Abstract
Both obesity and the metabolic syndrome are risk factors for type 2 diabetes and cardiovascular disease. Identification of novel biomarkers are needed to distinguish metabolic syndrome from equally obese individuals in order to direct them to early interventions that reduce their risk of developing further health problems. We utilized mass spectrometry-based targeted metabolic profiling of 221 metabolites to evaluate the associations between metabolite profiles and established metabolic syndrome criteria (i.e. elevated waist circumference, hypertension, elevated fasting glucose, elevated triglycerides, and low high-density lipoprotein cholesterol) in plasma samples from obese men (n = 29; BMI = 35.5 ± 5.2 kg/m2) and women (n = 40; 34.9 ± 6.7 kg/m2), of which 26 met the criteria for metabolic syndrome (17 men and 9 women). Compared to obese individuals without metabolic syndrome, univariate statistical analysis and partial least squares discriminant analysis showed that a specific group of metabolites from multiple metabolic pathways (i.e. purine metabolism, valine, leucine and isoleucine degradation, and tryptophan metabolism) were associated with the presence of metabolic syndrome. Receiver operating characteristic curves generated based on the PLS–DA models showed excellent areas under the curve (0.85 and 0.96, for metabolites only model and enhanced metabolites model, respectively), high specificities (0.86 and 0.93), and good sensitivities (0.71 and 0.91). Moreover, principal component analysis revealed that metabolic profiles can be used to further differentiate metabolic syndrome with 3 versus 4–5 metabolic syndrome criteria. Collectively, these findings support targeted metabolomics approaches to distinguish metabolic syndrome from obesity alone, and to stratify metabolic syndrome status based on the number of criteria met.
Impact statement
We utilized mass spectrometry-based targeted metabolic profiling of 221 metabolites to evaluate the associations between metabolite profiles and established MetS criteria. To our best knowledge, the findings of this study provide the first evidence that metabolic profiles can be used to differentiate participants with MetS from similarly obese individuals who do not meet established criteria of MetS. Furthermore, the study demonstrated that within MetS participants, their unique metabolic profiles correlated to the number of criteria used for MetS determination. Taken together, this metabolic profiling approach can potentially serve as a novel tool for MetS detection and monitoring, and provide useful metabolic information for future interventions targeting obesity and MetS.
Keywords: HPLC-MS/MS, targeted metabolic profiling, metabolic syndrome, obesity, metabolic pathways
Introduction
Metabolic syndrome (MetS) is a growing public health challenge worldwide, primarily due to the increasing rates of urbanization, excess energy intake, obesity, and sedentary lifestyles.1,2 MetS is defined by the presence of ≥3 cardiometabolic risk factors, including abdominal (central) obesity, hypertension, elevated fasting glucose, and dyslipidemia.3 Tragically, ∼34% of the American population and ∼47% of those ≥60 y of age meet the criteria for MetS,2 which increases the risk of developing type 2 diabetes mellitus (T2DM) by 5-fold and cardiovascular disease (CVD) by 2-fold over the next 5–10 years.4
MetS is a multifactorial disease. Therefore, individuals afflicted with MetS will present with varying combinations and degrees of metabolic abnormalities. Not all obese individuals will develop MetS and those with MetS will differ in the pathology they develop (i.e. CVD, T2DM, or both).1 Therefore, it is important to enhance the prognostic value of established MetS criteria (i.e. waist circumference, blood pressure, and blood glucose and lipid levels) by identifying novel metabolic markers to differentiate MetS from obese non-MetS individuals and to stratify the risk of MetS status based on the incremental number of clinical criteria fulfilled.
Metabolic profiling techniques, such as nuclear magnetic resonance spectroscopy (NMR)- and mass spectrometry (MS)-based metabolomics, have been used to differentiate between MetS/diabetes and healthy populations by measuring alterations of tens to hundreds of metabolites.5–7 Studies that define differential metabolic profiles of obese individuals compared with those equally obese but meeting MetS criteria have not been investigated, but are warranted in light of the continued growth of the prevalence of MetS.2 Due to the physiological heterogeneity inherent to obesity and MetS, not all obese individuals eventually develop MetS. Moreover, the degree of obesity is not necessarily linked with metabolic complication.1 Therefore, approaches that can differentiate MetS from equally obese non-MetS individuals using a cluster of metabolite biomarkers from multiple biochemical pathways are needed to provide better health status monitoring and to potentially direct individuals to early intervention strategies. In the present study, targeted metabolic profiling by HPLC-MS/MS was conducted to detect metabolic profile differences between obese individuals with MetS and equally obese individuals not meeting the MetS criteria (obese non-MetS group). The targeted metabolic profiling approach provided specific screening of 221 metabolites from most relevant metabolic pathways (e.g. citric acid cycle, glycolysis and amino acids metabolism). We hypothesized that metabolite biomarkers identified using a targeted metabolic profiling approach would differentiate a group of heterogeneous MetS participants from obese non-MetS participants, by utilizing their metabolic profiles to build statistical diagnostic models, and that differences in metabolic profiles would predict the incremental number of clinical criteria used for MetS determination.
