Abstract
The c-Myc (MYC) transcription factor is a major cancer driver and a well-validated therapeutic target. However, directly targeting MYC has been challenging. Thus, identifying proteins that interact with and regulate MYC may provide alternative strategies to inhibit its oncogenic activity. In this study, we report the development of a NanoLuc-based protein-fragment complementation assay (NanoPCA) and mapping of the MYC protein interaction hub in live mammalian cells. The NanoPCA system was configured to enable detection of protein–protein interactions (PPI) at the endogenous level, as shown with PRAS40 dimerization, and detection of weak interactions, such as PINCH1-NCK2. Importantly, NanoPCA allows the study of PPI dynamics with reversible interactions. To demonstrate its utility for large-scale PPI detection in mammalian intracellular environment, we have used NanoPCA to examine MYC interaction with 83 cancer-associated proteins in live cancer cell lines. Our new MYC PPI data confirmed known MYC-interacting proteins, such as MAX, GSK3A, and SMARCA4, and revealed a panel of novel MYC interaction partners, such as RAC-α serine/threonine-protein kinase (AKT)1, liver kinase B (LKB)1, and Yes-associated protein (YAP)1. The MYC interactions with AKT1, LKB1, and YAP1 were confirmed by coimmunoprecipitation of endogenous proteins. Importantly, AKT1, LKB1, and YAP1 were able to activate MYC in a transcriptional reporter assay. Thus, these vital growth control proteins may represent promising MYC regulators, suggesting new mechanisms that couple energetic and metabolic pathways and developmental signaling to MYC-regulated cellular programs.
Introduction
The c-Myc (MYC) transcription factor, identified three decades ago, is encoded by the proto-oncogene MYC and is highly deregulated in a wide range of human malignancies (Vennstrom et al., 1982; Meyer and Penn, 2008). MYC is a master regulator that integrates signals from numerous pathways to modulate diverse cellular processes, such as proliferation, cell growth, apoptosis, differentiation, transformation, and metabolism (Meyer and Penn, 2008; Tu et al., 2015). To understand MYC function in normal cell biology and tumor progression, it is crucial to establish clear relationships between MYC status and cellular regulatory networks.
Characterization of the MYC protein–protein interaction (PPI) network is one of the major efforts to systematically interrogate MYC function through establishing detailed molecular connectivity. A large set of the MYC interactome data has become available through the application of a number of PPI detection technologies, such as yeast two-hybrid (Rolland et al., 2014) and affinity purification-mass spectrometry–based methods (Ewing et al., 2007; Agrawal et al., 2010; Dingar et al., 2015). Due to the unstable nature and strong chromatin association of MYC protein in cells, capturing MYC-mediated PPIs in a live cellular context has the potential to provide novel insights into the MYC regulatory system.
Protein-fragment complementation assays (PCAs) provide an efficient way for the detection of PPIs both in vitro and in vivo (Michnick et al., 2000, 2007; Michnick, 2001; Kodama and Hu, 2012). In this system (Fig. 1A), two proteins of interest, X and Y, are genetically fused with complementary N- and C-terminal fragments of a reporter protein, respectively. Upon the interaction of X and Y, the complementary fusion fragments are brought into close proximity, reconstituting the PCA reporter protein activities (Fig. 1A).
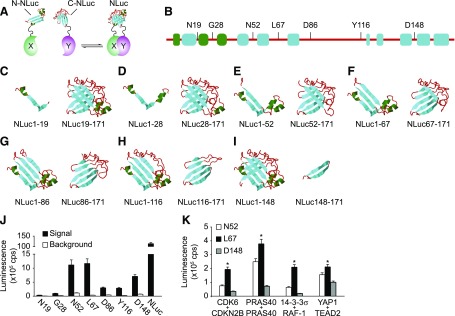
Fig. 1.
Design and development of NanoPCA. (A) NanoPCA design. N- or C-NLuc fragments are genetically fused to the N-terminal end of each protein of interest, X and Y, respectively. PCA signal can be detected when N- and C-NLuc fragments reconstitute into active full-length NLuc upon interaction of proteins X and Y. (B) Overall predicted secondary structure and selection of the splitting site of NLuc. (C–I) Schematic illustration of the predicted secondary structure of seven pairs of NLuc fragments. Secondary structural elements are colored as following: random coil with red, helix with green, and sheet with cyan. (J) Identification of the potential splitting site of NLuc using 14-3-3σ dimerization as a model. Luminescence signal was measured from live H1299 cells expressing the indicated NanoPCA 14-3-3σ constructs, respectively. H1299 cells expressing full-length NLuc were used as positive control. (K) Identification of the optimized splitting site of NLuc using four additional known PPIs as indicated. Error bars represent S.D. from three independent experiments. *P < 0.01.
The applicability of PCA has been significantly expanded by employing various reporter proteins, such as β-galactosidase (Rossi et al., 1997), fluorescent proteins (Hu et al., 2002; Zhang et al., 2002; Kodama and Hu, 2012; Tchekanda et al., 2014), and luciferases (Paulmurugan et al., 2002; Paulmurugan and Gambhir, 2003; Remy and Michnick, 2006). The poor reversibility of fluorescent protein-based PCAs and the weak optical intensity of conventional luciferase-based PCAs largely hinder their broad application (Magliery et al., 2005), particularly for high-throughput PPI screening assays. A PCA platform featuring robust sensitivity in detecting PPI at physiologic levels of expression that allows miniaturization in an ultrahigh-throughput screening (uHTS) format is highly desirable.
NanoLuc luciferase (NLuc) (Hall et al., 2012) is the smallest (19 kDa), ATP-independent monomeric luminescent protein, engineered from the luciferase of a luminous deep-sea shrimp, Oplophorus gracilirostris. Together with the novel imidazopyrazinone substrate, furimazine, NLuc generates glow-type luminescence with a specific activity about 100-fold greater than that of the Firefly or Renilla luciferases (Hall et al., 2012). In addition, NLuc is stable over a wide range of environmental conditions, such as temperature, pH, urea, and ionic strength (Hall et al., 2012). Thus, the relatively small size, bright signal, and physical stability make NLuc a candidate PCA reporter protein that may offer many advantages over conventional approaches for biochemical and pharmacological investigations.
