Abstract
The clinical benefits of HIV-1 non-nucleoside reverse transcriptase (RT) inhibitors (NNRTIs) are hindered by their unsatisfactory pharmacokinetic (PK) properties along with the rapid development of drug-resistant variants. However, the clinical efficacy of these inhibitors can be improved by developing compounds with enhanced pharmacological profiles and heightened antiviral activity. We used computational and structure-guided design to develop two next-generation NNRTI drug candidates, compounds I and II, which are members of a class of catechol diethers. We evaluated the preclinical potential of these compounds in BALB/c mice because of their high solubility (510 µg/ml for compound I and 82.9 µg/ml for compound II), low cytotoxicity, and enhanced antiviral activity against wild-type (WT) HIV-1 RT and resistant variants. Additionally, crystal structures of compounds I and II with WT RT suggested an optimal binding to the NNRTI binding pocket favoring the high anti-viral potency. A single intraperitoneal dose of compounds I and II exhibited a prolonged serum residence time of 48 hours and concentration maximum (Cmax) of 4000- to 15,000-fold higher than their therapeutic/effective concentrations. These Cmax values were 4- to 15-fold lower than their cytotoxic concentrations observed in MT-2 cells. Compound II showed an enhanced area under the curve (0–last) and decreased plasma clearance over compound I and efavirenz, the standard of care NNRTI. Hence, the overall (PK) profile of compound II was excellent compared with that of compound I and efavirenz. Furthermore, both compounds were very well tolerated in BALB/c mice without any detectable acute toxicity. Taken together, these data suggest that compounds I and II possess improved anti-HIV-1 potency, remarkable in vivo safety, and prolonged in vivo circulation time, suggesting strong potential for further development as new NNRTIs for the potential treatment of HIV infection.
Introduction
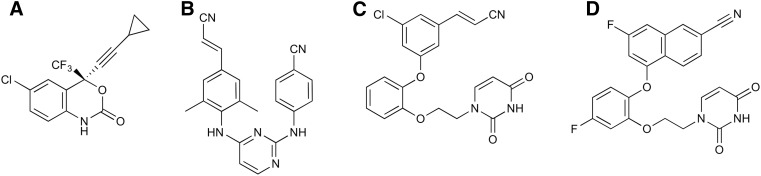
Globally, it is estimated that 36.7 million people are infected with HIV, and progression of the infection to AIDS causes over 1.1 million complications-related deaths per year. The current standard of care for therapeutically treating HIV infection is a minimum of three different antiretroviral drugs as a part of highly active antiretroviral therapy. Non-nucleoside reverse transcriptase (RT) inhibitors (NNRTIs) are major components of highly active antiretroviral therapy. These drugs significantly inhibit the catalytic function of HIV-RT by binding in an allosteric binding pocket 10 Å away from the RT polymerase active site and affect the chemical step of DNA synthesis (Spence et al., 1995, 1996; Fulco and McNicholl, 2009; de Béthune, 2010). Two of the five Food and Drug Administration–approved NNRTIs, efavirenz and rilpivirine (Fig. 1, A and B), are key components of the Food and Drug Administration–approved fixed-dose therapies Atripla, Complera, and Odefsey (Permpalung et al., 2012; Fellner, 2016).
Fig. 1.
Chemical structures of (A) efavirenz, (B) rilpivirine, (C) compound I, and (D) compound II.
In addition to the problem of promoting drug resistance, the clinical benefits of both of these drugs are hindered by their unsatisfactory pharmacokinetic (PK) properties. Their high cytotoxicity and poor aqueous solubility lead to low bioavailability and difficulties in formulation that require high dosage (Huang et al., 2015). Additionally, rapid blood clearance and insufficient drug concentration in vivo necessitates higher dosing, which results in high pill burden, lowered patient compliance, and development of cross-resistance between drug classes (Delaugerre et al., 2001). Furthermore, efavirenz’s low genetic barrier to resistance requires high-level adherence to therapy for efficacy (Delaugerre et al., 2001; Riddler et al., 2008). HIV-infected patients receiving efavirenz also suffer from side effects such as depression, insomnia, skin rash, lipodystrophy, and lipoatrophy (Riddler et al., 2008; de Waal et al., 2013). On the other hand, rilpivirine prescription requires a heightened vigilance in clinical practice due to its unfavorable interaction with acid suppressant medication (Janssen et al., 2005; Rathbun and Liedtke, 2011). Furthermore, as a substrate of CYP3A4, rilpivirine cannot be coadministered with drugs that are CYP3A4 inducers (Janssen et al., 2005; Riddler et al., 2008). Moreover, preclinical studies with rilpivirine (Edurant) reveal inhibition of the hERG ion channel, and clinical trials in Canada demonstrating prolongation of QT intervals in patients at higher dosing have prompted concerns of cardiotoxicity (http://www.hc-sc.gc.ca/dhp-mps/prodpharma/sbd-smd/drug-med/sbd_smd_2011_edurant_137484-eng.php; Summary Basis of Decision, 137484, Health Canada, Ottawa, Ontario, Canada). Therefore, development of alternative NNRTIs may prove beneficial for HIV-infected individuals, especially those who have experienced virologic failure.
To address the poor PK properties and issues of toxicity with currently approved NNRTIs, using computational and structure-guided design we successfully developed two next-generation NNRTI drug candidates, referred to as compounds I and II (Fig. 1, C and D). These compounds belong to a potent class of inhibitors known as catechol diethers, which possess better aqueous solubility, low cytotoxicity, and increased potency toward wild-type (WT) and resistant strains of HIV-1 (Lee et al., 2013, 2014). Compound I demonstrates picomolar activity (310 pM) against WT RT and low nanomolar activity toward clinically challenging variants (Lee et al., 2014), likely resulting from its molecular flexibility at the NNRTI binding pocket (Frey et al., 2015). Compound II possesses low nanomolar potency (1.9 nM) for WT RT and has very low cytotoxicity (>100 µM) (Lee et al., 2014). These encouraging findings prompted us to further investigate compounds I and II as potential anti-HIV drug candidates. We explored the PK profiles, single-dose safety, and tolerance of both compounds in BALB/c mice. In addition, the compounds were evaluated for potential off-target effects on a panel of 34 molecular targets including ion channels, receptors, and cytochrome P450 enzymes. Our pharmacological goal was to achieve serum concentrations that are expected to provide complete inhibition of RT based on a cell culture model of HIV inhibition. Indeed, PK analyses showed that the Cmax values for both compounds in BALB/c mice were several-fold higher than those required for inhibition of WT and resistant strains of HIV virus in cellular assays. Additionally, both compounds demonstrated longer serum residence times, suggesting that a once daily dosing might be sufficient to achieve the desired therapeutic serum concentration. The PK data were further supported by crystallographic studies with compound II and WT RT. Similar to compound I (Frey et al., 2015), compound II makes extensive contacts with residues within the non-nucleoside binding pocket (NNBP), further supporting its high potency and suggesting that compound II could be active against a range of clinically challenging variants. Hence, this study provides strong evidence that compounds I and II are promising anti-HIV drug candidates and should be tested further to determine their efficacy in animal models of HIV infection.