Methods
Participants
The protocol for this study was approved by the Institutional Review Boards at the University of Connecticut and The Ohio State University. Written informed consent was obtained from all participants before enrolling, and plasma samples were completely de-identified prior to analysis. Plasma samples were obtained after an overnight fast from individuals who underwent screening for inclusion into a previously published clinical study.8 Participants were non-diabetic, non-smokers, and were not using any vasoactive medications (e.g. blood pressure medications, statins) or dietary supplements. Obese (body mass index (BMI) ≥30 kg/m2) men and pre-menopausal women were screened for the presence of ≥3 of the following established risk factors for MetS3: waist circumference ≥102 cm for men and ≥88 cm for women, fasting triglycerides ≥150 mg/dL, fasting glucose ≥100 mg/dL, resting systolic (≥130 mmHg) and diastolic (≥85 mmHg) blood pressure, and HDL-cholesterol <40 mg/dL for men and <50 mg/dL for women. Of the 69 participants screened, 26 were classified as MetS and 43 as obese non-MetS (Table 1).
Table 1.
Participant characteristics
| Obese non-MetS (n = 43 (12 men)) | MetS (n = 26 (17 men)) | |
|---|---|---|
| Age (y) | 29.3 ± 10.3 | 27.4 ± 9.8 |
| BMI (kg/m2) | 35.2 ± 6.8 | 35.6 ± 4.5 |
| Waist circumference (cm) | 109.1 ± 15.9 | 112.1 ± 9.2 |
| SBP (mmHg) | 115.7 ± 11.4 | 122.7 ± 11.9* |
| DBP (mmHg) | 78.6 ± 7.4 | 82.6 ± 7.7* |
| MAP (mmHg) | 90.9 ± 8.2 | 96.1 ± 8.2* |
| HR (bpm) | 70.0 ± 12.4 | 71.1 ± 10.4 |
| Glucose (mg/dL) | 94.7 ± 8.4 | 106.9 ± 11** |
| TG (mg/dL) | 76.9 ± 29.4 | 154.2 ± 61.3** |
| Total cholesterol (mg/dL) | 160.5 ± 27.7 | 165.5 ± 39.4 |
| HDL-C (mg/dL) | 52.1 ± 10.2 | 38.0 ± 9.6** |
| LDL-C (mg/dL) | 93.0 ± 24.8 | 96.8 ± 34.8 |
Note: Data are means ± SD. DBP: diastolic blood pressure; HDL-C: high density lipoprotein-cholesterol; LDL-C: low density lipoprotein-cholesterol; MAP: mean arterial pressure; SBP: systolic blood pressure; TG: triglyceride.
P value < 0.05.
P value < 0.005.
Reagents
Authentic standards corresponding to the measured metabolites were purchased from Sigma-Aldrich (Saint Louis, MO, USA) or IROA Technologies (Boston, MA, USA). Stable isotope-labeled amino acid mix (20 AA U-13C, 97–99%; U-15 N, 97–99%) was purchased from Cambridge Isotope Laboratories (Tewksbury, MA, USA). HPLC-MS grade acetonitrile, ammonium acetate, and acetic acid were all purchased from Fisher Scientific (Pittsburgh, PA, USA).
Clinical chemistries
Plasma triglycerides, total and HDL-cholesterol, and glucose were measured as previously described8 using commercially available clinical assays (Pointe Scientific, Canton, MI, USA). Plasma LDL-cholesterol was calculated.9
Sample preparation
Plasma samples were prepared following the same protocol from our previous work.10,11 In brief, samples were kept at −80℃ until sample preparation steps. Prior to sample preparation, samples were randomized for analysis to ensure no batch effects and prevent experimental bias. Samples were thawed at 4℃, vortexed, and 50 µL was aliquot to a tube prior to adding 50 µL spiking solution (stable isotope-labeled amino acid mix, used for quality control purpose during mass spectrometry runs) and 250 µL methanol that contained a mix of 21 internal standards. After vortexing for 2 min, and incubating at −20℃ for 20 min, samples were centrifuged (22024.6 g for 20 min. The supernatant (150 µL) was collected and dried at 30℃ using a speedvac system. Samples were then reconstituted with in a 1:1 mixture of mobile phase A and B, and injected on the HPLC-MS/MS system for analysis.
