Abstract
During this decade, breakthrough conceptual shifts have commenced to emerge in the field of Alzheimer’s disease (AD) recognizing risk factors and the non-linear dynamic continuum of complex pathophysiologies amongst a wide dimensional spectrum of multi-factorial brain proteinopathies/neurodegenerative diseases. As is the case in most fields of medicine, substantial advancements in detecting, treating and preventing AD will likely evolve from the generation and implementation of a systematic precision medicine strategy. This approach will likely be based on the success found from more advanced research fields, such as oncology.
Precision medicine will require integration and transfertilization across fragmented specialities of medicine and direct reintegration of Neuroscience, Neurology and Psychiatry into a continuum of medical sciences away from the silo approach. Precision medicine is biomarker-guided medicine on systems-levels that takes into account methodological advancements and discoveries of the comprehensive pathophysiological profiles of complex multi-factorial neurodegenerative diseases, such as late-onset sporadic AD. This will allow identifying and characterizing the disease processes at the asymptomatic preclinical stage, where pathophysiological and topographical abnormalities precede overt clinical symptoms by many years to decades. In this respect, the uncharted territory of the AD preclinical stage has become a major research challenge as the field postulates that early biomarker guided customized interventions may offer the best chance of therapeutic success. Clarification and practical operationalization is needed for comprehensive dissection and classification of interacting and converging disease mechanisms, description of genomic and epigenetic drivers, natural history trajectories through space and time, surrogate biomarkers and indicators of risk and progression, as well as considerations about the regulatory, ethical, political and societal consequences of early detection at asymptomatic stages. In this scenario, the integrated roles of genome sequencing, investigations of comprehensive fluid-based biomarkers and multimodal neuroimaging will be of key importance for the identification of distinct molecular mechanisms and signaling pathways in subsets of asymptomatic people at greatest risk for progression to clinical milestones due to those specific pathways. The precision medicine strategy facilitates a paradigm shift in Neuroscience and AD research and development away from the classical “one-size-fits-all” approach in drug discovery towards biomarker guided “molecularly” tailored therapy for truly effective treatment and prevention options. After the long and winding decade of failed therapy trials progress towards the holistic systems-based strategy of precision medicine may finally turn into the new age of scientific and medical success curbing the global AD epidemic.
Keywords: Alzheimer’s disease, biomarkers, systems biology, precision medicine, precision medicine initiative
Introduction
The emerging paradigm of precision medicine (1) aims at optimizing the effectiveness of disease prevention and therapy, by considering an individual’s specific biological makeup (e.g. genetic, epigenetic, biomarker, phenotypic, lifestyle and psychosocial characteristics) that recognizes and embraces the heterogeneity of disease for targeted interventions aimed at specific biological subsets rather than the traditional concept of neurodegenerative diseases or brain proteinopathies, such as Alzheimer’s disease (AD) as homogenous clinicopathological or clinicobiological entities. Therefore, drug discovery and development in precision medicine starkly contrast with the classical “one-drug-fits-all” approach. This precision medicine, biomarker guided, therapy strategy is what has led to drastically improved treatment success in oncology. Historically and persistent to date, medicines are developed for categorical, typically clinically defined, “neurodegenerative disease” entities representing advanced late stages of biological dysfunction converging into clinical symptomatic phenotypes. This historically developed “one-size-fits-all” approach for treating biologically heterogeneous groups of clinical phenotypes continues to be utilized for the development of early detection, intervention and prevention options as well, where biomarker candidates are being validated against the plethora of heterogeneous clinical operationalized syndromes, rather than against genetically (risk profile) and biologically (molecular mechanisms and cellular pathways) determined entities.
We hypothesize that with the introduction of precision medicine into Neurology, Psychiatry and Neuroscience, these specialties will be reintegrated into the broader medical and scientific spectrum facilitating a comprehensive holistic systems model of disease aimed at effectively detecting, treating and preventing neurodegenerative diseases, such as AD beginning with the primary care provider and integration of lessons learned from the oncology, infectious disease and cardiovascular spaces. Additionally, the research and development community also integrate broader disciplines to overcome the challenge of both precise early preclinical detection and effective prevention and disease modification. This scientific revolution will only be possible due to the ever increasing array of customized mechanistic compounds and advancing technologies for more precise molecular targeting underlying specific biological dysfunction (pathophysiology) in order to curb the global epidemic of age-related sporadic neurodegenerative diseases, such as AD.
Besides other areas of substantial progress in medicine, the theoretically and scientifically matured translational research field of oncology has already initiated implementation and is stepping up practical progress of precision medicine, primarily due to the identification of the genomic nature of the malignant pathophysiology driven through individual patterns of oncogenes in affected patients and patient subgroups. We propose to learn from these lessons and allow unrestrictive and undogmatic exploration with transfertilization into neuroscience, neurology and psychiatry. Currently, precision medicine is in the process to be applied broadly across an ever increasing number of diseases, thanks to the implementation of large-scale biological databases and the development of high-throughput screening methods – the “omic” tools – discovering and characterizing disease mechanism related biomarkers. This methodologically exploratory, integrative and interdisciplinary approach, underlying precision medicine, is referred to as systems biology (SB) based on systems theory (2, 3).
A plethora of molecular alterations have been described in AD brain pathophysiology including, but not restricted to, modifications in amyloid precursor protein metabolism (4), tau phosphorylation (5), lipid alterations (6), membrane lipid dysregulation (7), mitochondrial dysfunction, amplified oxidative stress, activation of immunological and neuroinflammatory pathways (8), and the anomalous interplay of brain neurotransmitter systems (9). Given that these perturbations are reciprocally interrelated, a comprehensive exploratory systemic approach seems necessary in order to shed sufficient light on the decade long non-linear dynamic pathogenesis, particularly of polygenic late-onset sporadic AD across time and space and systems, including a final scientific frontier, the complex neural networks (10, 11).
The objective of precision medicine is to decipher the specific biological and molecular perturbations associated with disease among specific sub-populations. This approach helps understand the final diagnostic dilemma of clinical heterogeneity by identifying a person’s comprehensive and characteristic pattern of risk factors and biological dysfunction, as reflected by genomic and genetic variants, neuroimaging indicators (structural, functional, metabolic) as well as fluid-based biological markers (cerebrospinal fluid [CSF], blood, urine, saliva). By understanding the complexity of these alterations and identifying the importance of specific alterations among groups of patients, a refined preventive or therapeutic approach that is specifically personalized (i.e. “customized”) to the individual can be applied.
Different categories and methodological modalities of indicators will serve as innovative molecular mechanistic biomarkers providing the in vivo measurement of specific pathophysiological and topographic features in AD. These genetic drivers and related encoding and expression products, such as fluid biomarkers will foster the selection of the most beneficial treatment regime for individual patients by making through assessment of the molecular pathophysiological events responsible for the patient’s progression to clinical symptoms at different disease stages. Thus, effective targeted drugs as focused therapeutic strategies – i.e. “molecularly” targeted therapies for precise treatment of molecular pathophysiological pathways associated with AD – will be developed and/or improved (1,12). In this respect, the future neurologist and psychiatrist, as the oncologist today, will be able to deliver optimally targeted and timed interventions tailored to the definite biological profiles of patients. Notably, the Institute of Medicine (IOM) has summoned a number of boards of experts to examine key issues related to biomarkers, biomarker testing, genomics, and correlated disciplines. These efforts have emphasized the need for an efficient investigation of all the opportunities and challenges related to biomarker assays for “molecularly” targeted therapies. Particularly, in recent times, the IOM summoned a Committee on Policy Issues in the Clinical Development and Use of Biomarkers for Molecularly Targeted Therapies in order to provide suggestions on key clinical practice, regulatory, and reimbursement issues. Particularly, this led to the conceptualization of ten comprehensive recommendations – namely, the IOM Committee Recommendations for Advancing Appropriate Use of Biomarker Tests for Molecularly Targeted Therapies – based on the idea that properly validated and implemented biomarker tests and targeted therapies hold substantial ability to improve the quality of patient care and ameliorate significant clinical outcomes (13). These steps are supposed to allow precision medicine to express its potential for improving patient care and clinical outcomes (13).
The systems biology (SB) paradigm for complex multifactorial diseases: from systems theory to precision medicine
Next-generation molecular and high-throughput techniques are opening new avenues of research towards the discovery of mechanisms and networks underlying complex multifactorial diseases (14–19). These networks enhance progress towards new molecular signatures, comprehensive risk classification and translational (directly applicable to patient) targeted interventions leading to the conceptualization of the precision medicine paradigm (1,20–24). The most influential methodological and technical advancements for precision medicine are innovations in genome sequencing, which has led to several In Vitro Diagnostics (IVDs) in the cancer field. However, recent advancements in whole-genome sequencing (WGS) and screening of individuals’ sequences, copy number variants and structural rearrangements, candidate pathogenic or protective) are likely to reach the clinic as a routine procedure in the next 5–10 years (25).
Next-generation sequencing (NGS) technologies are already delivering in terms of both the detection and treatment of diseases with a basic genomic component (e.g., Mendelian and, as yet, uncharacterized diseases) (14,15,26). However, many complex diseases – including diabetes, neurodegenerative diseases, and most cancers will require a SB-based approach to identify effective interventions. A comprehensive understanding of the full pathophysiological spectrum of dimensional (not categorical) neurodegenerative diseases (proteinopathies), such as AD in precision medicine will require several advancements:
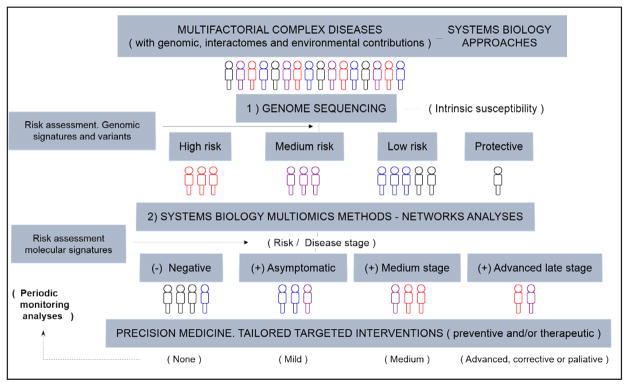
I) understanding of the multifactorial nature of the disease (i.e., involving a combination of genomic, epigenomic, interactomic, and environmental factors); II) resolution of the “altered networks”, affecting essential modules and interactomes; III) realization of the nonlinear dynamic aspect of the disease translated through its array of independent and interrelated mechanisms, with a fine balance, interplay with and between impaired complex networks and homeostatic defense mechanisms (14,15). For multifactorial diseases like AD, comprehensive holistic systems-level approaches are necessary, which is a strength of the SB paradigm, which aims at understanding the genotype-phenotype relationships and mechanisms at the levels of genome/epigenome, transcripts (RNAs), proteins/peptides, metabolites, interactomes, and environmental factors participating in complex cellular networks. SB is not so much concerned with inventories of working parts but, rather, with how those parts interact to produce working units of biological organization whose properties are much greater than the sum of their parts. Additionally, SB seeks to understand what makes complex networks and systems sustainable and viable, and how complex diseases can arise from “altered networks states”. Understanding these systems and networks in their functional and dysfunctional states can reveal characteristic molecular signatures and candidates for tailored interventions, according to the precision medicine paradigm (14–17,19,27–29). On this basis, the conceptualization of the SB paradigm for multifactorial diseases is presented in Figure 1. Briefly, comprehensive assessments of candidate groups of individuals should start with genome sequencing to reveal genomic signatures, for basic risk assessment at the genomic level (i.e. intrinsic susceptibility to disease). From here, further comprehensive systems-level analyses, with SB multi-omics methods (experimental and computational) are needed (14–16). These advanced methods, with incorporation of new standardized techniques and guidelines, continuously updated and refined, are needed to reveal specific molecular signatures and biomarkers, the underlying mechanisms and actual disease risk and disease stage, towards mechanistically-based targeted interventions (preventive and/or therapeutic); this represents the “true precision medicine” paradigm (14–15). Definitively, the key message is that for precision medicine-based strategies of complex multifactorial diseases to succeed, systems-level approaches are absolutely necessary (Figure 1).
Figure 1.