Materials and Methods
Synthesis of Compounds I and II.
The synthetic procedures for preparation of compounds I and II have been previously described in detail (Bollini et al., 2011; Lee et al., 2014).
Animals.
Six- to eight-week-old female BALB/c mice from Jackson Laboratories (Jackson Laboratories, Farmington, CT) were used. All mice were housed in cages containing water, food, and bedding. All procedures were approved by the Yale University Animal Care and Use Committee.
Inhibition Assay with Pico Green and Solubility Measurements for Compounds I and II.
Inhibition assays were carried out with the PicoGreen Enzcheck Reverse Transcriptase Assay Kit (Life Technologies, Eugene, OR) as described previously (Gray et al., 2015). The compound solubility was measured using a shake-flask procedure as described previously (Bollini et al., 2013; Lee et al., 2013)
Anti-HIV Activity in Infected TZM-bl Cells.
The in vitro anti-HIV-1 activity of compound II was determined using a luciferase reporter gene assay (Montefiori, 2009). Twenty-four hours prior to infection, 20,000 TZM-bl cells were seeded per well in a 48-well plate. Two hours before infection, the cells were incubated with medium containing compound II at concentrations ranging from 0.1 pM to 1 µM at 37°C. The cells were infected with HIV-1 JR-CSF (multiplicity of infection, 0.1) 2 hours later and cultured for 48 hours. Luciferase activity in cell lysates was measured using a luciferase assay kit, following the manufacturer’s protocol (Promega, Madison, WI).
In Vitro Pharmacological Profiling.
In vitro pharmacological profiling of efavirenz and compounds I and II were carried out by the Eurofins Groups (Cerep and Eurofins Panlabs; Taipei, Taiwan) in a panel of enzyme and radio-ligand assays with 34 targets. The detailed information of the assays can be obtained by clicking on the direct link provided in Supplemental Table 1. These links will direct the readers to the Eurofins Panlabs web page, where information about each assay has been provided in detail.
Crystallization, Data Collection, Structure Determination, and Refinement.
Recombinant RT52A enzyme was expressed and purified to homogeneity using methods described previously (Das et al., 2008; Frey et al., 2012). Crystals of RT52A in the complex with compound II were prepared using similar methods as for catechol diether complexes (Frey et al., 2012). The final optimized condition for crystal growth consisted of 18% (w/v) polyethylene glycol 8000, 100 mM ammonium sulfate, 15 mM magnesium sulfate, 5 mM spermine, and 50 mM 4-morpholineethanesulfonic acid (pH 6.0). Crystals were transferred to a cryo-solution containing 27% (v/v) ethylene glycol and flash cooled with liquid nitrogen.
Diffraction data for the best crystals were collected at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL) on beamline 24-ID-E through the Northeastern Collaborative Access Team. High-resolution data sets for the best diffracting crystals were processed with HKL2000 (Otwinowski and Minor, 1997). To obtain phases, molecular replacement was performed with Phaser (McCoy et al., 2007) using a previously determined RT:NNRTI complex (Protein Data Bank code: 4WE1) as the search model (Lee et al., 2014). The software program Coot (Emsley et al., 2010) was used for model building into the electron density. Structure was refined using Phenix (Adams et al., 2010) until acceptable R-factors, geometry statistics (ideal root-mean-square deviation for bonds and angles), and Ramachandran statistics were achieved. Iterative build, composite omit electron density maps were generated using Phenix Autobuild (Terwilliger et al., 2008). The PyMOL molecular viewer (DeLano, 2009) was used to visualize and analyze the structure and generate figures. Crystallography programs were compiled by SBGrid (Morin et al., 2013). The atomic coordinates and structure factors are deposited in the Protein Data Bank with codes 5TW3.
Pharmacokinetic Studies of Compounds I and II in BALB/c Mice.
Stock solutions of compounds I and II were diluted in phosphate-buffered saline containing 10% Tween 80 and administered i.p. at doses of 20 and 100 mg/kg. A cohort of three mice per group was used for PK measurements. A sample size of three animals was used based on previously published PK studies with NNRTIs. Additionally, lack of mortality or morbidity in our pilot experiments conducted with different doses (5, 10, 20, and 100 mg/kg) of compounds I and II suggested that n = 3 would be sufficient to obtain statistically significant data without using a higher number of animals. Serum samples were obtained from blood collected from the ocular venous plexus by retro-orbital venipuncture at various time points after i.p. injection and used for analysis as detailed subsequently.
Toxicity Studies of Compound II in BALB/c Mice.
In acute toxicity studies, three mice were treated with single i.p bolus injection of vehicle (dimethylsulfoxide) or compound II at 100 mg/kg. Mice were allowed free access to food and water throughout the experiments and were monitored daily for 3 days for morbidity and mortality.
Determination of Serum Compound I and II Concentrations by Quantitative High-Performance Liquid Chromatography (HPLC).
Efavirenz (5 µg/ml) and nevirapine (1 µg/ml) were used as internal standards for the analysis of compounds I and II, respectively. The compounds were extracted following previously published extraction protocols with modifications (Weller et al., 2007). Next, 200 µl of 0.01 M sodium hydroxide was added to all samples and vortexed briefly followed by the addition of 1 ml of methyl-tert-butyl-ether. The samples were then vortexed briefly and placed on ice for 5 minutes to separate the methyl-tert-butyl-ether layer. The methyl-tert-butyl-ether layer was collected in a glass tube and dried under nitrogen at room temperature. The residue was reconstituted in 50 µl of acetonitrile and 25 µl of the reconstituted sample was subjected to analytical HPLC.
The serum levels of compounds I and II were determined using the 210 Varian Prostar HPLC system (Agilent Technologies) (Andover, MA) equipped with 218/SD-1 pumps, a PS 325 UV-visible detector, a fraction collector (440 LC), and a computer with the Open Laboratory CDS software (ChemStation edition) for data analysis (Agilent Technologies). The C-18 analytical column (Varian Polaris 5, 150 × 4.6 mm) was equilibrated prior to data collection with 1% A and 99% B (where A is HPLC grade water and B is HPLC grade acetonitrile). The employed linear gradient mobile phase (flow rate = 1.0 ml/min) was as follows: 95% A/5% B at 0 minutes, 1% A/99% B at 30 minutes, and 95% A/5% B at 35 minutes. The detection wavelengths were 280 and 272 nm for compounds I and II, respectively, and the response time was set at 2.0 seconds. The lowest limits of detection were 25 and 50 ng/ml for compounds I and II, respectively. Good linearity was observed between concentrations ranging from 50 ng/ml to 100 µg/ml (for compound I) and from 25 ng/ml to 100 µg/ml (for compound II) in serum. The calculated extraction efficiency was 65%–70% for both compounds.