Targeted HPLC-MS/MS metabolic profiling
Targeted metabolomics of plasma samples was performed according to validated procedures,10,11 with minor modifications. In brief, chromatographic separation and analyte detection was performed using a Thermo Fisher Scientific Dionex Ultimate 3000 HPLC system with inline TSQ-Quantiva triple quadrupole tandem mass spectrometer (MS/MS) equipped with an electrospray ionization (ESI) source. Each sample was injected twice to perform detection in both negative and positive ionization modes. Regardless of ionization mode, chromatographic separations of primarily polar metabolites were performed on a Xbridge BEH hydrophilic interaction chromatography column (Waters Corporation, Milford, MA, 150 × 2.1 mm, 3.0 µm). HPLC separation was performed (0.30 mL/min) with the autosampler thermostatted to 4℃, and the column compartment to 40℃. Mobile phase A consisted of 5 mM ammonium acetate prepared in 10% acetonitrile containing 0.2% acetic acid and mobile phase B was 5 mM ammonium acetate prepared in 90% acetonitrile containing 0.2% acetic acid. Total running time for both ionization modes was 20 min with chromatographic gradient separation (0–2 min, 70% B; 5 min, 30% B; 9 min, 30% B; 11 min, 70% B; 20 min, 70% B). The HPLC-MS/MS was controlled by Xcalliber version 2.0 (Thermo Fisher Scientific). The targeted metabolic profiling was performed in selected-reaction-monitoring (SRM) mode, established by running multiple authentic chemical standards first, and then using the obtained retention time and SRM transition information to identify metabolites from samples. The 221 metabolites were selected according to our published work,10–12 and consistent with previous studies that these represent key metabolites of interest from relevant metabolic pathways.13,14 The detection parameters for these metabolites are listed in supplementary Table S1. The detection ability and measurement reliability for each of the metabolites using our mass spectrometry system were also factors in determining which metabolites to include in the analysis. The average inter-assay coefficient of variation for the quality control samples was below 15%, and indicated excellent reproducibility of our targeted metabolic profiling approach.
Data analyses
A Student’s paired t-test was used to evaluate participant characteristic data between groups. All raw metabolomics data were manually processed by the Quanbrowser module of Xcalibur version 2.0 (Thermo Fisher Scientific). Mass spectral data were normalized using pooled human serum quality control samples (QC). The coefficient of variation values were calculated for every metabolite. To search for potential metabolite biomarkers of MetS (metabolite selection), model building was performed using SPSS Version 22.0 (IBM Analytics, Armonk, NY, USA) and JMP Pro12 (SAS Institute, Cary, NC, USA). The statistical methods applied in this study are Mann–Whitney U test, principle components analysis (PCA), and generation of receiver operating characteristic (ROC) curve. Partial least squares discriminant analysis (PLS-DA) and Monte Carlo cross validation (MCCV, developed using in-house scripts) were also performed using Mathlab software (Mathworks, Natick, MA, USA) installed with the PLS toolbox. MCCV was applied to prevent the over-optimization of statistical model. It was conducted with 500 iterations, using data from 70% of the samples (randomly selected) as the training set while the remaining 30% served as the testing set for each iteration. Three specificities, 0.95, 0.85, and 0.75, for the training sets were used to determine the thresholds of PLS-DA-predicted Y values. The same thresholds were then applied to the test set to determine sensitivities and specificities. The sample classification can be correctly assigned, termed “true class,” or the sample class information can be randomly permuted, which is referred to as “random permutation.” The PLS-DA models can only be considered validated if the MCCV tests turn out that true model did not overlap with random permutation model. Metabolic pathway analysis was performed using the online tool MetaboAnalyst 3.0 (http://www.metaboanalyst.ca/).15
Results
There were 69 obese individuals screened for inclusion into the previously complete clinical study,8 of which 26 met the criteria for MetS (Table 1). MetS participants had three (n = 11), four (n = 11), or all five MetS criteria (n = 4), whereas obese non-MetS participants had ≤2 criteria. Obesity status was similar between groups on the basis of BMI and waist circumference. However, SBP, DBP, MAP, plasma glucose and triglycerides were higher, and plasma HDL-C lower, in MetS versus obese non-MetS participants (P < 0.05).