The systems biology (SB) paradigm for complex multifactorial diseases: from SB-based approaches to precision medicine. Pipeline. Multifactorial diseases involve genomics, interactomes, and environmental contributions for which SB-based approaches are needed. Comprehensive screenings of individuals, groups, and subgroups need to start with advanced genome sequencing methods in order to unveil specific variants and genomic signatures for basic risk assessment at genomic level (i.e. intrinsic susceptibility to disease) (point 1). From here, further comprehensive systems-level analyses, with SB multi-omics networks methods, both experimental and computational, are needed (point 2) (14–16). These advanced methods, with incorporation of new standardized techniques and guidelines, are expected to reveal specific molecular signatures and biomarker patterns in time and space, underlying mechanisms and actual disease risk and disease stage, towards mechanistically-based, rational-tailored interventions, preventive and/or therapeutic (i.e. “true precision medicine” paradigm) (14–15)
SB-based methods in precision medicine: next-generation molecular, high-throughput “omics” and computational methods
Advanced SB-based methods, including next-generation molecular, high-throughput “omics” approaches and computational methods are continuously advancing. Comprehensive SB experiments studying transcriptome, proteome/peptidome, and metabolome patterns and interactions were first achieved in yeast, a reference “model eukaryote” (30), with most relevant approaches being compiled in standardized protocols and databases (19). This opened the way to investigations in other organisms and, finally, in humans. Thus, Snyder and coworkers (2012) performed multi-omics analyses in longitudinal studies in humans by using integrative personal “omics” profile (iPOP), monitoring panels of biomarkers and patterns towards personalized diagnosis and personalized medicine (18). While still expensive, these approaches are progressively becoming more affordable. Examples of recent advanced studies using SB-based approaches and methods applied to multifactorial diseases include: I) integrative genomic approaches including transcriptomics (RNA sequencing) for tumor profiling towards personalized cancer therapy (31); II) identification of key regulators of pancreatic cancer progression through multidimensional systems-level analysis (32); III) integrative transcriptomics, proteomics, and network analyses to reveal candidate targeted therapies in chronic myeloid leukemia (33); IV) systems-level studies including Sequential Window Acquisition of all THeoretical Mass Spectra (SWATH-MS) proteomics, together with genomics, transcriptomics, metabolomics and trans-omic data for the discovery of mechanisms and signatures in liver disease (34); V) network-level analyses of transcriptome data integrated with genome-scale biological networks (protein-protein interaction, transcriptional regulatory and metabolic) to unveil molecular signatures of ovarian diseases (35); VI) integration of NGS technologies, SB and networks approaches to identify convergent patterns in autism (36); VII) integrative systems genomics, transcriptomics and proteomics studies for evaluation of a neuroblastoma cell lines as models for Parkinson’s disease (37); VIII) systems-based approaches to neurodegenerative diseases and biomarker discovery (3, 14, 15, 38, 39). More omics-driven studies and initiatives are in progress to revolutionize the healthcare system in the direction of the precision medicine paradigm (Figure 1) (40).
The SB paradigm applied to AD
Despite the huge potential of advanced systems-level approaches applied to multifactorial diseases, the reality is that evolving precision medicine initiatives for AD and other neurodegenerative diseases (1, 20–22, 41) are still missing the essential SB framework, which is key for the successful implementation and operationalization of precision medicine strategies (Figure 1). This critical issue needs to be fully corrected in order to advance healthcare towards “true precision medicine”.
In addition to a re-orientation away from the traditional “one-size-fits-all” approach, additional work is needed to establish a precision medicine neurodegenerative disease and AD initiative, supported by consequently applied SB-based methods (Figure 1). In particular, advanced molecular and high-throughput technologies, in conjunction with computational and integrative networks tools, need to be incorporated within the same study populations/cohorts. Additionally, rigorously defined methods, guidelines and standards are needed to move from “Research Use Only” (RUO) tools to laboratory developed tests (LDTs) or IVDs, which are required for biomarker-guided precision medicine. Raw data and integrative approaches will have to be deposited in protocols series, databases, and data repositories, with essential metadata (e.g., conditions and techniques used) to support the identification of real comparable datasets, for solid analytical studies. Even though with intrinsic limitations (14, 15, 27), relevant efforts are being made and advances are steadily achieved in both the experimental and computational areas of SB (3, 14, 15, 17, 19, 39, 42).
It should be noticed that system-level methods in neurodegenerative disease and AD research present obstacles. One significant hurtle is infancy of the stage of early stage molecular diagnostics as well as the reliance solely on advanced positron emission tomography (PET) or lumbar puncture modalities. Until reliable physiological and molecular signatures and validated (i.e. IVD, LDT) biomarkers (and companion diagnostics) are available, the diagnosis often relies on the occurrence of different clinical signs, symptoms and patterns, often detected only at an advanced late stage of the disease. Another key issue is the need for standardization of methods and data records. In this regard, newly developed systems-level methodologies and protocols need to be tested and compared with previously established procedures. Once validated, they lead to the establishment of new standards and guidelines towards reliable methods and specific disease signatures (43). This represents a continuous process until reliable and affordable methods and biomarkers are approved by medicine agencies, such as the US Food and Drug Administration (FDA) clinical trials guidance with adherence to the principles of good clinical practice (GCP) (44), the European Medicine Agency (EMA), and the European Clinical Trials Database (EudraCT) (45). This process entails the development of validation procedures and guidelines for: I) collection, pre-treatment, manipulation, and preservation of samples and data records, II) analytical methods, molecular and high-throughput “omics” techniques, statistical and bioinformatics approaches for analysis and integration of truly comparable datasets, and III) reliable molecular signatures, profiles and biomarkers of disease and disease stage, together with metadata from longitudinal studies and clinical trials, in continuous refinement. More specifically, the primary advances in terms of standardization and guidelines in AD originate from global efforts performed in the area of neuroimaging and biomarkers including the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (46) and the Dominantly Inherited Alzheimer Network (DIAN) (47) (14, 15, 48, 49). To date, much of the work in molecular signatures of AD remain in the discovery or very early validation phase rather than LDT or IVD stage.
Even though precision medicine-based strategies for AD are being the subject of increasing attention and investigations (1, 20–22, 41), the requirement to apply systems-level methods and approaches to AD (Figure 1) is mostly underestimated or even overlooked. SB approaches towards precision medicine paradigm are urgently needed; to this aim, multidisciplinary worldwide collaborations will be required to progress from SB to translational systems medicine and public health. Relevant examples of systems-level approaches in AD have been recently produced (3, 14, 15, 39). Finally, latest initiatives showing a promising future for SB and precision medicine are currently being developed: I) the European Association of Systems Medicine (EASyM) (50), II) the European Prevention of Alzheimer’s disease Consortium (EPAD) (51), and III) the Precision Medicine Initiative (PMI) at the National Institutes of Health (NIH) (24) with the recent approval of a $400 million increase in AD research funding (52).
The role of Genetics in precision medicine for AD – From genomic medicine to precision medicine: personal genomics profiles
It is well-known that AD is a multi-factorial genetically complex multi-factorial disease with heritability estimates between 58–79% (53, 54). Given the increase in average life expectancy and the subsequent rise of AD prevalence, the identification of subjects at high risk of developing AD is key for prognosis and early intervention. Thus, genetics can provide a valuable starting point for advancement. AD is a heterogeneous disease caused by a combination of environmental and genetic factors. Early-onset AD (EOAD) is caused by highly penetrant variants, the majority of which are attributable to mutations in one of three genes, amyloid precursor protein (APP, located at chromosome region 21q21.2) (55), presenilin 1 (PSEN1, located at 14q24.3) (56), and presenilin 2 (PSEN2, located at 1q42.13) (57). However, late-onset AD (LOAD) accounts for more than 95% of AD cases and is caused by a more complex underlying genetic architecture. To date, along with the polymorphism in the apolipoprotein E gene (APOE, chromosome 19q13.2) (58), a number of common and moderately rare genome-wide significant (GWS) susceptibility loci are associated with LOAD (59–66). Despite enormous efforts across the research community and the successful identification of those loci, the understanding of the aetiology of non-Mendelian forms of neurodegenerative diseases remains limited and the pressure to identify subjects at high risk of AD increases.
To date, however, the vast majority of genetic work in AD has been the search for individual genes or combinations of genes associated with a dichotomous outcome of an AD diagnosis. To compound this, the vast majority of these AD diagnoses were made solely based on clinical phenotypes rather than any indication of underlying biological signatures reflected by mechanism-specific biomarker quantification. This would be akin to using genetics to determine “cancer” presence instead of specific genetic profiles of individual subtypes of cancers. From a precision medicine standpoint, what is needed in AD science is the identification of genetic signatures of specific subtypes of individuals most likely to benefit (or not benefit) from targeted therapies. Even further, combination of genomic and proteomic, metabolomics, lipidomic signatures can further refine these models as well as identify modifiable biological pathways.
In addition to GWS loci, significant evidence (p = 4.9×10–26) for a polygenic component enriched in AD has recently been reported (67). This implies that the genetic architecture of AD includes many common variants of small effect that is likely to reflect a large number of susceptibility genes contributing to a complex set of biological pathways related to disease. The polygenic scoring approach is of utility for calculating an individual level genetic risk profile that can predict disease development. The APOE ε4 allele is the strongest known genetic risk factor for AD. In the presence of APOE ε4 alleles, the area under the ROC curve (AUC) is 67.8% (95% C.I. = 66–69%). Inclusion of the numbers of APOE ε2 alleles in the logistic regression model slightly increases prediction accuracy values; in particular, the AUC increases to 68.8% (95% C.I. = 67–70%). Prediction accuracy is further enhanced (AUC = 72% (95% C.I. = 70–73%), model improvement over APOE p = 2.7×10–12) when the variable based upon GWS single nucleotide polymorphisms (SNPs) or their proxies is added.
The addition of the polygenic component based upon about 87,600 SNPs further improves the prediction accuracy: AUC = 74.5% (95% C.I. = 73–76%, model improvement over and above APOE and GWS loci is significant (p = 1.3×10–11)). Remarkably, this actual AUC value is quite close to the upper limit AUCmax = 82% (95% C.I. = 78–85%) (68) that could be achieved given the genetic epidemiology of the disease, namely disease prevalence (2%) and SNP-heritability (24%) (69,70), indicating that the polygenic risk profiling captures the SNP-heritability very well and is quite suitable for AD genetic risk prediction.
Since AD is largely a disease of older people and the prevalence of AD escalates rapidly with age, then age needs to be taken into account in the context of practical application, such as in experimental designs comparing cases with high or low polygenic risk of AD. It is known that, given the same heritability, genetic liability is a better predictor of disease status for diseases with smaller prevalence, because “a higher proportion of those with high genetic liability are actually diseased” (70). The results of the analyses stratified by age confirm this finding and show the highest AUC value in the 60–69 age group (AUC = 79.2%). In this age group, AD prevalence is about 2–3% (71) and the maximum AUCmax estimate is 82% (95% C.I. = 78–86%), which is very close and, in fact, not significantly different (p = 0.08) from the actual one.
In summary, our analyses suggest that, while as yet unknown, the majority of the remaining common variant susceptibility loci are captured, either directly or indirectly, within the polygenic risk score model and this is quite suitable for AD genetic risk prediction. This analysis also indicates that the contribution of any new findings, not already captured by polygenic risk score, to the overall prediction of AD risk is likely to be small and attributed to rare variants, since the linkage disequilibrium between low frequency causal variants and commonly genotyped SNPs is low (72). One can further enhance the prediction accuracy by adding more environmental and/or clinical information. For example, the addition of age and sex to the prediction model, increases the AUC value from 75% to 78% (95% C.I. = 77–80%) (67). As demonstrated in other complex diseases, future polygenic score analysis of variants identified by exome/genome sequencing are expected to further inform our differentiated understanding of the genetic underpinnings of AD (73).
It is now possible to add to the genomic profile non-heritable genetic variants, such as de novo copy number variants or DNA methylation status. These variables do not contribute to SNP-heritability and, therefore, such genomic profiles could exceed the AUCmax previously shown. Other potential sources of increasing prediction accuracy are gene-gene/gene-environment interactions. Further analysis clarifying the significance of loci that do not currently reach genome-wide significance in the biological pathways established as being important in disease will refine and improve the prediction accuracy.
The concept of genomic profiling is largely understood as a possibility to determine the risk of disease for an individual given their genetic variants. Comprehensive genomic profiling is essential to set the stage for targeted therapies based on the patient’s unique disease risk profile. We advocate that genomic profiling is a promising tool for predicting the risk of future progression to AD among specific subsets of early symptomatic or pre-symptomatic individuals and should be investigated. The clinical value of the current AD genomic profiling for matching patients to targeted therapies needs to be further investigated; from this viewpoint, genomic profiling based upon biological pathways (74) could be a first step. As AD is caused by a complex interplay of genetic, lifestyle, and environment factors, epidemiologic studies should be used to examine interplay of these different factors among subpopulations of AD patients, as well as pre-clinical asymptomatic AD cohorts, to determine the clinical validity, clinical utility, and public health utility.