Pharmacokinetic Analyses.
The data were plotted as a concentration-time curve using PRISM 7.0 (GraphPad Software Inc, LaJolla, CA). The predicted area under the curve (AUC) of the concentration of drug in blood plasma against time (AUCpredicted) was calculated based on the linear trapezoid method (Bourget and Delouis, 1993). The maximum concentration (Cmax) of a drug observed in serum after its administration and the maximum time taken to reach Cmax were also calculated from the concentration-time curve. The total body clearance (CL) value was obtained by the following equation (Ratain and Plunkett, 2003):
| (1) |
Results
HIV-1 Inhibitory Activity and Solubility Measurements for Compounds I and II.
The intrinsic enzymatic inhibitory activities of compounds I and II against WT RT are compared with rilpivirine and efavirenz in Table 1. Consistent with their high potency observed in the cell-based assays (Lee et al., 2014), both compounds I and II showed single digit nanomolar IC50 values against WT RT. Table 1 also compares the solubility values of compounds I and II with that of rilpivirine and efavirenz. The solubility of rilpivirine was below 1 µg/ml, which is well outside the normal range of 4–4,000 µg/ml (Jorgensen and Duffy, 2002; Bollini et al., 2013). Compound I had a much higher solubility of 510 µg/ml, whereas the solubility value for compound II was slightly better than efavirenz, 82.9 versus 68 µg/ml. The C log P values for compounds I and II were below 4 and in the normal range of 0–5 for oral drugs (Jorgensen, 2009), while efavirenz was 4.6 and rilpivirine was above 5 (Lee et al., 2013, 2014).
TABLE 1.
HIV-RT inhibitory activity (IC50 in nM), experimental aqueous solubility (in µg/ml) at pH 6.5, and computed C log P
Data are the mean ± S.D. values from three different experiments involving triplicate measurements.
Binding Mode of Compound II in WT (RT).
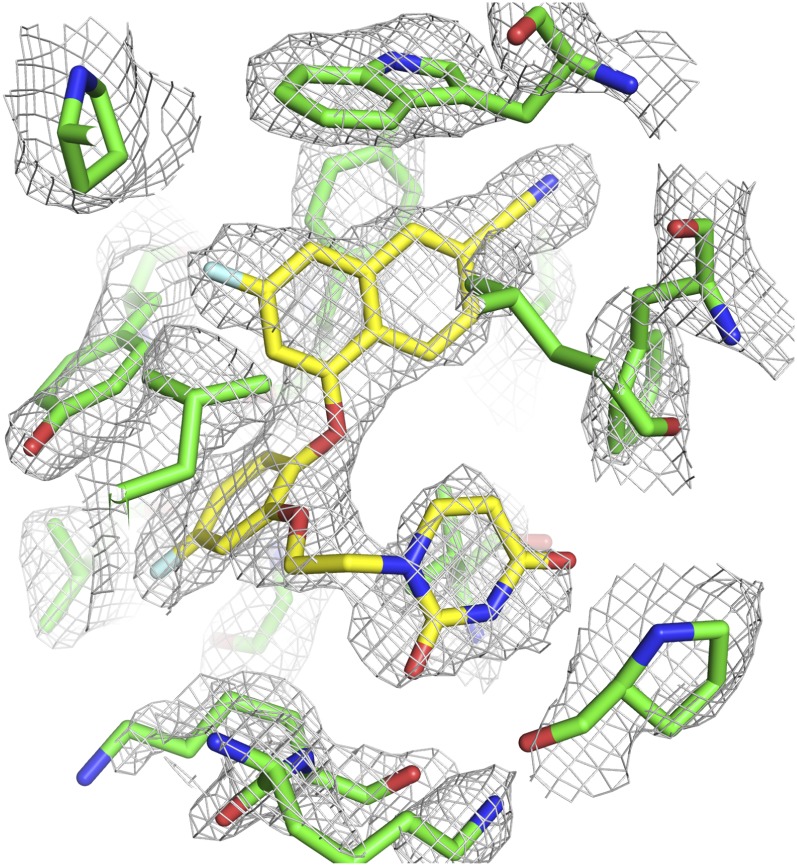
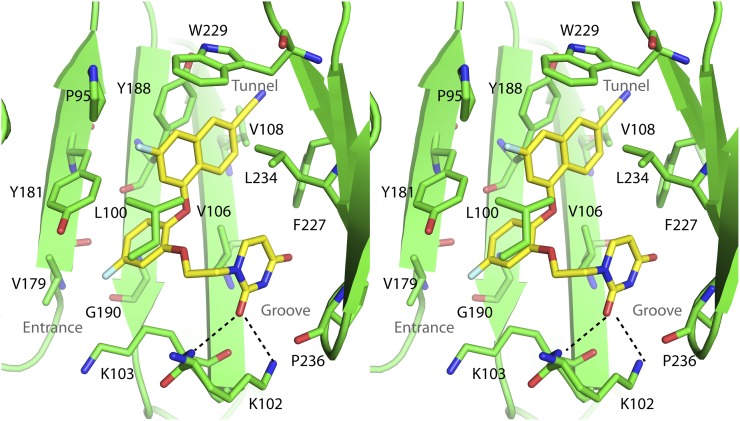
To elucidate the specific interactions between compound II and residues in the NNBP, the crystal structure of the RT:compound II complex was determined. The best crystal diffracted to amplitudes extending to a resolution of 2.85 Å, and phases were obtained via molecular replacement using the structure of RT in the complex with the nonhalogenated version of compound II (Protein Data Bank code: 4WE1) as a search model (Lee et al., 2014). Data collection and refinement statistics are listed in Table 2. The electron density of compound II can be clearly seen in the NNBP (Fig. 2). The overall conformation of compound II and the binding pocket is very similar to what we have observed previously with other RT:catechol diether complexes (Frey et al., 2012, 2014, 2015; Lee et al., 2013, 2014) (Fig. 3). The NNBP adopts an open-cleft conformation and features three channels described as entrance, groove, and tunnel (Ekkati et al., 2012). The naphthyl moiety of compound II forms extensive van der Waals interactions with P95, L100, V108, Y188, W229, and L234. In particular, the naphthyl ring forms an offset face-to-face π-π interaction with Y188 and a face-to-edge π-π interaction with W229. The cyano group attached to the naphthyl resides in the tunnel region protruding to the polymerase active site. The central catechol ring forms van der Waals interactions with K103 and Y181 and with the backbone of K101, Y188, and G190. The ring forms an offset face-to-face π-π interaction with Y181. The F on the catechol ring protrudes into the entrance site and contacts K103 as well as V179. The uracil moiety resides in the groove region and contacts K102, K103, F227, L234, H235, P236, and Y318. The C2 carbonyl forms weak hydrogen bonds with the side chain amino group of K102 (3.5 Å N–O distance) and backbone amide of K103 (3.3 Å N–O distance).
