Abstract
The intermediate-conductance Ca2+-activated K+ channel (KCa3.1) constitutes an attractive pharmacological target for immunosuppression, fibroproliferative disorders, atherosclerosis, and stroke. However, there currently is no available crystal structure of this medically relevant channel that could be used for structure-assisted drug design. Using the Rosetta molecular modeling suite we generated a molecular model of the KCa3.1 pore and tested the model by first confirming previously mapped binding sites and visualizing the mechanism of TRAM-34 (1-[(2-chlorophenyl)diphenylmethyl]-1H-pyrazole), senicapoc (2,2-bis-(4-fluorophenyl)-2-phenylacetamide), and NS6180 (4-[[3-(trifluoromethyl)phenyl]methyl]-2H-1,4-benzothiazin-3(4H)-one) inhibition at the atomistic level. All three compounds block ion conduction directly by fully or partially occupying the site that would normally be occupied by K+ before it enters the selectivity filter. We then challenged the model to predict the receptor sites and mechanisms of action of the dihydropyridine nifedipine and an isosteric 4-phenyl-pyran. Rosetta predicted receptor sites for nifedipine in the fenestration region and for the 4-phenyl-pyran in the pore lumen, which could both be confirmed by site-directed mutagenesis and electrophysiology. While nifedipine is thus not a pore blocker and might be stabilizing the channel in a nonconducting conformation or interfere with gating, the 4-phenyl-pyran was found to be a classical pore blocker that directly inhibits ion conduction similar to the triarylmethanes TRAM-34 and senicapoc. The Rosetta KCa3.1 pore model explains the mechanism of action of several KCa3.1 blockers at the molecular level and could be used for structure-assisted drug design.
Introduction
The intermediate-conductance Ca2+-activated K+ (KCa3.1) channel (KCNN4) was cloned independently by three groups in 1997 from pancreas (Ishii et al., 1997), placenta (Joiner et al., 1997), or lymph node (Logsdon et al., 1997). Similar to the related small-conductance Ca2+-activated K+ channel or KCa2 channels, KCa3.1 channels have fewer charges in their S4 segment than voltage-gated K+ (Kv) channels (Wei et al., 2005); therefore, they do not respond to changes in membrane voltage but instead are activated by Ca2+ binding to calmodulin, which functions as their β-subunit and induces Ca2+-dependent channel opening (Fanger et al., 1999). KCa3.1 channels are accordingly able to hyperpolarize the membrane toward the K+ equilibrium potential and modulate Ca2+ influx during cellular activation and proliferation by sustaining Ca2+ entry through Ca2+-release activated Ca2+ or transient receptor potential channels.
KCa3.1 is widely expressed in cells of the immune system such as T- and B-lymphocytes (Ghanshani et al., 2000; Wulff et al., 2004), mast cells (Shumilina et al., 2008), macrophages, and microglia (Chen et al., 2016), where it plays an important role in cellular activation, migration, and cytokine production (Feske et al., 2015). KCa3.1 is further important for volume regulation in erythrocytes (Vandorpe et al., 1998), which is why the channel is often referred to as the Gárdos channel after the Hungarian scientist, who first described a Ca2+-activated K+ efflux leading to erythrocyte dehydration (Gardos, 1958). More recently, missense mutations in KCa3.1 have been linked to hereditary stomatocytosis (xerocytosis), an autosomal dominant congenital hemolytic anemia. In addition to red and white blood cells, KCa3.1 is also expressed in dedifferentiated vascular smooth muscle cells (Köhler et al., 2003; Toyama et al., 2008); fibroblast (Peña and Rane, 1999); the vascular endothelium, where the channel is involved in the so-called endothelium-derived hyperpolarization response (Si et al., 2006; Edwards et al., 2010); and in secretory epithelia of the lung and the gastrointestinal tract (Heitzmann and Warth, 2008), where KCa3.1 participates in fluid and electrolyte movement across the epithelium. Based on this expression pattern, KCa3.1 inhibitors constitute very attractive drug candidates for reducing inflammatory cytokine production in autoimmune disorders (Feske et al., 2015), microglia activation in stroke (Chen et al., 2016), and Alzheimer’s disease (Maezawa et al., 2012), and for inhibiting smooth muscle cell and fibroblast proliferation in atherosclerosis (Toyama et al., 2008), asthma (Girodet et al., 2013), vascular restenosis (Köhler et al., 2003), and transplant rejection (Chen et al., 2013). Other potential indications for KCa3.1 inhibition are sickle cell disease (Brugnara, 2003) and secretory diarrhea in humans and farm animals (Rufo et al., 1997). Research groups in both the pharmaceutical industry and academia have identified several classes of potent and selective KCa3.1 blockers including the triarylmethanes senicapoc (triarylmethanes 2,2-bis-(4-fluorophenyl)-2-phenylacetamide) (McNaughton-Smith et al., 2008) and TRAM- 34 (1-[(2-chlorophenyl)diphenylmethyl]-1H-pyrazole) (Wulff et al., 2000), the benzothiazione NS6180 (4-[[3-(trifluoromethyl)phenyl]methyl]-2H-1,4-benzothiazin-3(4H)-one) (Strøbæk et al., 2013), and a series of dihydropyridine isosteric 4-phenyl-pyrans (Urbahns et al., 2003) and cyclohexadienes (Urbahns et al., 2005). Senicapoc, which was in phase-3 clinical trials for sickle cell anemia, where it failed for lack of efficacy in reducing the number of painful sickling attacks (Ataga et al., 2011), demonstrated that KCa3.1 inhibitors are safe and well tolerated in humans. However, based on issues such as short remaining patent lives or low oral availability, none of these compounds are currently suitable for further clinical development and there is a need for identifying novel KCa3.1 inhibitors.
Since structure-based drug design approaches have not previously been attempted for KCa3.1 due to the absence of x-ray crystal structures, we here applied the Rosetta molecular modeling suite (https://www.rosettacommons.org/) with membrane environment specific energy functions to generate structural models of the KCa3.1 pore and tested our predictions by site-directed mutagenesis and electrophysiology. Rosetta has previously been successfully used to calculate gating charges (Khalili-Araghi et al., 2010), simulate voltage-sensor movements (Decaen et al., 2008, 2009, 2011; Yarov-Yarovoy et al., 2012; Tuluc et al., 2016), construct open and closed state conformations of Kv channels (Yarov-Yarovoy et al., 2006a; Pathak et al., 2007), and model the interactions of scorpion toxins with voltage-gated Na+ channels (Cestèle et al., 2006; Wang et al., 2011; Zhang et al., 2011, 2012). More recently, Rosetta has been used to simulate the access and binding of local anesthetics and anticonvulsants to Na+ channels (Boiteux et al., 2014), study the interaction of capsaicin with the TRPV1 channel (Yang et al., 2015), and rationally design a vanilloid-sensitive TRPV2 channel (Yang et al., 2016). Similarly, the KCa3.1 structural models we developed in our current study accurately identified key residues for binding of several small molecule probes and explain their atomistic mechanisms of action. We propose that our models could be useful as tools for future structure-assisted KCa3.1 modulator design.
Material and Methods
RosettaMembrane Modeling of the KCa3.1 Channel Pore-Forming Domain.
Homology, de novo, and full-atom modeling of the KCa3.1 channel pore was performed using the Rosetta molecular modeling suite (Rohl et al., 2004) using membrane environment specific energy functions (Yarov-Yarovoy et al., 2006b, 2012; Barth et al., 2007) and loop modeling applications (Wang et al., 2007; Mandell et al., 2009). The pore-forming domain structure of the Kv1.2-Kv2.1 chimeric channel [Protein Data Bank (PDB) ID: 2R9R] (Long et al., 2007) and the open KcsA structure (PDB ID: 3FB5) (Cuello et al., 2010) were used as templates. The HHpred (Homology detection and Structure Prediction Toolkit, Max-Planck Institute for Developmental Biology, https://toolkit.tuebingen.mpg.de/hhpred) server-based (Soding et al., 2005; Hildebrand et al., 2009) alignment between KCa3.1, Kv1.2, and KcsA used for homology modeling is shown in Fig. 1, in which 10,000 models were generated and the 1000 lowest-energy models were clustered with a root-mean-square deviation threshold of 2.0 Å between Cα atoms. The top cluster homology models of the Kv1.2- and the KcsA-based KCa3.1 channel (from the loop modeling round) were used for docking of small molecules. All structural illustrations in this paper were generated using the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera/) (Pettersen et al., 2004). PDB format files of the empty Kv1.2- and KcsA-based KCa3.1 channel models are provided in the Supplemental Material; PDB files of all other models containing docked compounds are available upon request.
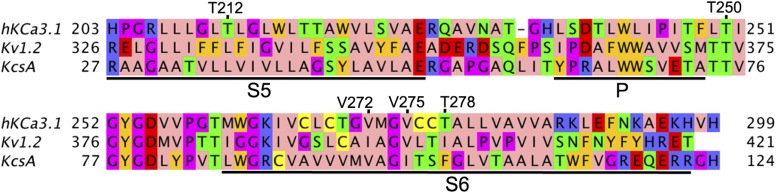
Fig. 1.
Sequence alignment between the KCa3.1, Kv2.1, and KcsA channel pore-forming domains. Transmembrane regions S5 and S6 and the pore P-helix are underlined by black bars and labeled. Positions of residues forming a part of the drug receptor sites discussed in this paper are marked and labeled. Amino acids were colored with the Jalview program (http://www.ebi.ac.uk/∼michele/jalview) (Clamp et al., 2004; Waterhouse et al., 2009) using the Zappo color scheme, where hydrophobic residues (I, L, V, A, and M) are colored pink; aromatic residues (F, W, and Y) are colored orange; positively charged residues (K, R, and H) are colored blue; negatively charged residues (D and E) are colored red; hydrophilic residues (S, T, N, and Q) are colored green; P and G colored magenta; and C is colored yellow.