In this work, we describe the development and validation of an NLuc-based PCA system that we termed NanoLuc-based PCA (NanoPCA). We demonstrate the following: 1) its enhanced sensitivity that enables the detection of PPI at various endogenous expression levels and weak interactions; 2) its reversibility that allows detection of protein dissociation upon inhibitory peptide or small-molecule PPI inhibitor treatment; and 3) its adaptability for large-scale uHTS PPI mapping of c-MYC interactors. With NanoPCA in H1299, a human nonsmall cell lung carcinoma cell line, and HCT116, a human colon carcinoma cell line, we successfully identified a MYC PPI hub by examining the PPIs between protein products of c-MYC and 83 cancer-associated genes. This study identified RAC-α serine/threonine-protein kinase (AKT)1, liver kinase B (LKB)1, and Yes-associated protein (YAP)1 as candidate MYC regulators, suggesting potential mechanisms through which MYC is coupled to cellular metabolic and developmental pathways.
Materials and Methods
Construction of Plasmids.
The N- and C-terminal fragments of the Nluc and RLuc were amplified by polymerase chain reaction from pNL1.1[Nluc] and pRL-null vector, respectively (Promega, Madison, WI). The fragments were cloned into the HindIII and NheI sites of the mutated pcDNA3.2/V5-DEST vector (g894t, t908g, a3182c, and c3243g) to generate Gateway cloning-friendly NLuc-PCA vectors. The mutated pcDNA3.2/V5-DEST vector was generated via mutagenesis using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). Gateway cloning system (Invitrogen, Carlsbad, CA) was used to generate the mammalian expression plasmid containing the gene cDNA with Nluc fragment tagged at its 5′ end. In all cases, a 10-amino-acid flexible linker sequence [(Gly4Ser)2] was inserted between the Nluc fragment and the partner gene cDNA. The pDONR vector containing the gene cDNA was either purchased from DNASU or cloned by polymerase chain reaction. All plasmids generated were confirmed by sequencing.
Cell Culture and Transfection.
Lung cancer H1299 cells, breast MCF7, pancreatic PANC-1, glioblastoma LN229 cells, colorectal carcinoma HCT116, prostate carcinoma DU145, skin melanoma A375, and liver hepatocellular carcinoma HepG2 cells were obtained from American Type Culture Collection (Manassas, VA). All cells maintained their appropriate culture medium. Cells were incubated at 37°C in humidified conditions with 5% CO2. Linear polyethylenimines (Polysciences, Warrington, PA; catalogue 23966) were used as the transfection reagent throughout the study.
PCA Measurements.
PCA signal measurements were performed 48 hours after transfection. N- and C-NLuc or N- and C-RLuc-tagged plasmids were cotransfected, as indicated. After 48-hour incubation, furimazine (NanoGlo; Promega, Madison, WI) or ViviRen (Promega) was added to the cells directly for live cell–based NanoPCA or Renilla luciferase-based PCA (RLuc-PCA) measurement, respectively. Furimazine or Coelenterazine-h (Promega) was used for cell lysate–based NanoPCA or RLuc-PCA measurement. The luminescence signals were measured immediately using an Envision Multilabel plate reader (PerkinElmer, Waltham, MA). Luminescence measured from PPI pairs and empty vector control pairs was defined as PCA signal and background, respectively.
Glutathione S-Transferase Pull-Down and Coimmunoprecipitation.
Glutathione S-transferase (GST) pull-down and coimmunoprecipitation assays were performed essentially as described previously (Havel et al., 2015). For Western blotting, cells were lysed and centrifuged for 15 minutes at 4°C. Total protein was quantified using the Bradford reagent, and equal amounts of total protein were mixed with 4× SDS sample buffer, incubated at 95°C for 5 minutes, and separated by SDS-PAGE. After electrophoresis, proteins were transferred to a nitrocellulose filter membrane (Bio-Rad, Hercules, CA) and blocked for 1 hour at room temperature. Each membrane was incubated with the appropriate primary antibody at 4°C overnight. The blots were then incubated with horseradish peroxidase–conjugated secondary antibodies for 1 hour, washed three times with PBST (1X Phosphate Buffered Saline Tween-20), and visualized using the Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore, Billerica, MA). The following primary antibodies were used: rabbit anti-PRAS40 (IBL, Minneapolis, MN); rabbit anti-GST (Santa Cruz Biotechnology, Santa Cruz, CA; sc-459); mouse anti-Flag (Sigma-Aldrich, St. Louis, MO; M2); mouse anti-LKB1 (Santa Cruz Biotechnology; sc-32245); mouse anti-AKT1 (Santa Cruz Biotechnology; sc-5298); rabbit anti-P65 (Santa Cruz Biotechnology; sc-372); rabbit anti-YAP (Santa Cruz Biotechnology; sc-15407); mouse anti-MYC (Santa Cruz Biotechnology; sc-40); and rabbit anti-MYC (Santa Cruz Biotechnology; sc-764).
Time-Resolved Föster (Fluorescence) Energy Transfer Assay.
Time-resolved (TR)–Föster/fluorescence energy transfer (FRET) assay was performed to monitor the small-molecule inhibitory effect, as described before (Du et al., 2013). Briefly, GST- and Venus-tagged PRAS40 were cotransfected and expressed in HEK293T cells, and cells were lysed with 1% Nonidet P-40 lysis buffer [20 mM Tris-HCl (pH 7.0), 50 mM NaCl, 1% Nonidet P-40]. Lysates were serially diluted in TR-FRET buffer [20 mM Tris-HCl (pH 7.0), 50 mM NaCl, 0.01% Nonidet P-40] to obtain the optimal signal. Terbium-conjugated GST antibody was used to couple GST-tagged protein as a FRET donor. Venus-tagged proteins served as a FRET acceptor. The FRET signal was measured on the Envision spectrophotometer (excitation wavelength, 337 nm; emission wavelength, 520 nm).
Myc Reporter Assay.