TABLE 2.
Data collection and refinement statistics for RT (WT) in the complex with compound II. For details on MolProbity, see Chen et al., 2010.
| Data Collection and Refinement Statistic | RT (Compound II) |
|---|---|
| PDB code | 5TW3 |
| Resolution limit (Å) | 2.85 |
| X-ray source | APS |
| X-ray source | 24ID-E |
| Wavelength (Å) | 0.97915 |
| Space group | C2 |
| Number of molecules in the asymmetric unit | 1 |
| Unit cell (Å) | a = 224.4, b = 69.5, c = 104.5 |
| Unit cell (°) | α = 90, β = 106.0, γ = 90 |
| Resolution range (Å) | 50.0–2.85 |
| Last shell (Å) | 2.90–2.85 |
| R-sym (last shell) | 0.069 (0.510) |
| Completeness (last shell) (%) | 99.4 (99.0) |
| Number of reflections (unique reflections) | 137490 (36173) |
| Redundancy (last shell) | 3.8 (3.8) |
| Average I/σ (last shell) | 24.2 (3.3) |
| Total number of atoms (protein, inhibitor, solvent, ions) | 7757, 32, 17, 1 |
| R-free | 0.2725 |
| R-factor | 0.2270 |
| Root-mean-square deviation bond length (Å) | 0.003 |
| Root-mean-square deviation bond angle (°) | 0.631 |
| Average B-factor (protein, inhibitor, solvent, ions) | 69.4, 53.9, 54.1, 87.3 |
| Ramachandran favored (MolProbity) (%) | 96.62 |
| Ramachandran allowed (MolProbity) (%) | 3.38 |
| Ramachandran outliers (MolProbity) (%) | 0 |
APS, Advanced Photon Source (Argonne National Laboratory, Argonne, IL); PDB, Protein Data Bank.
Fig. 2.
Omit, σA-weighted 2mFo-Fc electron density contoured to 1.0 σ for the RT:compound II. Compound II was omitted from the model to generate an iterative-build omit map using the original structure factors.
Fig. 3.
Stereo view of the crystal structure for compound II complexes with HIV-RT. Residues that interact with the inhibitor are shown as green sticks. Compound II is represented by yellow sticks. Black dotted lines indicate hydrogen bonds.
In Vitro Pharmacological Profiling.
In vitro pharmacological profiling was carried out to identify off-target effects responsible for high attrition rate in the drug discovery and development process. Our compounds along with efavirenz were subjected to a panel of 34 targets, which included various receptors, ion channels, enzymes, and hormones. A complete list of the targets evaluated for potential off-target effects is given in Supplemental Table 1.
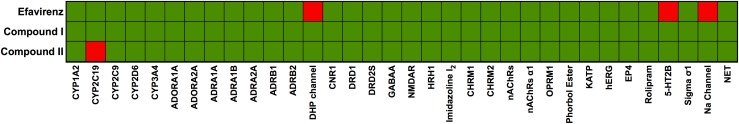
A heat map was generated based on the significant response obtained from these assays, as shown in Fig. 4, where the green squares represent less than 50% binding or inhibition and the red squares represent more than 50% inhibition. As shown in Fig. 4, in the case of efavirenz, significant response of more than 50% was noted for calcium channel L-Type, dihydropyridine (the DHP channel), serotonin (5-hydroxytryptamine) (5-HT2B), and sodium channel, site 2 (Na channel) assays, whereas compound I showed no adverse response to all targets tested. Similar to compound I, compound II also had no adverse response to any of the targets tested except for cytochrome P450 2C19 (CYP2C19) where a little over 50% response was seen.
Fig. 4.
In vitro pharmacological profiling of efavirenz, compound I, and compound II against targets for adverse drug reactions as described in Materials and Methods. Rows represent compounds tested and columns represent targets. Percentage inhibition at 10 µM concentration of the compounds is color -coded: <50% inhibition of target is labeled in green and >50% inhibition is labeled in red. Abbreviations: CYP, cytochrome P450; ADOR, adenosine receptor; ADR, adrenergic receptor; DHP, dihydropyridine or calcium channel L-type; CNR1, cannabinoid receptor; DRD, dopamine receptor; NMDAR, glutamate receptor; HRH1, histamine receptor; imidazole I2, imidazole I2 receptor; CHRM, muscarinic; nACHR, nicotinic acetylcholine; OPRM, opiate µ phorbol ester receptor, KATP, potassium channel; hERG, human ether-à-go-go-related gene; EP4, prostanoid receptor; rolipram, phosphodiesterase-4 inhibitor; 5-HT2B, serotonin receptor 2B; sigma σ1, sigma σ1receptor; Na channel, sodium channel; NET, norepinephrine transporter.
Pharmacokinetics of Compound I in BALB/c Mice.
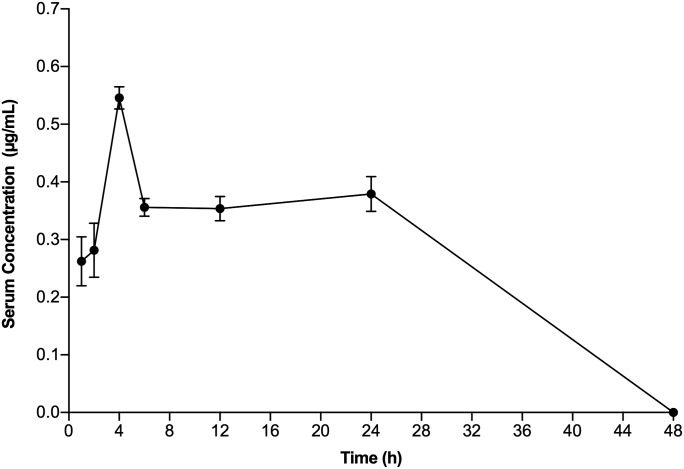
The serum concentration-time profile showed that the maximum concentration of compound I in BALB/c mice administered a single dose of 20 mg/kg by the i.p. route resulted in Cmax values of 0.54 ± 0.02 µg/ml at 4 hours that persisted for up to 24 hours and then slowly waned in the next 24 hours (Fig. 5; Table 3). An AUC0-last value of 13.1 ± 0.4 µg/h/ml was obtained for compound I by the linear trapezoid method. The CL level calculated using eq. 1 was 25.4 min/ml/kg. Thus, a 4000-fold higher serum concentration of compound I compared with its EC50 value in MTT (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide) cells was rapidly achieved and maintained for more than 24 hours after i.p. administration of the 20 mg/kg dose of compound I.
Fig. 5.
Pharmacokinetics of compound I in BALB/c mice. Serum levels of compound I were monitored after i.p. administration of 20 mg/kg compound I in three BALB/c mice. Blood samples were collected at 0.5, 1, 2, 4, 6, 12, 24, and 48 hours and analyzed as described in Materials and Methods. Data points represent mean ± S.D.