Discovery, development, and validation of pathophysiological biomarker candidates in AD
Despite the anticipation following genome-wide association studies (GWAS) and WGS capacity, the full power of precision medicine has yet to be realized and the prescription of several drugs, outside the cancer arena, is largely based on trials and errors (75). Precision medicine is widely considered to be the “Holy Grail” of the next era of medicine where specific patients are treated with specific interventions – medication, behavioral, environmental, etc. – based on the biology of their disease. In order for precision medicine to be fully realized, a full-spectrum of mechanism-specific biomarkers must be developed, validated and integrated into clinical medicine. In this respect, the potential and key significance for blood-based biomarkers in precision medicine for AD needs to be elaborated.
Biomarker-based (guided) stratification of therapeutic intervention is the key to precision medicine, with drugs such as trastuzumab (Herceptin) and imatinib (Gleevec) being the reference model after demonstrating remarkable efficacy for specific patients (76). While the cancer arena has generated numerous companion diagnostics to guide therapy (available at http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm301431.htm), few other areas of medicine have kept pace. However, recent work points towards the use of blood-based biomarkers for realizing the potential of precision medicine paradigms for numerous disease states including, but not limited to, multiple sclerosis (MS) (77), hypertension (78), idiopathic pulmonary fibrosis (IPF) (79), allergic diseases (76), and diabetes (80).
Blood-based biomarkers are ideal complementary biomarkers to genetic, cerebrospinal (CSF), and neuroimaging biomarkers as they are time- and cost-effective and can, therefore, provide and define restricted access to more advanced and invasive biomarkers in a multi-staged diagnostic process (81). Blood-based biomarkers have been studied extensively in AD with regards to diagnostics (81–84), risk prediction (85), understanding the complexity of the pathobiology (86, 87). Within the precision medicine area, the primary need is for companion diagnostic assays (CDx) that not only aid in the identification of which patients are most likely to respond to specific interventions, but also to rule out those patients who may suffer from safety and tolerability issues (75). While most current CDx’s approved by the FDA are single-assay (and single-gene), the tremendous advancements in “omics” technologies and analytic power open novel opportunities for the development of CDx’s for a range of disease states.
How can blood-based biomarkers aid in the development of precision medicine for AD? AD involves a broad range of pathophysiological processes – including immunological mechanisms, inflammation, metabolic dysfunction, neurotrophic dysfunction, and oxidative stress – in addition to the well-characterized amyloid beta (Aβ) and tau pathophysiologies. In fact, Aβ and tau related mechanisms do not seem to occur in isolation and without interaction with other intra or extracellular mechanisms and pathways in sporadic late-onset AD. This complex web of pathophysiologies through time, space and systems dimensions of disease in the brain clashes with the “one-drug-fits-all” belief as was the case prior to the biomarker-based guided therapies in cancer (75). Therefore, profiling biological pathways associated with AD may highlight novel opportunities for therapeutics (88, 89); notably, inflammatory pathways may represent such targets (90, 91). Using inflammation as an example, there are numerous studies linking inflammation to AD pathophysiology (92–94). Inflammation and immune system alterations have been reported to be associated with AD pathophysiology and risk (95–99) and long-term use of non-steroidal anti-inflammatory drugs (NSAIDs) is related to a reduced risk of developing AD (100–102). Based on pathobiological, clinical, and epidemiological data, multiple clinical trials have been completed utilizing NSAIDs for the treatment or prevention of AD (103–106); nevertheless, all failed to meet clinical trial endpoints. However, molecular markers have the potential for the identification of specific subgroups (i.e. endophenotypes) of disease state (78) that may be more likely to benefit from specific interventions. In fact, a preliminary work shows that, by profiling the inflammatory system, it is possible to identify a specific subgroup of AD patients with detectable alterations in the inflammatory system treated in the Alzheimer’s Disease Cooperative Study (ADCS) trial who benefitted from that “failed” trial (107). Additionally, another subset of cases most likely not benefitting and suffering from adverse responses (i.e. worsening of cognition) was identified by this approach. Therefore, as demonstrated by cancer research, molecular markers may be utilized in AD and neurodegenerative diseases to identify specific subsets of patients that most likely benefit from specific interventions and differentiate them from patients who likely not benefit using the same compound (78). Additionally, given the rapidly growing graveyard of failed AD therapeutics that never made it past Phase III trials, these novel blood-based patterns can be applied to the biorepository samples from these failed trials in order to (I) demonstrate proof-of-concept and (II) provide the requisite information for novel CDx-driven clinical trials.
Additionally, it is widely believed that novel disease-modifying candidate drugs will likely succeed in Phase III AD trials in the upcoming future. However, without a precision medicine-guided approach, the field will be in a similar dilemma facing the prescription of disease-modifying anti-rheumatic drugs in rheumatoid arthritis where these drugs are prescribed essentially by trial and error (74). This approach is inefficient with regards to cost and patient outcomes. However, if blood-based profiles (i.e. algorithms) of underlying biological disturbances can be generated to identify those patients most likely to respond or even respond adversely (e.g. inflammatory events) to these disease-modifying therapies, besides and in addition of exploiting the options of pharmacogenomics, these novel drugs will have a substantially increased impact in terms of patient outcomes and medical costs. In fact, these CDx assays are currently being developed in the area of rheumatoid arthritis. Additionally, at this point, these disease-modifying therapies remove soluble or aggregated forms of “amyloid” or “tau” from the brain; however, it remains possible that other specific forms of amyloid and/or tau are more relevant to pathophysiology and progression among specific sub-populations of individuals. At this point, the molecular detection strategies are not sufficiently advanced to generate IVDs (or LDTs) for sufficiently broad numbers of forms of amyloid or tau and the disease-modifying drugs are not sufficiently tailored. However, based on the advances in cancer research, one can easily envision a near future where interventions are CDx-guided to specific forms of amyloid for specific subsets of individuals. Finally, as has been clearly demonstrated from the cancer space, the development of CDx for specific molecules must begin early in the process, ideally in co-development beginning in preclinical development and the CDx should inform the design of Phase 2 and 3 trials. As the precision medicine model advances in AD, the clinical trial design and design of CDx for molecules will also evolve.
The use of profiles and algorithms to generate precision-medicine paradigms and CDx for AD introduces a number of challenges and new obstacles not yet adequately addressed. The international blood-based biomarker working group has recently generated the first-ever pre-analytical guidelines for blood-based biomarkers in AD (108); however, CDx introduce the need to fully understand the analytical accuracy and precision on treatment effects, variances, and other aspects of the device performance itself (109). A tremendous effort has been undertaken to move CSF-based AD biomarkers from RUO towards LDTs and IVDs, in recent years. However, in the blood-based biomarker area, the vast majority of work continues to be conducted on discovery-based platforms that will likely never transition to LDTs, much less to IVDs. Moreover, there are statistical issues to consider when transitioning from a clinical trial assay and bridging from a clinical trial assay and the CDx to be utilized for implementing FDA-approved drug (110). Despite these challenges, blood-based biomarkers offer an attractive and substantial window of opportunity for the development of a precision-medicine paradigm in AD.
Evolving conception of targeted therapeutic strategies in the field of AD
High-throughput molecular profiling (“omics” techniques) and SB are currently expanding our understanding of the molecular pathophysiology of neurodegenerative diseases. This increasing knowledge holds the promise for precision medicine to be fully realized both in patients who progress to first prodromal symptoms and to the late-stage dementia syndrome (to improve symptom control and/or decrease the rate of cognitive decay) and asymptomatic subjects at risk of dementia (to implement primary or secondary prevention programs). Unfortunately, there have been historical barriers for the implementation of precision medicine trials for dementia. Accordingly, despite a continuous and devastatingly low success rate of clinical trials in AD (phase 3 trials below 1.8%), the reductionistic “one-drug-fits-all” approach has continued to be adopted to date, whereby patients are all treated according to the severity of their cognitive decline – generally measured with the Mini Mental State Examination (MMSE) – (111) and their diagnostic classification, solely based on clinical criteria, e.g., the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRA) criteria (112). As a result of this traditional strategy, “symptomatic” treatments for AD have been developed and marketed even within an unselected and largely heterogeneous patient population (113), which provide minimal benefit even at the group level. However, the well-known heterogeneity of the involved pathophysiological processes coupled with the increasing number of putative biomarkers render the current AD and neurodegenerative diseases clinical trial paradigm reductionistic and inefficient. Innovative study designs are needed to facilitate the successful clinical development of targeted agents within specific molecular phenotypes of neurodegenerative diseases. In this scenario, new transformative clinical trial designs are warranted. In such innovative “disease-modifying” trials, eligibility should be based on an accurate CDx’s rather than the classic cognitive profile and the diagnostic characteristics of patients. Importantly, these study designs hold the promise to reduce the rates of unexpected events seen in drug development against cognitive decline (e.g., the unexpected absence of benefits – and even potential harms (114) – of cholinesterase inhibitors in subjects with mild cognitive impairment), as well as to keep the number of patients recruited in the respective trials at reasonable levels. For example, the success of agents targeting Aβ formation and/or aggregations for patients with AD is critically dependent on I) the presence of amyloid pathology assessed by CSF or amyloid imaging, and II) the evidence that such pathology is having an adverse impact on cognition (particularly in the prodromal phase of illness). However, a significant barrier when implementing this approach is the limited availability of reproducible, specific, sensitive as well as cost-effective biomarkers (115,116). New study designs are needed to facilitate the successful clinical development of targeted agents, specifically requiring three critical steps (as outlined below).
There are three steps critical to the advance of precision medicine in the field of neurodegenerative diseases and dementia disorders. The first step is the identification of “at risk” individuals in a preclinical phase. This can be achieved through the assessment of I) modifiable and non-modifiable risk factors, II) cognitive profile, III) biomarker proof of disease, and IV) changes of these factors over time. All of these variables can be combined in a probabilistic model for developing AD dementia over a defined time period, as described in the previously mentioned EPAD project (51,117). Such approach may be helpful not only to define the risk at the individual level but also to moving into individual contributor roles to the global risk. In the second step, it is important to tailor treatment based on the information gathered from the first step. This goal may be achieved by I) controlling modifiable risk factors, II) enhancing resilience, and III) modifying disease course (if necessary) through specific pharmacological interventions (Figure 2). The third step consists in assessing the outcomes of the first two phases. For example, the success of a tailored intervention in a preclinical population (118) may consist in improvements in cognition (or less-than-expected declines) and normalization of pathophysiological and topographic biomarkers. In general, this new paradigm states that risk of neurodegenerative disease at the individual level declines when any given contributory factor has been blunted.
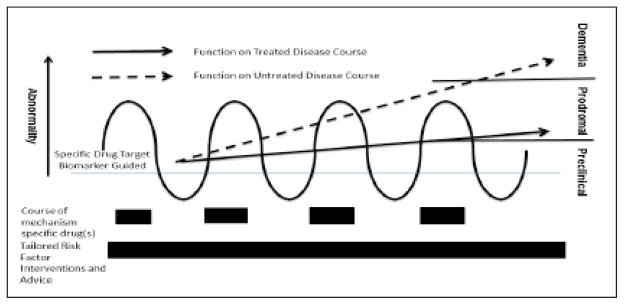
Figure 2.
Intervention paradigm for future secondary prevention of neurodegenerative diseases and dementia disorders at the preclinical level according to specific biomarker abnormalities, with the goal of allowing treatment decisions on the use of specific disease-modifying drugs. Concurrent tailored risk factor modification can optimize outcomes (as measured by cognitive and functional preclinical measures). Such approach can lead to a reduced incidence of neurodegenerative diseases and dementia disorders or a delayed progression from the preclinical stage to the full-blown clinical syndrome
Additionally, it is important to note that tailored interventions in neurodegenerative diseases and dementia disorders must not be restricted to pharmacological interventions but should also be aimed at reducing controllable risk factors and enhancing resilience. In this scenario, the future of prevention relies on evidence for individually tailored, effective, and safe interventions probably consisting of combined pharmacological and non-pharmacological approaches. The focus on non-pharmacological approaches is warranted in the earliest disease phases (generally occurring in the middle to late adulthood). This goal can be attained through the implementation of lifestyle interventions and control for known modifiable risk factors. Conversely, the use of pharmacological strategies should be limited to cases in whom the specific underlying pathological changes that precede disease phenotypes could be targeted through drugs only.
