) and cleaved adamantane-doxorubicin showed equivalent capacity to kill human U-87 glioblastoma cells in vitro as unmodified doxorubicin. Taken together, these data demonstrate our ability to load high levels of modified chemotherapeutic drugs into our affinity-based delivery platform and deliver these drugs almost exclusively in the acidic microenvironments, such as those surrounding the tumor tissue via pH-cleavable bond while minimizing drug delivery in neutral pH tissue, with the ultimate goal of reducing systemic through better local delivery.
Impact statement
Doxorubicin (DOX) is especially cytotoxic to the heart, liver, kidneys, and healthy tissues surrounding the tumor microenvironment. This systemic toxicity can be partially addressed by local, tumor-specific drug delivery systems. While pH-sensitive DOX delivery systems have been developed by several other groups, many lack a prolonged and consistent release profile required to successfully treat heterogeneous tumors. Our system of a chemically modified form of DOX combined with an affinity-based cyclodextrin delivery system is capable of delivering DOX for 87 days while maintaining its the drug cytotoxicity. This finding is particularly relevant to improving cancer treatments because it enables regulated local delivery of DOX specifically to tumor tissue and allows the drug to be continuously delivered over a therapeutically relevant amount of time.
Keywords: Doxorubicin, pH-sensitive, adamantane, cyclodextrin, hydrazone
Introduction
Doxorubicin (DOX) is a hydrophobic chemotherapeutic drug that is frequently administered as a treatment for more than 10 types of cancerous tumors, including breast, lung, and pancreatic cancer.1–3 In order to effectively treat many aggressive cases of cancer, it is often necessary to treat the patient with a sustained dosage of chemotherapeutics over an extended period of time. However, DOX is particularly cytotoxic to the heart but also has shown to have toxicity in brain, liver, and kidneys as well as being harmful to healthy tissues surrounding the tumor environment when available in high doses.1–4 We have previously shown DOX can undergo high loading into cyclodextrin-based polymers (pCD) and release at levels capable of clearing subcutaneous glioblastoma tumors.5 This work demonstrated the capacity for affinity-based release to deliver DOX at therapeutically relevant rates, and that a slow, steady rate was able to clear tumors where more rapid release rate such as from non-affinity, diffusion only controls was not capable of clearing the same tumors. While this work demonstrated the high potential of providing drug at appropriate rates, it did not address the toxicity issues of DOX to non-cancerous tissues. We therefore set out to develop a chemically modified form of DOX that can undergo the same high loading into our affinity-based polymers but which will be released only in response to the low pH surrounding tumor tissue. We hypothesize that our system has the potential to reduce systemic toxicity by accomplishing a therapeutically relevant local dose, without the need for a high systemic dose, and by reducing non-specific delivery in the absence of a tumor’s low pH environment.
The use of pH-sensitive drug delivery systems has been explored by several groups in order to enable a site-specific delivery of a variety of therapeutic drugs.6–11 Specifically, several groups have worked with pH-sensitive DOX conjugates3,8 or pH-sensitive polymers12–16 in order to overcome DOX’s toxicity and to create highly selective delivery systems for chemotherapeutics. However, the majority of these delivery systems provide a drug which is instantly available upon chemical cleavage and therefore lack the prolonged and consistent release profile that we and others have shown as necessary for successful treatment of cancerous tissue.4 There are multiple theories behind the need for sustained delivery, but they center around drug sensitivities during different stages of the cell cycle17,18 and in vivo heterogeneity of tumors due to cancer stem cells.19,20
Luo et al.21 previously synthesized and characterized the AD-DOX molecule used in this work, however they too were evaluating this for instantaneous release, rather than slow, sustained release from a polymeric implant. Briefly, in the synthesis of AD-DOX, the C-13 carbonyl group of DOX is bound to the hydrazide group of adamantane-1-carbohydrazide to form a hydrazone bond, which is cleavable in slightly acidic environments. Tumor tissue has pH range of 5.8–7.2, which is slightly lower than standard physiological pH 7.4.6 The difference in acidity is attributed to elevated cellular respiration activity within the growing tumor.7 The slightly acidic environment of the tumor is capable of cleaving the hydrazone bond of AD-DOX, enabling DOX to be selectively released into the tumor tissue.21 Similar conjugates have been synthesized using hydrazides with DOX and pullulan,2,22 amphiphilic multi-arm block copolymers,23 and single walled carbon nanotubes24 as well as others, to create pH-sensitive hydrazone bonds for DOX release.
