Abstract
Effective therapeutics for ovarian cancer continue to be urgently needed, particularly for chemotherapy-resistant cases. Here we present both a 3D-Matrigel culture-based expansion of our directed evolution method for generation of oncolytic virotherapies and two promising ovarian-cancer targeted oncolytic viruses, OvAd1 and OvAd2. OvAd1 was developed using Matrigel cell cultures, whereas OvAd2 was developed in parallel using traditional monolayer tissue culture methods. Both viruses are potent against a panel of platinum-resistant ovarian cancer cell lines and are attenuated on normal cells in vitro, resulting in therapeutic windows of ∼200-fold. We observed two benefits of the use of Matrigel-based cultures for directed evolution of these oncolytics: (1) use of Matrigel generated a bioselected pool that was more strongly attenuated on normal cells while retaining its potency against ovarian cancer cells, and (2) in an ovarian carcinomatosis model, the Matrigel-derived virus OvAd1 suppressed all tumor growth while the non-Matrigel-derived virus was 50% effective. Neither virus stimulated formation of peritoneal adhesions as seen for Ad5-based therapies. Consequently, these viruses are novel candidates for development as new effective treatments for aggressive ovarian cancer.
Keywords: 3D cell culture, Matrigel, oncolytic, virotherapy, ovarian cancer, directed evolution, adenovirus, replicating
Introduction
Ovarian cancer has the highest mortality rate of all gynecological malignancies. Although the overall 5-year survival rate is ∼40%, the survival rate drops markedly to ∼10% with increasing age and stage, largely because of treatment-resistant recurrence.1 Of the gynecological cancers, ovarian cancer has the highest recurrence rate, with over 60% recurring as therapeutically resistant after optimal de-bulking surgery and frontline treatment with paclitaxel in combination with platinum-based chemotherapy (American Cancer Society, http://www.cancer.org/). These patients have very poor prognoses and few therapeutic options.2
Most ovarian cancer patients die from intraperitoneal (i.p.) organ failure, and this has led to trials of i.p.-delivered therapies. The need for effective treatment for frontline-therapy-resistant ovarian patients, combined with the potential efficacy of body cavity-restricted treatment has stimulated the development of ovarian-directed oncolytic viruses, several of which have entered clinical testing.3, 4, 5, 6, 7, 8 Select oncolytic adenoviruses have demonstrated adequate safety profiles in patients along with some encouraging signs of activity,9 but overall, oncolytic viruses to date have lacked sufficient potency for treatment of ovarian cancer patients. Because oncolytic adenoviruses replicate selectively in and lyse tumor cells, thus releasing an amplified dose of virus that then infects and similarly kills remaining tumor cells, these biotherapeutic agents have the potential, as yet unrealized, to yield complete therapeutic responses in patients while generating minimal side effects.
To improve the therapeutic utility of oncolytic viruses, we have developed a method termed “directed evolution” that selects for potency from a starting library or pool of diverse adenoviral serotypes or recombinants thereof.10 This approach is unbiased for serotype or mechanism of action in achieving the desired selection criteria and is not limited by the extent of our existing knowledge of oncolytic mutations; this is in contrast with the conventional approach of designed or engineered oncolytic viruses. The first therapeutic virus generated by this method, ColoAd1, was isolated for its rapid and lethal replication in colon cancer cells.10 Successful application of directed evolution is dependent both on maximizing the biodiversity of the initial viral pool and on carrying out the selection process on cell cultures that recreate the target tissue as accurately as possible.