The neuroimaging perspective on targeted precision therapies for AD
Imaging pathological findings like fibrillar Aβ and neurofibrillary tangles (NFTs) in vivo may enable targeting therapy to the stage of AD based on the results of clinical trials of therapeutics in which patients have been stratified by quantitative analysis of binding by PET agents specific to the therapeutic intervention, such as anti-amyloid or anti-tau. Quantitative analysis has enabled validation of cognitive assessments with the sensitivity necessary for secondary prevention strategies during the preclinical phase of the disease, when the outcome of therapeutic intervention is more likely to yield greater benefit. Imaging of Aβ has pushed traditional AD clinical trial measures to a new limit; imaging of tau may enable development of other metrics for risk stratification, or even “liquid biopsy”, as the basis for screening patients into still earlier interventions targeting the tau infrastructure. As stated above, it remains to be known if tailored amyloid or tau interventions will be needed for specific forms of these proteins among specific sub-populations. If that becomes the case, PET imaging can enable identification of presence/absence and then CSF assays will be needed to guide specific interventions to the given patient.
The identification, localization, and quantification of typical neuropathologic changes in the post-mortem brain tissue have long been regarded as the definitive diagnosis of AD. [18F]Fluorodeoxyglucose-PET (FDG-PET) and volumetric magnetic resonance imaging (MRI) approaches aiming to support the diagnostic workup in clinical practice are currently applicable only after the onset of clinical symptoms, which usually reflects considerable progression of disease (119).
New diagnostic solutions that allow non-invasive neuroimaging of pathological and pathophysiological findings in AD could support not only the evaluation of disease processes, but also help in the development of therapies targeting either Aβ or tau (120) and/or other mechanisms. Distinctive neuropathological findings include extracellular Aβ plaques and intracellular tau-associated NFTs. These plaques are predominantly found in the precuneus, anterior and posterior cingulate, parietal, frontal and lateral temporal cortices, a characteristic distribution which can be used for visual reading of PET scans. The visual cortex and the primary sensorimotor cortex are spared of Aβ deposits until very late in the course of the AD, consistent with the sequence of subsequent clinical symptomatology (121). The utility of pattern-based analyses of presence and progression of cognitive loss and potential for precision medicine approaches remains unknown.
In healthy control subjects, the cortical uptake of Aβ agents is low in comparison with patients suffering from prodromal AD of the hippocampal type/MCI-due-to-AD or fully developed AD dementia. However, a significant proportion of cognitively healthy elderly show increased cortical Aβ binding. This finding is supported by postmortem histopathological data showing Aβ plaques upwards of 30% of the non-demented elderly population above 75 years of age, likely representing preclinical AD. However, population-based studies have yet to be conducted to inform such base rates at the individual or population level. Ongoing trials are testing the hypothesis that removal of this amyloid among cognitively normal elders will reduce risk for development of AD. Given the significant pathological comorbidity associated with presence of amyloid and tau, it is likely that a precision medicine approach of combination therapy consisting of disease-modifying therapies (at some point targeted to the specific type of amyloid and/or tau protein) with medications targeting other systems of dysfunction (e.g., inflammation, metabolic dysfunction, neurotrophic dysfunction, oxidative stress) will achieve substantially greater clinical impact than the single-drug approach.
Deposition of Aβ can be detected by amyloid-specific imaging agents for positron emission tomography – computed tomography (PET/CT) as early as fifteen years before the onset of AD symptoms whereas the next most sensitive metric, cerebral hypometabolism (FDG-PET/CT) is detectable only 10 years prior to symptom onset. Aβ PET/CT is thought to precede by 10 years the declines in even the most sensitive cognitive metrics including episodic memory (122).
Although studies have demonstrated that a negative Aβ scan indicates absence of AD with a high level of accuracy (high negative predictive value), and therefore can be used to stratify patients for trials of anti-amyloid therapy (123), for the purpose of therapeutic decision-making in clinical practice it will not be practical to screen for AD using PET scans except as the population has been stratified for risk using another method with high sensitivity (124), albeit low specificity or prognostic quality (125). To date, CSF tests are also reproducible and, in classical Kaplan-Meier curves (126) as well as models of disease progression (127), stratify to poor prognosis and seem to detect declines in the Aβ1-42 peptide approximately twenty-five years before onset of symptoms. Therefore, given the prevalence of AD at the age of 75 and the high cost of early-onset dementia, CSF could be used to stratify people beginning at the age of 40 to be referred for screening with PET/CT. More desirable and generalizable for baseline screening of larger populations for more specific secondary investigations using CSF and/or imaging methods would clearly be blood-based biomarker technology.
Cortico/cerebellar standardized uptake value ratio (SUVr) is used as an index of pathologic Aβ deposits in patients with AD, comparisons and controls in research. Other cortical regions, like pons or centrum semiovale, are being evaluated as alternative or additional reference regions. The SUVr calculates the ratio between selected cortical regions-of-interest (ROI) and the cerebellum as a reference. Quantitative evaluations using global and regional cortico-cerebellar SUVr may allow an objective, quantitative value to enhance the visual read to discern cognitively healthy individuals from AD patients (128, 129).
Quantitation also enables increased sensitivity in early stages of AD. Rodrique and colleagues performed Aβ PET studies of 137 cognitively healthy adults. Eight cortical regions were created for each hemisphere along with a cerebellar hemisphere reference region excluding the cerebral peduncles, and segmental and global cortico-cerebellar SUVr were derived. There was a progressive increase in cortical Aβ deposition with increasing age. Twenty percent of subjects older than 60 years showed increased Aβ deposition with the cut-off SUVr of 1.22 for optimum differentiation of clinically significant versus insignificant Aβ deposition. Direct correlations were demonstrated with increasing age, APOE ε4 carrier status, and inverse correlations with cognitive performance for processing speed, working memory, and reasoning ability. Episodic memory showed no correlation with Aβ uptake. These findings led the authors to conclude that detectable cognitive deficits may commence even in earlier stages of AD in patients with elevated cortical Aβ (130).
SUVr was the key measurement; visual estimation of grey-white matter differentiation may result to be complicated by a lack of consistency among different PET or PET/CT systems that may have different resolutions, image noise levels, and reconstruction algorithms (131).
Significant difference in SUVr was seen between MCI and AD subjects in precuneus, posterior cingulate, and frontal median segments. Visual evaluation of the PET scans showed a good sensitivity of 84.6% and a low specificity of 38.1% for discriminating AD patients from control subjects. On the other hand, quantitative assessment of the global cortico-cerebellar SUVr showed a very good sensitivity of 92.3% and specificity of 90.5% with a cut-off SUVr value of 1.122. The lower specificity (38.1%) of visual assessment demonstrates the difficulty of differentiating patients from healthy controls based on grey-white matter differentiation alone. The authors conclude that this lower specificity was due to the variability among scanner performance, resolution and image noise among the three participating PET/CT systems (132).
Quantitation also facilitates longitudinal assessment which can be more discerning of early disease, document the natural history of disease, serve as a biomarker in clinical trials of targeted therapy, and, then, assist in clinical practice in the individualization of therapeutic regiments. SUVr has been used to quantify changes in Aβ deposition in 49 cognitively healthy elderly subjects (MMSE > 29) and 36 subjects with MCI (MMSE > 24) who underwent Aβ PET imaging (133). Comparison of baseline and two-year follow-up SUVr levels revealed that subjects who were Aβ positive at baseline showed a significant increase in SUVr, thus suggesting progressive deposition of Aβ. On the other hand, subjects negative for Aβ at baseline did not show increase in SUVr after two years, thus suggesting lack of progression of Aβ deposition. Out of 59 Aβ negative subjects at baseline, there was transformation to Aβ positive SUVr levels only in four subjects. The authors concluded that SUVr may be a reliable and reproducible indicator for monitoring changes in Aβ deposition. Subsequent work has provided confirmatory evidence in support of this conclusion (134,135) and similar results are being obtained with PET agents imaging tau (136,137), relevant to the precision administration of therapeutic interventions targeting tau. It is possible that, like in cancer, positive biomarker may not be the indication, but rather a notion that the biomarker is dynamically changing (biomarker trajectories in a patient) becomes the indication for intervention. As such, it is possible that some amyloid “positive” cognitively normal older adults will be monitored over time to determine if amyloid levels change and, at that point, intervention begins to maximize the clinical impact of the drugs.
Accuracy in quantitation requires co-registration of the nuclear image with an anatomic image like CT or MRI. Most patients undergoing PET for neuropsychiatric diseases will also receive an MRI scan as part of their routine care, for instance to identify micro-hemorrhages. Integrated MRI and PET scanners (bi-modal or multimodal high to ultra-high field hybrid scanners) allow optimal co-registration of PET and MRI data for correction for atrophy and partial volume effects, and correction of patient motion (138). This combination of biomarkers may also be useful at identifying combination therapies. The technical development of scanner hardware and integrated analysis tools (diagnostic packages) is ever advancing.
Whether monitoring changes in Aβ deposition, or tau in NFTs, or even inflammation can be useful in assessing the efficacy of therapeutic regimens remains to be proven in clinical trials of novel therapeutics but would enable precision medicine by tailoring regimens per patient, minimizing the risks of adverse effects, and mitigating impact on healthcare budgets.
The regulatory perspective on precision medicine
Precision medicine offers a promising vision on the development of new drugs in areas with a high medical need. Assuming that some drugs may act differently in different patients, precision medicine is searching for a relevant interaction between patient and treatment leading to an improved efficacy or improved patient safety in a given subgroup of patients, thus resulting in a certain degree of treatment personalization. The investigation of new treatments in a biomarker-defined subpopulation has gained considerable attention during the last decade. Whereas the expectations regarding tailor-made medicines are high in many therapeutic areas, essentially cancer drugs have been successfully approved in biomarker defined (guided) subgroups.
Precision medicine usually involves the exploration of a predictive biomarker implying a positive treatment-by-subgroup interaction. This interaction is usually suggested by drug action and investigated in pre-clinical research or in surrogate endpoints in early clinical phases with the hope that biological and statistical interaction are interrelated. Demonstration of a true (and relevant) interaction with respect to the clinically relevant endpoint, however, often remains a difficult task. On the other hand, interaction on a specific statistical scale does not necessarily imply that there is a biological interaction but it may just be induced by the choice of the scale.
According to the usual regulatory paradigm, an independent confirmation of a medicine’s efficacy in the population to be treated in a generally large Phase III trial, not relying on historical data, is essential for drug approval. In that sense, drug approval calls for the effectiveness and tolerability in the biomarker-defined subgroup but not necessarily on a full proof of the usefulness of the restriction to a limited population. Even in drugs that have been approved in a biomarker-restricted population, evidence of a truly predictive biomarker that is capable to discriminate between the group of patients benefitting from the drug and those who do not benefit is still scarce. Often, clinical trials with hard clinical endpoints are not powered to detect a significant treatment-by-subpopulation interaction, which is further complicated by the different additional sources of variability. The presence of a variation from occasion to occasion within a patient can hardly be identified if multiple measurements per patient are difficult or impossible to perform and may be confounded with a patient-by-treatment interaction. The desired setting implies patients with a high probability to respond to treatment opposed to patients with a low probability to respond. This setting, however, is not easy to be distinguished from that of patients that all have an intermediate probability to respond.
Therefore, much work is required to explore and confirm reasonable predictive biomarkers. Validation of predictive biomarkers on the basis of clinically relevant endpoints may be rather challenging in AD. On the other hand, early surrogate endpoints that could be used for the evaluation of the biomarker-based patient selection and are capable to predict the treatment effect in clinically relevant endpoints, are not yet established. Thus, the investigation of predictive biomarkers intended to define a receptive population remains a challenge for future research.
As in other therapeutic areas, the promise of developing drugs that are highly effective in a well-defined part of the respective patient population is of critical need; however, justification of the selection is challenging, requiring more evidence and a good understanding of the underlying sources of variability. However, the biomarker-guided therapy approach to treating sub-populations of patients is well-established in the cancer field. Additionally, there are many examples in the cancer field where this approach drastically reduced the time from drug development to clinical use, which was completely due to the biomarker-based implementation throughout the trial process. Therefore, while challenging, there is an established model to regulatory approval that can be followed.