RosettaLigand Docking of Small Molecules to the hKCa3.1 Channel.
Ligand docking was performed using the RosettaLigand application (Meiler and Baker, 2006; Davis and Baker, 2009), which is comprised of three stages that progress from low-resolution conformational sampling and scoring to full-atom optimization and all-atom energy function. In the first, low-resolution stage, the ligand is placed randomly within the binding site and its center of mass is constrained to move within a 10-Å-diameter sphere. Hundreds of ligand conformers are generated using OEChem, version 1.7.4 (OpenEye Scientific Software, Inc., Santa Fe, NM; www.eyesopen.com) (Hawkins et al., 2010; Hawkins and Nicholls, 2012) and are then randomly rotated as a rigid body and scored for shape compatibility with the target protein. The best-scoring models are filtered by root-mean-square deviation to eliminate near duplicates and one of the remaining models is selected at random to continue to the next stage. The second, high-resolution stage employs the Monte Carlo minimization protocol in which the ligand position and orientation are randomly perturbed by a small deviation (0.1 Å and 3°); receptor side chains are repacked using a rotamer library; the ligand position, orientation, and torsions and protein side-chain torsions are simultaneously optimized using quasi-Newton minimization and the end result is accepted or rejected based on the Metropolis criterion. Scoring uses the full-atom Rosetta energy function with softened van der Waals repulsion. The side-chain rotamers are searched simultaneously during full repack cycles and one at a time in the rotamer trials cycles. The full repack makes ∼106 random rotamer substitutions at random positions and accepts or rejects each on the Metropolis criterion. Rotamer trials choose the single best rotamer at a random position in the context of the current state of the rest of the system, with the positions visited once each in random order. The ligand is treated as a single residue and its input conformers serve as rotamers during this stage. During the energy minimization step, the finely sampled rotamer library and soft-repulsive energy function allow access to off-rotamer conformations. The third and final stage is a more stringent gradient-based minimization of the ligand position, orientation, and torsions and receptor torsions for both side chains and backbone. Scoring uses the same Rosetta energy function, but with a hard-repulsive van der Waals potential, which creates a more rugged energy landscape that is better at discriminating native from non-native binding modes. Twenty thousand docking trajectories were generated for each channel–ligand pair and the top structures were selected according to the Rosetta energy.
Site-Directed Mutagenesis and Electrophysiology.
The human wild-type KCa3.1 cDNA coding region was originally cloned in the pEGFP-C1 plasmid vector (Wulff et al., 2001). To construct the mutations, polymerase chain reaction–based site-directed mutagenesis was performed directly on the wild-type plasmid and the desired mutations were confirmed by sequencing. COS-7 cells were obtained from the American Type Culture Collection (Manassas, VA). Cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and were used for expression of KCa3.1 and mutant channels using FuGENE 6 transfection reagent (Promega, Madison, WI) in Opti-MEM reduced serum medium (Life Technologies, Benicia, CA). Transfected cells were cultured in 24-well plates for 24 or 48 hours, detached by trypsinization, washed, attached to poly-L-lysine-coated glass cover slips, and then studied in the whole-cell mode of the patch-clamp technique with an EPC-10 amplifier (HEKA Elektronik, Lambrecht, Germany). Patch pipettes were pulled from soda lime glass micro-hematocrit tubes (Kimble Chase, Rochester, NY) to resistances of 2 to 3 MΩ to achieve good access and efficient and complete cell dialysis for internals with high free Ca2+ concentrations. The pipette solution contained 145 mM K+ aspartate, 2 mM MgCl2, 10 mM HEPES, 10 mM K2EGTA, and 8.5 mM CaCl2 (1 μM free Ca2+), pH 7.2, 290 mOsm. To reduce chloride leak currents endogenous to COS-7 cells, which can contaminate the K+ current at depolarized potentials, we used a Na+ aspartate external solution containing 160 mM Na+ aspartate, 4.5 mM KCl, 2 CaCl2, 1 mM MgCl2, and 5 mM HEPES, pH 7.4, 300 mOsm. The K+ currents were elicited with voltage ramps from −120 to 40 mV of 200-millisecond duration applied every 10 seconds. Whole-cell KCa3.1 current amplitude was measured at 40 mV in the control external solution or inhibitor-containing external solutions. TRAM-34, nifedipine, and 4-phenyl-puran stocks were prepared in dimethylsulfoxide and drug dilutions were freshly prepared before each recording. Data analysis was performed using IgorPro (WaveMetrics, Inc., Portland, OR) or Microsoft Excel (Microsoft, Seattle, WA). Concentration-response curve fitting to the Hill equation to obtain the IC50 values and Hill coefficients was performed with Origin 9.0 (OriginLab, Northampton, MA). All data are expressed as mean ± S.D.
Results
Atomistic Structural Model of the KCa3.1 Pore Domain.
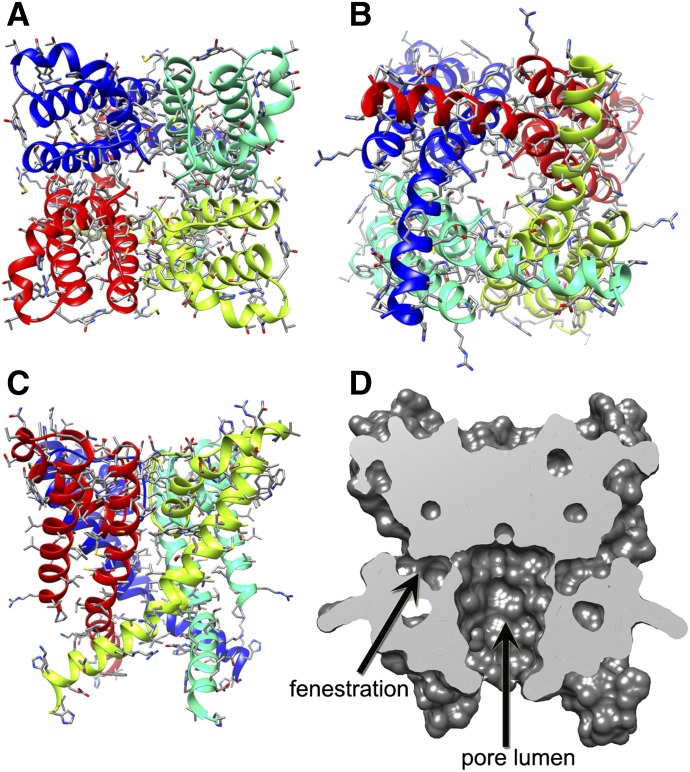
We applied Rosetta modeling to study the molecular mechanism of action of multiple small molecules interacting with multiple receptor sites within the KCa3.1 channel pore. Out of the available open state K+ channel structures, such as KcsA, MthK, KvAP, and Kv1.2, we chose the Kv1.2 channel X-ray crystal structure (Long et al., 2007) as a template for homology modeling, because several of the compounds used in our study cross react to Kv1.2 at higher concentrations (Wulff et al., 2000; Strøbæk et al., 2013). To begin with, we built a homology model of the human KCa3.1 channel pore-forming domain as described in Materials and Methods. Due to significant sequence differences between KCa3.1 and Kv1.2 channels in the loop between the S5 and P helix in the selectivity filter region (Fig. 1), we predicted the structure of this loop de novo. The most frequently sampled and lowest-energy KCa3.1 channel models converged on the model, as presented in Fig. 2, A–D and provided as a PDB file in the Supplemental Material. Key residues forming possible receptor sites for small molecule drugs in our study were positioned within the pore lumen or fenestration regions of the KCa3.1 pore structure (Fig. 2D).
Fig. 2.
Kv1.2-based homology model of the KCa3.1 channel pore-forming domain. (A) View of the ribbon representation of the homology model of the KCa3.1 channel pore-forming domain from the extracellular side of the membrane. Each subunit is colored individually. Side chains are shown in stick representation. (B) View of the model shown in (A) from the intracellular side of the membrane. (C) Transmembrane view of the model shown in (A). (D) Surface representation of the transmembrane view of the model shown in (A). The pore has been sliced in the middle to highlight the pore lumen and fenestration regions of the structure.