A natural progression of this approach is to perform the iterative rounds of selection in more sophisticated 3D cultures. Seminal work from the Bissell laboratory demonstrated that basement membrane proteins, and the architecture they induce in cells cultured in their presence, are critical for reproduction in culture of physiological patterns of cellular polarization, growth regulation, apoptotic susceptibility, chemotherapeutic resistance, adhesion, tissue specific function, cytoskeletal organization, signal transduction and gene expression, morphogenesis, and differentiation in cultures of both normal and transformed cells.11, 12, 13, 14 Others have corroborated and extended these early studies, reporting that culturing cells such as ovarian cells in gels whose physical and chemical properties closely resemble human tissues yields data that are more clinically relevant than that obtained from monolayer (traditional) cell cultures. Such biomimetic cultures are commonly referred to as three-dimensional (3D) to distinguish them from monolayer or two-dimensional (2D) cultures.15, 16
Because our goal is the development of oncolytic viruses effective against high-grade, chemoresistant ovarian primary tumors and metastases, we chose SKOV3 cells to model this target tissue. SKOV3 cells were derived from platinum-therapy-resistant ascites associated with an ovarian serous adenocarcinoma, the most prevalent (>50%) ovarian tumor type.17 The biodiversity of the initial viral pool was increased relative to our prior work by the inclusion of ColoAd1 (along with representative serotypes from all the non-oncogenic adenovirus groups). Through iterative passaging of the initial adenoviral pool on two types of SKOV3 cultures and subsequent cloning, we developed two novel ovarian oncolytic viruses, termed OvAd1 and OvAd2, generated on Matrigel versus on monolayer cultures, respectively. In vitro and in vivo comparisons of these two viruses indicate that 3D culture methods may be superior to monolayer culture methods in generating oncolytic viruses that are both potent in vivo and attenuated on non-malignant normal cells.
Results
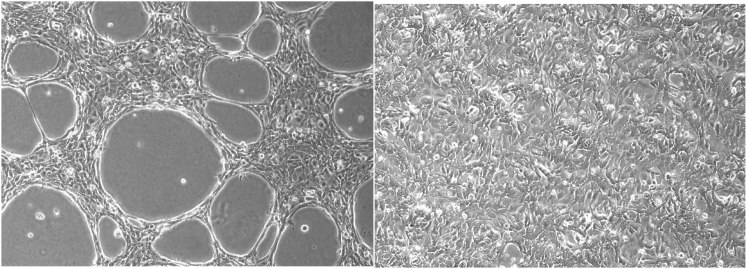
Directed evolution of an ovarian oncolytic virus was carried out on the ovarian cancer cell line SKOV3 because these cells are representative of the poor-prognosis patient population for which this virotherapy was intended. SKOV3 cells form lacy, three-dimensional structures when cultured on growth-factor reduced Matrigel allowing visual verification that this basement membrane extract had induced a change in the morphology of these cells (Figure 1). SKOV3 cells seeded onto Matrigel initially grow into multilayered rope-like strands (Figure S1) that widen into the lacy morphology presented in Figure 1. Growth in 3D can be observed by focusing the microscope in the vertical direction (Figure S1). Our observations are consistent with previously reported morphology of SKOV3 cells grown on Matrigel.18
Figure 1.
SKOV3 Cells Grown on Matrigel Acquire Substratum-Dependent Morphology
SKOV3 cells grown on GFR-Matrigel (left) compared with those grown by traditional method on tissue culture plastic (right) display distinct morphologies.
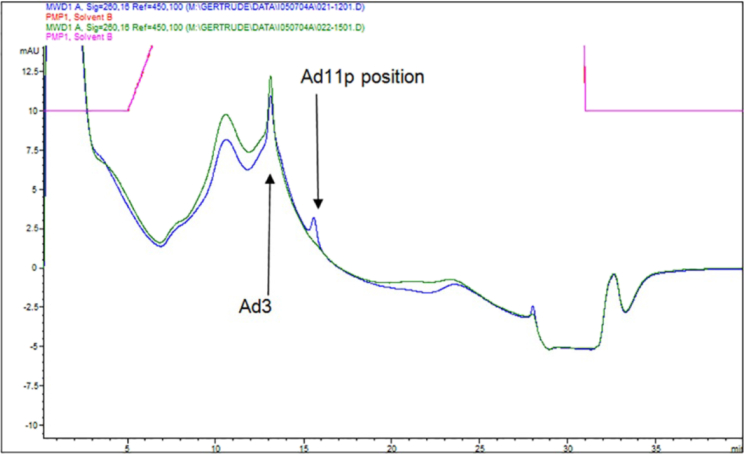
To determine whether a basement membrane substratum underlying the ovarian cell cultures would affect the nature of the oncolytic viruses selected by directed evolution, the process was performed in parallel using SKOV3 cells grown on Matrigel, a commercial basement membrane gel substance, or using SKOV3 cells grown as monolayers (i.e., by traditional method on tissue culture plastic). The two resultant selected viral pools, SM10 (passaged on Matrigel) and SP10 (passaged on 2D monolayers), were titered by anion-exchange (AIEX) chromatography,19 an analysis that also revealed that the SP10 pool predominantly contained viruses related to Ad3, whereas the SM10 pool contained viruses related to Ad3 and Ad11p or Ad35 (the AIEX retention characteristics of Ad11p and Ad35 are indistinguishable [Figure 2; Figure S2]). Note that by AIEX neither SM10 nor SP10 contained viruses with the characteristics of Ad5, the serotype on which almost all previous oncolytic viruses have been based. The fact that no Ad5-related viruses had been selected during the directed evolution of both viral pools supports our previous suggestion10 that Ad5 is likely not the most effective or promising human adenovirus (Ad) serotype basis for all oncolytic virus development, a suggestion subsequently supported by others developing ovarian oncolytic viruses.20 Accurate viral particle counts for each stock, whether purified or a crude cell lysate, were derived from the AIEX results as viral particles per milliliter (vp/mL), enabling direct comparison of the potency of each stock on the various cell lines by MTS assay on a “viral particle per cell” (vppc) basis.19
Figure 2.