Ethical and societal considerations regarding precision medicine
Precision medicine considers the impact of individual variation at the level of genomics/epigenomics, pharmacogenomics, transcriptomics, proteomics/peptidomics, metabolomics/lipidomics, and neural network systems on the differences in predisposition to disease, pathophysiological mechanisms, and response to drugs (139). For the benefit of patients with cognitive decline and dementia disorders, it is hoped that precision medicine in the field of neuroscience and neurodegenerative diseases will at some point practically deliver its’ groundbreaking assumptions and promises (aimed at both prevention and treatment of disease). However, the practical implementation of a precision medicine-based approach for both pre-dementia and dementia patients, raises – besides medical, scientific, and organizational challenges – important ethical, legal, political and social issues. All things considered, the public acceptance of a new medical approach will be clearly influenced by recipients’ estimation of benefits and costs or risks involved.
First, the use of precision medicine in the field of neurodegenerative diseases will fundamentally change our approach of “taxonomizing” simplified (reductionistic) theoretical disease categories (ultimately challenging a so far largely unchallenged and uncontroversial definition of what is “normal” versus “pathological”) (140) into a much more differentiated and dynamic dimensional concept of genetically and biological diverse subsets of defined pathophysiologies. In this scenario, the classical categorical diagnosis based on clinical late-stage phenotypes will likely shift to the biomarker-based (guided) dimensional approach, which will likely move healthcare solutions and spending from inefficient “one-size-fits-all” treatment to more effective and less risky and more economic customized (tailored) personalized therapy and prevention (140, 141). However, the patient might choose to have genetic and/or biomarker testings for early risk assessment and detection of a potentially untreatable disease – like some forms of neurodegenerative diseases and dementia disorders – but, subsequently, decline to be informed of the test’s results, ultimately posing serious ethical decisions and more demanding and complex physician-patient communication and agreement processes (139). A second ethical issue raised by precision medicine in the field of dementia is the potential disclosure of individually sensitive information and data to employers, banks, and insurance companies, possibly leading to “genetic or biological” discrimination (139,142). Another question concerns the issue of informed consent and data rights in order to store and to make use of patients’ data in large-scale databases (143) in relation to confidentiality, security, privacy and constitutional or legal personality rights. To this aim, political stance and legal regulations will orient the way privacy issue is addressed, towards a selective limited access to anonymized data secured by a gatekeeper and protected against re-identification (139) or towards an Open Data model sustained by cryptographic techniques as, for instance, the differential privacy technique (144). Practices of security, transparency, and accountability will take on extraordinary importance in the implementation of precision medicine in the field of neurodegenerative diseases and dementia disorders. Altogether, major challenges of precision medicine encompass scientific and technological issues, security, and benefits of “omics” testing, development of new technologies and assessment methods, related ethical considerations, and socially related (economic, educational, lifestyle) data collection and practice (140). Paradoxically, the implementation of an individual-centered model is clearly dependent on a large international collective effort (140) involving various stakeholders (researchers, caregivers, payers, regulators, policy makers, governments, and citizens in general). Unfortunately and inevitably in the beginning stage, the variety of stakeholders having differentiated goals and interests as well as different levels of scientific literacy (145,146) may lead to conflicts of interest and misunderstandings in a complex societal and multidisciplinary arena blurring boundaries between research, healthcare, politics, and society. Further advances in data analysis and interpretation tools are necessary as well, so that information obtained through tests and technologies can be properly transferred and translated understandably to the primary care physicians and the public. In the future application of precision medicine, increasing attention needs to be given to these diverse ethical issues, including cost-effectiveness and social acceptability. An overriding concern remains, however, that political, ethical, and legal regulations are not being established, thus leaving problematic grey zones (Table 2). A concerted effort is needed to provide broad societal support for studies (including studies in ethics), study participation and ultimately implementation of precision medicine in the field of neurodegenerative diseases, to finally enable universal and personalized applications. Indeed, precision medicine in neuroscience, neurology and psychiatry is still a bold vision beyond the horizon of current perception, it will require to integrate genomic and biological data with phenotypical, social, cultural, and personal preferences and lifestyles to provide a more individualized prevention and treatment of biological mechanisms ultimately progressing to cognitive decline and dementia but crucially considering ethical, societal, political and public health perspectives (147).
Table 2.
Issues related to precision medicine in which political, ethical and legal regulation needs to be achieved
| Societal concerns raised by precision medicine | Ethical principles challenged by precision medicine | Requirements for societal acceptance |
|---|---|---|
| ▪ Data governance - Who will make use of the data? - How the data will be used? ▪ Data security How to prevent the disclosure of individually sensitive information leading to “genetic or biological” discrimination? |
Privacy Confidentiality | * Data protection Need for developing and improving anonymization methods including protection against re-identification, safeguards and gatekeepers, and cryptographic techniques in a privacy by design approach * Accountability in case of data leak Need for developing response plans for cases of privacy breach |
| ▪ Blurred boundaries between politics and society How to prevent misunderstandings due to conflicts of interest between various stakeholders? |
Communication | * Science and health literacy Need for accessible communication tools for all stakeholders to discuss goals and means on a “meta-level”, without excluding professional and cultural (including legal) backgrounds (e.g. scientists vs. non-scientists) * Transparency Need for willingness in the establishment of a transparency culture |
| ▪ Blurred boundaries between research and healthcare How to adapt the physician-patient communication to novel goals and means of precision medicine? |
Informed consent | * Science and health literacy Need for suitable communication tools for physicians to adapt communication to the understanding ability of the patient (particularly on individual results, secondary and incidental findings) * Transparency Need for information tools on:
|
Directions for AD biomarker research
Presently, considerable advances in discovery, development, and validation of AD-mechanism related biomarkers have paved the way for the novel era of multimodal investigations integrating modalities and different biological fluids (2,148,149). They result from neurogenetics (150–152), structural/functional/metabolic neuroimaging as well as neurophysiology (153, 154), neurochemistry using biological fluids (155–157), namely CSF (158–160) and blood (plasma/serum) (161–165). The longitudinal dynamics and predictive performance of this multimodal approach is not definitely established and should be examined according to our expectation, in terms of sensitivity and/or specificity, and their condition, i.e. in combination or isolated (166, 167). Moreover, opinions of regulatory agencies and industry stakeholders in AD biomarker discovery area are constantly in discussion and development (168–170).
Future perspectives – It is time to facilitate precision medicine in neuroscience, neurology and psychiatry
In order to swiftly advance the application of the precision medicine paradigm (1) to a broader spectrum of complex diseases (171), various governments around the world are supporting the promotion of the Precision Medicine Initiatives (PMIs). These are substantial efforts that aim at generating the extensive scientific knowledge needed to facilitate breakthrough progress in early detection, prevention and therapy and integrate and successfully utilize the model of precision medicine into every day clinical practice (172, 173).
On January 20, 2015, U.S. President Barack Obama announced a research initiative aiming at accelerating the progress toward a new era of Precision Medicine, the Precision Medicine Initiative Cohort Program (PMI-CP) (available at https://www.whitehouse.gov/precision-medicine) that is estimated to recruit a research cohort of over one million U.S. citizens. This population will be requested to give consent for extensive characterization of biological specimens and behavioral data, all interconnected to electronic health records. The systematic collection of deep, big and complex data will enable to perform observational studies of drugs and devices and, potentially, facilitate more rigorous interventional studies addressing specific questions (172,173). Although focused at the first stage on a number of important disease areas, such as cancer, this approach is explicitly expected to target brain diseases such as AD as well.
Cross-sectoral collaborations and interdisciplinary research are supposed to elucidate our understanding of how neurodegenerative diseases develop and to translate emerging knowledge more efficiently into preventive and therapeutic approaches. Some of the PMIs have provision for work in the social and regulatory sciences, and aim at engaging governments and broader publics around technological development. The concept of open and responsible innovation balances mandates for regulation, investment, and promoting access to future diagnostics and therapies in neurodegenerative diseases. The successful development of the PMI-CP will need the combination of well-established and innovative technologies for both gathering and managing big, deep and complex data (174). This will be accomplished thanks to advances in information technology that have provided significant reductions in the cost of data storage as well as comparable increases in analytic capabilities, thus allowing the assembly and analysis of massive clinical databases in biomedicine.
Finally, in addition to technological advances, individuals and patients are now on the table with scientists and clinicians, they have become active participants and have increasingly become more engaged in healthcare and health research, more connected and organized through social media, and even more “impatient” as they are eagerly looking for better treatments for both themselves and people they care. Researcher engagement with patients, caregivers, and advocacy groups to collect patient-related genomic and SB information (Figure 1) and outcome measures can be employed to increase the success of clinical trials, to enable a proactive discussion with regulatory agencies, to help the definition of therapeutic value, and to ensure that PMIs can address patients’ needs. These aspects indicate a cultural, ethical and conceptual shift critically important for the success of precision medicine. However, coverage and payment decisions for any advanced diagnostic and therapy for neurodegenerative diseases, such as AD, need to be based on available medical evidence of positive health outcomes and relative costs (175). Definitively, the use of adequate resources and a sustained commitment of time, energy, knowledge, and expertise from the scientific and biomedical communities will allow to progressively embrace the full potential of precision medicine.
Table 1.
Terminology and evolving lexicon of precision medicine
| PMI | Precision Medicine Initiative |
| PMI-CP | Precision Medicine Initiative Cohort Program |
| SB | Systems Biology |
| GWAS | Genome-Wide Association Studies |
| NGS | Next-Generation Sequencing |
| WGS | Whole-Genome Sequencing |
| iPOP | Integrative Personal “Omics” Profile |
| CDx | Companion Diagnostic Assays |
| IOM | Institute of Medicine |
| EASyM | European Association of Systems Medicine |
| EPAD | European Prevention of Alzheimer’s disease Consortium |
Acknowledgments
Funding: HH is supported by the AXA Research Fund, the Fondation Université Pierre et Marie Curie and the “Fondation pour la Recherche sur Alzheimer”, Paris, France. The research leading to these results has received funding from the program “Investissements d’avenir” ANR-10-IAIHU-06 (Agence Nationale de la Recherche-10-IA Agence Institut Hospitalo-Universitaire-6). SEO is supported by grants from the NIH/NIA (R56AG054073 and U01AG051412).