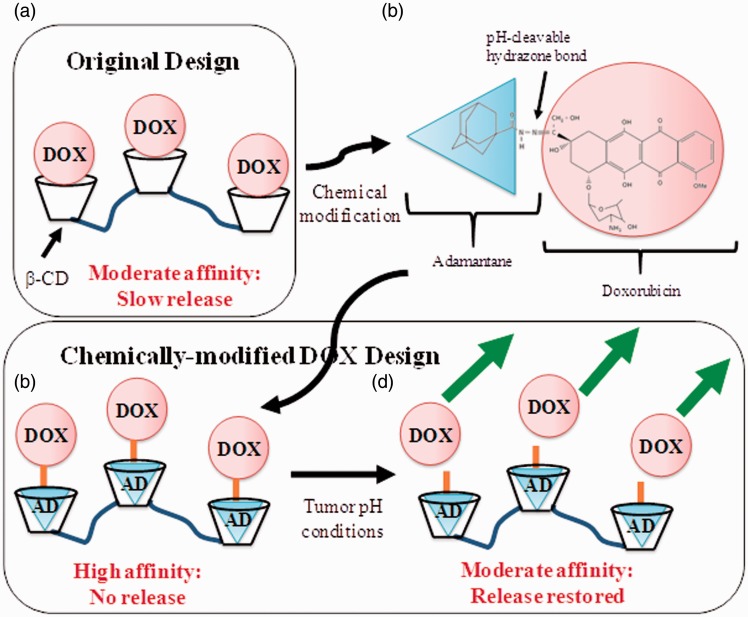
While the hydrazone bond provides pH sensitivity required for release only in or near the tumor tissue, the AD-group in our system provides an affinity moiety that is known to bind β-cyclodextrin (β-CD) with a high affinity association equilibrium constant (KA) of 5.2 × 104 M−1.25 Our previous work showed that DOX alone has a modest affinity to β-CD polymers, resulting in steady release,5 and that multiplexing interactions between drug and CD polymers would geometrically decrease the delivery rate until almost no release is detected.26 The cyclic structure of CD provides a “pocket” into which drugs such as DOX can bind. Soluble CD is routinely used to increase the solubility of hydrophobic drugs.27–29 We turn that observation on its head by making insoluble forms of CD (CD polymers or pCD) to entrap rather than solubilize drug. With these CD polymers, we then show high loading levels and slow, sustained release of many different drugs.27–31 While unmodified DOX is capable of binding pCD, we hypothesized that by adding additional affinity groups we can substantially reduce the rate of release to almost none, and through the use of a pH-sensitive linkage enabling tumor-dependent cleavage of the extra affinity moiety, bringing the release rate back up to therapeutically relevant rates. In this paper, AD-DOX was synthesized according to previously published work.21 Nuclear magnetic resonance (NMR) and Fourier transform infrared spectroscopy (FTIR) confirmed the successful synthesis of AD-DOX. A cytotoxicity study involving AD-DOX and unmodified DOX as a control using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cell proliferation assay (MTT) confirmed that AD-DOX had an approximately equivalent capacity for cancer cell killing as unmodified DOX. Lastly, an in vitro drug release study using pCD disks releasing AD-DOX into neutral phosphate-buffered saline (PBS), pH 7.4, and slightly acidic acetate buffer, pH 5.0, indicated a rapid release profile in acidic conditions and a slow, sustained release of two orders of magnitude lower dose in neutral conditions due to the effects of the AD linker. Figure 1 shows the expected strategy, where DOX alone (Figure 1(a)) shows only modest affinity; incorporation of a high affinity adamantane (Figure 1(b)) results in a bound drug (Figure 1(c)) which can be cleaved at low pH, resulting in the previously observed, slow sustained release rates (Figure 1(d)). Release of DOX from chemically similar, but non-affinity dextran control polymers showed rapid release in both neutral and acidic conditions due to the lack of specific affinity to DOX.
Figure 1.
Schematic depiction of the drug delivery systems. (a) The previous drug delivery system, where a moderate affinity between DOX and pCD is capable of controlling the rate of release suitable for tumor treatment. (b) Chemical diagram and schematic depiction of DOX modified with an adamantane through a pH-degradable hydrazone linkage. (c and d) The drug delivery system developed in this study, where (c) high affinity AD group attached by pH-cleavable hydrazone bond reduces the rate of background drug release to as little as possible, whereas in (d) the low pH tumor microenvironments, the bond to the AD group is cleaved, restoring it to the moderate affinity, slow release system. (A color version of this figure is available in the online journal.)
DOX: doxorubicin; AD: adamantane; β-CD: β-cyclodextrin
Materials and methods
Materials
Lightly epichlorohydrin cross-linked β-cyclodextrin prepolymer was purchased from CycloLab, Budapest, Hungary. Low-molecular weight dextran prepolymer was obtained from Polysciences Inc. (Warrington, PA). Hexamethylene diisocyanate (HDI) is from Sigma-Aldrich (St. Louis, MO). Adamantane-1-carbohydrazide is from Matrix Scientific (Columbia, SC). Doxorubicin hydrochloride is from LC Laboratories (Woburn, MA). U-87 MG cells (ATCC® HTB-14™) are a human-derived glioblastoma cell line, obtained from ATCC® Microbiology (Manassas, VA). Dulbecco’s Modified Eagle’s Medium (DMEM) and fetal bovine serum (FBS) are obtained from Gibco (Carlsbad, CA). All other reagents, solvent, chemicals, etc. were purchased from Fisher Scientific in the highest grade available.
Synthesis of polymerized dextran (pDextran) and cyclodextrin (pCD) disks
Affinity-based pCD and non-affinity, diffusion-only pDextran control polymers were synthesized according to a previously published protocol, where the respective prepolymers were cross-linked with HDI in a ratio of 1:0.16 (glucose residue:HDI).27,28 Briefly, 1 g of dried pCD or pDextran was dissolved in 4 mL dimethylformamide (pCD) or dimethylsulfoxide (pDextran), cross-linked, cured at 70℃ for 45 min (pCD) or 120 min (pDextran), and punched into 8 mm disks. All disks were washed in solutions of 100% solvent, 50:50 solvent:water, and 100% water over several days.