Myc reporter assay was performed by transfection of HEK293T, H1299, and HCT116 cells with the Myc Reporter (luc) plasmids. Forty-eight hours after transfection, luciferase activities were determined using Dual-Glo luciferase assay system (Promega). Relative Myc activity was expressed by normalizing the luciferase activity (Firefly/Renilla) to the empty vector control.
Data Analysis and Statistics.
For uHTS data, all analyses were performed using the MatLab package (MathWorks, Natick, MA). The statistical significance of the differences between NanoPCA PPI and control signals was calculated with the one-way analysis of variance, which can be used to compare the differences in means of two or more groups of data points (Heiman, 2010; http://www.stat.cmu.edu/∼hseltman/309/Book/Book.pdf). In particular, for every PPI, two statistical groups were generated. The first group contained the NanoPCA signals obtained in quadruplicate for the PPI (e.g., N-Nluc-MYC + C-Nluc-gene X), and the second group included NanoPCA signals obtained in quadruplicate for two empty vector controls (e.g., control 1: N-Nluc-MYC + C-Nluc and control 2: N-Nluc + C-Nluc-gene X). The P values less than 0.05 were considered significant. Because only two groups of data were subjected to analysis of variance analysis, no additional tests (e.g., post hoc tests) are required to confirm the difference between the PPI and control groups. However, to ensure that the PPI signals are stronger than the signals of the empty vector controls, the signal/background ratios were calculated as a ratio of averaged PPI signal to the highest of averaged signals obtained for the empty vector controls: signal over background (S/B) = PPIAVR/max(control 1AVR/control 2AVR). Accordingly, PPIs that demonstrate P values less than 0.05 and S/B > 1 were considered as positive. For other two groups of data, the significance of differences was analyzed using paired two-tailed Student's t test in Prism 6.0 software.
Results
For efficient detection of PPIs for protein network analysis and pharmacological studies in a live cell environment, we sought to develop a sensitive assay system that permits the implementation of ultrahigh-throughput screening applications. We selected the highly versatile PCA system (Michnick et al., 2000, 2007; Michnick, 2001; Kodama and Hu, 2012) and investigated the feasibility of adapting NLuc for enhanced performance in a PCA configuration.
Selection of a Nanoluc Protein Complementation Pair.
To develop the NanoPCA, we first examined potential fragmentation sites and generated a library of seven pairs of NLuc fragments (Fig. 1, B–I). To minimize the structural perturbation, the cleavage sites were localized within seven unstructured domains (Fig. 1, B–I), according to a predicted secondary structure of NLuc using the I-TASSER suite (Yang et al., 2015). We then genetically fused the corresponding N- and C-terminal NLuc fragment (N-NLuc and C-NLuc) to a test protein, 14-3-3σ (Fu et al., 2000). Coexpression of the N- and C-NLuc-14-3-3σ constructs in H1299 cells led to robust luciferase activity, suggesting that constitutive 14-3-3σ dimerization leads to reconstitution of the inactive N- and C-NLuc fragments into active NLuc (Fig. 1J). Live cells expressing three potential pairs of NLuc fragments, with fragmentation sites at Asn52 (N52), Leu67 (L67), and Asp148 (D148), reached about 10% of the full-length luminescence signal (Fig. 1J). To eliminate the bias of using a single 14-3-3 PPI example and to further narrow down the fragmentation site among N52, L67, and D148, four additional known PPIs were examined, CDK6/CDKN2B (Hannon and Beach, 1994), PRAS40/PRAS40 (Havel et al., 2015; Mo et al., 2016), 14-3-3σ/RAF-1 (Fu et al., 2000), and YAP1/TEAD2 (Vassilev et al., 2001; Zhao et al., 2008). As shown in Fig. 1K, these four PPIs showed the highest luminescence signal at fragmentation site at L67, which was selected for NanoPCA in the following studies.
Comparative Sensitivity of NanoPCA for PPI Detection.
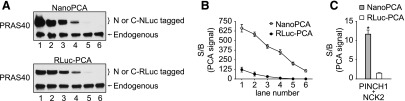
The RLuc-PCA has been widely used for detection of PPIs (Paulmurugan and Gambhir, 2003). Therefore, RLuc-PCA was used as a benchmark for the development of NanoPCA using PRAS40 dimerization as a model system (Havel et al., 2015; Mo et al., 2016). We first generated a dose-response curve using a range of PRAS40 protein expression levels and measured PCA signals for PRAS40 dimerization. As revealed by Western blot analysis (Fig. 2A), PRAS40 proteins as the N- and C-NLuc–tagged, or N- and C-RLuc–tagged fusions were exogenously expressed at levels ranging from less to greater than endogenous PRAS40 expression. At expression levels that were comparable to or even lower than the endogenous PRAS40 (Fig. 2A, lanes 4–6), PRAS40 dimerization could be detected using NanoPCA with S/B up to 350-fold. At such endogenous levels, the PRAS40 interaction could not be detected with RLuc-PCA (Fig. 2B). Thus, the increased sensitivity of NanoPCA compared with that of the RLuc-PCA platform suggests a particular advantage of the NanoPCA in the setting of low protein expression. Moreover, this enhanced sensitivity allows detection of PPIs under physiologic expression conditions.
Fig. 2.
Characterization of the sensitivity of NanoPCA. (A) Expression of N- and C-NLuc or RLuc-tagged PRAS40 compared with endogenous PRAS40. HEK293T cells were cotransfected with various amounts of NanoPCA or RLuc-PCA PRAS40 construct. Whole-cell lysate was applied to Western blot analysis with anti-PRAS40 antibodies. (B) PCA analysis. Whole-cell lysate from the same sample as indicated in (A) used for Western blot analysis was added into the 1536-well plate for PCA detection. (C) Comparison between the NanoPCA and RLuc-PCA for measuring the extremely weak PPI, PINCH1/NCK2, in live H1299 cells. The data are expressed as mean ± S.D. from three independent experiments. *P < 0.01.