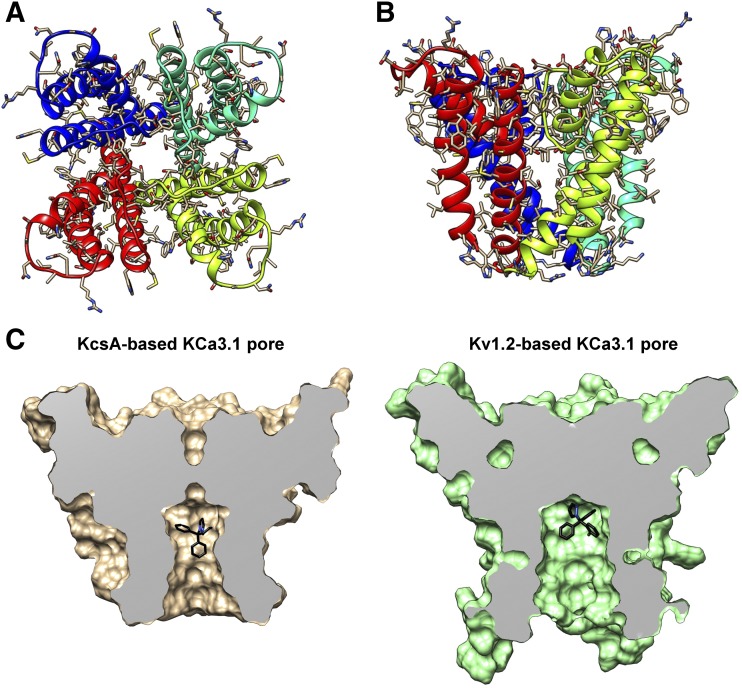
However, because the Kv1.2 channel contains a proline-valine-proline sequence motif that forms a bend in the S6 helix—and therefore might not be the most appropriate template for the corresponding leucine-valine-alanine sequence in KCa3.1 despite the similarities in pharmacology between KCa3.1 and Kv1.2—we also built a second KCa3.1 pore model using the KcsA structure in an open-conductive state (Cuello et al., 2010) as a template (Fig. 3; PDB file provided in the Supplemental Material). The KcsA-based KCa3.1 model reveals a relatively smaller pore lumen volume compared with the Kv1.2-based KCa3.1 model (Fig. 3C), which originates from differences in relative positions of the pore lining helices in the original KcsA and Kv1.2 structures.
Fig. 3.
KcsA-based homology model of the KCa3.1 channel pore-forming domain and comparison of the TRAM-34 position in the KcsA- and the Kv1.2-based models. (A) View of the ribbon representation of the homology model of the KCa3.1 channel pore-forming domain from the extracellular side of the membrane. Each subunit is colored individually. (B) Transmembrane view of the model shown in (A). (C) Surface representations of the transmembrane views of TRAM-34 (black) energy minimized in the KcsA- and Kv1.2-based model. The pore has been sliced in the middle to highlight the pore lumen and fenestration regions of the structure.
Model of the Triarylmethane Receptor Site in the Pore Lumen.
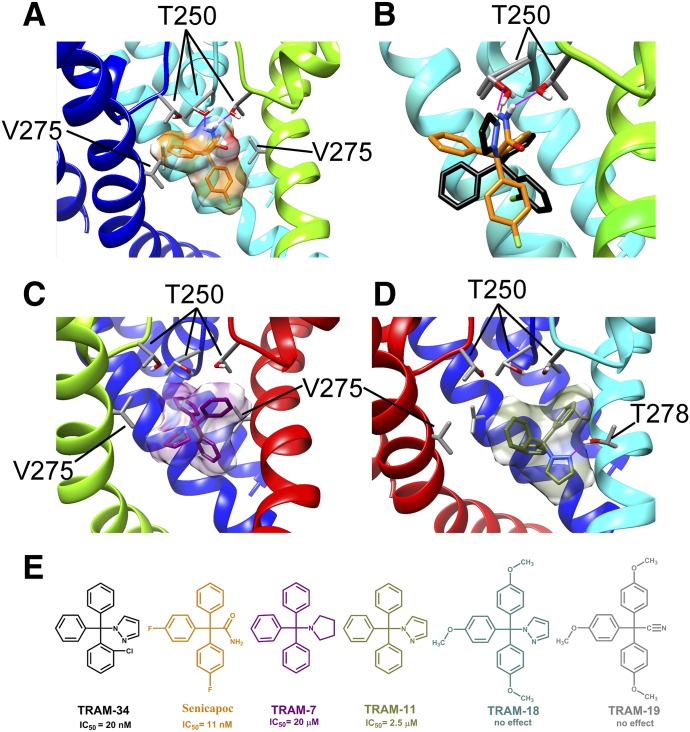
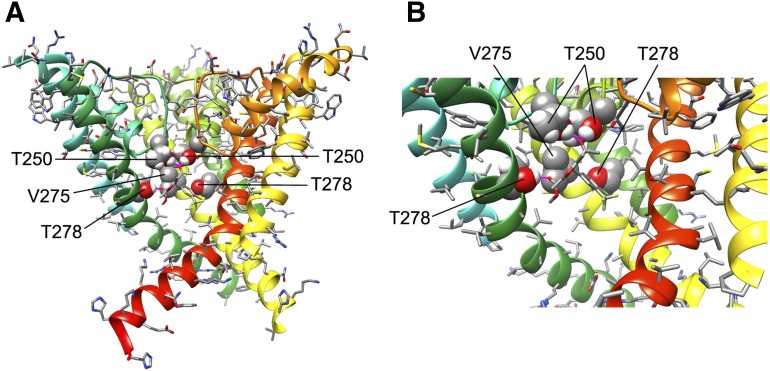
Based on mutagenesis studies, the pore lumen of KCa3.1 is known to be the target for both triarylmethane-type KCa3.1 blockers such as clotrimazole and TRAM-34 as well as the benzothiazinone NS6180 (Wulff et al., 2001; Strøbæk et al., 2013). Both TRAM-34 and NS6180 inhibit KCa3.1 with similar potencies (IC50 value of 10–20 nM) and are 200- to 1000-fold selective over other voltage-gated cation channels (Strøbæk et al., 2013). Our Kv1.2-based open state KCa3.1 model (Fig. 4) shows that the side chains of T250 (in the selectivity filter region) and V275 (in transmembrane segment S6) project directly toward the pore lumen environment and are in a position to potentially interact with small molecule blockers. We used the RosettaLigand application, which has been developed by the Meiler and Baker groups to predict small molecule/protein interactions (Meiler and Baker, 2006; Davis and Baker, 2009; Davis et al., 2009; Lemmon and Meiler, 2013; Tinberg et al., 2013; DeLuca et al., 2015), in order to generate preliminary models of TRAM-34 (Fig. 4), several other structurally related triarylmethanes (Fig. 5), and NS6180 (Fig. 6) binding to KCa3.1 in an open state as described in Materials and Methods.
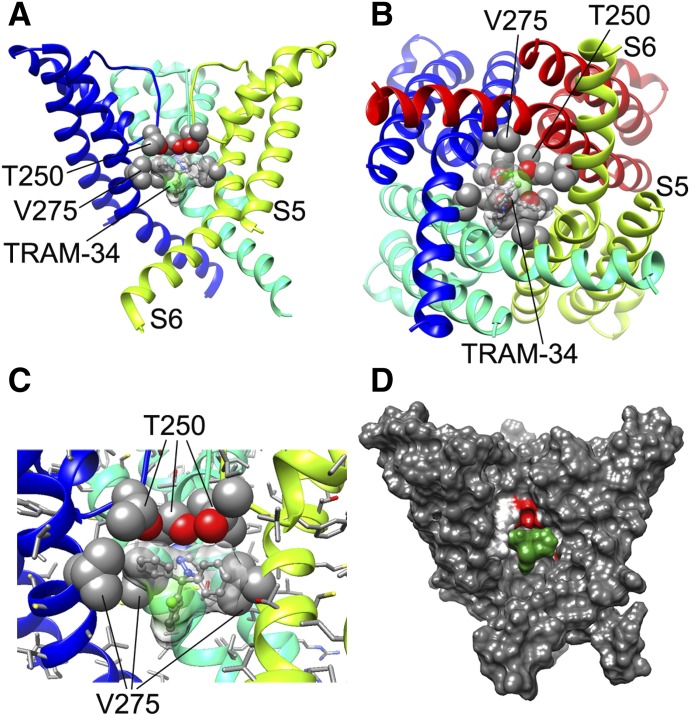
Fig. 4.
TRAM-34 interaction with the KCa3.1 channel pore. (A) Transmembrane view of the ribbon representation of one of the lowest-energy models of the KCa3.1 channel complex with TRAM-34. Only three subunits are shown for clarity and each subunit is colored individually. Side chains of T250 and V275 are shown in space-filling representation and labeled. TRAM-34 is shown in ball and stick representation with a transparent molecular surface. Transmembrane segments S5 and S6 are labeled for one of the subunits. (B) Intracellular view of the model shown in (A) with all four subunits shown. (C) Close-up of the transmembrane model shown in (A). (D) Surface representation of the transmembrane view of the model shown in (A). Only three subunits are shown for clarity. T250 and V275 side chain atoms are colored light gray (carbon), white (hydrogen), and red (oxygen). TRAM-34 is colored green.
Fig. 5.
Interaction of several TRAM-34 derivatives with the KCa3.1 channel pore. (A) Transmembrane view of the ribbon representation of one of the lowest-energy models of the KCa3.1 channel complex with senicapoc. Only three subunits are shown for clarity and each subunit is colored individually. T250 and V275 side chain atoms are colored light gray (carbon), white (hydrogen), and red (oxygen). Senicapoc is shown in stick representation with a transparent molecular surface. Hydrogen bonds are shown in pink. (B) Overlay of the lowest-energy TRAM-34 (black) and senicapoc (orange) models. Hydrogen bonds between the NH2 group of senicapoc and two T250 residues are shown in pink. The hydrogen bond between one T250 residue and the pyrazole ring of TRAM-34 is shown in green. (C) Representative binding pose of 1-tritylpyrrolodine (TRAM-7) (shown in stick presentation with a transparent molecular surface). (D) One of two dominant binding poses of TRAM-11. (E) Structures of TRAM-34, senicapoc, TRAM-11, TRAM-7, 2,2,2-tris(4-methoxyphenyl)acetonitrile (TRAM-18), and 1-[tris(4-methoxyphenyl)methyl]-1H-pyrazole (TRAM-19).
Fig. 6.