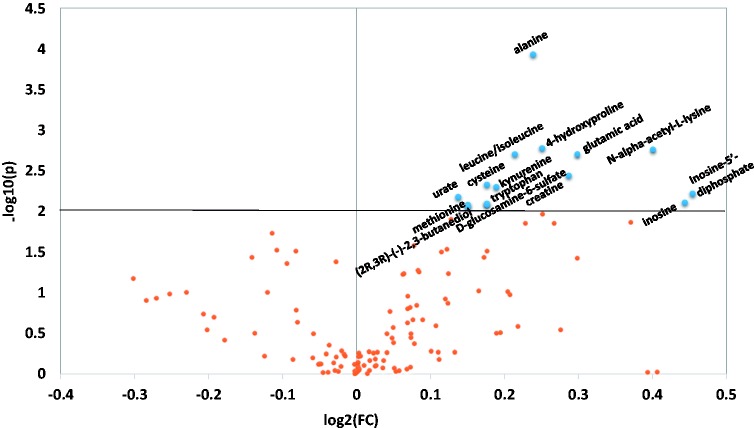
To differentiate the metabolic profile between obese non-MetS and MetS participants, a recently developed HPLC-MS/MS approach was used.12 Of the 221 metabolite targets considered for analysis, 144 metabolites from more than 20 metabolic pathways were identified. Mann–Whitney U test P value together with the fold changes between the two groups is plotted in Figure 1. Several cutoff values were applied using the U test, and the cutoff P value of 0.01 was chosen to limit the number of metabolites to be used in subsequent multivariate statistical modelling. Fifteen metabolites were identified to be differentially regulated in MetS compared with obese non-MetS individuals (Table 2). Variable importance in projection (VIP) analysis, which estimates the contribution of each variable to the classification, is reported in Table 2. Variables with a VIP scores >1 can be considered important for the separation of the two groups of participants.4 The VIP value of the 15 metabolites identified ranged from 1.59 to 2.43, which suggested their potential use in a multivariate statistical model that differentiates MetS and obese non-MetS status. The fold change and effect size of each metabolite between the two groups of study participants was also calculated and reported in Table 2. Interestingly, all 15 significantly changed metabolites from the MetS group were detected at a higher level than in the obese non-MetS group. The effect size was at medium to large level (0.57–0.87) for all significantly changed metabolites, which takes the group size into consideration, and again indicated the strong differences of these metabolites between these two groups.
Figure 1.
Volcano plot showing the P value (y-axis) and the fold changes (x-axis) of the metabolites detected in this study. Cutoff P value of 0.01 was used. The metabolites above the horizontal line with significant P value are alanine, trans-4-hydroxyproline, N-alpha-acetyl-l-lysine, leucine/isoleucine, glutamic acid, creatine, cysteine, kynurenine, inosine-5′-diphosphate, urate, inosine, tryptophan, methionine, d-glucosamine-6-sulfate, (2R, 3R) – (−)-2, 3-butanediol. (A color version of this figure is available in the online journal.)
Table 2.
Metabolites with P value less than 0.01 in comparison of MetS versus Obese non-MetS group
| Metabolite | P value | VIP value | Fold changes | Effect size |
|---|---|---|---|---|
| Alanine | 1.17E-04 | 2.43 | 1.18 | 0.87 |
| Trans-4- hydroxyproline | 1.67E-03 | 2.06 | 1.19 | 0.73 |
| N-alpha-acetyl- L-lysine | 1.73E-03 | 1.94 | 1.32 | 0.69 |
| Leucine/isoleucine | 1.98E-03 | 1.86 | 1.16 | 0.67 |
| Glutamic acid | 1.98E-03 | 1.81 | 1.23 | 0.65 |
| Creatine | 3.64E-03 | 1.93 | 1.22 | 0.69 |
| Cysteine | 4.71E-03 | 1.59 | 1.13 | 0.57 |
| Kynurenine | 5.02E-03 | 1.78 | 1.14 | 0.64 |
| Inosine-5′- diphosphate | 6.07E-03 | 1.94 | 1.37 | 0.69 |
| Urate | 6.66E-03 | 1.89 | 1.10 | 0.68 |
| Inosine | 7.77E-03 | 1.88 | 1.36 | 0.67 |
| Tryptophan | 8.01E-03 | 1.69 | 1.13 | 0.60 |
| Methionine | 8.25E-03 | 1.72 | 1.11 | 0.61 |
| d-glucosamine-6- sulfate | 8.25E-03 | 1.77 | 1.13 | 0.63 |
| (2R,3R)-(-)-2,3- butanediol | 8.77E-03 | 1.75 | 1.11 | 0.62 |
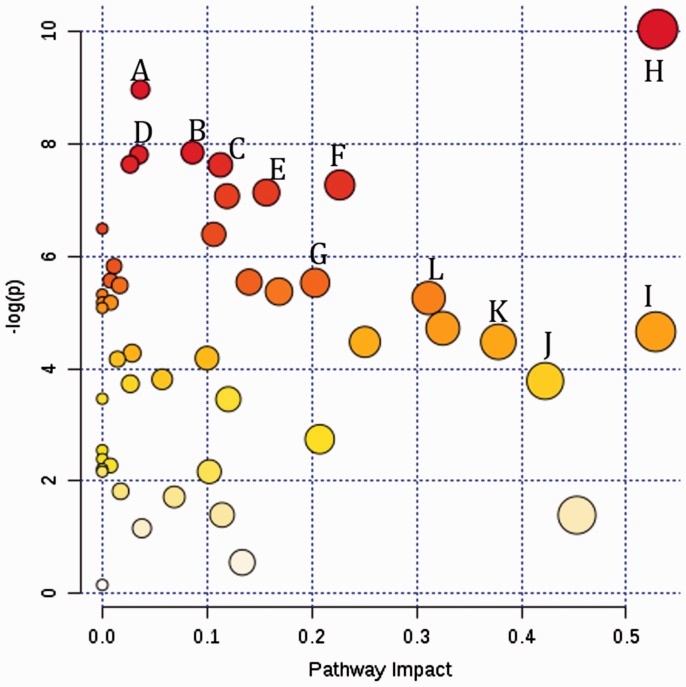