Footnotes
Conflict of interest disclosure: HH declares no competing financial interests related to the present article. He serves as Senior Associate Editor for the journal Alzheimer’s & Dementia®; he has been a scientific consultant and/or speaker and/or attended scientific advisory boards of Axovant, Anavex, Eli Lilly and company, GE Healthcare, Cytox, Jung Diagnostics, Roche, Biogen Idec, Takeda-Zinfandel, Oryzon Genomics; and receives research support from the Association for Alzheimer Research (Paris), Pierre and Marie Curie University (Paris), Pfizer & Avid (paid to institution); and has patents as co-inventor, but received no royalties: A patent in vitro multiparameter determination method for the Diagnosis and early diagnosis of neurodegenerative disorders. Patent number: 8916388 Issued. A patent in vitro procedure for diagnosis and early diagnosis of neurodegenerative diseases. Patent number: 8298784 Issued. A patent Neurodegenerative Markers for Psychiatric Conditions. Publication number: 20120196300 Issued. A patent IN VITRO MULTIPARAMETER DETERMINATION METHOD FOR THE DIAGNOSIS AND EARLY DIAGNOSIS OF NEURODEGENERATIVE DISORDERS. Publication number: 20100062463 Issued. A patent IN VITRO METHOD FOR THE DIAGNOSIS AND EARLY DIAGNOSIS OF NEURODEGENERATIVE DISORDERS. Publication number: 20100035286 Issued. A patent In vitro Procedure for Diagnosis and Early Diagnosis of Neurodegenerative Diseases. Publication number: 20090263822 Issued. A patent in vitro method for the diagnosis of neurodegenerative diseases. Patent number: 7547553 Issued. A patent CSF Diagnostic in Vitro Method for Diagnosis of Dementias and Neuroinflammatory Diseases. Publication number: 20080206797 Issued. A patent in vitro Method For the Diagnosis of Neurodegenerative Diseases. Publication number: 20080199966 Issued. A patent Neurodegenerative Markers for Psychiatric Conditions. Publication number: 20080131921 Issued. SEO has the following patents pending related to precision medicine: PCT/US2011/036496 and PCT/US2014/067562 (additional patent filed). SEO has served on an advisory board for and received honoraria from Roche and has equity in Cx Precision Medicine, Inc. RF is a salaried employee (Chief Medical Officer) of Siemens Healthineers; he is a non-voting (innovator manufacturer representative) member of FDA’s MIDAC (Medical Imaging Drugs Advisory Committee) and CMS’ MEDCAC (Medicare Evidence Development & Coverage Advisory Committee). BD reports personal fees from Eli Lilly. VEP reports personal fees from Cytox Ltd. SL has received lecture honoraria from Roche. JIC, CR, KR, KB, NB, RN have no conflict of interest to disclose
References
- 1.Reitz C. Toward precision medicine in Alzheimer’s disease. Ann Transl Med. 2016;4:107. doi: 10.21037/atm.2016.03.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hampel H, Lista S, Khachaturian ZS. Development of biomarkers to chart all Alzheimer’s disease stages: the royal road to cutting the therapeutic Gordian Knot. Alzheimers Dement. 2012;8:312–336. doi: 10.1016/j.jalz.2012.05.2116. [DOI] [PubMed] [Google Scholar]
- 3.Lista S, Khachaturian ZS, Rujescu D, Garaci F, Dubois B, Hampel H. Application of Systems Theory in Longitudinal Studies on the Origin and Progression of Alzheimer’s Disease. Methods Mol Biol. 2016;1303:49–67. doi: 10.1007/978-1-4939-2627-5_2. [DOI] [PubMed] [Google Scholar]
- 4.O’Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer’s disease. Annu Rev Neurosci. 2011;34:185–204. doi: 10.1146/annurev-neuro-061010-113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gendron TF, Petrucelli L. The role of tau in neurodegeneration. Mol Neurodegener. 2009;4:13. doi: 10.1186/1750-1326-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giannopoulos PF, Joshi YB, Praticò D. Novel lipid signaling pathways in Alzheimer’s disease pathogenesis. Biochem Pharmacol. 2014;88:560–564. doi: 10.1016/j.bcp.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Paolo G, Kim TW. Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat Rev Neurosci. 2011;12:284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verri M, Pastoris O, Dossena M, et al. Mitochondrial alterations, oxidative stress and neuroinflammation in Alzheimer’s disease. Int J Immunopathol Pharmacol. 2012;25:345–353. doi: 10.1177/039463201202500204. [DOI] [PubMed] [Google Scholar]
- 9.Francis PT. The interplay of neurotransmitters in Alzheimer’s disease. CNS Spectr. 2005;10:6–9. doi: 10.1017/s1092852900014164. [DOI] [PubMed] [Google Scholar]
- 10.Kaddurah-Daouk R, Zhu H, Sharma S, et al. Alterations in metabolic pathways and networks in Alzheimer’s disease. Transl Psychiatry. 2013;3:e244. doi: 10.1038/tp.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noorbakhsh F, Overall CM, Power C. Deciphering complex mechanisms in neurodegenerative diseases: the advent of systems biology. Trends Neurosci. 2009;32:88–100. doi: 10.1016/j.tins.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Tan L, Jiang T, Tan L, Yu JT. Toward precision medicine in neurological diseases. Ann Transl Med. 2016;4:104. doi: 10.21037/atm.2016.03.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyman GH, Moses HL. Biomarker Tests for Molecularly Targeted Therapies--The Key to Unlocking Precision Medicine. N Engl J Med. 2016;375:4–6. doi: 10.1056/NEJMp1604033. [DOI] [PubMed] [Google Scholar]
- 14.Castrillo JI, Oliver SG. Methods Mol Biol (MIMB) Series. Humana Press. Springer Science+Business Media; New York: 2016. Systems Biology of Alzheimer’s disease. [Google Scholar]
- 15.Castrillo JI, Oliver SG. Alzheimer’s as a systems-level disease involving the interplay of multiple cellular networks. In: Castrillo JI, Oliver SG, editors. Systems Biology of Alzheimer’s disease. Methods Mol Biol (MIMB) Humana Press. Springer Science+Business Media; New York: 2016. pp. 1303pp. 3–48. [DOI] [PubMed] [Google Scholar]
- 16.Castrillo JI, Pir P, Oliver SG. Yeast Systems Biology: towards a systems understanding of regulation of eukaryotic networks in complex diseases and biotechnology. In: Walhout M, Vidal M, Dekker J, editors. Handbook of Systems Biology. Elsevier; New York: 2013. pp. 343–365. [Google Scholar]
- 17.Walhout M, Vidal M, Dekker J. Handbook of Systems Biology. Elsevier; New York: 2013. [Google Scholar]
- 18.Chen R, Mias GI, Li-Pook-Than J, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castrillo JI, Oliver SG. Methods in Molecular Biology. Humana Press, Springer; New York: 2011. Yeast Systems Biology. Methods and Protocols; p. 759. [DOI] [PubMed] [Google Scholar]
- 20.Kosik KS. Personalized medicine for effective Alzheimer disease treatment. JAMA Neurol. 2015;72:497–498. doi: 10.1001/jamaneurol.2014.3445. [DOI] [PubMed] [Google Scholar]
- 21.Montine TJ, Montine KS. Precision medicine: Clarity for the clinical and biological complexity of Alzheimer’s and Parkinson’s diseases. J Exp Med. 2015;212:601–605. doi: 10.1084/jem.20150656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovacs GG. Molecular Pathological Classification of Neurodegenerative Diseases: Turning towards Precision Medicine. Int J Mol Sci. 2016;17(2) doi: 10.3390/ijms17020189. pii: E189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanton C, Soria JC, Bardelli A, et al. Consensus on precision medicine for metastatic cancers: a report from the MAP conference. Ann Oncol. 2016 doi: 10.1093/annonc/mdw192. pii: mdw192. [DOI] [PubMed] [Google Scholar]
- 24.National Institute of Health (NIH) [Accessed 20 July 2016];The Precision Medicine Initiative Cohort Program. https://www.nih.gov/precision-medicine-initiative-cohort-program.
- 25. [Accessed 20 July 2016];Illumina’s executive chairman Jat Flatley speaks of genomic sequencing future. 2016 Jun 20; http://www.sandiegouniontribune.com/news/2016/jun/20/jay-flatley-illumina-innovation/
- 26.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 28.Castrillo JI, Oliver SG. Metabolomics and systems biology in Saccharomyces cerevisiae. In: Esser K, editor. The Mycota. Vol. XIII. A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research. Fungal Genomics. Springer; New York: 2006. pp. 3–18. [Google Scholar]
- 29.Castrillo JI, Oliver SG. Yeast as a model for systems Biology studies on complex Diseases. In: Nowrousian M, editor. Fungal Genomics. 2. Springer-Verlag; Berlin: 2014. pp. 3–30. The Mycota. A Comprehensive Treatise on Fungi as Experimental Systems for Basic and Applied Research. [Google Scholar]
- 30.Castrillo JI, Zeef LA, Hoyle DC, et al. Growth control of the eukaryote cell: a systems biology study in yeast. J Biol. 2007;6:4. doi: 10.1186/jbiol54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uzilov AV, Ding W, Fink MY, et al. Development and clinical application of an integrative genomic approach to personalized cancer therapy. Genome Med. 2016;8:62. doi: 10.1186/s13073-016-0313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajamani D, Bhasin MK. Identification of key regulators of pancreatic cancer progression through multidimensional systems-level analysis. Genome Med. 2016;8:38. doi: 10.1186/s13073-016-0282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abraham SA, Hopcroft LE, Carrick E, et al. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature. 2016;534:341–346. doi: 10.1038/nature18288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams EG, Wu Y, Jha P, et al. Systems proteomics of liver mitochondria function. Science. 2016;352:aad0189. doi: 10.1126/science.aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kori M, Gov E, Arga KY. Molecular signatures of ovarian diseases: Insights from network medicine perspective. Syst Biol Reprod Med. 2016;62:266–282. doi: 10.1080/19396368.2016.1197982. [DOI] [PubMed] [Google Scholar]
- 36.An JY, Claudianos C. Genetic heterogeneity in autism: From single gene to a pathway perspective. Neurosci Biobehav Rev. 2016 Jun 16;68:442–453. doi: 10.1016/j.neubiorev.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 37.Krishna A, Biryukov M, Trefois C, et al. Systems genomics evaluation of the SH-SY5Y neuroblastoma cell line as a model for Parkinson’s disease. BMC Genomics. 2014;15:1154. doi: 10.1186/1471-2164-15-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lausted C, Lee I, Zhou Y, et al. Systems approach to neurodegenerative disease biomarker discovery. Annu Rev Pharmacol Toxicol. 2014;54:457–481. doi: 10.1146/annurev-pharmtox-011613-135928. [DOI] [PubMed] [Google Scholar]
- 39.Rollo JL, Banihashemi N, Vafaee F, et al. Unraveling the mechanistic complexity of Alzheimer’s disease through systems biology. Alzheimers Dement. 2016;12:708–718. doi: 10.1016/j.jalz.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Sheridan C. Omics-driven startups challenge healthcare model. Nat Biotechnol. 2015;33:887–889. doi: 10.1038/nbt0915-887. [DOI] [PubMed] [Google Scholar]
- 41.Cholerton B, Larson EB, Quinn JF, et al. Precision medicine: clarity for the complexity of dementia. Am J Pathol. 2016;186:500–506. doi: 10.1016/j.ajpath.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sannerud R, Esselens C, Ejsmont P, et al. Restricted Location of PSEN2/γ-Secretase Determines Substrate Specificity and Generates an Intracellular Aβ Pool. Cell. 2016;166:193–208. doi: 10.1016/j.cell.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.FitzGerald GA. Measure for measure: biomarker standards and transparency. Sci Transl Med. 2016;8:343fs10. doi: 10.1126/scitranslmed.aaf8590. [DOI] [PubMed] [Google Scholar]
- 44.U.S. Food and Drug Administration. [Accessed 20 July 2016];Clinical Trials Guidance Documents. http://www.fda.gov/regulatoryinformation/guidances/ucm122046.htm.
- 45.EudraCT. [Accessed 20 July 2016]; https://eudract.ema.europa.eu/
- 46.Alzheimer’s Disease Neuroimaging Initiative (ADNI) [Accessed 20 July 2016]; http://www.adni-info.org/
- 47.DIAN Observational Study. [Accessed 20 July 2016]; http://dian-info.org/
- 48.Burton A. Kaj Blennow: the route to biomarkers and the Söderberg prize. Lancet Neurol. 2016 doi: 10.1016/S1474-4422(16)30097-7. [DOI] [PubMed] [Google Scholar]
- 49.Zwan MD, Rinne JO, Hasselbalch SG, et al. Use of amyloid-PET to determine cutpoints for CSF markers: A multicenter study. Neurology. 2016;86:50–58. doi: 10.1212/WNL.0000000000002081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.1st Conference of the European Association of Systems Medicine (EASYM); 26th - 28th October 2016; Berlin. [Accessed 20 July 2016]. https://www.easym.eu/ [Google Scholar]
- 51.European Prevention of Alzheimer’s Dementia consortium. [Accessed 20 July 2016]; http://ep-ad.org/
- 52.Senate Makes Monumental Commitment To Care And Support. [Accessed 20 July 2016];Alzheimer’s Association Celebrates inclusion of the HOPE for Alzheimer’s Act in Senate Funding Bill. 2016 Jun 7; http://www.alz.org/documents_custom/060716-HOPE.pdf.