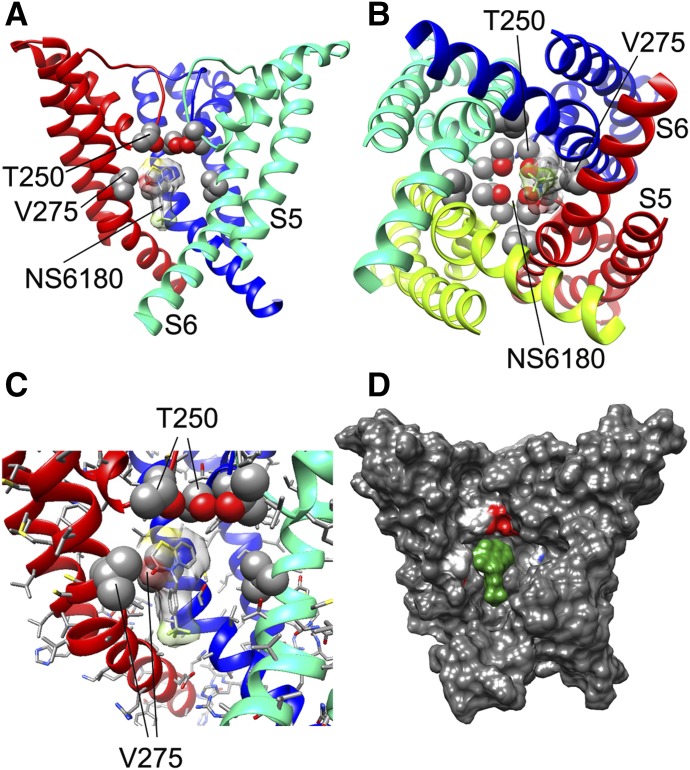
NS6180 interaction with the KCa3.1 channel pore. (A) Transmembrane view of the ribbon representation of one of the lowest-energy models of the KCa3.1 channel complex with NS6180. Only three subunits are shown for clarity and each subunit is colored individually. Side chains of T250 and V275 are shown in space-filling representation and labeled. NS6180 is shown in stick representation with a transparent molecular surface. Transmembrane segments S5 and S6 are labeled for one of the subunits. (B) Intracellular view of the model shown in (A) with all four subunits shown. (C) Close-up of the transmembrane view of the model shown in (A). (D) Surface representation of the transmembrane view of the model shown in (A). Only three subunits are shown for clarity. T250 and V275 side chain atoms are colored light gray (carbon), white (hydrogen), and red (oxygen). NS6180 is colored green.
TRAM-34 was placed into 20,000 random starting positions in the outer vestibule, the fenestration region, and the inner pore. While minimization in the outer vestibule or the fenestration region did not converge on any energetically favorable binding poses (Supplemental Fig. 1), TRAM-34 converged in the pore. Our most energetically favorable model of the KCa3.1-TRAM-34 complex suggests that TRAM-34 interacts with T250 and V275 in the pore-lumen (Fig. 4, A–C; PDB file provided in the Supplemental Material), two residues that have previously been shown to completely abolish triarylmethane sensitivity when mutated to the corresponding residues in KCa2.3 (T250S and V275A), which is not sensitive to TRAM-34 (Wulff et al., 2001). Conversely, introduction of T and V into the corresponding positions in KCa2.3 renders this channel sensitive to triarylmethanes (Wulff et al., 2001). The T250 side chain in each subunit forms part of the ion binding site in the potassium channel selectivity filter (Zhou et al., 2001). Based on our model, TRAM-34 is situated near the T250 side chains from all four subunits and blocks ion conduction directly by occupying the site that would normally be occupied by potassium before it enters the selectivity filter. The pyrazole ring of TRAM-34 is positioned between the V275 side chains from adjacent subunits with the pyrazole nitrogen forming a hydrogen bond with the T250 side chain from one of the subunits (Fig. 4, A–C). In contrast to the Kv1.2-based KCa3.1 model, TRAM-34 failed to converge in the KcsA-based KCa3.1 model (Supplemental Fig. 2). We suggest that the narrower pore lumen below the selectivity filter in the KcsA-based KCa3.1 model (Fig. 3C) does not allow shape complementarity between TRAM-34 and its receptor site near V275 and formation of a hydrogen bond with T250.
The binding orientation of TRAM-34 in the Kv1.2-based KCa3.1 model agrees well with the structure activity relationship observations that were previously made when testing 83 triarylmethanes (Wulff et al., 2000). TRAM-34’s pyrazole ring could be replaced by other hydrogen bond–accepting heterocycles such as imidazole or tetrazole but not by pyrrole or other substituents lacking a hydrogen bond acceptor without at least a 500-fold drop in potency. The molecular dimensions of the binding site are also in line with our previous estimation of the molecular dimensions of the triarylmethane pharmacophore (Wulff et al., 2000) of roughly 9.5 Å × 9.5 Å × 8.6 Å. To determine if the Rosetta KCa3.1 pore model would support these structure-activity relationships we docked a sample set of triarylmethanes (Fig. 5E) consisting of the high-affinity blocker senicapoc (IC50 value of 11 nM), 1-tritylpyrrolodine and 1-tritryl-1H-pyrazole (TRAM-11), two compounds blocking KCa3.1 with micromolar affinity, and two inactive compounds, 2,2,2-tris(4-methoxyphenyl)acetonitrile and 1-[tris(4-methoxyphenyl)methyl]-1H-pyrazole (Wulff et al., 2000). For senicapoc the 50 lowest-energy binding poses all showed two hydrogen bonds between the amino group of the ligand and the oxygen atoms of the hydroxyl groups of T250 residues from adjacent subunits (Fig. 5A). Similar to TRAM-34 (black in Fig. 5B), senicapoc (orange in Fig. 5B) is occupying the potassium binding site below the selectivity filter and is thus physically obstructing permeation. However, instead of acting as a hydrogen bond acceptor, senicapoc acts as a hydrogen bond donor (Fig. 5B). For the less potent TRAM-11 (IC50 value of 2.5 μM) Rosetta converged on two preferred binding poses that both show this compound lower down in the cavity, where despite having the same pyrazole substituent as TRAM-34 (which could theoretically form a hydrogen bond with the T250 side chain) it is out of range for a hydrogen bond (∼2.1 Å) with T250 and instead forms a hydrogen bond with T278 (Fig. 5D) and rotates around this hydrogen bond in its different poses. Unfortunately, we could not confirm this binding pose experimentally. Mutating T278 to alanine and valine resulted in nonfunctional channels, while mutating T278 to a large, aromatic phenylalanine did not significantly change TRAM-11 activity (see the Supplemental Material). Rosetta modeling of the T278F mutant explained these somewhat puzzling results by showing 278F facing away from the cavity and TRAM-11 positioned higher up and then forming a hydrogen bond with T250 (Supplemental Fig. 3). 1-Tritylpyrrolodine, a triarylmethane blocking KCa3.1 around 20 μM, which is missing the chloro-substituent and bears an aliphatic pyrrolodine ring instead of TRAM-34’s pyrazole ring, also was modeled lower down in the cavity by Rosetta (Fig. 5C), where it rotated around itself without forming any detectable hydrogen bonds or other stable interactions. Interestingly, when either one of the two inactive TRAMs, 2,2,2-tris(4-methoxyphenyl)acetonitrile or 1-[tris(4-methoxyphenyl)methyl]-1H-pyrazole (Fig. 5E), were energy minimized in the pore lumen, Rosetta did not converge on any particular binding pose and the compounds were often ejected from the cavity during energy minimization in keeping with their experimentally observed inability to block KCa3.1.
We also generated a structural model of the benzothiazinone NS6180 (Fig. 6), another small molecule that has been shown by electrophysiology and site-directed mutagenesis to interact with T250 and V275 in the KCa3.1 pore lumen (Strøbæk et al., 2013). In contrast to TRAM-34, which is positioned directly under the selectivity filter region and interacts with residues from all four subunits, NS6180 interacts with the T250 and V275 side chains from only two adjacent subunits and its elongated structure is oriented along the S6 helices (Fig. 6, A–C). Based on this model, we suggest that NS6180, although sitting differently from TRAM-34, blocks potassium conduction in a similar fashion, namely, by overlapping with the potassium binding site within the pore lumen (Zhou et al., 2001) and thus preventing ion permeation. This binding model is in agreement with electrophysiological experiments showing that TRAM-34 and NS6180 have nearly identical potencies and are very hard to wash out (Strøbæk et al., 2013).
Structural Modeling of the Unknown KCa3.1 Binding Site of the Dihydropyridine Nifedipine.