To put individual metabolite into the context of connected metabolic pathways network, and to understand the potential impact to these pathways from MetS, we constructed a metabolic pathways impact map using MetaboAnalyst 3.0 (Figure 2). From the metabolome view of all metabolites detected, 11 metabolic pathways showed significant P value following exposure (top section with y-axis higher or equal to 6), and seven metabolic pathways showed great pathway impact (x-axis larger or equal to 0.3). The major pathways that were increased by the presence of MetS include purine metabolism (i.e. urate); valine, leucine and isoleucine degradation; aminoacyl-tRNA biosynthesis; tryptophan metabolism; cysteine and methionine metabolism; lysine degradation; pyrimidine metabolism; arginine and proline metabolism; glycine, serine and threonine metabolism; taurine and hypotaurine metabolism; alanine, aspartate and glutamate metabolism; pantothenate and CoA biosynthesis.
Figure 2.
A metabolome view showing all impacted metabolic pathways in comparison between MetS and Obese non-MetS groups in this study. (a) Purine metabolism; (b) valine, leucine and isoleucine degradation; (c) aminoacyl-tRNA biosynthesis; (d) tryptophan metabolism; (e) cysteine and methionine metabolism; (f) lysine degradation; (g) pyrimidine metabolism; (h) arginine and proline metabolism; (i) glycine, serine and threonine metabolism; (j) taurine and hypotaurine metabolism; (k) alanine, aspartate and glutamate metabolism; (l) pantothenate and CoA biosynthesis. (A color version of this figure is available in the online journal.)
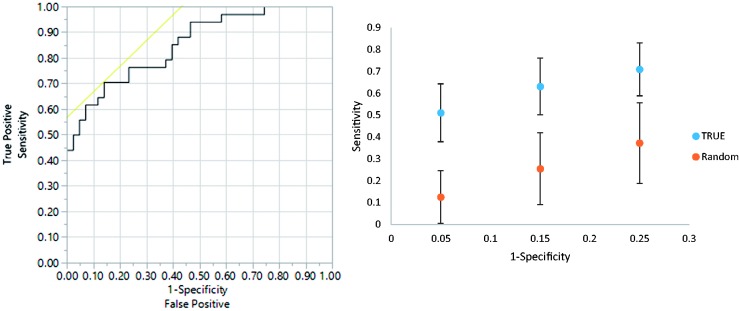
Multivariate statistical analysis was performed to determine the extent to which metabolic profiles could differentiate MetS from obese non-MetS individuals. Metabolites with p < 0.005 were applied for initial PLS-DA analysis (Figure 3(a)), specifically isoleucine/leucine, cysteine, 4-hydroxyproline, alanine, N-alpha-acetyl-L-lysine, creatine, and glutamic acid. As shown in Figure 3(a), PLS-DA model-derived ROC curve is generated to distinguish the MetS group from obese non-MetS group using metabolic profiles detected in this study and AUROC of 0.85, sensitivity of 0.71, and specificity of 0.86 were obtained, which indicated good diagnostic power of this metabolite model for detecting MetS individuals and discriminating against those who were only obese. Monte Carlo cross-validation (MCCV) was then applied (Figure 3(b)) with 70% samples used as training set and the remaining 30% were used as testing set. Sensitivities were calculated for test specificities of 0.95, 0.85, and 0.75. The sample classification can be correctly assigned, termed “true class,” or the sample class information can be randomly permuted, which is referred to as a “random permutation.” Three groups of error bar from true class to random permutation group did not have any overlap, which indicates that clear separation between the obese non-MetS and MetS groups can be achieved and validated.
Figure 3.
ROC (upper panel) and MCCV (lower panel) using metabolites with p < 0.005 in comparison of MetS group versus Obese non-MetS in the metabolites only model. Seven metabolites are used, AUROC = 0.85, sensitivity = 0.71 and specificity = 0.86. True: true class model; random: random permutation model. (A color version of this figure is available in the online journal.)