Synthesis of AD-DOX
The synthesis of AD-DOX was carried out according to a published protocol.21 Briefly, 67 mg of adamantane-1-carbohydrazide and 100 mg doxorubicin hydrochloride were dissolved in 50 mL of dry methanol with 50 µL of trifluoroacetic acid as a catalyst. The mixture was refluxed at 50℃ for 48 h in the dark, concentrated by rotary evaporation and precipitated three times in ethyl acetate. The product was collected by centrifugation and dried under vacuum. Figure 1(b) displays the expected final chemical structure of the modified drug.
1H NMR spectroscopy
A small sample of AD-DOX (<1 mg/mL) was dissolved in deuterated dimethylsulfoxide and evaluated using a 400 MHz Varian Inova NMR (Varian Inc., Palo Alto, CA) from the Instrumentation Core in the Case Western Reserve University, Department of Chemistry.
FTIR
Small samples of dried AD-DOX, DOX, and AD were each ground into a fine powder with a mortar and pestle. Dried potassium bromide (KBr, 0.1 g) was added to the powdered drug sample and further ground and mixed. Each sample was placed in a 13 mm die and pressed with a force of 10 metric tons for 10 min. Each pellet was removed from the die and scanned in an Excalibur Series-BioRad FTS3000MX spectrometer (Hercules, CA). Two hundred background and 400 sample scans were collected from 500 to 4000 cm−1. Varian Resolution Pro Software (version 5) was used to transform the data using ratio and transmittance transformations. A boxcar function was used to smooth out each plot.
Cytotoxicity MTT assay with AD-DOX
U-87 MG human glioblastoma cells were grown in DMEM with 10% FBS, resuspended in a flask and incubated for five days at 37℃ and 5% CO2. The cell culture medium was removed and the cells were washed with PBS. The cells were trypsinized, counted by hemocytometer, and the 7500 cells were pipetted into each well of a 96-well plate and incubated overnight; 24 h later, the cells were treated with DOX or AD-DOX with 36 μM concentration with three 10-fold serial diluted solutions and a 10 μM concentration with two 10-fold serial diluted solutions. The total volume of solution in each well was 100 µL. The cells were incubated for 24 h. Twenty microliters of 5 mg/mL MTT solution was added to each well.32 One row of the tray was left as a negative control of MTT solution with no cells. The cells were incubated for 3.5 h at 37℃. The cell culture medium was removed and 150 µL of dimethylsulfoxide was added to the cells.32 The cells were covered in foil and placed on an agitator for 15 min. The samples were then read on a Bio-Tek 96-well plate reader (H1Winooski, VT) with 590 nm detection and with a reference filter wavelength of 620 nm, in order to quantify the number of cells killed in each well.
Drug loading
Two 3 mg samples of AD-DOX were each dissolved in 80 µL dimethylsulfoxide. Each drug solution was diluted to a volume of 4.4 mL with PBS. Six pCD disks were added to one of the solutions and six pDextran disks were added to the other. Both samples were covered in foil and placed on an agitator for three days. Each loaded disk was washed with distilled and deionized water and dried at room temperature overnight. The dried disks were transferred into an individual vial containing either 1 mL PBS (pH 7.4) or in acetate buffer (pH 5.0). All four conditions (two polymers, two pHs) were in triplicate. The initial (available) drug mass loaded was determined by long-term drug release followed by a final solvent extraction (in DMSO).
AD-DOX release
Every 48 h during the release of the loaded disks, all of the solution surrounding each disk was removed and replenished with 1 mL fresh PBS or acetate buffer in order to maintain infinite sink conditions. Two hundred microliters of the drug release aliquots (diluted as necessary) were stored in a 96-well plate, and then quantified by the plate reader, calculating the drug amount in each sample using calibration curves generated from known amounts of drug. Since drug was appropriately diluted to avoid high concentration quenching, fluorescence spectroscopy was used for quantification with a Biotek™ microplate reader using excitation and emission wavelengths of 498 and 590 nm. Release was followed for 87 days, at which point the control samples had mostly plateaued by the affinity-based polymer in neutral conditions continued to show release.
Statistical analysis
All data are displayed as the mean of each condition tested in triplicate with the standard deviation as the error bars. One way ANOVA statistics was performed on cytotoxicity data with Microsoft Excel 2013. A P-value of 0.019 was obtained and a value < 0.05 was determined to be statistically significant. A two-tailed t-test assuming unequal variances was performed at each concentration of the cytotoxicity data. A t-test value <0.01 was determined to be statistically significant.