- 53.Gatz M, Pedersen NL, Berg S, et al. Heritability for Alzheimer’s disease: the study of dementia in Swedish twins. J Gerontol A Biol Sci Med Sci. 1997;52:M117–M125. doi: 10.1093/gerona/52a.2.m117. [DOI] [PubMed] [Google Scholar]
- 54.Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 55.Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 56.Sherrington R, Rogaev EI, Liang Y, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 57.Levy-Lahad E, Wasco W, Poorkaj P, et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 58.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 59.Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 62.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Escott-Price V, Bellenguez C, Wang LS, et al. Gene-wide analysis detects two new susceptibility genes for Alzheimer’s disease. PloS One. 2014;9:e94661. doi: 10.1371/journal.pone.0094661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guerreiro R, Wojtas A, Bras J, et al. TREM2 Variants in Alzheimer’s Disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Escott-Price V, Sims R, Bannister C, et al. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain. 2015;138:3673–3684. doi: 10.1093/brain/awv268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Escott-Price V, Shoai M, Pither R, Williams J, Hardy J. Polygenic score prediction captures nearly all common genetic risk for Alzheimer’s disease. Neurobiol Aging. 2016 doi: 10.1016/j.neurobiolaging.2016.07.018. In press. [DOI] [PubMed] [Google Scholar]
- 69.Lee SH, Harold D, Nyholt DR, et al. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer’s disease, multiple sclerosis and endometriosis. Hum Mol Genet. 2013;22:832–841. doi: 10.1093/hmg/dds491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liesi E, Hebert LE, Weuve J, Scherr PA, Denis A, Evans DA. Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology. 2013;80:1778–1783. doi: 10.1212/WNL.0b013e31828726f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wray NR. Allele frequencies and the r2 measure of linkage disequilibrium: impact on design and interpretation of association studies. Twin Res Hum Genet. 2005;8:87–94. doi: 10.1375/1832427053738827. [DOI] [PubMed] [Google Scholar]
- 73.Purcell SM, Moran JL, Fromer M, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones L, Holmans PA, Hamshere ML, et al. Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer’s disease. PLoS One. 2010;5:e13950. doi: 10.1371/journal.pone.0013950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jorgensen JT. Companion diagnostics: the key to personalized medicine. Foreword Expert Rev Mol Diagn. 2015;15:153–156. doi: 10.1586/14737159.2015.1002470. [DOI] [PubMed] [Google Scholar]
- 76.Moingeon P. Biomarkers for Allergen Immunotherapy: A «Panoromic» View. Immunol Allergy Clin North Am. 2016;36:161–179. doi: 10.1016/j.iac.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 77.Comabella M, Sastre-Garriga J, Montalban X. Precision medicine in multiple sclerosis: biomarkers for diagnosis, prognosis, and treatment response. Curr Opin Neurol. 2016;29:254–262. doi: 10.1097/WCO.0000000000000336. [DOI] [PubMed] [Google Scholar]
- 78.Currie G, Delles C. Use of Biomarkers in the Evaluation and Treatment of Hypertensive Patients. Curr Hypertens Rep. 2016;18:54. doi: 10.1007/s11906-016-0661-6. [DOI] [PubMed] [Google Scholar]
- 79.Hambly N, Shimbori C, Kolb M. Molecular classification of idiopathic pulmonary fibrosis: personalized medicine, genetics and biomarkers. Respirology. 2015;20:1010–1022. doi: 10.1111/resp.12569. [DOI] [PubMed] [Google Scholar]
- 80.Pearson ER. Personalized medicine in diabetes: the role of ‘omics’ and biomarkers. Diabet Med. 2016;33:712–717. doi: 10.1111/dme.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Bryant SE, Edwards M, Johnson LA, et al. A blood screening test for Alzheimer’s disease. Alzheimers Dement (Amst) 2016;3:83–90. doi: 10.1016/j.dadm.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O’Bryant SE, Xiao G, Zhang F, et al. Validation of a serum screen for Alzheimer’s disease across assay platforms, species, and tissues. J Alzheimers Dis. 2014;42:1325–1335. doi: 10.3233/JAD-141041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DeMarshall CA, Nagele EP, Sarkar A, et al. Detection of Alzheimer’s disease at mild cognitive impairment and disease progression using autoantibodies as blood-based biomarkers. Alzheimers Dement (Amst) 2016;3:51–62. doi: 10.1016/j.dadm.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gupta VB, Doecke JD, Hone E, et al. Plasma apolipoprotein J as a potential biomarker for Alzheimer’s disease: Australian Imaging, Biomarkers and Lifestyle study of aging. Alzheimers Dement (Amst) 2015;3:18–26. doi: 10.1016/j.dadm.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Winston CN, Goetzl EJ, Akers JC, et al. Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimers Dement (Amst) 2016;3:63–72. doi: 10.1016/j.dadm.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tampubolon G. Repeated systemic inflammation was associated with cognitive deficits in older Britons. Alzheimers Dement (Amst) 2015;3:1–6. doi: 10.1016/j.dadm.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guedes JR, Santana I, Cunha C, et al. MicroRNA deregulation and chemotaxis and phagocytosis impairment in Alzheimer’s disease. Alzheimers Dement (Amst) 2015;3:7–17. doi: 10.1016/j.dadm.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Henchcliffe C, Dodel R, Beal MF. Biomarkers of Parkinson’s disease and Dementia with Lewy bodies. Prog Neurobiol. 2011;95:601–613. doi: 10.1016/j.pneurobio.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 89.Hu WT, Chen-Plotkin A, Arnold SE, et al. Biomarker discovery for Alzheimer’s disease, frontotemporal lobar degeneration, and Parkinson’s disease. Acta Neuropathol. 2010;120:385–399. doi: 10.1007/s00401-010-0723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Durrenberger PF, Fernando FS, Kashefi SN, et al. Common mechanisms in neurodegeneration and neuroinflammation: a BrainNet Europe gene expression microarray study. J Neural Transm (Vienna) 2015;122:1055–1068. doi: 10.1007/s00702-014-1293-0. [DOI] [PubMed] [Google Scholar]
- 91.Heneka MT, Kummer MP, Latz E. Innate immune activation in neurodegenerative disease. Nat Rev Immunol. 2014;14:463–477. doi: 10.1038/nri3705. [DOI] [PubMed] [Google Scholar]
- 92.O’Bryant SE, Johnson L, Edwards M, et al. The link between C-reactive protein and Alzheimer’s disease among Mexican Americans. J Alzheimers Dis. 2013;34:701–706. doi: 10.3233/JAD-122071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duong T, Nikolaeva M, Acton PJ. C-reactive protein-like immunoreactivity in the neurofibrillary tangles of Alzheimer’s disease. Brain Res. 1997;749:152–156. doi: 10.1016/s0006-8993(96)01359-5. [DOI] [PubMed] [Google Scholar]
- 94.Iwamoto N, Nishiyama E, Ohwada J, Arai H. Demonstration of CRP immunoreactivity in brains of Alzheimer’s disease: immunohistochemical study using formic acid pretreatment of tissue sections. Neurosci Lett. 1994;177:23–26. doi: 10.1016/0304-3940(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 95.Heneka MT, Carson MJ, El Khoury J, et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14:388–405. doi: 10.1016/S1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wyss-Coray T, Rogers J. Inflammation in Alzheimer disease-a brief review of the basic science and clinical literature. Cold Spring Harb Perspect Med. 2012;2:a006346. doi: 10.1101/cshperspect.a006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schmidt R, Schmidt H, Curb JD, Masaki K, White LR, Launer LJ. Early inflammation and dementia: a 25-year follow-up of the Honolulu-Asia Aging Study. Ann Neurol. 2002;52:168–174. doi: 10.1002/ana.10265. [DOI] [PubMed] [Google Scholar]
- 99.Akiyama H, Barger S, Barnum S, et al. Inflammation and Alzheimer’s disease. Neurobiol Aging. 2000;21:383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.in t’ Veld BA, Ruitenberg A, Hofman A, et al. Nonsteroidal antiinflammatory drugs and the risk of Alzheimer’s disease. N Engl J Med. 2001;345:1515–1521. doi: 10.1056/NEJMoa010178. [DOI] [PubMed] [Google Scholar]
- 101.Anthony JC, Breitner JC, Zandi PP, et al. Reduced prevalence of AD in users of NSAIDs and H2 receptor antagonists: the Cache County study. Neurology. 2000;54:2066–2071. doi: 10.1212/wnl.54.11.2066. [DOI] [PubMed] [Google Scholar]
- 102.Etminan M, Gill S, Samii A. Effect of non-steroidal anti-inflammatory drugs on risk of Alzheimer’s disease: systematic review and meta-analysis of observational studies. BMJ. 2003;327:128. doi: 10.1136/bmj.327.7407.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Aisen PS, Schafer KA, Grundman M, et al. Effects of rofecoxib or naproxen vs placebo on Alzheimer disease progression: a randomized controlled trial. JAMA. 2003;289:2819–2826. doi: 10.1001/jama.289.21.2819. [DOI] [PubMed] [Google Scholar]
- 104.Thal LJ, Ferris SH, Kirby L, et al. A randomized, double-blind, study of rofecoxib in patients with mild cognitive impairment. Neuropsychopharmacology. 2005;30:1204–1215. doi: 10.1038/sj.npp.1300690. [DOI] [PubMed] [Google Scholar]
- 105.Adapt_Research_Group. Lyketsos CG, Breitner JC, et al. Naproxen and celecoxib do not prevent AD in early results from a randomized controlled trial. Neurology. 2007;68:1800–1808. doi: 10.1212/01.wnl.0000260269.93245.d2. [DOI] [PubMed] [Google Scholar]
- 106.Breitner JC, Baker LD, Montine TJ, et al. Extended results of the Alzheimer’s disease anti-inflammatory prevention trial. Alzheimers Dement. 2011;7:402–411. doi: 10.1016/j.jalz.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.O’Bryant SE, Rissman RA, Lyketsos C. A proinflammatory endophenotype predicts treatment response in a multicenter trial of NSAIDs in AD. Alzheimers Dement. 2014;10:P273–P274. [Google Scholar]
- 108.O’Bryant SE, Gupta V, Henriksen K, et al. Guidelines for the standardization of preanalytic variables for blood-based biomarker studies in Alzheimer’s disease research. Alzheimers Dement. 2015;11:549–560. doi: 10.1016/j.jalz.2014.08.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li M, Yu T, Hu YF. The impact of companion diagnostic device measurement performance on clinical validation of personalized medicine. Stat Med. 2015;34:2222–2234. doi: 10.1002/sim.6476. [DOI] [PubMed] [Google Scholar]
- 110.Li M. Statistical consideration and challenges in bridging study of personalized medicine. J Biopharm Stat. 2015;25(3):397–407. doi: 10.1080/10543406.2014.920340. [DOI] [PubMed] [Google Scholar]
- 111.Folstein M, Folstein S. «Mini-mental state» A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 112.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 113.Ritchie CW, Ames D, Clayton T, Lai R. A meta-analysis of randomised trials for the efficacy and safety of donepezil, galantamine and rivastigmine for the treatment of Alzheimer’s disease. Am J Geriatr Psychiatry. 2004;12:358–369. doi: 10.1176/appi.ajgp.12.4.358. [DOI] [PubMed] [Google Scholar]
- 114.Ritchie CW, Zhinchin G. Low dose, high dose, or no dose: better prescribing of cholinesterase inhibitors for Alzheimer’s disease. Int Psychogeriatr. 2013;25:511–515. doi: 10.1017/S1041610212002414. [DOI] [PubMed] [Google Scholar]
- 115.Vellas B, Hampel H, Rougé-Bugat ME, et al. Alzheimer’s disease therapeutic trials: EU/US Task Force report on recruitment, retention, and methodology. J Nutr Health Aging. 2012;16:339–345. doi: 10.1007/s12603-012-0044-x. [DOI] [PubMed] [Google Scholar]
- 116.Aisen P, Touchon J, Andrieu S, et al. Registries and Cohorts to Accelerate Early Phase Alzheimer’s Trials. A report from the E.U./U.S. Clinical Trials in Alzheimer’s Disease Task Force. J Prev Alz Dis. 2016;3:68–74. doi: 10.14283/jpad.2016.97. [DOI] [PubMed] [Google Scholar]
- 117.Ritchie CW, Molinuevo JL, Truyen L, Satlin A, Van der Geyten S, Lovestone S European Prevention of Alzheimer’s Dementia (EPAD) Consortium. Development of interventions for the secondary prevention of Alzheimer’s dementia: the European Prevention of Alzheimer’s Dementia (EPAD) project. Lancet Psychiatry. 2016;3:179–86. doi: 10.1016/S2215-0366(15)00454-X. [DOI] [PubMed] [Google Scholar]
- 118.Dubois B, Hampel H, Feldman HH, et al. Preclinical Alzheimer’s disease: Definition, natural history, and diagnostic criteria. Alzheimers Dement. 2016;12:292–323. doi: 10.1016/j.jalz.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Urbanelli L, Magini A, Ciccarone V, et al. New perspectives for the diagnosis of Alzheimer’s disease. Recent Pat CNS Drug Discov. 2009;4:160–181. doi: 10.2174/157488909789104839. [DOI] [PubMed] [Google Scholar]