AIEX Analysis of SP10 and SM10 Identifies Ad Serotypes in Each Pool and Allows Accurate Viral Particle Counts
The bioselected pools, SM10 and SP10, were analyzed by AIEX chromatography. The elution positions of adenoviral peaks are characteristic for each adenoviral serotype (Figure S2) and are evident in the chromatograms as the narrow peaks superimposed on the broader protein peaks (discussed in Kuhn et al.19) contributed by the SKOV3 culture medium. The position of each narrow peak in the chromatograms of the passaged viral pools indicate that SP10 (dark green chromatogram) is predominantly composed of Ad3-related viruses, whereas SM10 (blue chromatogram) contains both Ad3 and Ad11p-related viruses.
To test whether our viral selection method had increased the potency of the viral pools against the SKOV3 cognate cells, MTS assays were used. The MTS data showed that the two bioselected pools, SM10 and SP10, had very similar potencies on SKOV3 (Figures 3A and 3C), and that both selected pools had increased several logs in potency relative to viruses in the initial pool (Tables 1 and 3; Figure S3). This result encouraged us to plaque-purify 12 viruses from each pool. Stocks of these 24 viral isolates were then titered and characterized by AIEX chromatography prior to MTS assay. The AIEX chromatograms showed the majority of the viral isolates from each pool had column retention characteristics indicating they were related to Ad3, while a few of the isolates from the SM10 pool were related to Ad11p or Ad35, consistent with the composition of the SM10 and SP10 pools shown in Figure 2.
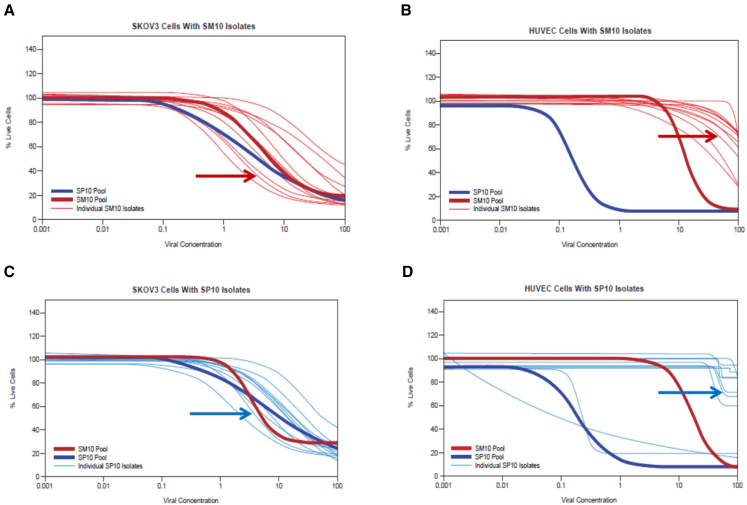
Figure 3.