TABLE 3.
Pharmacokinetic parameters of compounds I and II in comparison with efavirenz in BALB/c mice
Data are the mean ± S.D. values of triplicate measurements; n represents the number of mice.
| Pharmacokinetic Parameter | Compound I (n = 3) | Compound II (n =3) | Efavirenza (n = 3) |
|---|---|---|---|
| Dose (mg/kg) | 20 | 20; 100 | 20 |
| Cmax (µg/ml) | 0.54 ± 0.02 | 11.8 ± 0.7; 52.3 ± 5.7 | 1.8 ± 0.6 |
| Tmax (hour) | 4 | 4; 4 | 4 |
| AUC0–last (µg/h/ml) | 13.1 ± 0.4 | 201.7 ± 23.4; 896.4 ± 50.71 | 20.4 ± 21.1 |
| CL (ml/min/kg) | 25.4 | 1.6; 1.8 | 16.3 |
Tmax, time taken to reach Cmax.
Pharmacokinetics and Toxicity of Compound II in BALB/c Mice.
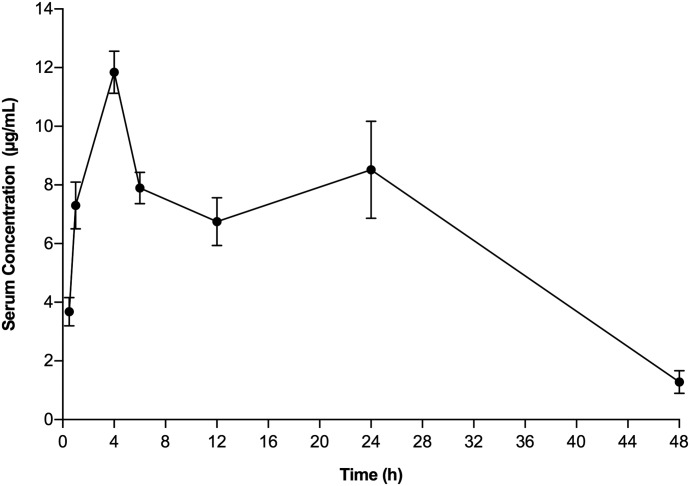
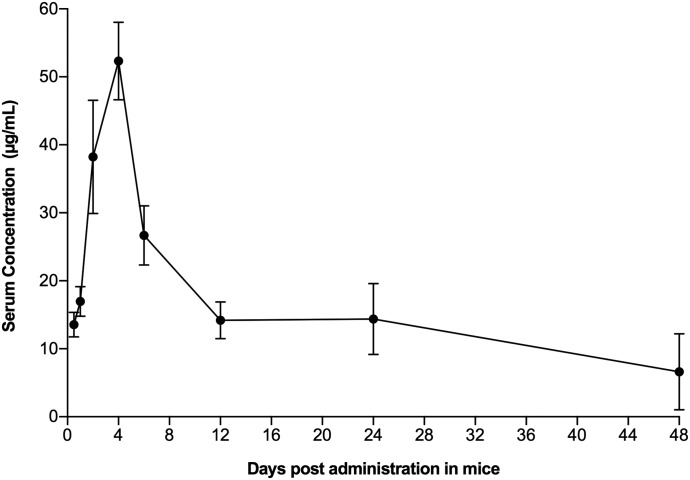
The serum concentration-time curve for compound II determined at the 20 mg/kg dose showed Cmax values of 11.8 ± 0.7 µg/ml at 4 hours (Fig. 6; Table 3). The AUC0–last values of 201.7 ± 23.4 µg/h/ml and CL of 1.6 min/ml/kg were calculated similarly. Thus, higher levels of compound II in serum were rapidly achieved and maintained for almost 48 hours after i.p. administration of the 20 mg/kg dose of compound II. Additionally, we also looked at the PK of a single high dose of 100 mg/kg of compound II in BALB/c mice (Fig. 7), where Cmax values of 52.3 ± 5.3 µg/ml at 4 hours and AUC0–last values of 896.4 ± 50.71 µg/h/ml were obtained. A CL value of 1.8 min/ml/kg was observed, which was similar to that seen with the 20 mg/kg dose. These mice were also observed for 96 hours for any sign of toxicity in terms of morbidity or mortality. All of the treated mice remained healthy throughout the 96-hour observation period, with no evidence of morbidity or clinical distress, suggesting that the single-bolus dose of compound II was nontoxic to BALB/c mice at dose levels of 100 mg/kg.
Fig. 6.
Pharmacokinetics of compound II in BALB/c mice. Serum level of compound II was monitored after i.p. administration of 20 mg/kg compound I in three BALB/c mice. The blood samples were collected at 0.5, 1, 2, 4, 6, 12, 24, and 48 hours and analyzed as described in Materials and Methods. Data points represent mean ± S.D.
Fig. 7.
Pharmacokinetics of compound II in BALB/c mice administered at 100 mg/kg dose. Serum level of compound I was monitored after i.p. administration of 100 mg/kg compound I in three BALB/c mice. The blood samples were collected at 0.5, 1, 2, 4, 6, 12, 24, and 48 hours and analyzed as described in Materials and Methods. Data points represent mean ± S.D.
Anti-HIV Activity of Compound II in Infected TZM-bl Cells.
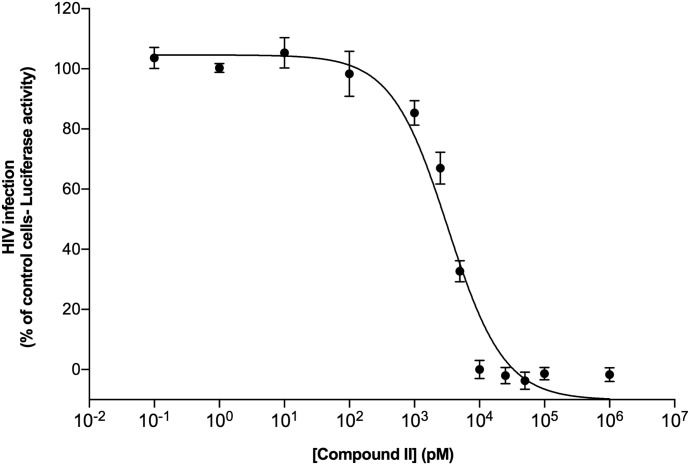
Our previous assay for HIV infection was based on cell death induced by HIV replication in MT2 cells and cell viability measured in MTT assays as readout for antiviral potency (Lee et al., 2013, 2014). We also performed antiviral assays in a TZM-bl indicator cell line that expresses a luciferase reporter driven by the HIV long terminal repeats and thus a linear measure for HIV infection. TZM-bl cells were preincubated with compound II for 2 hours followed by infection with the HIV-1 JR-CSF strain at a multiplicity of infection of 0.1 to assess the antiviral potency of compound II. Figure 8 depicts the concentration-dependent inhibition of HIV-1 infection and an EC50 value of 3.5 nM for compound II, comparable to the low EC50 value observed in the MTT assays (Lee et al., 2013). Effective inhibition of HIV JR-CSF infected cells reinforced the fact that compound II has potent inhibitory activity against HIV-1. These results encourage animal infection experiments, given the favorable toxicity profile and PK of compound II.