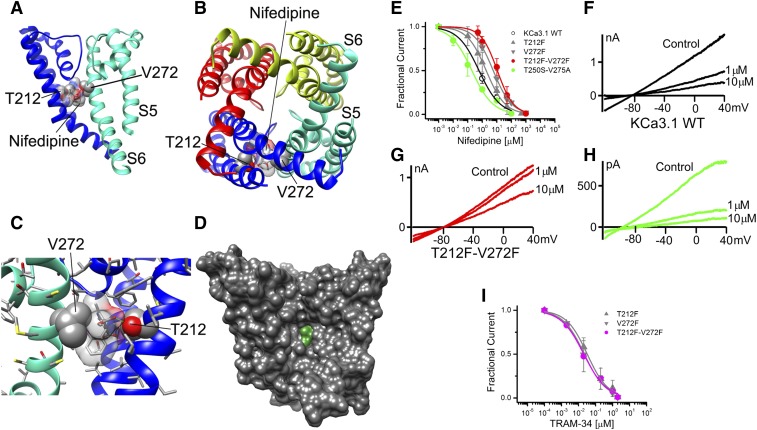
To be useful for structure-assisted design efforts, a model should also be able to correctly predict binding site locations and the molecular mechanism of action of small molecules that have not been previously mapped by electrophysiology and site-directed mutagenesis. Therefore, we decided to challenge the KCa3.1 pore model with two compounds whose binding sites are unidentified. Nifedipine, which is a potent calcium channel blocker, has long been known to also inhibit KCa3.1 at low micromolar concentrations (Jensen et al., 1998; Wulff et al., 2001). It more recently served as the template for medicinal chemists at Bayer (Leverkusen, Germany), who isosterically replaced the N in the dihydropyridine ring with an O or C atom and generated several phenyl-pyrans (Urbahns et al., 2003) and cyclohexadienes (Urbahns et al., 2005), which inhibit KCa3.1 at low nanomolar concentrations without affecting L-type calcium channels. We used the RosettaLigand method to generate structural models of nifedipine binding to the KCa3.1 pore lumen and fenestration regions. Modeling of nifedipine to the pore lumen region did not converge on any particular model (data not shown),which is in agreement with the finding that the T250S-V275A double mutant in the pore lumen does not reduce the potency of nifedipine inhibition of KCa3.1 but rather enhances nifedipine activity (Fig. 7, E–H). In contrast, modeling of nifedipine to the fenestration region converged on three groups of models with the most frequently sampled binding pose of the drug shown in Fig. 7 (PDB file provided in the Supplemental Material). This model suggests that nifedipine may fit within the fenestration of the KCa3.1 pore between the side chains of T212 (in S5) and V272 (in S6) from adjacent subunits (Fig. 7, A–C) and could be stabilizing the channel in a nonconducting conformation without directly occluding the pore.
Fig. 7.
Nifedipine interaction with the KCa3.1 channel pore. (A) Transmembrane view of the ribbon representation of one of the lowest-energy models of the KCa3.1 channel complex with nifedipine. Only two adjacent subunits are shown for clarity and each subunit is colored individually. Side chains of T212 and V272 are shown in space-filling representation and labeled. Nifedipine is shown in stick representation with a transparent molecular surface. Transmembrane segments S5 and S6 are labeled for one of the subunits. (B) Intracellular view of the model shown in (A) with all four subunits shown. (C) Close-up and 90° rotated transmembrane view of the model shown in (B) with only two subunits shown. Hydrogen bonds are shown in green. (D) Surface representation of the transmembrane view of the model shown in (B). All four subunits are shown. Nifedipine is colored green. (E) Concentration-dependent inhibition of wild-type (WT) and mutant KCa3.1 channels by nifedipine. WT KCa3.1 channels are inhibited by nifedipine with an IC50 concentration of 0.74 ± 0.18 µM (h = 0.82 ± 0.28; n = 5). While the fenestration mutants T212F (IC50 = 2.70 ± 1.98 µM; h = 0.71 ± 0.14; n = 11), V272F (IC50 = 5.86 ± 2.12 µM; h = 0.75 ± 0.06; n = 4), and T212F-V272F (10.28 ± 5.18 µM; h = 1.38 ± 0.58; n = 12) decrease nifedipine block by 4-, 8-, and 14-fold, respectively, the pore double mutant T250S-V275A (IC50 = 0.16 ± 0.15 µM; h = 0.73 ± 0.26; n = 4) does not reduce the nifedipine block. (F–H), Individual current traces showing inhibition of KCa3.1 currents in the presence 1 and 10 µM nifedipine. (I) Concentration-dependent relationship of inhibition for the fenestration mutants by TRAM-34 demonstrating the lack of an effect of the fenestration mutants on the action of this pore-selective blocker: T212F (IC50 = 29.0 ± 14.5 nM; h = 0.76 ± 0.26; n = 5), V272F (IC50 = 23.0 ± 11.5 nM; h = 0.64 ± 0.13; n = 4), and T212F-V272F (IC50 = 28.5 ± 6.3 nM; h = 0.89 ± 0.11; n = 6). Data points are expressed as mean ± S.D.; n = number of independent cells used to construct the concentration-response curves.
We tested this prediction by mutating T212 and V272 to phenylalanine, reasoning that the bulky phenylalanine side chains will overlap and close off the fenestration binding site for nifedipine, thus reducing its affinity to the channel. The T212F and T272F single mutants reduced nifedipine’s IC50 value by approximately 4- and 8-fold, respectively (Fig. 7E), while the T212F-V272F double mutant reduced the IC50 value by 14-fold (Fig. 7E). Interestingly, all three fenestration mutants retained the same high sensitivity to TRAM-34 (IC50 value of ∼20 nM) as the wild-type KCa3.1 channel, suggesting that the mutations had not significantly altered the pore lumen structure (Fig. 7I).
Structural Modeling of the Unknown Binding Sites of a 4-Phenyl-Pyran.
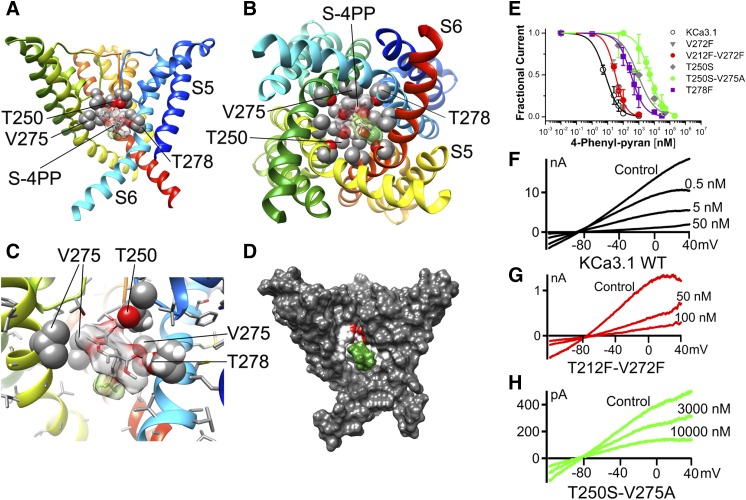
We next resynthesized an exemplary 4-phenyl-pyran from the Bayer KCa3.1 study (Urbahns et al., 2003) using an alternative synthetic route (see Supplemental Material) since we wanted to further probe the KCa3.1 model with a more potent nifedipine derivative. In electrophysiological experiments we found that the 4-phenyl-pyran 3 blocked KCa3.1 current with an IC50 value of 7.7 nM (Fig. 8E), which is in agreement with the previously reported IC50 value of 8 nM (Urbahns et al., 2003). Based on the structural similarity between nifedipine and the 4-phenyl-pyran we expected that this compound would also bind in the fenestration region. However, to our surprise, we found that the RosettaLigand method failed to converge on any energetically favorable binding pose for the R enantiomer of the 4-phenyl-pyran in the fenestration region and only predicted one hydrogen bonding interaction with threonine 212 for the S enantiomer. In contrast, modeling of both the (S)- and (R)-4-phenyl-pyran in the inner pore converged on the very energetically favorable binding pose shown for the S enantiomer in Fig. 8, A–D and for the R enantiomer in Fig. 9. Both the (S)- and (R)-4-phenyl-pyran are positioned near the side chains of T250 in the selectivity filter region, V275 in S6, and T278 in S6 from three adjacent subunits and block ion conduction directly by undergoing three hydrogen bonds and three van der Waals interactions. Subsequent electrophysiological experiments with the fenestration mutants confirmed this prediction and showed that the V272F and V212F-V272V mutants were still remarkably sensitive (IC50 value of ∼30 nM), while mutations in the pore lumen dramatically reduced potency with the T250S-V275A mutant, being more than 500-fold less sensitive (IC50 value of ∼4.4 μM). The T250 side chains from two adjacent subunits form hydrogen bonds with the carbonyl-oxygen of the acetyl substituent in the 3-position of the pyran ring (Fig. 8, A and C), while T278 from one of the subunits forms a hydrogen bond with the carbonyl-oxygen of the methyl-ester in the 5-position (Fig. 8, A and C). This observation is in agreement with the finding that mutating T278 to phenylalanine reduced the IC50 value by 40-fold. Additionally, V275 side chains from three adjacent subunits have van der Waals interactions with the (S)-4-phenyl-pyran (Fig. 8, B and C). The para-chloro atom on the phenyl ring of the 4-phenyl-pyran system is positioned between side chains of T278 and A279 in S6 (Fig. 8B), while the meta-CF3 group is oriented down toward the intracellular side of the membrane (Fig. 8C). The (R)-enantiomer is flipped but is undergoing the same interactions with T250, V275, and T278 from different subunits because of the overall symmetry of the tetrameric channel (Fig. 9). Overall, this binding orientation of the 4-phenyl-pyran and the contacts it is making to the channel through its two carbonyl oxygens, and the chloro- and trifluoromethyl substituents on the phenyl ring, are consistent with the published structure-activity relationship findings that these four moieties are essential for high-potency KCa3.1 inhibition in the phenyl-pyran series of compounds (Urbahns et al., 2003).
Fig. 8.