Results
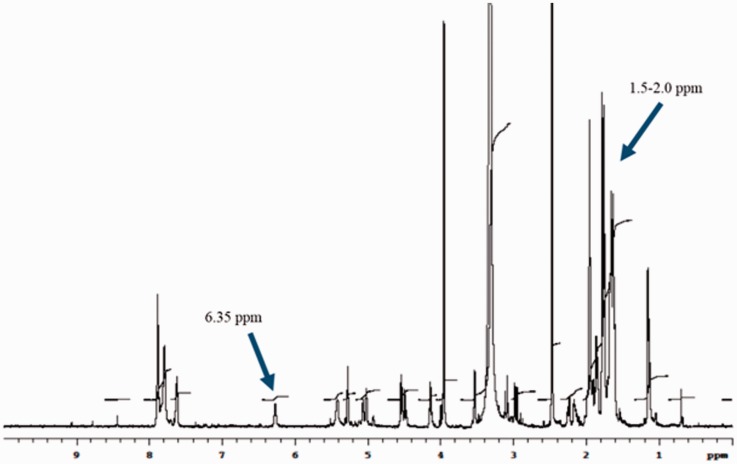
NMR spectrum of AD-DOX
We confirmed that the AD-DOX synthesis resulted in the expected product by H1 NMR spectroscopy. Specifically, the peak at 6.35 ppm indicates the protons in the amine group of the AD hydrazide, while the increased signal at 1.5–2.0 ppm is representative of AD protons (Figure 2). These peaks are consistent with that detailed by Luo et al.21 and confirmed the presence of the hydrazone bond between AD hydrazide and DOX and the successful synthesis of AD-DOX.
Figure 2.
1H NMR spectrum of AD-modified DOX confirms hydrazone bond. Successful synthesis of AD-DOX is indicated by the presence of amine protons at 6.35 ppm and AD protons of the hydrazone bond form the increased signal at 1.5–2.0 ppm. (A color version of this figure is available in the online journal.)
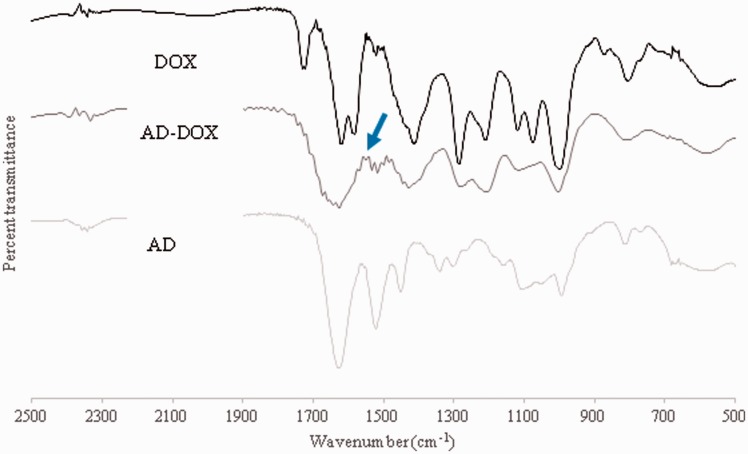
FTIR spectrum of AD, DOX, and AD-DOX
Further confirmation of the correct chemical structure was performed by FTIR spectroscopy. Specifically, FTIR spectra were generated for AD, DOX, and AD-DOX from 500 to 4000 cm−1 (Figure 3). The spectra were superimposed upon one another in order to evaluate if the hydrazone bond was successfully created between AD and DOX. The formation of the hydrazone bond is indicated by a peak over the span of 1560–1570 cm−1 on the AD-DOX spectrum that is not present on the other spectra.33
Figure 3.
FTIR spectrum confirms the drug modification. Spectra were obtained from AD, DOX, and AD-DOX from 500 to 4000 cm−1. The peak at 1560–1570 cm−1 on the AD-DOX spectrum indicates the C = N (hydrazone) bond that was formed during the synthesis of AD-DOX and that is not on either the AD or DOX spectra. Arrow on figure signifies the location of the hydrazone bond associated peak. (A color version of this figure is available in the online journal.)
DOX: doxorubicin; AD: adamantane; AD-DOX: adamantane-modified doxorubicin
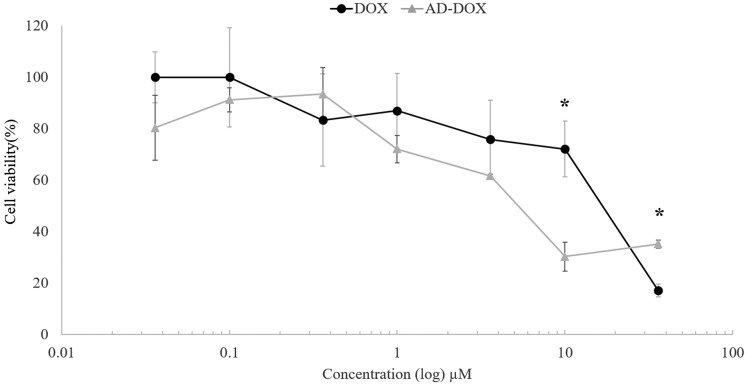
Cytotoxicity MTT assay of DOX and AD-DOX
Since the drug delivery system could theoretically either deliver normal DOX upon pH cleavage or AD-DOX uncleaved (albeit most likely quite slowly), we wanted to compare the bioactivity of both molecules. AD-DOX and normal DOX were evaluated with U-87 human glioblastoma cells using an MTT assay of mitochondrial activity as an indirect measure of cell death. The cytotoxicity assay results indicated that the modified AD-DOX drug had approximately the same intrinsic cell killing capability as unmodified DOX (Figure 4). There were statistically significant differences calculated at high concentrations, with regular DOX more toxic at 36 μM, but with AD-DOX more toxic at 10 μM. Specifically, at 10 μM, AD-DOX killed 69.7% of the cells compared with DOX killing 27.9%. The results from the ANOVA test suggested that the data are statistically significant with a P-value of 0.019. At concentrations of 36 and 10 μM, the t-test values were 9.6 × 10−5 and 1.6 × 10−3, which demonstrated statistically significant data at those specific concentrations. At other tested concentrations, the t-test value was >0.01 and was considered statistically insignificant. Within the lower concentration ranges tested, AD-DOX has an approximately equivalent cytotoxicity as DOX. Using the % viability plot (Figure 4), the IC 50 values for each drug were estimated to be 6 µM for the uncleaved (AD-DOX) and 20 µM (DOX).