Assessing Tumor Specificity: Potency of the SM10 and SP10 Pools and Viral Isolates on SKOV3 versus HUVEC Monolayers
(A) Potency of SM10 pool isolates assessed on monolayers of SKOV3 cells. The SP10 and SM10 pools were included in all MTS analyses as references to allow comparison between assays. Results at 6 dpi are presented. The SM10 and SP10 pools as well as the 12 isolates of the SM10 pool had similar potencies (within a 1-log range) against SKOV3 cells. The red arrow indicates the clonal isolate of SM10 that was renamed OvAd1. (B) The same viral isolates and pools as in (A) were assessed in parallel on HUVEC monolayers; endpoints read at 10 dpi (the HUVEC assay required more days than the SKOV3 assay to reveal any killing of HUVECs by the 12 SM10 isolates). The SM10 pool was about 2 logs more attenuated on HUVECs than was the SP10 pool, and the SM10 pool isolates were 2–3 logs more attenuated on HUVECs than was the SP10 pool. The red arrow indicates the clonal isolate of SM10 that was renamed OvAd1. (C) Potency of SP10 pool isolates assessed on monolayers of SKOV3 cells, along with the SM10 and the SP10 pools; results at 6 dpi are presented. As in (A), SM10 and SP10 pools had very similar potencies on SKOV3, and the SP10 isolates were within about a 1-log range of one another and of the pools. The blue arrow indicates the clonal isolate of SP10 that was renamed OvAd2. (D) The same viral isolates and pools as in (C) were analyzed in parallel on HUVECs. Results at 7 dpi are presented. The SP10 pool and two SP10 isolates are 3 or more logs more potent against HUVEC than the SM10 pool, indicating that the SP10 pool and these two isolates are at best weakly tumor selective. The remaining 10 isolates of the SP10 pool appear to be tumor selective. The blue arrow indicates the clonal isolate of SP10 that was renamed OvAd2.
Table 1.
IC50 of Each Virus Determined by MTS Assay on Four Cell Lines of Non-ovarian Tumor Types
| Virus | HT-29 | MDA-231 | PC-3 | Panc-1 |
|---|---|---|---|---|
| OvAd1 and 2 | 1 | 20 | 0.05 | 0.7 |
| ColoAd1 | <0.1 | 0.5 | 0.02 | 2–3 |
| Ad3 | >200 | 10 | 4 | 30 |
| Ad5 | 20 | 0.2 | 40 | 0.02 |
| ONYX-015 | >1000 | 10 | >200 | 2–3 |
Table 3.
Tumor Specificity of Each Virus as Indicated by IC50 Values at 7 dpi on Monolayers of Malignant as Compared with Normal Primary Human Cells
| SKOV3 | OVCAR3 | HUVEC | |
|---|---|---|---|
| OvAd1 | 1 | 1 | ∼200 |
| OvAd2 | 1 | 1 | ∼200 |
| Ad3 | 50 | 200 | ∼200 |
| ONYX-015 | >400 | 60 | 100 |
| Ad5 | >400 | 100 | 8 |
| Ad11p | 100 | ND | ∼150 |
| ColoAd1 | 15 | 10 | 17 |
To identify the most desirable isolate for further development from among the 12 isolates of each pool, we next measured the tumor selectivity of each of the 24 isolates by MTS assay. Tumor selectivity was defined as potency (indicated by the IC50) on human normal primary cells such as human umbilical vein endothelial cells (HUVECs), divided by potency on malignant cells such as SKOV3. The desired oncolytic virus would thus have the highest tumor selectivity score or ratio, indicating that it would require a very large number of viral particles to kill normal human cells, while very few viral particles (small viral concentration) would be required to kill tumor cells.
Results represented in Figures 3A and 3C demonstrated that the potency against SKOV3 cells was similar for the SM10 and SP10 pools, and the range of potencies of the 12 isolates of each pool was also similar. Potencies on HUVECs, however, were distinct. About 2 logs more virus were required for SM10 to kill HUVECs as compared to SP10, indicating a much wider therapeutic window for the Matrigel-derived pool as compared to the 2D-derived pool. Consistent with this, all 12 isolates of the SM10 pool were tumor selective in that they were attenuated on HUVECs, whereas 2 of the isolates of the SP10 pool, and the SP10 pool itself, were not tumor selective because they efficiently killed the normal HUVECs (Figures 3B and 3D).Taken together, it appears that culturing cells on basement membrane gel, as opposed to on tissue culture plastic, does increase the probability of generating a tumor selective oncolytic, in this case by attenuating the potency of the selected pool against normal cells while retaining potency against the target cancer cells.
The viral isolate with the greatest tumor selectivity was chosen from each selected pool for further characterization. The Matrigel-derived viral isolate was named OvAd1 (indicated by red arrows in Figures 3A and 3B), whereas the monolayer-derived viral isolate was named OvAd2 (indicated by blue arrows in Figures 3C and 3D).