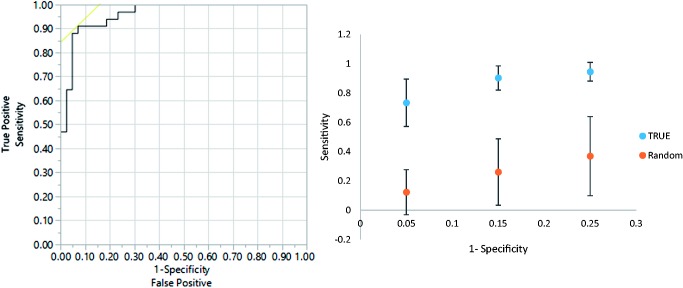
Additionally, an enhanced PLS-DA model was established by using these seven metabolites and combining three clinical characteristics (plasma glucose, plasma TG and HDL) that also have P values less than 0.005 in comparison to the two groups to test the possibility of model performance enhancement. As shown in Figure 4, an ROC curve is again generated, and the enhanced metabolite mode showed excellent AUROC of 0.96 for differentiating MetS participants from obese non-MetS, with sensitivity of 0.91 and specificity of 0.93, which is superior to the metabolites only model shown in Figure 3. MCCV was again performed and indicated that robust detection of MetS participants can be achieved by this enhanced PLS-DA model. The successful establishment of these PLS-DA models proved that shared changes of metabolites from a group of heterogeneous MetS patients can be detected and used for the differentiation of MetS from obese non-MetS population.
Figure 4.
ROC (upper panel) and MCCV (lower panel) using p < 0.005 in comparison of MetS group versus Obese non-MetS, metabolites and clinical characteristics (plasma glucose, plasma TG, HDL) combined model. AUROC = 0.96, sensitivity = 0.91, specificity = 0.93. True: true class model; random: random permutation model. (A color version of this figure is available in the online journal.)
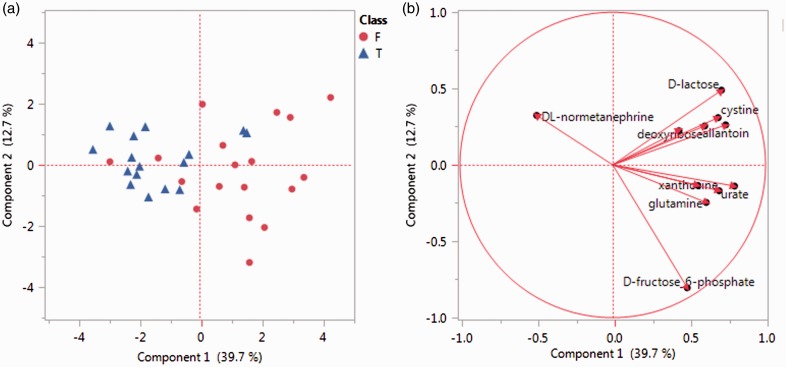
Based on the findings of metabolic profiles differentiating obese non-MetS individuals from MetS participants in this study, we considered that metabolite profiles would also differ between MetS participants meeting 3 criteria compared with those meeting ≥4 criteria. Metabolic profile comparison of these two subgroups was conducted and principal component analysis (PCA) was performed using 11 significantly changed metabolites (Table 3). Based on the metabolic profiles, good separation between the subgroup of 3 criteria and the subgroup of ≥4 criteria can be observed in the PCA score plot (Figure 5(a)) with only a few exceptions. The loading plot (Figure 5(b)) showed the different contribution of these 11 significantly different metabolites to the separation of these two subgroups of MetS participants.
Table 3.
Metabolites with P value less than 0.05 in comparison of subgroups of MetS participants (3 factors versus 4 and 5 factors)
| Metabolites | P value | VIP value | Fold changes | Effect size |
|---|---|---|---|---|
| Deoxyribose | 1.81E-02 | 1.71 | 0.82 | −0.73 |
| Lactose | 4.73E-02 | 1.60 | 0.83 | −0.69 |
| Mesoxalate | 1.65E-02 | 1.80 | 0.90 | −0.77 |
| normetanephrine | 3.55E-04 | 2.92 | 1.51 | 1.25 |
| Hydroxybutyric acid | 3.10E-02 | 1.61 | 0.98 | −0.69 |
| Glutamine | 2.18E-02 | 1.85 | 0.89 | −0.79 |
| Cystine | 1.81E-02 | 2.22 | 0.67 | −0.95 |
| Allantoin | 4.01E-02 | 1.59 | 0.90 | −0.68 |
| Xanthosine | 1.12E-02 | 2.00 | 0.78 | −0.86 |
| Urate | 1.50E-02 | 2.16 | 0.88 | −0.92 |
| d-fructose-6- phosphate | 4.73E-02 | 1.83 | 0.75 | −0.79 |
Figure 5.