- 120.Brier MR, Gordon B, Friedrichsen K, et al. Tau and Aβ imaging, CSF measures, and cognition in Alzheimer’s disease. Sci Transl Med. 2016;8:338ra66. doi: 10.1126/scitranslmed.aaf2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ferreira LK, Busatto GF. Neuroimaging in Alzheimer’s disease: current role in clinical practice and potential future applications. Clinics (Sao Paulo) 2011;66:19–24. doi: 10.1590/S1807-59322011001300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Herholz K, Ebmeier K. Clinical amyloid imaging in Alzheimer’s disease. Lancet Neurol. 2011;10:667–670. doi: 10.1016/S1474-4422(11)70123-5. [DOI] [PubMed] [Google Scholar]
- 123.Isaac M, Vamvakas S, Abadie E, Jonsson B, Gispen C, Pani L. Qualification opinion of novel methodologies in the predementia stage of Alzheimer’s disease: cerebrospinal-fluid related biomarkers for drugs affecting amyloid burden--regulatory considerations by European Medicines Agency focusing in improving benefit/risk in regulatory trials. Eur Neuropsychopharmacol. 2011;21:781–788. doi: 10.1016/j.euroneuro.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 124.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mulder C, Verwey NA, van der Flier WM, et al. Amyloid-beta(1-42), total tau, and phosphorylated tau as cerebrospinal fluid biomarkers for the diagnosis of Alzheimer disease. Clin Chem. 2010;56:248–253. doi: 10.1373/clinchem.2009.130518. [DOI] [PubMed] [Google Scholar]
- 126.Buchhave P, Minthon L, Zetterberg H, Wallin AK, Blennow K, Hansson O. Cerebrospinal fluid levels of β-amyloid 1-42, but not of tau, are fully changed already 5 to 10 years before the onset of Alzheimer dementia. Arch Gen Psychiatry. 2012;69:98–106. doi: 10.1001/archgenpsychiatry.2011.155. [DOI] [PubMed] [Google Scholar]
- 127.Bateman RJ, Xiong C, Benzinger TL, et al. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fleisher AS, Chen K, Liu X, et al. Using positron emission tomography and florbetapir F18 to image cortical amyloid in patients with mild cognitive impairment or dementia due to Alzheimer disease. Arch Neurol. 2011;68:1404–1411. doi: 10.1001/archneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 129.Wong DF, Rosenberg PB, Zhou Y, et al. In vivo imaging of amyloid deposition in Alzheimer disease using the radioligand 18F-AV-45 (florbetapir [corrected] F 18) J Nucl Med. 2010;51:913–920. doi: 10.2967/jnumed.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rodrique KM, Kennedy KM, Devous MD, Sr, et al. β-Amyloid burden in healthy aging: regional distribution and cognitive consequences. Neurology. 2012;78:387–395. doi: 10.1212/WNL.0b013e318245d295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Quigley H, Colloby SJ, O’Brien JT. PET imaging of brain amyloid in dementia: a review. Int J Geriatr Psychiatry. 2011;26:991–999. doi: 10.1002/gps.2640. [DOI] [PubMed] [Google Scholar]
- 132.Camus V, Payoux P, Barré L, et al. Using PET with 18F-AV-45 (florbetapir) to quantify brain amyloid load in a clinical environment. Eur J Nucl Med Mol Imaging. 2012;39:621–631. doi: 10.1007/s00259-011-2021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Joshi A, Pontecorvo M, Breault C, Lu M, Mintun M, Carpenter A, Skorvonsky D. Use of Florbetapir-PET to Assess Progression of Amyloid Burden over Time. Human Amyloid Imaging Abstract. J Nucl Med. 2012;53:89. [Google Scholar]
- 134.Doraiswamy PM, Sperling RA, Johnson K, et al. Florbetapir F 18 amyloid PET and 36-month cognitive decline: a prospective multicenter study. Mol Psychiatry. 2014;19:1044–1051. doi: 10.1038/mp.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Grundman M, Pontecorvo MJ, Salloway SP, et al. Potential impact of amyloid imaging on diagnosis and intended management in patients with progressive cognitive decline. Alzheimer Dis Assoc Disord. 2013;27:4–15. doi: 10.1097/WAD.0b013e318279d02a. [DOI] [PubMed] [Google Scholar]
- 136.Johnson KA, Schultz A, Betensky RA, et al. Tau positron emission tomographic imaging in aging and early Alzheimer disease. Ann Neurol. 2016;79:110–119. doi: 10.1002/ana.24546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schwarz AJ, Yu P, Miller BB, et al. Regional profiles of the candidate tau PET ligand 18F-AV-1451 recapitulate key features of Braak histopathological stages. Brain. 2016;139:1539–1550. doi: 10.1093/brain/aww023. [DOI] [PubMed] [Google Scholar]
- 138.Hutton C, Sibille L, Bullich S, et al. Comparison of two methods, with and without MRI, for quantification of florbetaben (18F) PET. Poster PW024 EANM. 2014 [Google Scholar]
- 139.Cuticchia AJ. Existing Ethical Principles and their Application to Personal Medicine. Open Ethics J. 2008;2:29–33. [Google Scholar]
- 140.McGonigle IV. The collective nature of personalized medicine. Genet Res. 2016;98:e3. doi: 10.1017/S0016672315000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shomron N. Prioritizing personalized medicine. Genet Res. 2014;96:e007. doi: 10.1017/S0016672314000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Thomas SM. Society and ethics - the genetics of disease. Curr Opin Genet Dev. 2004;14:287–291. doi: 10.1016/j.gde.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 143.Gurwitz D, Lunshof JE, Altman RB. A call for the creation of personalized medicine databases. Nat Rev Drug Discov. 2006;5:23–26. doi: 10.1038/nrd1931. [DOI] [PubMed] [Google Scholar]
- 144.Simmons S, Sahinalp C, Berger B. Enabling Privacy-Preserving GWASs in Heterogeneous Human Populations. Cell Syst. 2016;3:54–61. doi: 10.1016/j.cels.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lea DH, Kaphingst KA, Bowen D, Lipkus I, Hadley DW. Communicating Genetic and Genomic Information: Health Literacy and Numeracy Considerations. Public Health Genomics. 2011;14:279–289. doi: 10.1159/000294191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zarcadoolas C, Pleasant A, Greer DS. Understanding health literacy: an expanded model. Health Promot Int. 2005;20:195–203. doi: 10.1093/heapro/dah609. [DOI] [PubMed] [Google Scholar]
- 147.Budin-Ljøsne I, Harris JR. Ask not what personalized medicine can do for you--ask what you can do for personalized medicine. Public Health Genomics. 2015;18:131–138. doi: 10.1159/000373919. [DOI] [PubMed] [Google Scholar]
- 148.Hampel H, Lista S, Teipel SJ, et al. Perspective on future role of biological markers in clinical therapy trials of Alzheimer’s disease: a long-range point of view beyond 2020. Biochem Pharmacol. 2014;88:426–449. doi: 10.1016/j.bcp.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 149.Hampel H, Lista S. Use of biomarkers and imaging to assess pathophysiology, mechanisms of action and target engagement. J Nutr Health Aging. 2013;17:54–63. doi: 10.1007/s12603-013-0003-1. [DOI] [PubMed] [Google Scholar]
- 150.Hampel H, Lista S. Alzheimer disease: from inherited to sporadic AD-crossing the biomarker bridge. Nat Rev Neurol. 2012;8:598–600. doi: 10.1038/nrneurol.2012.202. [DOI] [PubMed] [Google Scholar]
- 151.Bertram L, Hampel H. The role of genetics for biomarker development in neurodegeneration. Prog Neurobiol. 2011;95:501–504. doi: 10.1016/j.pneurobio.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 152.Zetzsche T, Rujescu D, Hardy J, Hampel H. Advances and perspectives from genetic research: development of biological markers in Alzheimer’s disease. Expert Rev Mol Diagn. 2010;10:667–690. doi: 10.1586/erm.10.48. [DOI] [PubMed] [Google Scholar]
- 153.Teipel SJ, Grothe M, Lista S, Toschi N, Garaci FG, Hampel H. Relevance of magnetic resonance imaging for early detection and diagnosis of Alzheimer disease. Med Clin North Am. 2013;97:399–424. doi: 10.1016/j.mcna.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 154.Ewers M, Sperling RA, Klunk WE, Weiner MW, Hampel H. Neuroimaging markers for the prediction and early diagnosis of Alzheimer’s disease dementia. Trends Neurosci. 2011;34:430–442. doi: 10.1016/j.tins.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lista S, O’Bryant SE, Blennow K, et al. Biomarkers in sporadic and familial Alzheimer’s Disease. J Alzheimers Dis. 2015;47:291–317. doi: 10.3233/JAD-143006. [DOI] [PubMed] [Google Scholar]
- 156.Rosen C, Hansson O, Blennow K, Zetterberg H. Fluid biomarkers in Alzheimer’s disease - current concepts. Mol Neurodegener. 2013;8:20. doi: 10.1186/1750-1326-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Blennow K, Zetterberg H, Fagan AM. Fluid biomarkers in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a006221. doi: 10.1101/cshperspect.a006221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Blennow K, Hampel H, Weiner M, Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol. 2010;6:131–44. doi: 10.1038/nrneurol.2010.4. [DOI] [PubMed] [Google Scholar]
- 159.Hampel H, Shen Y, Walsh DM, et al. Biological markers of amyloid beta-related mechanisms in Alzheimer’s disease. Exp Neurol. 2010;223:334–346. doi: 10.1016/j.expneurol.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hampel H, Blennow K, Shaw LM, Hoessler YC, Zetterberg H, Trojanowski JQ. Total and phosphorylated tau protein as biological markers of Alzheimer’s disease. Exp Gerontol. 2010;45:30–40. doi: 10.1016/j.exger.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.O’Bryant SE, Lista S, Rissman RA, et al. Comparing biological markers of Alzheimer’s disease across blood fraction and platforms: Comparing apples to oranges. Alzheimers Dement (Amst) 2016;3:27–34. doi: 10.1016/j.dadm.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.O’Bryant SE, Gupta V, Henriksen K, et al. Guidelines for the standardization of preanalytic variables for blood-based biomarker studies in Alzheimer’s disease research. Alzheimers Dement. 2015;11:549–560. doi: 10.1016/j.jalz.2014.08.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Henriksen K, O’Bryant SE, Hampel H, et al. The future of blood-based biomarkers for Alzheimer’s disease. Alzheimers Dement. 2014;10:115–131. doi: 10.1016/j.jalz.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Snyder HM, Carrillo MC, Grodstein F, et al. Developing novel blood-based biomarkers for Alzheimer’s disease. Alzheimers Dement. 2014;10:109–114. doi: 10.1016/j.jalz.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gupta VB, Sundaram R, Martins RN. Multiplex biomarkers in blood. Alzheimers Res Ther. 2013;5:31. doi: 10.1186/alzrt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Edwards M, Balldin VH, Hall J, O’Bryant S. Combining select neuropsychological assessment with blood-based biomarkers to detect mild Alzheimer’s disease: a molecular neuropsychology approach. J Alzheimers Dis. 2014;42:635–640. doi: 10.3233/JAD-140852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lista S, Garaci FG, Ewers M, et al. CSF Abeta1-42 combined with neuroimaging biomarkers in the early detection, diagnosis and prediction of Alzheimer’s disease. Alzheimers Dement. 2014;10:381–392. doi: 10.1016/j.jalz.2013.04.506. [DOI] [PubMed] [Google Scholar]
- 168.Hampel H, Frank R, Broich K, et al. Biomarkers for Alzheimer’s disease: academic, industry and regulatory perspectives. Nat Rev Drug Discov. 2010;9:560–574. doi: 10.1038/nrd3115. [DOI] [PubMed] [Google Scholar]
- 169.Broich K, Weiergraber M, Hampel H. Biomarkers in clinical trials for neurodegenerative diseases: regulatory perspectives and requirements. Prog Neurobiol. 2011;95:498–500. doi: 10.1016/j.pneurobio.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 170.Hampel H, Lista S. The rising global tide of cognitive impairment. Nat Rev Neurol. 2016;12:131–132. doi: 10.1038/nrneurol.2015.250. [DOI] [PubMed] [Google Scholar]
- 171.Baldacci F, Lista S, Garaci F, Bonuccelli U, Toschi N, Hampel H. Biomarker Guided Classification Scheme of Neurodegenerative Diseases. J Sport Health Sci. 2016 doi: 10.1016/j.jshs.2016.08.007. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fox JL. Obama catapults patient-empowered Precision Medicine. Nat Biotechnol. 2015;33:325. doi: 10.1038/nbt0415-325. [DOI] [PubMed] [Google Scholar]
- 174.OECD. Can Big Data Help? OECD Publishing; 2015. [Accessed 19 July 2016]. Dementia Research and Care. http://dx.doi.org/10.1787/9789264228429-en. [Google Scholar]
- 175.OECD. OECD Science, Technology and Industry Policy Papers. OECD Publishing; 2015. [Accessed 19 July 2016]. Enhancing Translational Research and Clinical Development for Alzheimer’s Disease and other Dementias. http://dx.doi.org/10.1787/5js1t57jts44-en. [Google Scholar]