OvAd1 and OvAd2 Are Two Different Chimeras of ColoAd1 and Ad3
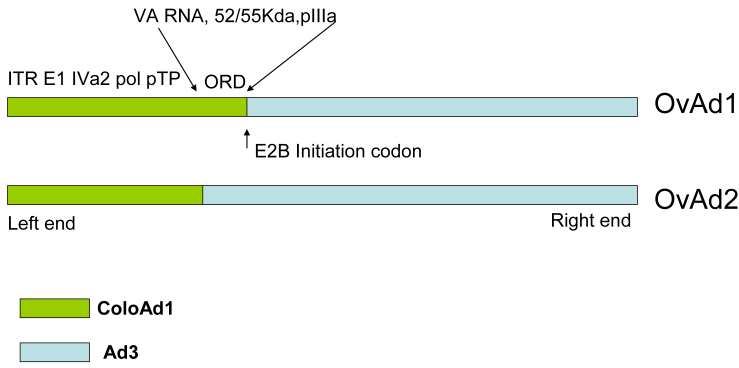
DNA sequence analysis of OvAd1 and OvAd2 revealed that these two viruses are both chimeras of wild-type Ad3 and ColoAd1, the oncolytic virus previously developed in this laboratory.10 The first ∼13,000 bp of OvAd1 and the first ∼10,000 bp of OvAd2 are identical to ColoAd1, with the remainder of each virus identical to Ad3 (Figure 4). Interestingly, we did not find any point mutations in OvAd1 or OvAd2, similar to our previous observation10 that no point mutations had been incorporated during the directed evolution of ColoAd1. It is of note that the E2B region of ColoAd1, which is a recombinant product of the E2B regions of Ad11p and Ad3,10 was selected for in both OvAd1 and OvAd2. The repeated selection of this chimeric E2B region suggests its importance for oncolytic potency.
Figure 4.
OvAd1 and OvAd2 Are Chimeras of ColoAd1 and Ad3
OvAd1 and OvAd2 differ in a 3-kb region between bp 10,150 and 13,060. We have termed this region the ORD, or OvAd Region of Difference. OvAd1 is identical to ColoAd1 in the ORD, whereas OvAd2 is identical to Ad3. Because this region of ColoAd1 was derived from Ad11p, OvAd1 is also identical to Ad11p in the ORD. The ORD encodes Viral Associated RNA, and L1 proteins 55 Kda and pIIIa. Additionally, the E2B transcription unit spans the ORD. A number of splicing events control E2B functions, and the localization of the ORD within the E2B transcription unit predicts that E2B functions may differ between OvAd1 and OvAd2.
OvAd1 and OvAd2 Exhibit Mixed Potency on Broad Panel of Tumor Types
Having shown that OvAd1 and OvAd2 have acquired enhanced potency (i.e., ability to kill) SKOV3 ovarian cells, we asked whether these viruses had also acquired enhanced potency against other tumor types. Potencies of OvAd1 and OvAd2 were compared with those of the parental viruses, ColoAd1 and Ad3, on Panc-1, PC-3, MDA-231-mt1, and HT-29 cells representing pancreatic, prostate, breast, and colon cancers, respectively. ONYX-015, the most clinically advanced adenovirus virotherapy, and Ad5, the serotype basis for ONYX-015 and all other adenoviral virotherapies clinically tested to date, were included for reference. The IC50 values, or potency, derived from the MTS assays on this broad panel of cancer types are presented in Table 1. The potencies of OvAd1 and OvAd2 were very similar to one another on each of these cancer lines. The potencies of the OvAds are in a range similar to but are not of greater potency than ColoAd1, indicating that increased broad potency against this set of tumor types was not selected for in our method. The other OvAd parental, Ad3, is not as potent as either ColoAd1 or the OvAds on any cell line in this set, suggesting that the Ad3 components of the OvAd viruses are probably not the source of OvAd potency on these (non-ovarian) cancer lines. In summary, the OvAds do not demonstrate increased potency against all the tested tumor types but perhaps do warrant further investigation for use in prostate and pancreatic cancer.
OvAd and OvAd2 Are Broadly Potent against High-Grade Serous Platinum-Resistant Ovarian Cell Lines and Exhibit Potential for Clinical Efficacy
OvAd1 and OvAd2 were selected for enhanced potency against the ovarian cancer line SKOV3, but it was not yet clear whether these viruses had acquired broader potency against an array of ovarian cancer tumor types. To test their wider ovarian efficacy, we analyzed their potency against a panel of five ovarian cell lines representative of the tumor types found in our advanced-disease, platinum-resistant target patient population as well as one cell line that is platinum sensitive.