Such a sensitivity of NanoPCA may allow the detection of PPIs with low binding affinity. To test this notion, we examined the performance of NanoPCA with the PINCH1/NCK2 pair, a known PPI with a low binding affinity (KD ∼3 × 10−3 M) (Vaynberg et al., 2005). We observed significant PCA signal upon expression of N-NLuc-PINCH1 and C-NLuc-NCK2 with S/B of more than 10-fold (Fig. 2C). No significant signals were detected for PINCH1/NCK2 with the conventional RLuc-PCA. Our results demonstrate that NanoPCA has enhanced detection sensitivity compared with RLuc-PCA, enabling the detection of PPIs at low expression levels of tested proteins, as well as PPIs with low binding affinity.
NanoPCA for Monitoring PPI Dynamics.
Irreversibility is a perceived limitation of PCA, particularly PCAs based on fluorescence proteins (Hu et al., 2002; Magliery et al., 2005; Kodama and Hu, 2012). Poor reversibility often results in high false-positive discovery rate in PPI detection, and also largely hinders the further application of PCA for monitoring protein dynamics.
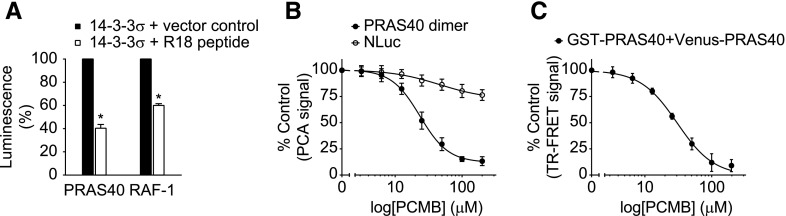
To test the reversibility of NanoPCA in live cells, we first evaluated the reversible interactions between 14-3-3σ and its binding partners, such as PRAS40 and RAF1, by coexpressing a disruptive peptide, R18, known to effectively disrupt the binding of 14-3-3 with client proteins (Masters and Fu, 2001). When the R18 disruptive peptide was coexpressed in cells with 14-3-3σ and PRAS40 or RAF1, we observed 60% and 40% less luminescence from the interaction of 14-3-3σ with PRAS40 or RAF-1 than cells without R18 (Fig. 3A), suggesting the dissociation of the interactions in the NanoPCA settings.
Fig. 3.
Monitoring PPI dissociation using NanoPCA. (A) PCA signal for 14-3-3σ with PRAS40 or RAF-1 in the presence or absence of disruptive peptide R18. (B and C) The p-chloromercuribenzoic acid induced dissociation of PRAS40 dimer with NanoPCA (B) and TR-FRET (C). A dose-dependent decrease of PCA and TR-FRET signal for PRAS40 dimerization with increasing PCMB concentration is observed. The data are expressed as mean ± S.D. from three independent experiments. *P < 0.01.
We next examined the reversibility of NanoPCA in cell lysate using a small-molecule PPI inhibitor. We monitored the dissociation of PRAS40 dimer upon the treatment of cells with our previously described PRAS40 dimerization inhibitor, p-chloromercuribenzoic acid. With p-chloromercuribenzoic acid treatment, a dose-dependent decrease of luminescence signal from the PRAS40 dimer, but not from full-length NLuc, was observed with IC50 of 22 μM (Fig. 3B), which is consistent with the IC50 of 29.9 μM from TR-FRET assay (Fig. 3C), as well as our previously observed IC50 using GST pull-down experiments (Mo et al., 2016). These results suggest that reconstitution of the complementary fragments of NanoLuc is dependent on target protein interactions, and the associated N- and C-terminal NanoPCA subunits were able to dissociate reversibly.
Miniaturized NanoPCA Allows Rapid PPI Detection across Multiple Cell Lines.
The high sensitivity of the NanoPCA technology enables miniaturized configuration for uHTS, which permits a wide range of applications, such as uHTS PPI mapping. The uHTS assay platform in the 1536-well plate was developed similarly, as described previously (Mo and Fu, 2016; Mo et al., 2016). Briefly, cells were plated in a high-density 1536-well plate, and, after 24 hours, forward transfection was performed robotically from 384-well DNA plate in four replicates. To test the utility of uHTS NanoPCA technology for rapid PPI mapping, we focused on the oncogenic MYC interactome as an example. MYC is dysregulated in more than 50% of human cancers (Meyer and Penn, 2008). However, the molecular basis of the oncogenic activity of MYC under various physiologic contexts is still not completely understood.
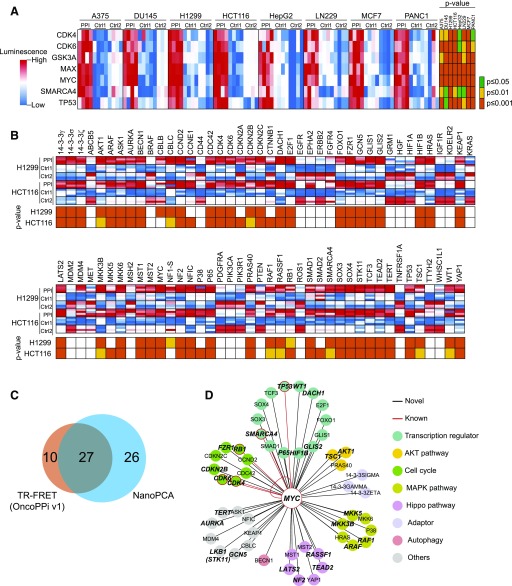
The complexity of PPI maps largely results from cellular context-dependent protein dynamics, and thus an experimental method allowing PPI detection in multiple cell lines is highly preferable (Schaefer et al., 2013). We tested the scalability of NanoPCA for PPI detection across multiple cell lines. We selected seven known MYC interaction partners, CDK4, CDK6, GSK3A, MAX, SMARCA4, TP53, and MYC homodimer, and eight different human cancer cell lines, A375, DU145, H1299, HCT116, HepG2, LN229, MCF7, and PANC1, from various tissues of origin. MYC and test partners were coexpressed in these cells. NanoPCA signals were recorded. A heat map was constructed using PPI signals compared with background from empty vector controls (Fig. 4A). Using the NanoPCA platform, we validated the selected known PPIs with significantly higher S/B > 1.0 and P < 0.05 in all of the cell lines tested (Fig. 4A). These results suggest that these known MYC interaction partners may play a generalized role in MYC signaling across tissues or cell lines of different origins.
Fig. 4.