PCA plots showing detailed comparison of MetS subgroups (participants with 3 factors versus 4 and 5 factors). F: participants with 4 and 5 risk factors; T: participants with only 3 risk factors. (a) Score plot showing separation of the two subgroups of MetS participants based on their metabolic profile of selected metabolites. (b) Loading plot showing contributions of each metabolites in the separation of two subgroups. (A color version of this figure is available in the online journal.)
Discussion
The findings of this cross-sectional study provide the first evidence that metabolic profiles can be used to differentiate participants with MetS from similarly obese individuals who do not meet the established criteria of MetS. Furthermore, the study demonstrated that within MetS participants, their unique metabolic profiles correlated to the number of criteria used for MetS classification.
In recent years, metabolomics has been broadly applied to study comprehensive characterization of the small molecule metabolites found in living organism.16,17 Since the metabolome investigated is closely tied to the phenotype of an organism, and is also sensitive to perturbation such as lifestyle and dietary changes, metabolomics provides a unique approach to examine the pathophysiological status of individuals, and provides immediate feedback and guidance for intervention and prevention. In this study, the metabolic profile of participants afflicted with MetS was particularly focused via a large-scale metabolomics investigation of 221 targeted metabolites from more than 20 metabolic pathways. The advantage of our targeted metabolic profiling approach was that it provided broad coverage of relevant metabolites while simultaneously enabling confident metabolite identification, thus avoiding tedious compound database search and confirmation steps required for untargeted metabolic profiling approaches.5 The metabolite targets were carefully selected from metabolomics studies10,11,13,14 in which their detectability and measurement reproducibility were validated. Thus, the metabolites reported from the present study are readily interpreted within their biological context to potentially address questions of public health and scientific significance.
In our study, 15 metabolites with statistical significance were detected when comparing MetS and obese non-MetS individuals, including nucleosides, amino acids and derivatives, amino sugars, purine derivatives and polyols. Amino acid levels have been reported to be associated with obesity. Specifically, blood levels of branched chain amino acids (BCAAs) are elevated in obese, insulin-resistant or T2DM subjects relative to healthy controls.18 It is also suggested that increased catabolism of BCAAs is induced by obesity, and are correlated with insulin resistance even in obese individuals that were deemed healthy but insulin resistant relative to lean controls.19 In our study, the amino acids alanine, trans-4-hydroxyproline, N-alpha-acetyl-l-lysine, leucine/isoleucine, glutamic acid, cysteine, kynurenine, tryptophan, and methionine were detected at higher levels in MetS compared with obese non-MetS individuals. MetS-mediated increases in these metabolites suggest altered amino acid metabolism, which may play a pathogenic role in the transition from obese non-MetS to MetS.
Glucose and energy metabolism are well established to be altered in obesity in both animal and human studies. Elevated level of free fatty acids and basal lipolysis has also been linked to obesity, and their alteration is closely related to CVD. Previous metabolomics studies have reported that metabolic dysregulation of glucose and lipids is the most relevant factor for the pathophysiology of T2DM.20 In our study, we did not observe many significantly altered glucose or lipid metabolites, which could potentially be explained by the similar obesity status of the MetS and obese non-MetS participants. As other studies have compared those with MetS versus healthy controls,7 we focused on two equally obese groups distinguished by the classification of MetS to provide alternative approach for the early diagnostics of MetS, and better understanding of their metabolic profile differences. Findings from our study provide novel evidence by identifying a group of altered metabolic pathways from MetS comparing to obese non-MetS, which could be used as potential intervention targets to prevent the transition from obesity to MetS in the future. Furthermore, we discovered a potential metabolic profile from MetS subgroups that are correlated to the number of MetS criteria present.
While our findings are potentially clinically impactful, many areas remain to be explored in future studies. First, while a group of fatty acids were included in our detection panel of targeted metabolites, other important lipid metabolites need to be investigated to advance an understanding of alterations in lipid metabolism during MetS. Moreover, linking the metabolic profiles to the heterogeneity of MetS, such as different combinations of MetS criteria, was not extensively explored in this study primarily due to limited number of participants in each subgroup of MetS. Thus, prospective studies are warranted to address these areas and advance our understanding of the utility of metabolic profiling to predict the risk for developing MetS.
Conclusion
Our results demonstrated a panel of plasma metabolites from numerous physiologically significant metabolic pathways (i.e. purine metabolism, valine, leucine and isoleucine degradation and tryptophan metabolism) can be integrated into a PLS-DA model for MetS detection. Furthermore, with the enhancement provided by the clinical criteria of MetS, this metabolic profiling approach can potentially serve as a novel tool for MetS detection and monitoring, and provide useful metabolic information for future interventions targeting obesity and MetS. While these findings from a small sample size are promising, further validation and prospective studies using a larger patient cohort will be needed to substantiate the results, verify the important biological changes of these key metabolites, and determine the association of pathophysiology of MetS to these metabolite biomarkers.