The IC50 of each virus on each ovarian cell line was derived from replicate MTS assays and used to calculate the potency of the OvAds relative to their oncolytic parental virus ColoAd1; these results are summarized in Table 2. OvAd1 and OvAd2 potencies were very similar to one another on each cell line. The OvAd viruses were 0.5–3 logs more potent than ColoAd1 on the three lines representative of the most prevalent types of high-grade serous (HGS-OvCa), platinum-resistant ovarian cancer and equivalently potent on platinum-resistant non-serous lines. The potency of the OvAds against all five of the platinum-resistant ovarian cancer cell lines, a broadly divergent group, bodes well for their potential efficacy against clinical tumors because ovarian tumors are highly heterogeneous.21
Table 2.
OvAd1 and OvAd2 Are Both Potent against HGS-OvCa Lines and Likely Clinically Efficacious as Evidenced by Comparison with Potency of ColoAd1
| Cell Line | Tumor Classification | Tumor Type Prevalence (%) | Platinum Sensitivity | Relative Potency: IC50 ColoAd1/OvAds |
|---|---|---|---|---|
| CaOV3 | papillary serous adenocarcinoma | 40–50 | R | 2,000 |
| OVCAR3 | ascites, serous progressive adenocarcinoma | 40–50 | R | 12.5 |
| SKOV3 | ascites, serous adenocarcinoma | 40–50 | R | 20 |
| BG-1 | unclassified carcinoma | unknown | R | 2 |
| ES-2 | poorly differentiated clear cell carcinoma | 6 | R | 1 |
| IGROV | endometroid carcinoma, untreated tumor | 15 | S | 0.01 |
The IC50s of OvAd1, OvAd2, and ColoAd1 were determined by MTS assay on monolayers of each cell line. The IC50s of OvAd1 and OvAd2 were equivalent on each line. Relative potency, presented here, is the ratio of the IC50 of ColoAd1 divided by IC50 of OvAd1&2 on the same cell line; thus, the higher the number the greater the potency of the OvAds relative to ColoAd1.
ONYX-01522 was also included in these MTS assays. ONYX-015 was less potent than ColoAd1, or the OvAds, on all lines in the ovarian cell line panel, most of which are p53(−) and therefore would not be expected to bias against ONYX-015. These data are consistent with the poor clinical efficacy of ONYX-015 for treatment of ovarian cancer.5, 23 Ad5, the industry standard, and Ad3, a major source of the OvAds’ genome, were similarly relatively attenuated on this panel of ovarian cell lines (data not shown).
As an indication of how safe these viruses might be in the clinic, we compared the potency of OvAd1 and OvAd2 to that of ONYX-015 on HUVECs, a primary normal human cell. Assays on HUVECs showed all three viruses, OvAd1, OvAd2, and ONYX-015, had very similar low potency (IC50 values ∼100–200 vp/cell) on these primary endothelial cells. These data, along with the reported clinical safety profile of ONYX015 and extensive testing of ColoAd1 on human primary epithelial and vascular cells (S. Illingworth, Y. Di, A. Bruno, I.K., M.B., J. Lei, M.R. Duffy, S. Alvis, Brian Champion, T.H., L.S., J. Beadle, and K.F., unpublished data), indicate the potential clinical utility of the OvAd viruses. Note also that for these experiments, normal HUVECs were cultured according to manufacturer’s instructions including the addition of growth factors and corticosteroids that have been reported to increase susceptibility of cells to virus infection.24, 25 As such, these assay results on primary normal cells may underestimate the true therapeutic index relative to polarized, contact-inhibited layers of cells in normal tissue.
The OvAds’ two parental viruses, ColoAd1 and Ad3, were also included in these MTS assays. Table 3 summarizes the IC50 values for these viruses on two platinum-resistant HG-OvCa lines as compared with HUVECs. The OvAds’ IC50 is 1 vppc on the tumor cells while being 200-fold more attenuated (less potent) on the HUVEC. Each parent virus is similar to the OvAds only on one cell type: Ad3 is similar to the OvAds in being attenuated on the HUVEC, and ColoAd1 is similar to the OvAds in being relatively (10-fold less) potent on the ovarian cancer cells. Although we do not yet know the mechanism(s) responsible for these distinct potencies on malignant versus normal cells, the data summarized in Table 3 suggest the mechanism of OvAd attenuation on HUVECs may derive from Ad3, while the mechanism of potency against ovarian tumor cells derives primarily from ColoAd1.