Figure 4.
Cytotoxicity studies show AD-DOX is comparable to unmodified DOX. DOX modified with an AD group attached by a pH-cleavable hydrazone bond was compared with regular, unmodified DOX and tested at 36, 10, 3.6, 1, 0.36, 0.1, and 0.036 μM. AD-DOX was observed to have an approximately equivalent cytotoxicity to DOX within most of tested concentration ranges. Statistically significant concentrations are labeled with asterisks where the t-value <0.01. At high concentrations, some differences at 10 μM AD-DOX were observed to be slightly more toxic, whereas at 36 μM DOX was observed to be slightly more toxic
Drug release
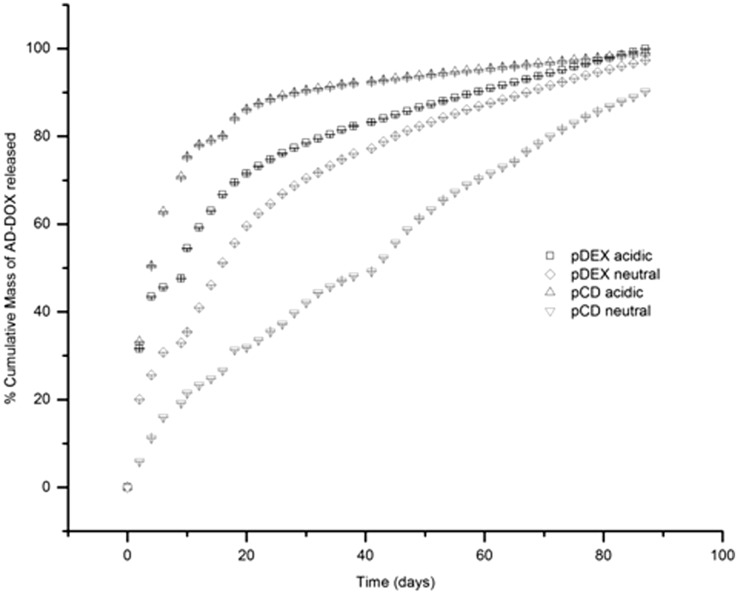
Drug release was evaluated for the two different polymers (affinity pCD and non-affinity pDextran control polymers) under neutral pH and acidic pH to determine whether cleaving the adamantane group would impact the delivery rate. Figure 5 demonstrates that drug release in the non-affinity pDextran control polymer is highly non-linear as expected from a drug delivery system relying on diffusion alone. The majority of the drug is released in a burst within the first few days, leaving less drug behind to be released at later time points. In the affinity-based pCD polymers studied under neutral conditions where the hydrazone bond is still intact, the release rate of drug is much slower, with no initial burst. However, a release rate change is observed in that same delivery system when examined at a low pH, where hydrazone cleavage and loss of the high-affinity adamantane group can occur.34 In lower pH (pH 5.0), the release rate from pCD polymers is comparable to that of the non-affinity control polymers. Specifically, at 30 days, approximately 90% of the loaded drug was released from the pCD disk in acidic conditions compared with only approximately 40% released in neutral conditions.
Figure 5.
Cumulative release studies show AD-DOX release is dependent on pH change. Due to the added affinity from the AD group, release from pCD polymers was shown to be very slow, with virtually no burst. Whereas at low, tumor pH the adamantane group is cleaved and the DOX is released at a much faster rate. The release observed is comparable to that seen by systems using chemically similar non-affinity pDextran polymers in either pH
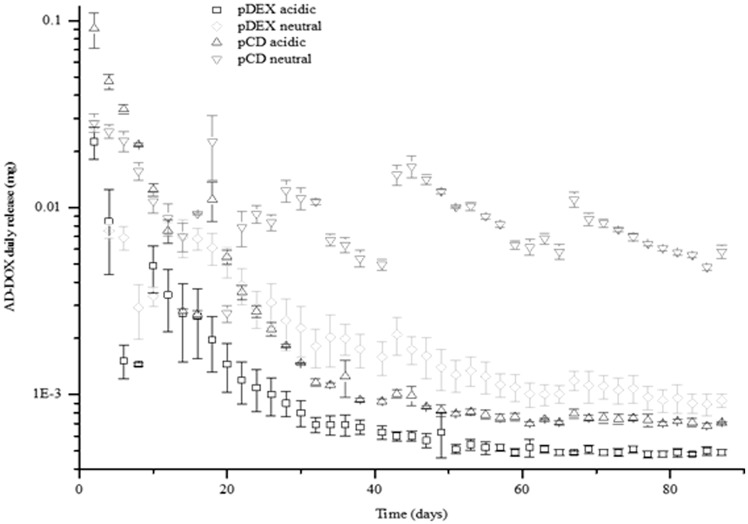
Figure 6 is a semi-log replot of the cumulative release data demonstrating the daily release quantities in the four different conditions (pCD neutral pH, pCD low pH, pDextran neutral pH, and pDextran low pH). One observation is that the total amount of drug released from the pCD systems is much higher than the total drug released from the chemically similar, but non-affinity pDextran polymers. There was almost a 4-fold difference in total drug loading detected between the two polymers. This is consistent with previous observations that more drug can be loaded into affinity-based polymers than their non-affinity controls.27,28 Specifically, we calculated that the average mass loaded into each pDextran disk was approximately 103 ± 45 µg AD-DOX/mg polymer and the average mass loaded into each pCD disk was approximately 378 ± 148 µg AD-DOX/mg polymer.