High-throughput PPI mapping of oncogenic MYC interactome. (A) Heat map of known MYC PPIs in eight cell lines showing four independent replicates. (B) Oncogenic PPI profiling of N-NLuc-MYC against C-NLuc–tagged genes library. Four independent replicates were shown in the heat map. (C) The Venn diagram of the distribution of positive PPIs identified in NanoPCA platform and TR-FRET–based OncoPPi v1 network (Li et al., 2016). (D) An oncogenic MYC PPI hub. PPIs that are positive with S/B > 1.0 and P < 0.05 in both cell lines were selected. Colors are assigned as indicated. PPIs that are double positive in both NanoPCA and TR-FRET were denoted in bold italic. MAPK, mitogen-activated protein kinase.
Identification of Novel MYC Partners with NanoPCA Profiling.
We explored the capability of NanoPCA for PPI profiling to discover new MYC interaction partners. As a proof of concept, we mapped the interactions of MYC with a focused set of human cancer-associated proteins using the OncoPPi library v.1 (Li et al., 2016) (Supplemental Table 1), expressed in H1299 and HCT116 cell lines. As shown in the heat map (Fig. 4B), 58 PPIs were considered as positive with S/B > 1.0 and P value < 0.05 in at least one cell line, and 53 of these 58 PPIs were positive in both cell lines. Using NanoPCA technology, 10 of 12 (about 83%) of known MYC-interacting partners (Chatr-Aryamontri et al., 2015) were successfully identified in at least one cancer cell line (Fig. 4, A and B). As shown in the Venn diagram (Fig. 4C), 27 of 53 NanoPCA-positive were also validated by TR-FRET, as described for the OncoPPi v1 network (Li et al., 2016).
To build a high-confidence MYC oncogenic PPI network, we selected 53 interactions, including 6 known (in red) and 49 novel (in black) interactions that were positive in both cell lines (Fig. 4D). These 49 novel MYC-interacting partners span across various groups of established oncogenic signaling pathways, including a panel of transcription regulators, AKT signaling, cell cycle regulators, mitogen-activated protein kinase signaling, Hippo pathway, adaptor proteins, autophagy, and other pathways.
Validation of AKT1, LKB1, P65, and YAP1 as MYC Interaction Partners.
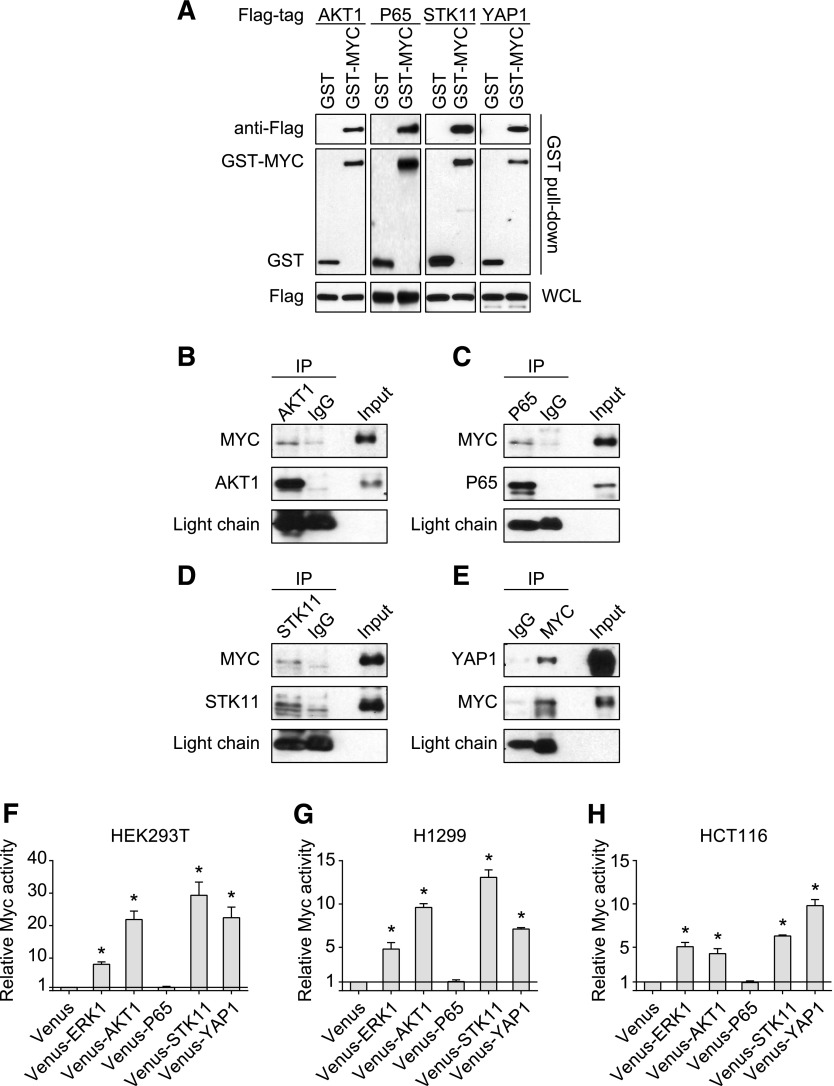
This oncogenic MYC PPI hub suggests several potential mechanisms underlying MYC regulation through interaction with several novel binding partners, such as AKT1, LKB1, P65, and YAP1. GST pull-down was employed as an orthogonal confirmatory method to validate these four novel PPIs. The GST-affinity chromatography gave rise to significantly higher signals for the PPI pairs than empty tag vector controls (Fig. 5A). To further validate the connectivity between these four genes with MYC, coimmunoprecipitation assay was used to examine interaction of endogenously expressed proteins. As shown in Fig. 5B, we successfully detected protein complexes of MYC with ATK1, LKB1, P65, and YAP1, respectively, with the indicated protein antibodies, but not with IgG controls.
Fig. 5.
Validation of novel PPI interactors. (A) GST pull-down and (B–E) endogenous coimmunoprecipitation validation of MYC interaction with (B) AKT1, (C) P65, (D) STK11, and (E) YAP1. (F–H) Functional validation of MYC activity in the presence of AKT1, STK11, P65, and YAP1 in (F) HEK293T, (G) H1299, and (H) HCT116 cell lines. The data are expressed as mean ± S.D. from three independent experiments. *P < 0.01. WCL, whole cell lysate.