Matrigel-Derived OvAd1 Is More Potent Than OvAd2 in an In Vivo Ovarian Model
Given their promising potency and safety profiles, OvAd1 and OvAd2 were tested for in vivo efficacy in an institutionally approved and supported SKOV3 intraperitoneal carcinomatosis model of ovarian cancer. This model was chosen not only to test potency but also to determine whether these viruses would cause peritoneal adhesions. Such adhesions had prevented continuation of the ovarian clinical trial of Schering-Plough’s Ad5-based p53-encoding gene therapy virus.3, 26 Similarly, inflammatory reactions associated with adhesions have marred clinical trials of ONYX0155 and other Ad5-derived agents for treatment of ovarian cancer.27
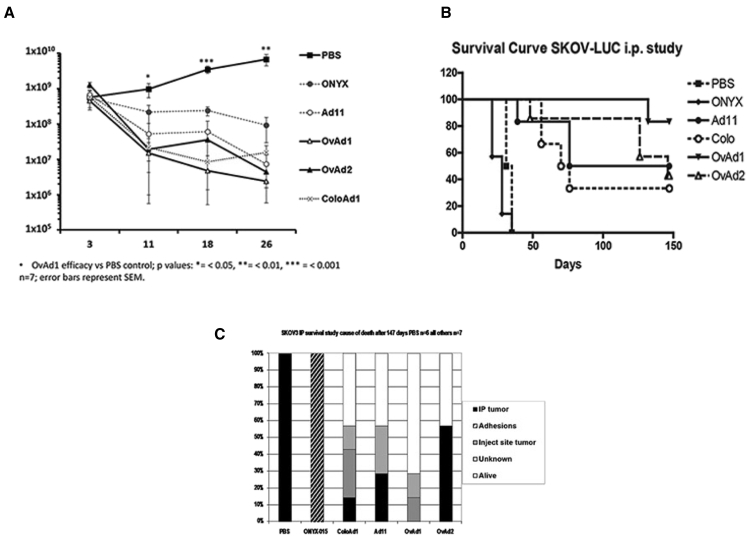
In the carcinomatosis SKOV3 model, all viruses caused a decrease of the luciferase signal over the imaging period of 26 days (Figure 5A) at which point some animals began to be removed from the study after reaching a humane endpoint (Figure 5B). On day 18, the last day on which all group members were still included, OvAd1 and OvAd2 appeared to have the most substantial impact on tumor burden as assessed by luciferase group averages. Because of variability in the imaging, the difference between the viruses OvAd1 and OvAd2 was not statistically significant.
Figure 5.
OvAd1 and OvAd2 Proved Effective in Mouse Model of Ovarian Cancer and Did Not Generate Peritoneal Adhesions
Ovarian carcinomatosis tumors were established by administration of 5 × 106 SKOV-3.Luc cells on day 0 (t0) into the peritoneal cavity and randomly assigned to seven groups of seven mice each. At days 3, 5, and 7, mice were administered PBS, ONYX-015, Ad11p, OvAd1, or OvAd2 via a 0.5-mL i.p. injection. All virus-treated mice received a dose of 5 × 1010 viral particles (vp) per injection for a total of 1.5 × 1011 vp/mouse. (A) During the first 26 days, tumor burden was followed by luciferase imaging, and (B) survival was followed out to 147 days. (C) Autopsy observations of pathological changes contributing to mouse mortality.
Mice treated with ONYX-015, an Ad5-based oncolytic virus, developed adhesions in the peritoneal cavity and so had to be culled during days 21–31, in contrast to untreated control mice which succumbed to tumor burden later, from days 31 to 34. None of the group B viruses, OvAd1, OvAd2, ColoAd1 or Ad11, caused peritoneal adhesions, and all had some surviving animals out to the end of the study (147 days). An autopsy recording the underlying cause for removing an animal from study revealed four of seven peritoneal tumors in OvAd2-treated mice and zero of seven tumors in the OvAd1-treated mice (Figure 5C). OvAd1-treated mice had just two events, both late in the study: one of these animals had an injection site tumor in the needle tract and was perfectly healthy but removed under local animal welfare rules due to the risk of ulceration; the other OvAd1-treated mouse had weight loss to the humane endpoint level (10%), but no tumor could be located. It is possible that a small tumor was present but not visible. Blood from each mouse in this study was analyzed for presence of adenovirus. In no case could virus be detected in the bloodstream, addressing the safety concern that oncolytic viruses may escape the peritoneal cavity.