(S)-4-Phenyl-pyran interaction with KCa3.1 channel pore. (A) Transmembrane view of the ribbon representation of one of the lowest-energy models of the KCa3.1 channel complex with (S)-4-phenyl-pyran (shown in stick representation with a transparent molecular surface). Only three subunits are shown for clarity and each subunit is colored individually. Side chains of T250, V275, and T278 are shown in space-filling representation and labeled. Transmembrane segments S5 and S6 are labeled for one of the subunits. (B) Intracellular view of the model shown in (A) with all four subunits shown. (C) Close-up of the transmembrane view shown in (A). (D) Surface representation of the transmembrane view of the model shown in (A). Only three subunits are shown for clarity. T250, V275, and T278 side chain atoms are colored light gray (carbon), white (hydrogen), and red (oxygen). S-4-phenyl-pyran is colored green. (E) Concentration-dependent inhibition of wild-type (WT) and mutant KCa3.1 channels by 4-phenyl-pyran. WT KCa3.1 is inhibited by 4-phenyl-pyran with an IC50 value of 7.7 ± 4.5 nM (h = 1.20 ± 0.27; n = 5). While the pore mutants T250S (IC50 = 969.9 ± 242.7 nM; h = 0.61 ± 0.16; n = 3), T250S-V275A (IC50 = 4.45 ± 1.63 µM; h = 1.02 ± 0.36; n = 7), and T278F (IC50 = 321.04 ± 130.82 nM; h = 0.97 ± 0.38; n = 11) decrease the 4-phenyl-pyran block by 125-, 575-, and 40-fold, respectively, neither of the fenestration mutants V272F (IC50 = 30.1 ± 2.4 nM; h = 1.65 ± 0.48; n = 3) or T212F-V272F (IC50 = 30.2± 4.5 nM; h = 1.54 ± 0.25; n = 5) significantly reduce the 4-phenyl-pyran block. Data points are expressed as mean ± S.D.; n = number of independent cells used to construct the concentration-response curves. (F–H) Current traces showing examples of inhibition of WT KCa3.1, T212-V272F, and T250S-T275A currents by 4-phenyl-pyran.
Fig. 9.
(R)-4-Phenyl-pyran interaction with KCa3.1 channel pore. (A) Transmembrane view of the ribbon representation of one of the lowest-energy models of the KCa3.1 channel complex with (R)-4-phenyl-pyran (shown in stick representation). Only three subunits are shown for clarity and each subunit is colored individually. Side chains of T250, V275, and T278 are shown in space-filling representation and labeled. Hydrogen bonds are shown in pink. (B) Close-up view of the model shown in (A).
Discussion
Using the Rosetta molecular modeling suite we generated a molecular model of the pore domain of the calcium-activated K+ channel KCa3.1 and tested the model by first confirming previously mapped binding sites and visualizing the mechanism of TRAM-34, senicapoc, and NS6180 inhibition at the atomistic level. All three compounds block ion conduction directly by fully or partially occupying the site that would normally be occupied by K+ before it enters the selectivity filter. We then tested if our KCa3.1 model would be useful for predicting the previously unknown binding sites of nifedipine and the isosteric 4-phenyl-pyran. Rosetta predicted receptor sites for nifedipine in the fenestration region and for the 4-phenyl-pyran in the pore lumen, which could both be confirmed by site-directed mutagenesis and electrophysiology. While nifedipine is thus not a pore blocker and might be stabilizing the channel in a nonconducting conformation or interfere with gating, the 4-phenyl-pyran was found to be a classical pore blocker that directly inhibits ion conduction similar to the triarylmethanes TRAM-34 and senicapoc.
The Kv1.2 channel structure (Long et al., 2007) was one of several possible templates for homology modeling of the KCa3.1 channel pore. Of the other available open state K+ channel structures, such as MthK (Jiang et al., 2002), KvAP (Jiang et al., 2003), and KcsA (Cuello et al., 2010), we also explored the open conductive state KcsA structure here and found it to be an inferior template since the high-affinity inner pore blocker TRAM-34 failed to converge in the KcsA-based KCa3.1 pore model. Therefore, we believe that based on the overall 27% sequence identity between KCa3.1 and Kv1.2 in the pore domain and the similarities in pharmacology, Kv1.2 is the most appropriate available template in the absence of a bona fide KCa3.1 structure. Several venom peptides such as charybdotoxin and maurotoxin inhibit both Kv1.2 and KCa3.1 with similar potencies (Sprunger et al., 1996; Castle et al., 2003), suggesting similarities in the outer vestibule. Similarly, several of the small molecule KCa3.1 blockers used in this study such as TRAM-34 and NS6180 inhibit Kv1.2 at low micromolar concentrations (Wulff et al., 2000; Strøbæk et al., 2013), again suggesting that Kv1.2 is a better template than the bacterial channels despite the fact that the proline-valine-proline sequence in the Kv1.2 channel S6 segments creates a significant bend downstream from the small molecule binding site in the inner pore that is unlikely to be present in the KCa3.1 channel structure. Another possible template would have been the recently solved structure of the sodium-activated K+ channel Slo2.2 in the closed state (Hite et al., 2015). However, although initially being classified as a Ca2+-activated K+ channel channel (KCa4.1), this channel has only 7% sequence identity to KCa1.1 and even less to KCa3.1, and has therefore just been renamed as KNa1.2 according to the new International Union of Basic and Clinical Pharmacology nomenclature (Alexander et al., 2015). The channel is also insensitive to both TRAM-34 and NS6180 (Strøbæk et al., 2013), making it an unattractive template.
Our new Rosetta model of the open state conformation of the KCa3.1 channel pore correctly identified T250 and V275, two residues previously suggested based on mutagenesis to form the triarylmethane (TRAM-34 and senicapoc) and benzothiazinone (NS6180) receptor sites, as key interacting residues. Although these findings are confirmatory in nature, the atomistic models provide new insights into the orientation and the chemical nature of the interactions of the two ligands with KCa3.1. While TRAM-34 is sitting directly under the selectivity filter with its pyrazole positioned between the V275 side chains from adjacent subunits and the pyrazole nitrogen forming a hydrogen bond with the T250 side chain from one of the subunits, NS6180 is oriented along S6 interacting with the T250 and V275 side chains from only two adjacent subunits. It should also be mentioned here that an older KCa3.1 pore model, which had been built to model the interaction of charybdotoxin with the KCa3.1 pore (Rauer et al., 1999) and which was based on the closed KcsA structure (Doyle et al., 1998), did not provide enough space in the inner pore to accommodate TRAM-34 and related triarylmethanes. As shown in this study, our new KCa3.1 model based on the open Kv1.2 structure now easily accommodates TRAM-34 and other triaylmethanes such as senicapoc (Fig. 4) in the inner cavity and is in agreement with our own previously performed mutagenesis studies (Wulff et al., 2001; Strøbæk et al., 2013). Other less-potent triarylmethanes were modeled with binding poses lower down in the cavity, where they were not in a position to effectively prevent K+ permeation, while inactive compounds did not converge and were ejected from the cavity during energy minimization.
In addition to the inner pore lumen, which contains the binding sites for TRAM-34 and NS6180, we identified the fenestration region in the KCa3.1 channel model as a key binding site for nifedipine in this study (see Fig. 7). This suggests a possible mechanism of KCa3.1 inhibition by nifedipine via stabilizing the channel in a nonconducting conformation without directly occluding the pore. The presence of nifedipine in the fenestration could, for example, allosterically affect the selectivity filter similar to what has recently been reported in a crystallographic study for the binding of several dihydropyridines to the homotetrameric CaVAb channel, a model for voltage-gated Ca2+ channels (Tang et al., 2016). The presence of one dihydropyridine molecule in the lipid-facing interface between two subunits was sufficient to induce an asymmetric, nonconducting conformation in the selectivity filter of the channel (Tang et al., 2016). Alternatively, the nifedipine molecule in the fenestration could prevent the gating movement that is translated from the C-terminal calmodulin to the gate, which for KCa3.1 has been proposed to be located at the level of the selectivity filter (Klein et al., 2007; Garneau et al., 2014). Since the T212F-V272F double mutant significantly reduced nifedipine binding, but did not eliminate it completely, we think it is possible that the fenestration region may be a dynamic part of the KCa3.1 channel structure and adopt conformations that may allow low-affinity nifedipine binding. It would, of course, also be possible that similar to CaVAb (Tang et al., 2016), KCa3.1 has a second, lower-affinity binding site for dihydropyridines in the inner cavity, which would be in line with the observation that the T250S-V275A mutant is 4.5-fold more sensitive to nifedipine than the wild-type channel.
One very interesting and in some ways unexpected observation from our study was that the nifedipine isosteric 4-phenyl-pyran is not binding to the same site in KCa3.1 and does not have the same atomistic mechanism of action as its template nifedipine. Medicinal chemists generally assume when they make isosteric replacements to improve potency and selectivity that the template and the derivatives bind to the same site when deducing structure-activity relationships. This is clearly not the case here and the findings certainly surprised us. However, the pore lumen binding site for the 4-phenyl-pyran that accommodates both the S and R enantiomers is perfectly consistent with the published structure-activity relationships of the phenyl-pyrans (Urbahns et al., 2003) and carba-analogous cyclohexadienes (Urbahns et al., 2005), and the fact that racemic compounds in these two series are equally or more potent than structurally closely related nonracemic compounds. In our model there is no contact to the pyran ring O (which was the pyran ring NH in nifedipine and is a simple aliphatic carbon in the equally potent cyclohexadienes), while the carbonyl groups of the ester and acetyl substituents are stabilized by hydrogen bonds in keeping with the observation that the presence and a relatively small size of the ester or acetyl side chains in the 3- and 5- positions is absolutely required for high-potency KCa3.1 inhibition (Urbahns et al., 2003, 2005). Our model further provides an explanation for the observation that para-chloro and meta-CF3 are the optimal substitutions of the 4-phenyl-pyran system by showing van der Waals interactions between T278 and A279 and the Cl atom and the bulky, lipophilic CF3 group.