Figure 6.
Periodic release aliquots confirm release low background release of AD-DOX. Replotting the cumulative release data by the individual release aliquots on a semi-log plot, it is apparent that release from the high affinity AD-DOX/pCD system is two orders of magnitude lower than either release after pH-dependent cleavage and the resulting DOX/pCD system at low pH, or release from either non-affinity pDextran control
Discussion
The aim of this study was to address the toxicity of systemically delivered DOX through two means. The first was to reduce the systemic dose by developing a low-dose, local, controlled release delivery system with a slow, steady rate, necessary to overcome limitations such as cell cycle stage and tumor stem cell heterogeneity. This slow steady rate is not possible by other, highly non-linear, “burst type” delivery systems. The second strategy was to further reduce the amount of drug available by adding a pH degradable linker to an affinity group. The affinity group, adamantane, binds the drug to the delivery system until the tumor pH cleaves the adamantane from the drug, allowing for the normal drug release. An adamantane-modified version of the drug DOX using a hydrazone bond had previously been published and demonstrated to cleave in a pH-sensitive fashion, restoring the original DOX. However, in that work, the modified drug was used in a small, soluble, instantly available nanoparticulate, whereas in this work, the drug modification is used to bind the drug to a delivery device, reducing available drug and providing it only when needed.
Previous work from our lab and others has shown that the use of molecular interactions between drug and polymer can delay the release of drug beyond that capable of diffusion alone, termed “affinity-based drug delivery.”27,28 In this work, we show that DOX, which has a moderate affinity to the affinity-based pCD, can be changed to have a high affinity by the addition of an adamantane group to DOX. The net result of combining AD-DOX with pCD is that there is virtually no release, other than that which could possibly be accounted for as unbound, unmodified, or prematurely cleaved drug. At tumor pH, previously demonstrated capable of removing the adamantane, we see a change in release rate back up to the moderate affinity condition, and comparable to the rate of non-affinity pDextran control polymers.
While delivery rates are close, some difference is observed between the low pH and neutral pH pDextran controls. This is possibly attributed to ionization of DOX upon pH change, impacting some very low affinity hydrogen bonding that the drug is capable of showing with the dextran hydroxyls. When compared with the release rates of other pH-dependent DOX delivery systems,2–4,10,14,35 our delivery system shows minimal (6%) background release of DOX in neutral (pH 7.4) conditions after two days compared with other pH-dependent DOX delivery systems that show ≥20% background release of DOX at pH 7.42,3,10,14 (See Supplementary Figure 1). Ideally, the background release at pH 7.4 would be 0%,4,35 in order to minimize the release and toxicity of DOX in healthy tissues. While we are able to achieve a relatively low background release of DOX at pH 7.4 with our delivery system, we were not able to achieve a 0% background release. Further reduction of background delivery is the topic of future research. Additionally, future work will entail a more in-depth study of the cytotoxicity of our AD-DOX delivery system to other non-cancerous cell lines in neutral conditions (i.e. cardiomyocytes, hepatocytes). Furthermore, when we compare the release kinetics in pH 5.0–5.5, our system demonstrates the most prolonged release profile, an order of magnitude longer than other pH-dependent DOX delivery systems. Supplementary Table 1 displays the times required for each pH-dependent DOX delivery system to release either 50% or 90% of DOX in pH 5.0–5.5 conditions.
In addition to changes in the release rate, the use of drug/polymer affinities has also been shown to increase the total drug loading,26,36 which is consistent with our observations; our system showed that both the moderate affinity DOX/pCD combination and the higher affinity AD-DOX/pCD have much higher maximum loading than the control DOX/pDextran and AD-DOX/pDextran systems. This total loading was more than 3-fold higher than the 10–12% maximum loading rule-of-thumb used in developing conventional drug delivery systems. The higher loading observed allows for the use of a smaller implant to deliver the same amount of drug and for an even longer time.
Additionally, recent data from our lab have shown that affinity-based drug delivery systems can even be reloaded with DOX in vivo for additional windows of therapeutic delivery.5 One concern of such delivery systems is that without high specificity, drug can reload or diffuse into off-target sites. This is the first work showing addition of further functionality to create a lock-and-key specificity between the polymer delivery system and the reloading drug.