To gain further functional insights into these new MYC interactions, we tested the effect of AKT1, P65, LKB1, and YAP1 on MYC-driven transcriptional activity using an E-box–based MYC reporter assay in three different cell lines, HEK293T, H1299, and HCT116. As shown in Fig. 5, F–H, in the presence of extracellular signal-regulated kinase 1, a known MYC interactor (Sears et al., 2000), as a positive control, MYC activity was increased compared with the empty vector control. Although P65 expression failed to enhance the MYC reporter activity, expression of AKT1, LKB1, and YAP1 led to increased reporter signals, suggesting their positive roles in modulating MYC activity.
Discussion
In the present study, we have demonstrated that NanoPCA is a highly sensitive technique with simple add-and-read mode (no washing step) that can be readily used to detect PPIs in a live cell environment at low protein expression level. The Gateway cloning friendly vector system developed in this study provides a convenient platform for cDNA library-based PPI network mapping. Additionally, the stable glow-type luminescence of NLuc (Hall et al., 2012) is well suited for high-throughput applications. In contrast, Gaussia luciferase-based PCA is limited by the fast flash kinetics of the Gaussia luciferase reaction (Tannous et al., 2005; Remy and Michnick, 2006; Tannous, 2009; Gilad et al., 2014). Because of its simplicity and sensitivity, NanoPCA can be miniaturized and automated for high-throughput applications. Moreover, NanoPCA is reversible and thus allows the study of PPI dynamics, such as PPI inhibitor discovery.
Design and selection of the optimal fragmentation site are critical for developing PCA technology with robust performance. In the NanoPCA settings, an optimal pair of NLuc fragments was identified as the splitting site at Leu67 located in an unstructured coil region from Ile62 to Gln74 (Fig. 1B). This fragmentation site is in agreement with another splitting position at N65, which is also in the Ile62-Gln74 region, identified by Zhao et al. (2016) using protein aggregation as a model system. In accordance with previous PCA technologies, the split NanoPCA protein achieves up to 10% reconstitution of activity compared with full-length NLuc (Fig. 1J). Further engineering the peptide fragments through amino acid substitution may further increase the reconstitution activity. For example, Dixon et al. (2016) recently developed a NanoLuc Binary Technology (NanoBiT), through engineering and substituting 16 amino acid residues of the native NLuc 1–156 peptide, and increased the reconstitution activity up to 37% of NLuc.
With the NanoPCA technology presented in this work, we have demonstrated its robust performance in a uHTS format and capability of detecting known and discovering novel MYC-interacting proteins in a live cancer cellular context. In contrast, previous MYC interactomes established using other high-throughput PPI detection platforms use nonhuman expression systems (such as yeast two-hybrid), or lack the ability to detect PPI signal in live cells without lysing and washing steps (such as affinity purification-coupled mass spectrometry). These limitations highlight the critical importance of developing a homogenous high-throughput PPI detection method with the capability of monitoring PPIs under physiologically relevant conditions in various cellular contexts, such as with NanoPCA.
MYC is a validated oncogene with pluripotent activities through engaging a wide spectrum of other proteins; to date, about 568 MYC-interacting proteins have been reported (Chatr-Aryamontri et al., 2015). In this study, we tested a total of 83 cancer-associated proteins for MYC binding, and 53 of these were positive. Such high connectivity (∼64%) partly results from the focused set of cancer-focused genes included in the screen (Barabási et al., 2011), and also probably reflects the intrinsic nature of MYC as an important signaling node (Chatr-Aryamontri et al., 2015). However, PCAs are prone to both false negatives and false positives. False positives could be in part due to overexpression-induced reconstitution of the luciferase activity. Thus, alternative experimental approaches without involving luciferase reporter are required for confirmation of positive interactions. The near endogenous level of fusion proteins required for detecting positive PPIs in our NanoPCA is expected to significantly reduce the potential false-positive rate. In this study, 27 of 53 positive PPIs have been confirmed in both NanoPCA and TR-FRET, whereas other 26 were solely positive in NanoPCA. These 26 PPIs will need further investigation to assess whether they are true or false positives. For example, MYC-YAP1 interaction, which was positive in NanoPCA, but not in TR-FRET, has been confirmed as true positive in alternative GST-pull-down and endogenous co-immunoprecipitation experiments. In contrast, false negatives could result from protein tag-mediated steric hindrance for certain PPIs. Using multiple fusion orientations for a particular PPI pair is expected to capture true positives, reducing false negatives.
The prominent role of MYC in tumorigenesis makes it an ideal candidate for targeted cancer therapy. Because numerous lines of evidence suggest that direct pharmacological inhibition of MYC is challenging due to its lack of enzymatic activity and binding pocket (Tu et al., 2015), a better understanding of MYC function and regulation through mapping of its PPI network may inform promising alternative strategies to inhibit the oncogenic activity of MYC (Hart et al., 2014). Interestingly, through this study, we have revealed a large number of potential MYC interactors and validated three promising MYC partners, including AKT1, LKB1, and YAP1. These novel MYC PPIs provide evidence for the previously described pluripotency of MYC in regulating these signaling pathways (Partanen et al., 2007; Clegg et al., 2011; Xiao et al., 2013). Although our current studies focused on the establishment of the novel NanoPCA platform and the demonstration of its utility for the rapid discovery of multiple novel MYC partners, functional roles of these newly revealed partners in the MYC regulatory system remain to be established.
The phosphatidylinositol 3-kinase/AKT/mTOR (mechanistic target of rapamycin) survival pathway, LKB1/AMPK metabolic pathway, and Hippo-YAP developmental pathway are three of frequently dysregulated cell signaling pathways involved in tumorigenesis and progression. Moreover, the functional relevance of MYC with AKT1, LKB1, and YAP1 has been suggested by previous studies (Partanen et al., 2007; Clegg et al., 2011; Xiao et al., 2013). For example, the association between MYC and AKT1 activity has been reported based on the evidence that several known MYC modulators, such as FOXO1, MAD1, and CDCA7, can be phosphorylated by AKT1 in vitro or in vivo (Bouchard et al., 2004; Zhu et al., 2008; Gill et al., 2013). The direct interaction of AKT1 and MYC discovered in our study, together with the MYC-TSC1 interaction (Fig. 4D), provides another layer of linkage between the phosphatidylinositol 3-kinase/AKT/mTOR (mechanistic target of rapamycin) survival pathway and the MYC transcriptional machinery.