Fig. 8.
HIV-1 inhibition by compound II in TZM-bl cells. TZM-bl cells presented with compound II at varying concentrations were infected with HIV-1 (JRCSF strain). Virus infection was measured 2 days later by measuring luciferase activity in the cells. The percentage of the luciferase activity was normalized relative to the HIV-1 infected control TZM-bl cells that were not exposed to compound II. The values are mean ± S.D. from three different experiments involving triplicate measurements.
Discussion
Compounds I and II were derived from a low-micromolar hit in a virtual screen by docking (Bollini et al., 2011; Lee et al., 2014). The hit, which contained a diphenylmethane substructure, was subjected to extensive analyses using free-energy perturbation calculations for model complexes of analogs of the hit with HIV-RT. The results led us to prepare substituted catechol diethers with a cyanovinyl group as in compound I, which yielded EC50 values as low as 55 pM in the infected T-cell assays. Subsequently, crystal structures were obtained that confirmed the correctness of the modeled complexes. Additional free-energy perturbation–guided efforts then addressed possible replacements of the cyanovinylphenyl substructure, which led to discovery of additional novel compounds, including compound II (Bollini et al., 2011; Lee et al., 2014).
These compounds are highly effective NNRTIs that have strong potential for development as new anti-HIV-1 drugs with improved antiviral efficacy and drug resistance profiles (Lee et al., 2013, 2014; Frey et al., 2014, 2015; Gray et al., 2015). As noted in Table 1, these compounds are suitable drug candidates for further preclinical studies due to their low effective intrinsic inhibitory activities at low nanomolar concentrations, better solubility profiles, and low C log P values compared with rilpivirine (Janssen et al., 2005; Sun et al., 2012) and efavirenz (Lee et al., 2013, 2014; Frey et al., 2014). Our in vitro pharmacological profiling of these compounds against a broad range of targets, which are associated with off-target effects in humans, has shown that these compounds exhibit little to no adverse effects on these targets (Fig. 4). Importantly, compounds I and II exhibited no adverse reaction to major targets such as the hERG channel, the blockade of which is responsible for major cardiac arrhythmias (Bowes et al., 2012). Furthermore, these compounds show no inhibition of CYP3A4, which is the major cytochrome P450 enzyme responsible for the metabolism of the majority of currently approved drugs. Even though compound II showed inhibition of CYP2C19, it was at a high concentration of 10 μM and CYP2C19 metabolizes only 10%–15% of the marketed drug in contrast to CYP3A4 (Nazir et al., 2016; Wang et al., 2016). In comparison, efavirenz in our panel showed inhibition of binding of ligands to dihydropyridine and Na channels and 5-hydroxytryptamine receptors, explaining some of the side effects seen with efavirenz treatment. The excellent in vitro pharmacological qualities of these compounds led us to evaluate the safety and toxicity profile, and the PK profiles, of these compounds in a detailed in vivo PK analysis in BALB/c mice.
The serum concentration-time profiles after intraperitoneal administration of compounds I and II showed that both compounds were rapidly absorbed, achieving maximum concentrations that were 4000- and 15,000-fold higher, respectively, than their therapeutic/effective concentrations (EC50) and 17- and 5-fold lower, respectively, than the cytotoxic concentration (CC50) (Lee et al., 2013, 2014). Interestingly, the serum concentration-time profiles of compounds I and II were notable in that concentrations of 2000- to 4000-fold above the required therapeutic range were achieved in serum as early as 0.5 hours postinjection and were maintained for more than 24 hours. The observed Cmax value of compound I was comparable to the Cmax value of efavirenz observed in a similar PK study in BALB/c mice conducted by Destache et al. (2010). Unlike compound I, the Cmax value of compound II was 10-fold higher compared with that of efavirenz (Destache et al., 2010) (Table 3). A plateau in serum concentration was observed for both compounds after 6 hours postinjection, which was sustained for a period of 24 hours (Figs. 5 and 6). The plateau was followed by a rapid decline to nondetectable levels by 48 hours for compound I. Interestingly, a concentration of 2.9 µg/ml was detected for compound II at 48 hours, which was ∼3-fold higher than the efavirenz level (Destache et al., 2010). Hence, the PK data suggest that the longer serum residence time of these compounds makes them amenable to a daily dose regimen since plasma concentrations of the drug would be maintained in the therapeutic range under these conditions. Additionally, no morbidities and mortalities were observed in the compound-treated mice, suggesting that the Cmax values achieved were nontoxic.
Further analyses of the PK data showed that the AUC0–last and total body CL values for compound I were similar to those of efavirenz (Table 3). However, for compound II a much higher AUC0–last value of 201.7 µg/h/ml and a much lower CL level of 1.09 ml/min/kg compared with the CL levels of compound I (25.4 ml/min/kg) and efavirenz (16.3 ml/min/kg) were noted in our PK analysis. The larger AUC0–last values and lower CL value could suggest that a higher volume of distribution can be achieved by compound II and also that compound II is subjected to slower metabolism, thereby achieving sustained levels for maintaining good virus control.
The PK data were further supported by crystallographic studies with RT:compound II, where compound II made extensive contacts with residues within the NNBP, further explaining its high potency and suggesting that compound II could be active against a range of NNBP mutants. Notably, the naphthyl group of compound II interacts with both P95 and W229, located at the p66/p51 dimerization interface and primer grip, respectively. Mutations with these two residues might destabilize the RT dimer or interfere with the correct placement of the primer in the active site, leading to a significant decrease in RT activity and therefore viral fitness. Indeed, clinical studies and in vitro mutational analysis have shown that both residues are immutable (Pelemans et al., 2000; Auwerx et al., 2005; Ceccherini-Silberstein et al., 2005), suggesting that compound II’s interactions with these two residues will be able to compensate for potential lost interactions due to other NNBP mutations. Moreover, although compound II forms a hydrogen bond with the frequently mutated K103, the hydrogen bond donor is the backbone amide of K103, suggesting that the interaction is likely maintained in a number of K103 mutations. The preliminary crystal structures of compound II in the complex with the Y181C mutant and K103N/Y181C double mutant show that compound II binds to these two mutants in an almost identical conformation as in WT RT (compound II and NNBP residues for WT and mutants align to root-mean-square deviation <0.4). In particular, the π-π interaction between the naphthyl group and W229, the van der Waals interaction with P95, and the hydrogen bond between the uracil group and K103 or N103 are maintained in the mutant structures (unpublished data). The previous report that compound II can inhibit common NNRTI-resistant mutants with a single mutation (Y181C) or double mutations (K103N and Y181C) at low nanomolar concentrations (Lee et al., 2014) supports the present hypothesis that compound II may be effective against a number of HIV strains harboring different NNBP mutations in RT.