Structure-based or structure-assisted design of ion channel modulators has long been a challenging endeavor. The field has recently advanced greatly with the increasing availability of both X-ray and cryo-EM crystal ion channel structures. However, cryo-EM structure resolution does not reveal atomistic details of small molecule/channel interactions and there still is an urgent need for co-crystals of medically relevant ion channels with small molecule drugs to truly enable structure-based drug design. This knowledge gap can be bridged by computational modeling approaches such as the Rosetta molecular modeling suite with membrane environment specific energy function. Our study has shown that Rosetta can generate atomistic models of small molecule/channel interactions that 1) confirm previously mapped binding sites; 2) accurately predict unknown binding sites; 3) suggest molecular mechanisms of small molecule action; and 4) explain structure-activity relationships for compounds belonging to different structural classes and binding to different sites in the pore domain.
Abbreviations
- KCa3.1
intermediate-conductance Ca2+-activated K+
- Kv
voltage-gated K+
- NS6180
4-[[3-(trifluoromethyl)phenyl]methyl]-2H-1,4-benzothiazin-3(4H)-one
- PDB
Protein Data Bank
- senicapoc
2,2-bis-(4-fluorophenyl)-2-phenylacetamide
- TRAM-11
1-tritryl-1H-pyrazole
- TRAM-34
1-[(2-chlorophenyl)diphenylmethyl]-1H-pyrazole
Authorship Contributions
Participated in research design: Nguyen, Wulff, Yarov-Yarovoy.
Conducted experiments: Nguyen, Pressly, Jenkins, Yarov-Yarovoy.
Contributed new reagents or analytic tools: Singh.
Performed data analysis: Nguyen, Singh, Pressly, Jenkins, Yarov-Yarovoy.
Wrote or contributed to the writing of the manuscript: Nguyen, Pressly, Wulff, Yarov-Yarovoy.
Footnotes
This work was supported by startup funds from the University of California and by the CounterACT Program, National Institutes of Health Office of the Director, and National Institutes of Health National Institute of Neurologic Disorders and Stroke [Grant U54NS079202]. B.P. was supported by the National Institutes of Health National Institute of General Medical Sciences funded Pharmacology Training Program [Grant T32GM099608].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Alexander SP, Kelly E, Marrion N, Peters JA, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Southan C, Buneman OP, et al. CGTP Collaborators (2015) The Concise Guide to PHARMACOLOGY 2015/16: Overview. Br J Pharmacol 172:5729–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataga KI, Reid M, Ballas SK, Yasin Z, Bigelow C, James LS, Smith WR, Galacteros F, Kutlar A, Hull JH, et al. ICA-17043-10 Study Investigators (2011) Improvements in haemolysis and indicators of erythrocyte survival do not correlate with acute vaso-occlusive crises in patients with sickle cell disease: a phase III randomized, placebo-controlled, double-blind study of the Gardos channel blocker senicapoc (ICA-17043). Br J Haematol 153:92–104. [DOI] [PubMed] [Google Scholar]
- Barth P, Schonbrun J, Baker D. (2007) Toward high-resolution prediction and design of transmembrane helical protein structures. Proc Natl Acad Sci USA 104:15682–15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux C, Vorobyov I, French RJ, French C, Yarov-Yarovoy V, Allen TW. (2014) Local anesthetic and antiepileptic drug access and binding to a bacterial voltage-gated sodium channel. Proc Natl Acad Sci USA 111:13057–13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugnara C. (2003) Sickle cell disease: from membrane pathophysiology to novel therapies for prevention of erythrocyte dehydration. J Pediatr Hematol Oncol 25:927–933. [DOI] [PubMed] [Google Scholar]
- Castle NA, London DO, Creech C, Fajloun Z, Stocker JW, Sabatier JM. (2003) Maurotoxin: a potent inhibitor of intermediate conductance Ca2+-activated potassium channels. Mol Pharmacol 63:409–418. [DOI] [PubMed] [Google Scholar]
- Cestèle S, Yarov-Yarovoy V, Qu Y, Sampieri F, Scheuer T, Catterall WA. (2006) Structure and function of the voltage sensor of sodium channels probed by a β-scorpion toxin. J Biol Chem 281:21332–21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Lam J, Gregory CR, Schrepfer S, Wulff H. (2013) The Ca2+-activated K+ channel KCa3.1 as a potential new target for the prevention of allograft vasculopathy. PLoS One 8:e81006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Nguyen HM, Maezawa I, Grössinger EM, Garing AL, Köhler R, Jin LW, Wulff H. (2016) The potassium channel KCa3.1 constitutes a pharmacological target for neuroinflammation associated with ischemia/reperfusion stroke. J Cereb Blood Flow Metab 36:2146–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. (2004) The Jalview Java alignment editor. Bioinformatics 20:426–427. [DOI] [PubMed] [Google Scholar]
- Cuello LG, Jogini V, Cortes DM, Pan AC, Gagnon DG, Dalmas O, Cordero-Morales JF, Chakrapani S, Roux B, Perozo E. (2010) Structural basis for the coupling between activation and inactivation gates in K+ channels. Nature 466:272–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Baker D. (2009) RosettaLigand docking with full ligand and receptor flexibility. J Mol Biol 385:381–392. [DOI] [PubMed] [Google Scholar]
- Davis IW, Raha K, Head MS, Baker D. (2009) Blind docking of pharmaceutically relevant compounds using RosettaLigand. Protein Sci 18:1998–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen PG, Yarov-Yarovoy V, Scheuer T, Catterall WA. (2011) Gating charge interactions with the S1 segment during activation of a Na+ channel voltage sensor. Proc Natl Acad Sci USA 108:18825–18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA. (2009) Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc Natl Acad Sci USA 106:22498–22503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA. (2008) Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc Natl Acad Sci USA 105:15142–15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca S, Khar K, Meiler J. (2015) Fully flexible docking of medium sized ligand libraries with RosettaLigand. PLoS One 10:e0132508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280:69–77. [DOI] [PubMed] [Google Scholar]
- Edwards G, Félétou M, Weston AH. (2010) Endothelium-derived hyperpolarising factors and associated pathways: a synopsis. Pflugers Arch 459:863–879. [DOI] [PubMed] [Google Scholar]
- Fanger CM, Ghanshani S, Logsdon NJ, Rauer H, Kalman K, Zhou J, Beckingham K, Chandy KG, Cahalan MD, Aiyar J. (1999) Calmodulin mediates calcium-dependent activation of the intermediate conductance KCa channel, IKCa1. J Biol Chem 274:5746–5754. [DOI] [PubMed] [Google Scholar]
- Feske S, Wulff H, Skolnik EY. (2015) Ion channels in innate and adaptive immunity. Annu Rev Immunol 33:291–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardos G. (1958) The function of calcium in the potassium permeability of human erythrocytes. Biochim Biophys Acta 30:653–654. [DOI] [PubMed] [Google Scholar]
- Garneau L, Klein H, Lavoie MF, Brochiero E, Parent L, Sauvé R. (2014) Aromatic-aromatic interactions between residues in KCa3.1 pore helix and S5 transmembrane segment control the channel gating process. J Gen Physiol 143:289–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanshani S, Wulff H, Miller MJ, Rohm H, Neben A, Gutman GA, Cahalan MD, Chandy KG. (2000) Up-regulation of the IKCa1 potassium channel during T-cell activation. Molecular mechanism and functional consequences. J Biol Chem 275:37137–37149. [DOI] [PubMed] [Google Scholar]
- Girodet PO, Ozier A, Carvalho G, Ilina O, Ousova O, Gadeau AP, Begueret H, Wulff H, Marthan R, Bradding P, et al. (2013) Ca2+-activated K+ channel-3.1 blocker TRAM-34 attenuates airway remodeling and eosinophilia in a murine asthma model. Am J Respir Cell Mol Biol 48:212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins PC, Nicholls A. (2012) Conformer generation with OMEGA: learning from the data set and the analysis of failures. J Chem Inf Model 52:2919–2936. [DOI] [PubMed] [Google Scholar]
- Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT. (2010) Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J Chem Inf Model 50:572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzmann D, Warth R. (2008) Physiology and pathophysiology of potassium channels in gastrointestinal epithelia. Physiol Rev 88:1119–1182. [DOI] [PubMed] [Google Scholar]
- Hildebrand A, Remmert M, Biegert A, Söding J. (2009) Fast and accurate automatic structure prediction with HHpred. Proteins 77 (Suppl 9):128–132. [DOI] [PubMed] [Google Scholar]
- Hite RK, Yuan P, Li Z, Hsuing Y, Walz T, MacKinnon R. (2015) Cryo-electron microscopy structure of the Slo2.2 Na+-activated K+ channel. Nature 527:198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii TM, Silvia C, Hirschberg B, Bond CT, Adelman JP, Maylie J. (1997) A human intermediate conductance calcium-activated potassium channel. Proc Natl Acad Sci USA 94:11651–11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen BS, Strobaek D, Christophersen P, Jorgensen TD, Hansen C, Silahtaroglu A, Olesen SP, Ahring PK. (1998) Characterization of the cloned human intermediate-conductance Ca2+-activated K+ channel. Am J Physiol 275:C848–C856. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. (2002) Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417:515–522. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. (2003) X-ray structure of a voltage-dependent K+ channel. Nature 423:33–41. [DOI] [PubMed] [Google Scholar]