Lastly, we explored whether the chemical modification of DOX impacts the bioactivity of the drug. Since presumably some drug will be released before cleavage occurs, it could be a concern that this lost drug is wasted if it has no capacity for tumor killing. Using cytotoxicity studies, we demonstrated that the AD-modified drug is comparable in its capacity to kill cancer cells as unmodified or cleaved drug. In fact at some, in high concentrations, the modified drug shows even slightly higher toxicity than the unmodified drug. These results indicate the possibility of using a lower concentration of AD-DOX to achieve a more significant cell killing than unconjugated DOX, further decreasing the risks of cytotoxicity associated with DOX.
Future work can explore the use of this AD-DOX/pCD system in animal models of cancer, as well as applying the same moderate affinity/high affinity strategy in other applications.
Supplementary Material
Acknowledgements
The authors would like to thank Dale Ray of the Case Western Reserve University, Department of Chemistry for assistance and use of NMR facilities; and Sean T. Zuckerman for technical and writing assistance. This work was supported by an NSF CAREER award, CBET-0954489, and an associated REU Supplement (E.C.).
Authors’ contributions
All authors participated in the design of the experiments, analysis of the data, and drafting of the manuscript; ELC and ASF conducted the experiments.
References
- 1.Tacar O, Sriamornsak P, Dass CR. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J Pharm Pharmacol 2013; 65: 157–70. [DOI] [PubMed] [Google Scholar]
- 2.Lu D, Wen X, Liang J, Gu Z, Zhang X, Fan Y. A pH-sensitive nano drug delivery system derived from pullulan/doxorubicin conjugate. J Biomed Res 2009; 89: 177–83. [DOI] [PubMed] [Google Scholar]
- 3.Du C, Deng D, Shan L, Wan S, Cao J, Tian J, Achilefu S, Gu Y. A pH-sensitive doxorubicin prodrug based on folate-conjugated BSA for tumor-targeted drug delivery. Biomaterials 2013; 34: 3087–97. [DOI] [PubMed] [Google Scholar]
- 4.Dong L, Xia S, Wu K, Huang Z, Chen H, Chen J, Zhang J. A pH/enzyme-responsive tumor-specific delivery system for doxorubicin. Biomaterials 2010; 31: 6309–16. [DOI] [PubMed] [Google Scholar]
- 5.Fu AS, von Recum HA. Affinity-based delivery and reloading of doxorubicin for treatment of glioblastoma multiforme, Cleveland, OH: Case Western Reserve University, 2013. [Google Scholar]
- 6.Chang G, Li C, Lu W, Ding J. N-Boc-histidine-capped PLGA-PEG-PLGA as a smart polymer for drug delivery sensitive to tumor extracellular pH. Macromol Biosci 2010; 10: 1248–56. [DOI] [PubMed] [Google Scholar]
- 7.Devalapally H, Shenoy D, Little S, Langer R, Amiji M. Poly(ethylene oxide)-modified poly(beta-amino ester) nanoparticles as a pH-sensitive system for tumor-targeted delivery of hydrophobic drugs: part 3. Therapeutic efficacy and safety studies in ovarian cancer xenograft model. Cancer Chemother Pharmacol 2007; 59: 477–84. [DOI] [PubMed] [Google Scholar]
- 8.Ulbrich K, Subr V. Polymeric anticancer drugs with pH-controlled activation. Adv Drug Deliv Rev 2004; 56: 1023–50. [DOI] [PubMed] [Google Scholar]
- 9.Bajpai SK, Saggu SPS. Controlled release of an anti-malarial drug from a pH-sensitive poly(methacrylamide-co-methacrylic acid) hydrogel system. Des Monomers Polym 2007; 10: 543–54. [Google Scholar]
- 10.Chen CY, Kim TH, Wu WC, Huang CM, Wei H, Mount CW, Tian Y, Jang SH, Pun SH, Jen AK. pH-dependent, thermosensitive polymeric nanocarriers for drug delivery to solid tumors. Biomaterials 2013; 34: 4501–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang T, Zhang Z, Zhang Y, Lu H, Zhou J, Li C, Hou L, Zhang Q. Dual-functional liposomes based on pH-responsive cell-penetrating peptide and hyaluronic acid for tumor-targeted anticancer drug delivery. Biomaterials 2012; 33: 9246–58. [DOI] [PubMed] [Google Scholar]
- 12.Qiu Y, Park K. Environment-sensitive hydrogels for drug delivery. Adv Drug Deliv Rev 2001; 53: 321–39. [DOI] [PubMed] [Google Scholar]
- 13.Mandracchia D, Pitarresi G, Palumbo FS, Carlisi B, Giammona G. pH-sensitive hydrogel based on a novel photocross-linkable copolymer. Biomacromolecules 2004; 5: 1973–82. [DOI] [PubMed] [Google Scholar]
- 14.Zheng H, Wang Y, Che S. Coordination bonding-based mesoporous silica for pH-responsive anticancer drug doxorubicin delivery. J Phys Chem C 2011; 115: 16803–13. [Google Scholar]