The good pharmacological profile of compound II compared with compound I and efavirenz makes it a great candidate for efficacy trials in animal models of HIV infection. Previously, it has been established that efficacy studies of various NNRTIs at 100 mg/kg produce more than 90% inhibition of the viral capsid core p24 protein (p24 antigen) production and higher AUC0–last values (Rabin et al., 1996; Stoddart et al., 2007). As such, we carried out a PK study for compound II at 100 mg/kg in BALB/c mice (Fig. 7). A 5-fold increase in the dose caused a Cmax increase of 4.4-fold and an AUC0–last increase of 4.4-fold (Table 3). These features coupled with constant plasma clearance (1.8 ml/min/kg) after high i.p. doses suggest that compound II follows linear PK. Since compound II was well tolerated by BALB/c mice over a period of 96 hours at this high concentration, the future efficacy studies could be carried out at a 100 mg/kg dose without any concern for toxicity. Additionally, compound II showed low nanomolar potency against JR-CSF strains of HIV-1 virus in the single-infectivity assay.
In summary, these findings suggest that nontoxic and therapeutic concentrations of compounds I and II can rapidly be achieved and maintained for prolonged periods in the serum of BALB/c mice. Additionally, these results strongly encourage further preclinical development of these potent catechol diether compounds and encourage studies of antiviral potency in animal models for HIV infection.
Abbreviations
- 5-HT2B
5-hydroxytryptamine
- AUC
area under the curve
- CL
clearance
- HPLC
high-performance liquid chromatography
- MTT
3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide
- NNBP
non-nucleoside binding pocket
- NNRTI
non-nucleoside reverse transcriptase inhibitor
- PK
pharmacokinetic
- RT
reverse transcriptase
- WT
wild type
Authorship Contributions
Participated in research design: Kudalkar, Beloor, Kumar, Anderson.
Conducted experiments: Kudalkar, Beloor, Chan.
Contributed new reagents or analytic tools: Lee, Jorgensen.
Performed data analysis: Kudalkar, Beloor, Chan.
Wrote or contributed to the writing of the manuscript: Kudalkar, Beloor, Chan, Jorgensen, Kumar, Anderson.
Footnotes
This work was supported in part by the National Institutes of Health [Grants AI44616, GM49551, AI112443, and AI122384] and the Ruth L. Kirschstein National Research Service Award Individual Postdoctoral Fellowship [AI122864]. This work is based on research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institutes of Health National Institute of General Medical Sciences [Grant P41 GM103403]. Crystal screening was conducted with support from the Yale Macromolecular X-ray Core Facility [1S10OD018007-01]. This research used resources from the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J, Van Nieuwenhove J, Rodríguez-Barrios F, de Castro S, Velázquez S, Ceccherini-Silberstein F, De Clercq E, Camarasa MJ, Perno CF, Gago F, et al. (2005) The N137 and P140 amino acids in the p51 and the P95 amino acid in the p66 subunit of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase are instrumental to maintain catalytic activity and to design new classes of anti-HIV-1 drugs. FEBS Lett 579:2294–2300. [DOI] [PubMed] [Google Scholar]
- Bollini M, Cisneros JA, Spasov KA, Anderson KS, Jorgensen WL. (2013) Optimization of diarylazines as anti-HIV agents with dramatically enhanced solubility. Bioorg Med Chem Lett 23:5213–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollini M, Domaoal RA, Thakur VV, Gallardo-Macias R, Spasov KA, Anderson KS, Jorgensen WL. (2011) Computationally-guided optimization of a docking hit to yield catechol diethers as potent anti-HIV agents. J Med Chem 54:8582–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourget P, Delouis JM. (1993) Review of a technic for the estimation of area under the concentration curve in pharmacokinetic analysis. Therapie 48:1–5. [PubMed] [Google Scholar]
- Bowes J, Brown AJ, Hamon J, Jarolimek W, Sridhar A, Waldron G, Whitebread S. (2012) Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nat Rev Drug Discov 11:909–922. [DOI] [PubMed] [Google Scholar]
- Ceccherini-Silberstein F, Gago F, Santoro M, Gori C, Svicher V, Rodríguez-Barrios F, d’Arrigo R, Ciccozzi M, Bertoli A, d’Arminio Monforte A, et al. (2005) High sequence conservation of human immunodeficiency virus type 1 reverse transcriptase under drug pressure despite the continuous appearance of mutations. J Virol 79:10718–10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Bauman JD, Clark AD, Jr, Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E. (2008) High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci USA 105:1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Béthune MP. (2010) Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: a review of the last 20 years (1989–2009). Antiviral Res 85:75–90. [DOI] [PubMed] [Google Scholar]
- DeLano WL (2009) PyMOL molecular viewer: Updates and refinements, in Proceedings of the 238th American Chemical Society National Meeting; 2009, Aug 16–20; Washington, DC.