Authors’ contributions
JZ, KDB and RSB designed the experiment, FZ, MX and JZ conducted the experiments, all authors participated in the interpretation of the studies, analysis of the data, and writing and review of the manuscript.
Supplementary Material
Acknowledgements
We'd like to extend our gratitude to the study participants.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This study was supported by Miami University (to JZ) and a grant from the National Dairy Council (to RSB).
References
- 1.Vidal-Puig A. The Metabolic Syndrome and its Complex Pathophysiology. In: Orešič M, Vidal-Puig A. (eds). A systems biology approach to study metabolic syndrome, Cham: Springer International Publishing, 2014, pp. 3–16. [Google Scholar]
- 2.Aguilar M, Bhuket T, Torres S, Liu B, Wong RJ. PRevalence of the metabolic syndrome in the United States, 2003–2012. JAMA 2015; 313: 1973–4. [DOI] [PubMed] [Google Scholar]
- 3.Alberti KGMM, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, Fruchart J-C, James WPT, Loria CM, Smith SC. Harmonizing the metabolic syndrome. A Joint Interim Statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 2009;120:1640–5. [DOI] [PubMed]
- 4.Kaur J. A comprehensive review on metabolic syndrome. Cardiol Res Practice 2014; 2014: 943162–943162. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Paredes RM, Quinones M, Marballi K, Gao X, Valdez C, Ahuja SS, Velligan D, Walss-Bass C. Metabolomic profiling of schizophrenia patients at risk for metabolic syndrome. Int J Neuropsychopharmacol 2014; 17: 1139–48. [DOI] [PubMed] [Google Scholar]
- 6.Suhre K, Meisinger C, Döring A, Altmaier E, Belcredi P, Gieger C, Chang D, Milburn MV, Gall WE, Weinberger KM, Mewes H-W, Hrabé de Angelis M, Wichmann HE, Kronenberg F, Adamski J, Illig T. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One 2010; 5: e13953–e13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hyötyläinen T. Metabolomics in the systems-level study of the metabolic syndrome. In: Orešič M, Vidal-Puig A. (eds). A systems biology approach to study metabolic syndrome, Cham: Springer International Publishing, 2014, pp. 213–36. [Google Scholar]
- 8.Ballard KD, Mah E, Guo Y, Pei R, Volek JS, Bruno RS. Low-fat milk ingestion prevents postprandial hyperglycemia-mediated impairments in vascular endothelial function in obese individuals with metabolic syndrome. J Nutr 2013; 143: 1602–10. [DOI] [PubMed] [Google Scholar]
- 9.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972; 18: 499–502. [PubMed] [Google Scholar]
- 10.Zhu J, Djukovic D, Deng L, Gu H, Himmati F, Chiorean EG, Raftery D. Colorectal cancer detection using targeted serum metabolic profiling. J Proteome Res 2014; 13: 4120–30. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J, Djukovic D, Deng L, Gu H, Himmati F, Abu Zaid M, Chiorean E, Raftery D. Targeted serum metabolite profiling and sequential metabolite ratio analysis for colorectal cancer progression monitoring. Anal Bioanal Chem 2015; 407: 7857–63. [DOI] [PubMed] [Google Scholar]
- 12.Schelli K, Rutowski J, Roubidoux J, Zhu J. Staphylococcus aureus methicillin resistance detected by HPLC-MS/MS targeted metabolic profiling. J Chromatogr B Epub ahead of print 3 June 2016. DOI: 10.1016/j.jchromb.2016.05.052. [DOI] [PubMed] [Google Scholar]
- 13.Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr 2006; 1125: 76–88. [DOI] [PubMed] [Google Scholar]
- 14.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion–switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc 2012; 7: 872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res 2015; 43: W251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larive CK, Barding GA, Dinges MM. NMR spectroscopy for metabolomics and metabolic profiling. Anal Chem 2015; 87: 133–46. [DOI] [PubMed] [Google Scholar]
- 17.Xiao JF, Zhou B, Ressom HW. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. TrAC, Trends Anal Chem 2012; 32: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felig P, Wahren J. Influence of endogenous insulin secretion on splanchnic glucose and amino acid metabolism in man. J Clin Invest 1971; 50: 1702–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O’Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med 2011; 17: 448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Xu Z, Lu X, Yang X, Yin P, Kong H, Yu Y, Xu G. Comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry for metabonomics: biomarker discovery for diabetes mellitus. Anal Chim Acta 2009; 633: 257–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.