- 15.Zhang H, Dong Y, Wang L, Wang G, Wu J, Zheng Y, Yang H, Zhu S. Low swelling hyperbranched poly(amine-ester) hydrogels for pH-modulated differential release of anticancer drugs. J Mater Chem 2011; 21: 13530–7. [Google Scholar]
- 16.Kim B, Shin Y. pH-sensitive swelling and release behaviors of anionic hydrogels for intelligent drug delivery system. J Appl Polym Sci 2007; 105: 3656–61. [Google Scholar]
- 17.Shah MA, Schwartz GK. Cell cycle-mediated drug resistance: an emerging concept in cancer therapy. Clin Cancer Res 2001; 7: 2168–81. [PubMed] [Google Scholar]
- 18.Deisseroth AB, DeVita VT., Jr The cell cycle: probing new molecular determinants of resistance and sensitivity to cytotoxic agents. Cancer J Sci Am 1995; 1: 15–21. [PubMed] [Google Scholar]
- 19.Sundar SJ, Hsieh JK, Manjila S, Lathia JD, Sloan A. The role of cancer stem cells in glioblastoma. Neurosurg Focus 2014; 37: E6–E6. [DOI] [PubMed] [Google Scholar]
- 20.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev 2015; 29: 1203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo GF, Xu XD, Zhang J, Yang J, Gong YH, Lei Q, Jia HZ, Li C, Zhuo RX, Zhang XZ. Encapsulation of an adamantane-doxorubicin prodrug in pH-responsive polysaccharide capsules for controlled release. ACS Appl Mater Interf 2012; 4: 5317–24. [DOI] [PubMed] [Google Scholar]
- 22.Lu D, Wen X, Liang J, Zhang X, Gu Z, Fan Y. Novel pH-sensitive drug delivery system based on natural polysaccharide for doxorubicin release. Chin J Polym Sci 2008; 26: 369–74. [Google Scholar]
- 23.Prabaharan M, Grailer JJ, Pilla S, Steeber DA, Gong S. Amphiphilic multi-arm-block copolymer conjugated with doxorubicin via pH-sensitive hydrazone bond for tumor-targeted drug delivery. Biomaterials 2009; 30: 5757–66. [DOI] [PubMed] [Google Scholar]
- 24.Gu YJ, Cheng J, Jin J, Cheng SH, Wong WT. Development and evaluation of pH-responsive single-walled carbon nanotube-doxorubicin complexes in cancer cells. Int J Nanomed 2011; 6: 2889–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Granadero D, Bordello J, Perez-Alvite MJ, Novo M, Al-Soufi W. Host-guest complexation studied by fluorescence correlation spectroscopy: adamantane-cyclodextrin inclusion. Int J Molecular Sci 2010; 11: 173–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thatiparti TR, Averell N, Overstreet D, von Recum HA. Multiplexing interactions to control antibiotic release from cyclodextrin hydrogels. Macromol Biosci 2011; 11: 1544–52. [PubMed] [Google Scholar]
- 27.Wang NX, von Recum HA. Affinity-based drug delivery. Macromol Biosci 2011; 11: 321–32. [DOI] [PubMed] [Google Scholar]
- 28.Thatiparti TR, von Recum HA. Cyclodextrin complexation for affinity-based antibiotic delivery. Macromol Biosci 2010; 10: 82–90. [DOI] [PubMed] [Google Scholar]
- 29.Fu AS, Thatiparti TR, Saidel GM, von Recum HA. Experimental studies and modeling of drug release from a tunable affinity-based drug delivery platform. Annals Biomed Eng 2011; 39: 2466–75. [DOI] [PubMed] [Google Scholar]
- 30.Halpern JM, Gormley CA, Keech MA, von Recum HA. Thermomechanical properties, antibiotic release, and bioactivity of a sterilized cyclodextrin drug delivery system. J Mater Chem 2014; 2: 2764–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thatiparti TR, Shoffstall AJ, von Recum HA. Cyclodextrin-based device coatings for affinity-based release of antibiotics. Biomaterials 2010; 31: 2335–47. [DOI] [PubMed] [Google Scholar]
- 32.van de Loosdrecht AA, Beelen RHJ, Ossenkoppele GJ, Broekhoven MG, Langenhuijsen MMAC. A tetrazolium-based colorimetric MTT assay to quantitate human monocyte mediated cytotoxicity against leukemic cells from cell lines and patients with acute myeloid leukemia. J Immunological Methods 1994; 174: 311–20. [DOI] [PubMed] [Google Scholar]
- 33.Belskaya NP, Dehaen W, Bakulev VA. Synthesis and properties of hydrazones bearing amide, thioamide and amidine functions. Online J Org Chem 2010; 1: 275–332. [Google Scholar]
- 34.Sanson C, Schatz C, Le Meins JF, Soum A, Thevenot J, Garanger E, Lecommandoux S. A simple method to achieve high doxorubicin loading in biodegradable polymersomes. J Control Release 2010; 147: 428–35. [DOI] [PubMed] [Google Scholar]
- 35.Low SA, Yang J, Kopeček Bone-targeted acid-sensitive doxorubicin conjugate micelles as potential osteosarcoma therapeutics. Bioconjugate Chem 2014; 25: 2012–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivera-Delgado E, Ward E, von Recum HA. Providing sustained transgene induction through affinity-based drug delivery. J Biomed Mater Res A 2016; 104: 1135–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.