The functional relevance of LKB1 for MYC function has been suggested recently based on the observation of indirect suppression of MYC activity by LKB1 in epithelial polarity regulation, as well as through MZF1, extracellular signal-regulated kinase 1/2, or signal transducer and activator of transcription 3 signaling in lung adenocarcinoma cell line models (Partanen et al., 2007; Liang et al., 2009; Tsai et al., 2015). The LKB1-MYC PPI revealed in this study indicates a potential mechanism of direct regulation of MYC by LKB1. It is noteworthy that we observed a significant increase of MYC reporter signal upon the overexpression of LKB1, which is unexpected based on previous data of LKB1-induced suppression of MYC (Partanen et al., 2007; Liang et al., 2009; Tsai et al., 2015). Further investigation, such as profiling of LKB1-dependent MYC target gene expression, is needed to examine how the LKB1-MYC PPI contribute to the process of tumorigenesis and progress.
Recently, the connection between the Hippo-YAP and MYC pathways has been explored, and a positive feedback loop linked by c-ABL has been suggested underlying YAP and MYC’s oncogenic activity in liver cancer (Xiao et al., 2013). Through our study, we identified and validated the YAP1-MYC PPI, suggesting a direct coupling of MYC to the Hippo developmental pathway. Moreover, the interactions between MYC and NF2, RASSF1, LATS2, and TEAD2 (Fig. 4D) identified in our study further suggest the involvement of MYC at different regulatory points in the Hippo pathway.
Conclusion
In summary, we have developed an NLuc-based PCA technology, termed NanoPCA. We have demonstrated robust performance of NanoPCA to study dynamic PPIs in live cells with high specificity, sensitivity, and reversibility. The significantly enhanced sensitivity of NanoPCA enabled the detection of PPIs at endogenous expression levels, or with low binding affinity, in 1536-well uHTS format. Application of NanoPCA allowed rapid mapping of the oncogenic MYC PPI hub across multiple cancer cell lines. We validated three promising MYC interactors (AKT1, LKB1, and YAP1), revealing their functional importance in the regulation of oncogenic MYC. The established NanoPCA PPI sensors will be readily available for protein interaction studies and chemical probe discovery.
Acknowledgments
The authors thank members of the Fu laboratory for helpful discussions, valuable comments, and editing and Dr. Edward Prochownik for MYC reporter plasmids.
Abbreviations
- AKT
RAC-α serine/threonine-protein kinase/protein kinase B
- FRET
Föster/fluorescence energy transfer
- GST
glutathione S-transferase
- LKB
liver kinase B
- MYC
c-Myc
- NanoPCA
NanoLuc-based PCA
- NLuc
Nanoluc luciferase
- PCA
protein-fragment complementation assay
- PPI
protein–protein interaction
- RLuc-PCA
Renilla luciferase-based PCA
- S/B
signal over background
- TR
time-resolved
- uHTS
ultrahigh-throughput screening
- YAP
Yes-associated protein
Authorship Contributions
Participated in research design: Mo, Qi, Havel, Goetze, Khuri, Du, Fu.
Conducted experiments: Mo, Qi, Niu, Luo, Havel, Bell, Goetze.
Performed data analysis: Mo, Ivanov, Fu.
Wrote or contributed to the writing of the manuscript: Mo, Qi, Ivanov, Moreno, Johns, Cooper, Khuri, Fu.
Footnotes
This work was supported in part by National Institutes of Health National Cancer Institute Cancer Target Discovery and Development Network [Grant U01CA168449] and Winship Cancer Institute of Emory University [Grant 5P30CA138292].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Agrawal P, Yu K, Salomon AR, Sedivy JM. (2010) Proteomic profiling of Myc-associated proteins. Cell Cycle 9:4908–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási AL, Gulbahce N, Loscalzo J. (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Marquardt J, Brás A, Medema RH, Eilers M. (2004) Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J 23:2830–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O’Donnell L, et al. (2015) The BioGRID interaction database: 2015 update. Nucleic Acids Res 43:D470–D478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg NJ, Couto SS, Wongvipat J, Hieronymus H, Carver BS, Taylor BS, Ellwood-Yen K, Gerald WL, Sander C, Sawyers CL. (2011) MYC cooperates with AKT in prostate tumorigenesis and alters sensitivity to mTOR inhibitors. PLoS One 6:e17449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingar D, Kalkat M, Chan PK, Srikumar T, Bailey SD, Tu WB, Coyaud E, Ponzielli R, Kolyar M, Jurisica I, et al. (2015) BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J Proteomics 118:95–111. [DOI] [PubMed] [Google Scholar]
- Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, et al. (2016) NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem Biol 11:400–408. [DOI] [PubMed] [Google Scholar]
- Du Y, Fu RW, Lou B, Zhao J, Qui M, Khuri FR, Fu H. (2013) A time-resolved fluorescence resonance energy transfer assay for high-throughput screening of 14-3-3 protein-protein interaction inhibitors. Assay Drug Dev Technol 11:367–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O’Connor L, Li M, et al. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 3:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. (2000) 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol 40:617–647. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Shiloh R, Ber Y, Bialik S, Kimchi A. (2014) Discovering protein-protein interactions within the programmed cell death network using a protein-fragment complementation screen. Cell Reports 8:909–921. [DOI] [PubMed] [Google Scholar]
- Gill RM, Gabor TV, Couzens AL, Scheid MP. (2013) The MYC-associated protein CDCA7 is phosphorylated by AKT to regulate MYC-dependent apoptosis and transformation. Mol Cell Biol 33:498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, et al. (2012) Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol 7:1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Beach D. (1994) p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 371:257–261. [DOI] [PubMed] [Google Scholar]