- Delaugerre C, Rohban R, Simon A, Mouroux M, Tricot C, Agher R, Huraux JM, Katlama C, Calvez V. (2001) Resistance profile and cross-resistance of HIV-1 among patients failing a non-nucleoside reverse transcriptase inhibitor-containing regimen. J Med Virol 65:445–448. [PubMed] [Google Scholar]
- Destache CJ, Belgum T, Goede M, Shibata A, Belshan MA. (2010) Antiretroviral release from poly(DL-lactide-co-glycolide) nanoparticles in mice. J Antimicrob Chemother 65:2183–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waal R, Cohen K, Maartens G. (2013) Systematic review of antiretroviral-associated lipodystrophy: lipoatrophy, but not central fat gain, is an antiretroviral adverse drug reaction. PLoS One 8:e63623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekkati AR, Bollini M, Domaoal RA, Spasov KA, Anderson KS, Jorgensen WL. (2012) Discovery of dimeric inhibitors by extension into the entrance channel of HIV-1 reverse transcriptase. Bioorg Med Chem Lett 22:1565–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellner C. (2016) Pharmaceutical approval update. PT 41:220–221. [PMC free article] [PubMed] [Google Scholar]
- Frey KM, Bollini M, Mislak AC, Cisneros JA, Gallardo-Macias R, Jorgensen WL, Anderson KS. (2012) Crystal structures of HIV-1 reverse transcriptase with picomolar inhibitors reveal key interactions for drug design. J Am Chem Soc 134:19501–19503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey KM, Gray WT, Spasov KA, Bollini M, Gallardo-Macias R, Jorgensen WL, Anderson KS. (2014) Structure-based evaluation of C5 derivatives in the catechol diether series targeting HIV-1 reverse transcriptase. Chem Biol Drug Des 83:541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey KM, Puleo DE, Spasov KA, Bollini M, Jorgensen WL, Anderson KS. (2015) Structure-based evaluation of non-nucleoside inhibitors with improved potency and solubility that target HIV reverse transcriptase variants. J Med Chem 58:2737–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco PP, McNicholl IR. (2009) Etravirine and rilpivirine: nonnucleoside reverse transcriptase inhibitors with activity against human immunodeficiency virus type 1 strains resistant to previous nonnucleoside agents. Pharmacotherapy 29:281–294. [DOI] [PubMed] [Google Scholar]
- Gray WT, Frey KM, Laskey SB, Mislak AC, Spasov KA, Lee WG, Bollini M, Siliciano RF, Jorgensen WL, Anderson KS. (2015) Potent inhibitors active against HIV reverse transcriptase with K101P, a mutation conferring rilpivirine resistance. ACS Med Chem Lett 6:1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Li C, Chen W, Liu T, Yu M, Fu L, Sun Y, Liu H, De Clercq E, Pannecouque C, et al. (2015) Fused heterocycles bearing bridgehead nitrogen as potent HIV-1 NNRTIs. Part 3: optimization of [1,2,4]triazolo[1,5-a]pyrimidine core via structure-based and physicochemical property-driven approaches. Eur J Med Chem 92:754–765. [DOI] [PubMed] [Google Scholar]
- Janssen PA, Lewi PJ, Arnold E, Daeyaert F, de Jonge M, Heeres J, Koymans L, Vinkers M, Guillemont J, Pasquier E, et al. (2005) In search of a novel anti-HIV drug: multidisciplinary coordination in the discovery of 4-[[4-[[4-[(1E)-2-cyanoethenyl]-2,6-dimethylphenyl]amino]-2- pyrimidinyl]amino]benzonitrile (R278474, rilpivirine). J Med Chem 48:1901–1909. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL. (2009) Efficient drug lead discovery and optimization. Acc Chem Res 42:724–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Duffy EM. (2002) Prediction of drug solubility from structure. Adv Drug Deliv Rev 54:355–366. [DOI] [PubMed] [Google Scholar]
- King RW, Klabe RM, Reid CD, Erickson-Viitanen SK. (2002) Potency of nonnucleoside reverse transcriptase inhibitors (NNRTIs) used in combination with other human immunodeficiency virus NNRTIs, NRTIs, or protease inhibitors. Antimicrob Agents Chemother 46:1640–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WG, Frey KM, Gallardo-Macias R, Spasov KA, Bollini M, Anderson KS, Jorgensen WL. (2014) Picomolar inhibitors of HIV-1 reverse transcriptase: design and crystallography of naphthyl phenyl ethers. ACS Med Chem Lett 5:1259–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WG, Gallardo-Macias R, Frey KM, Spasov KA, Bollini M, Anderson KS, Jorgensen WL. (2013) Picomolar inhibitors of HIV reverse transcriptase featuring bicyclic replacement of a cyanovinylphenyl group. J Am Chem Soc 135:16705–16713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. (2007) Phaser crystallographic software. J Appl Cryst 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC. (2009) Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 485:395–405. [DOI] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P. (2013) Collaboration gets the most out of software. eLife 2:e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir S, Iqbal Z, Ahmad L, Ahmad S. (2016) Variation in pharmacokinetics of omeprazole and its metabolites by gender and CYP2C19 genotype in Pakistani male and female subjects. Pak J Pharm Sci 29:887–894. [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed] [Google Scholar]
- Pelemans H, Esnouf R, De Clercq E, Balzarini J. (2000) Mutational analysis of Trp-229 of human immunodeficiency virus type 1 reverse transcriptase (RT) identifies this amino acid residue as a prime target for the rational design of new non-nucleoside RT inhibitors. Mol Pharmacol 57:954–960. [PubMed] [Google Scholar]
- Permpalung N, Putcharoen O, Avihingsanon A, Ruxrungtham K. (2012) Treatment of HIV infection with once-daily regimens. Expert Opin Pharmacother 13:2301–2317. [DOI] [PubMed] [Google Scholar]
- Rabin L, Hincenbergs M, Moreno MB, Warren S, Linquist V, Datema R, Charpiot B, Seifert J, Kaneshima H, McCune JM. (1996) Use of standardized SCID-hu Thy/Liv mouse model for preclinical efficacy testing of anti-human immunodeficiency virus type 1 compounds. Antimicrob Agents Chemother 40:755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratain MJ, Plunkett WK. (2003) Principles of pharmacokinetics, in Holland-Frei Cancer Medicine (Kufe DW, Pollock RE, Weichselbaum RR, Bast RC, Jr, Gansler TS, Holland JF, Frei E, III, eds) B.C. Decker Inc., Hamilton. [Google Scholar]
- Rathbun RC, Liedtke MD. (2011) Antiretroviral drug interactions: overview of interactions involving new and investigational agents and the role of therapeutic drug monitoring for management. Pharmaceutics 3:745–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddler SA, Haubrich R, DiRienzo AG, Peeples L, Powderly WG, Klingman KL, Garren KW, George T, Rooney JF, Brizz B, et al. AIDS Clinical Trials Group Study A5142 Team (2008) Class-sparing regimens for initial treatment of HIV-1 infection. N Engl J Med 358:2095–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence RA, Anderson KS, Johnson KA. (1996) HIV-1 reverse transcriptase resistance to nonnucleoside inhibitors. Biochemistry 35:1054–1063. [DOI] [PubMed] [Google Scholar]
- Spence RA, Kati WM, Anderson KS, Johnson KA. (1995) Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science 267:988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddart CA, Bales CA, Bare JC, Chkhenkeli G, Galkina SA, Kinkade AN, Moreno ME, Rivera JM, Ronquillo RE, Sloan B, et al. (2007) Validation of the SCID-hu Thy/Liv mouse model with four classes of licensed antiretrovirals. PLoS One 2:e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LQ, Qin B, Huang L, Qian K, Chen CH, Lee KH, Xie L. (2012) Optimization of 2,4-diarylanilines as non-nucleoside HIV-1 reverse transcriptase inhibitors. Bioorg Med Chem Lett 22:2376–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Adams PD, Read RJ, Zwart PH, Hung LW. (2008) Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias. Acta Crystallogr D Biol Crystallogr 64:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhao X, Lin J, Li H, Johnston SC, Lin Y, Pan Y, Liu L, Wang D, Wang C, et al. CHANCE investigators (2016) Association between CYP2C19 loss-of-function allele status and efficacy of clopidogrel for risk reduction among patients with minor stroke or transient ischemic attack. JAMA 316:70–78. [DOI] [PubMed] [Google Scholar]
- Weller DR, Brundage RC, Balfour HH, Jr, Vezina HE. (2007) An isocratic liquid chromatography method for determining HIV non-nucleoside reverse transcriptase inhibitor and protease inhibitor concentrations in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci 848:369–373. [DOI] [PubMed] [Google Scholar]