- Joiner WJ, Wang LY, Tang MD, Kaczmarek LK. (1997) hSK4, a member of a novel subfamily of calcium-activated potassium channels. Proc Natl Acad Sci USA 94:11013–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili-Araghi F, Jogini V, Yarov-Yarovoy V, Tajkhorshid E, Roux B, Schulten K. (2010) Calculation of the gating charge for the Kv1.2 voltage-activated potassium channel. Biophys J 98:2189–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H, Garneau L, Banderali U, Simoes M, Parent L, Sauvé R. (2007) Structural determinants of the closed KCa3.1 channel pore in relation to channel gating: results from a substituted cysteine accessibility analysis. J Gen Physiol 129:299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler R, Wulff H, Eichler I, Kneifel M, Neumann D, Knorr A, Grgic I, Kämpfe D, Si H, Wibawa J, et al. (2003) Blockade of the intermediate-conductance calcium-activated potassium channel as a new therapeutic strategy for restenosis. Circulation 108:1119–1125. [DOI] [PubMed] [Google Scholar]
- Lemmon G, Meiler J. (2013) Towards ligand docking including explicit interface water molecules. PLoS One 8:e67536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon NJ, Kang J, Togo JA, Christian EP, Aiyar J. (1997) A novel gene, hKCa4, encodes the calcium-activated potassium channel in human T lymphocytes. J Biol Chem 272:32723–32726. [DOI] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, MacKinnon R. (2007) Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450:376–382. [DOI] [PubMed] [Google Scholar]
- Maezawa I, Jenkins DP, Jin BE, Wulff H. (2012) Microglial KCa3.1 channels as a potential therapeutic target for Alzheimer’s disease. Int J Alzheimers Dis 2012:868972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell DJ, Coutsias EA, Kortemme T. (2009) Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods 6:551–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaughton-Smith GA, Burns JF, Stocker JW, Rigdon GC, Creech C, Arrington S, Shelton T, de Franceschi L. (2008) Novel inhibitors of the Gardos channel for the treatment of sickle cell disease. J Med Chem 51:976–982. [DOI] [PubMed] [Google Scholar]
- Meiler J, Baker D. (2006) ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins 65:538–548. [DOI] [PubMed] [Google Scholar]
- Pathak MM, Yarov-Yarovoy V, Agarwal G, Roux B, Barth P, Kohout S, Tombola F, Isacoff EY. (2007) Closing in on the resting state of the Shaker K+ channel. Neuron 56:124–140. [DOI] [PubMed] [Google Scholar]
- Peña TL, Rane SG. (1999) The fibroblast intermediate conductance KCa channel, FIK, as a prototype for the cell growth regulatory function of the IK channel family. J Membr Biol 172:249–257. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Rauer H, Pennington M, Cahalan M, Chandy KG. (1999) Structural conservation of the pores of calcium-activated and voltage-gated potassium channels determined by a sea anemone toxin. J Biol Chem 274:21885–21892. [DOI] [PubMed] [Google Scholar]
- Rohl CA, Strauss CE, Misura KM, Baker D. (2004) Protein structure prediction using Rosetta. Methods Enzymol 383:66–93. [DOI] [PubMed] [Google Scholar]
- Rufo PA, Merlin D, Riegler M, Ferguson-Maltzman MH, Dickinson BL, Brugnara C, Alper SL, Lencer WI. (1997) The antifungal antibiotic, clotrimazole, inhibits chloride secretion by human intestinal T84 cells via blockade of distinct basolateral K+ conductances. Demonstration of efficacy in intact rabbit colon and in an in vivo mouse model of cholera. J Clin Invest 100:3111–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumilina E, Lam RS, Wölbing F, Matzner N, Zemtsova IM, Sobiesiak M, Mahmud H, Sausbier U, Biedermann T, Ruth P, et al. (2008) Blunted IgE-mediated activation of mast cells in mice lacking the Ca2+-activated K+ channel KCa3.1. J Immunol 180:8040–8047. [DOI] [PubMed] [Google Scholar]
- Si H, Heyken WT, Wölfle SE, Tysiac M, Schubert R, Grgic I, Vilianovich L, Giebing G, Maier T, Gross V, et al. (2006) Impaired endothelium-derived hyperpolarizing factor-mediated dilations and increased blood pressure in mice deficient of the intermediate-conductance Ca2+-activated K+ channel. Circ Res 99:537–544. [DOI] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acid Res 33:W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprunger LK, Stewig NJ, O’Grady SM. (1996) Effects of charybdotoxin on K+ channel (KV1.2) deactivation and inactivation kinetics. Eur J Pharmacol 314:357–364. [DOI] [PubMed] [Google Scholar]
- Strøbæk D, Brown DT, Jenkins DP, Chen YJ, Coleman N, Ando Y, Chiu P, Jørgensen S, Demnitz J, Wulff H, et al. (2013) NS6180, a new KCa3.1 channel inhibitor prevents T-cell activation and inflammation in a rat model of inflammatory bowel disease. Br J Pharmacol 168:432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Gamal El-Din TM, Swanson TM, Pryde DC, Scheuer T, Zheng N, Catterall WA. (2016) Structural basis for inhibition of a voltage-gated Ca2+ channel by Ca2+ antagonist drugs. Nature 537:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinberg CE, Khare SD, Dou J, Doyle L, Nelson JW, Schena A, Jankowski W, Kalodimos CG, Johnsson K, Stoddard BL, et al. (2013) Computational design of ligand-binding proteins with high affinity and selectivity. Nature 501:212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama K, Wulff H, Chandy KG, Azam P, Raman G, Saito T, Fujiwara Y, Mattson DL, Das S, Melvin JE, et al. (2008) The intermediate-conductance calcium-activated potassium channel KCa3.1 contributes to atherogenesis in mice and humans. J Clin Invest 118:3025–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuluc P, Yarov-Yarovoy V, Benedetti B, Flucher BE. (2016) Molecular interactions in the voltage sensor controlling gating properties of CaV calcium channels. Structure 24:261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbahns K, Goldmann S, Krüger J, Horváth E, Schuhmacher J, Grosser R, Hinz V, Mauler F. (2005) IKCa-channel blockers. Part 2: discovery of cyclohexadienes. Bioorg Med Chem Lett 15:401–404. [DOI] [PubMed] [Google Scholar]
- Urbahns K, Horváth E, Stasch JP, Mauler F. (2003) 4-Phenyl-4H-pyrans as IKCa channel blockers. Bioorg Med Chem Lett 13:2637–2639. [DOI] [PubMed] [Google Scholar]
- Vandorpe DH, Shmukler BE, Jiang L, Lim B, Maylie J, Adelman JP, de Franceschi L, Cappellini MD, Brugnara C, Alper SL. (1998) cDNA cloning and functional characterization of the mouse Ca2+-gated K+ channel, mIK1. Roles in regulatory volume decrease and erythroid differentiation. J Biol Chem 273:21542–21553. [DOI] [PubMed] [Google Scholar]
- Wang C, Bradley P, Baker D. (2007) Protein-protein docking with backbone flexibility. J Mol Biol 373:503–519. [DOI] [PubMed] [Google Scholar]
- Wang J, Yarov-Yarovoy V, Kahn R, Gordon D, Gurevitz M, Scheuer T, Catterall WA. (2011) Mapping the receptor site for α-scorpion toxins on a Na+ channel voltage sensor. Proc Natl Acad Sci USA 108:15426–15431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei AD, Gutman GA, Aldrich R, Chandy KG, Grissmer S, Wulff H. (2005) International Union of Pharmacology. LII. Nomenclature and molecular relationships of calcium-activated potassium channels. Pharmacol Rev 57:463–472. [DOI] [PubMed] [Google Scholar]
- Wulff H, Gutman GA, Cahalan MD, Chandy KG. (2001) Delineation of the clotrimazole/TRAM-34 binding site on the intermediate conductance calcium-activated potassium channel, IKCa1. J Biol Chem 276:32040–32045. [DOI] [PubMed] [Google Scholar]
- Wulff H, Knaus HG, Pennington M, Chandy KG. (2004) K+ channel expression during B cell differentiation: implications for immunomodulation and autoimmunity. J Immunol 173:776–786. [DOI] [PubMed] [Google Scholar]
- Wulff H, Miller MJ, Hansel W, Grissmer S, Cahalan MD, Chandy KG. (2000) Design of a potent and selective inhibitor of the intermediate-conductance Ca2+-activated K+ channel, IKCa1: a potential immunosuppressant. Proc Natl Acad Sci USA 97:8151–8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Vu S, Yarov-Yarovoy V, Zheng J. (2016) Rational design and validation of a vanilloid-sensitive TRPV2 ion channel. Proc Natl Acad Sci USA 113:E3657–E3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Xiao X, Cheng W, Yang W, Yu P, Song Z, Yarov-Yarovoy V, Zheng J. (2015) Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat Chem Biol 11:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Baker D, Catterall WA. (2006a) Voltage sensor conformations in the open and closed states in ROSETTA structural models of K+ channels. Proc Natl Acad Sci USA 103:7292–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, DeCaen PG, Westenbroek RE, Pan CY, Scheuer T, Baker D, Catterall WA. (2012) Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci USA 109:E93–E102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Schonbrun J, Baker D. (2006b) Multipass membrane protein structure prediction using Rosetta. Proteins 62:1010–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Yarov-Yarovoy V, Scheuer T, Karbat I, Cohen L, Gordon D, Gurevitz M, Catterall WA. (2011) Structure-function map of the receptor site for β-scorpion toxins in domain II of voltage-gated sodium channels. J Biol Chem 286:33641–33651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Yarov-Yarovoy V, Scheuer T, Karbat I, Cohen L, Gordon D, Gurevitz M, Catterall WA. (2012) Mapping the interaction site for a β-scorpion toxin in the pore module of domain III of voltage-gated Na+ channels. J Biol Chem 287:30719–30728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. (2001) Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 414:43–48. [DOI] [PubMed] [Google Scholar]