- Hart JR, Garner AL, Yu J, Ito Y, Sun M, Ueno L, Rhee JK, Baksh MM, Stefan E, Hartl M, et al. (2014) Inhibitor of MYC identified in a Kröhnke pyridine library. Proc Natl Acad Sci USA 111:12556–12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havel JJ, Li Z, Cheng D, Peng J, Fu H. (2015) Nuclear PRAS40 couples the Akt/mTORC1 signaling axis to the RPL11-HDM2-p53 nucleolar stress response pathway. Oncogene 34:1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman GW. (2010) The one-way analysis of variance, in Basic Statistics for the Behavioral Sciences, 6th ed, pp 290–317, Wadsworth, Belmont, CA. [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9:789–798. [DOI] [PubMed] [Google Scholar]
- Kodama Y, Hu CD. (2012) Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. Biotechniques 53:285–298. [DOI] [PubMed] [Google Scholar]
- Li Z, Ivanov AA, Su R, Gonzalez PV, Qi Q, Liu S, Webber P, McMillan E, Rusnak LCP, Chen X, et al. (2016) The OncoPPi network of cancer-focused protein–protein interactions to inform biological insights and therapeutic strategies. Nat Commun, in press DOI: 10.1038/ncomms14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Nan KJ, Li ZL, Xu QZ. (2009) Overexpression of the LKB1 gene inhibits lung carcinoma cell proliferation partly through degradation of c-myc protein. Oncol Rep 21:925–931. [DOI] [PubMed] [Google Scholar]
- Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. (2005) Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc 127:146–157. [DOI] [PubMed] [Google Scholar]
- Masters SC, Fu H. (2001) 14-3-3 proteins mediate an essential anti-apoptotic signal. J Biol Chem 276:45193–45200. [DOI] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. (2008) Reflecting on 25 years with MYC. Nat Rev Cancer 8:976–990. [DOI] [PubMed] [Google Scholar]
- Michnick SW. (2001) Exploring protein interactions by interaction-induced folding of proteins from complementary peptide fragments. Curr Opin Struct Biol 11:472–477. [DOI] [PubMed] [Google Scholar]
- Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. (2007) Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov 6:569–582. [DOI] [PubMed] [Google Scholar]
- Michnick SW, Remy I, Campbell-Valois FX, Vallée-Bélisle A, Pelletier JN. (2000) Detection of protein-protein interactions by protein fragment complementation strategies. Methods Enzymol 328:208–230. [DOI] [PubMed] [Google Scholar]
- Mo XL, Fu H. (2016) BRET: NanoLuc-based bioluminescence resonance energy transfer platform to monitor protein-protein interactions in live cells. Methods Mol Biol 1439:263–271. [DOI] [PubMed] [Google Scholar]
- Mo XL, Luo Y, Ivanov AA, Su R, Havel JJ, Li Z, Khuri FR, Du Y, Fu H. (2016) Enabling systematic interrogation of protein-protein interactions in live cells with a versatile ultra-high-throughput biosensor platform. J Mol Cell Biol 8:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partanen JI, Nieminen AI, Mäkelä TP, Klefstrom J. (2007) Suppression of oncogenic properties of c-Myc by LKB1-controlled epithelial organization. Proc Natl Acad Sci USA 104:14694–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulmurugan R, Gambhir SS. (2003) Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem 75:1584–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulmurugan R, Umezawa Y, Gambhir SS. (2002) Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci USA 99:15608–15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I, Michnick SW. (2006) A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods 3:977–979. [DOI] [PubMed] [Google Scholar]
- Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. (2014) A proteome-scale map of the human interactome network. Cell 159:1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F, Charlton CA, Blau HM. (1997) Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation. Proc Natl Acad Sci USA 94:8405–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer MH, Lopes TJ, Mah N, Shoemaker JE, Matsuoka Y, Fontaine JF, Louis-Jeune C, Eisfeld AJ, Neumann G, Perez-Iratxeta C, et al. (2013) Adding protein context to the human protein-protein interaction network to reveal meaningful interactions. PLOS Comput Biol 9:e1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. (2000) Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14:2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous BA. (2009) Gaussia luciferase reporter assay for monitoring biological processes in culture and in vivo. Nat Protoc 4:582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. (2005) Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther 11:435–443. [DOI] [PubMed] [Google Scholar]
- Tchekanda E, Sivanesan D, Michnick SW. (2014) An infrared reporter to detect spatiotemporal dynamics of protein-protein interactions. Nat Methods 11:641–644. [DOI] [PubMed] [Google Scholar]
- Tsai LH, Wu JY, Cheng YW, Chen CY, Sheu GT, Wu TC, Lee H. (2015) The MZF1/c-MYC axis mediates lung adenocarcinoma progression caused by wild-type lkb1 loss. Oncogene 34:1641–1649. [DOI] [PubMed] [Google Scholar]
- Tu WB, Helander S, Pilstål R, Hickman KA, Lourenco C, Jurisica I, Raught B, Wallner B, Sunnerhagen M, Penn LZ. (2015) Myc and its interactors take shape. Biochim Biophys Acta 1849:469–483. [DOI] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. (2001) TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev 15:1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaynberg J, Fukuda T, Chen K, Vinogradova O, Velyvis A, Tu Y, Ng L, Wu C, Qin J. (2005) Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility. Mol Cell 17:513–523. [DOI] [PubMed] [Google Scholar]
- Vennstrom B, Sheiness D, Zabielski J, Bishop JM. (1982) Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J Virol 42:773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Wang J, Ou C, Zhang Y, Ma L, Weng W, Pan Q, Sun F. (2013) Mutual interaction between YAP and c-Myc is critical for carcinogenesis in liver cancer. Biochem Biophys Res Commun 439:167–172. [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Campbell RE, Ting AY, Tsien RY. (2002) Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol 3:906–918. [DOI] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22:1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Nelson TJ, Vu Q, Truong T, Stains CI. (2016) Self-assembling NanoLuc luciferase fragments as probes for protein aggregation in living cells. ACS Chem Biol 11:132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Blenis J, Yuan J. (2008) Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc Natl Acad Sci USA 105:6584–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]