Discussion
We have described two novel ovarian oncolytic viruses, OvAd1 and OvAd2, both products of our ongoing efforts to develop potent virotherapies. In the current work, we targeted platinum-resistant, poor-prognosis ovarian cancers. Our strategy was to select for viruses potent against the platinum-resistant, ovarian serous adenocarcinoma ascites-derived SKOV3 cell line, grown either as a 2D monolayer or as a 3D culture formed on the basement membrane extract Matrigel. Both 2D and 3D culture methods selected oncolytic viruses potent against a diverse panel of chemo-resistant ovarian lines while being at least as attenuated on normal human cells as the most clinically advanced oncolytic adenovirus, ONYX-015. Furthermore, both viruses were effective in an in vivo model, and neither induced peritoneal adhesions seen previously with Ad5-based treatments.5, 27 The 3D-derived OvAd1 was more potent in vivo than the 2D-derived OvAd2 (none of the mice treated with OvAd1 developed or had detectable tumors, whereas four of seven treated with OvAd2 did). High-grade serous ovarian carcinoma (HGS-OvCa) tumors are highly intratumorally heterogeneous, thus effective treatments will need to treat all the many subpopulations of these tumors (elimination of single tumor sub-populations is not effective treatment). The pronounced efficacy of the OvAd viruses against the three HGS-OvCa lines, given the distinct genotypes of these cell lines, lends hope that the OvAds will be effective against many ovarian tumor subpopulations,21 whether as monotherapies or in combination with compatible therapeutics.
OvAd1 and OvAd2 are replication-competent therapeutics, which combined with their efficacy against ovarian cancer cells of various genotypes, suggests their clinical potential. Replication-competent oncolytic viruses specifically infect malignant cells, replicate many fold, lyse the cancer cell, and ideally, go on to iteratively infect and kill surrounding tumor cells until all malignant cells have been killed. We previously reported development of the oncolytic virus ColoAd1 and the directed evolution method to identify potent viruses in an unbiased manner from a diverse pool of adenoviruses. In the present work, the method was extended to include selection on Matrigel-based 3D cultures. The decision to test the effects of Matrigel was stimulated by reports that such 3D cultures are more physiologically relevant than 2D cultures. That the use of Matrigel had affected the product of directed evolution became evident when the tumor selectivity of the Matrigel-derived selected viral pool SM10 was compared to that of the monolayer-derived selected viral pool SP10. The Matrigel-derived pool was 2 logs more attenuated than the monolayer-derived pool on primary normal HUVECs, while the two pools’ potencies on SKOV3 ovarian cells were similar. As a whole, therefore, the Matrigel-derived pool has greater tumor selectivity than the monolayer-derived pool.
The Matrigel-derived oncolytic OvAd1 was more effective in eliminating cancer cells than the monolayer-derived OvAd2 in an intraperitoneal model of ovarian cancer, suggesting that the Matrigel cultures may be better agents than monolayer cultures for selecting viruses potent against SKOV3 tumors, presumably because the in vitro 3D SKOV3 cultures were better mimics of in vivo SKOV3 tumor biology. However, without additional studies analyzing a much larger number of viral isolates derived by the Matrigel and monolayer methods, we cannot make a conclusive statement about the benefit of using Matrigel beyond the fact that viruses derived from the Matrigel arm were distinguishable from those derived from the monolayer arm. Additionally, it is not yet clear whether OvAd1 will be similarly more potent than OvAd2 against other in vivo models.
In considering the genetic bases of these attributes of potency and selectivity, there are significant similarities and differences between OvAd1 and OvAd2. These two viruses were generated from two distinct series of selective steps; therefore, their similarity is striking. Both OvAd1 and OvAd2 carry over 20 kb of the right end of Ad3, and AIEX chromatography of SM10 and SP10 indicated that most of the bioselected viruses were related to Ad3; this Ad3 prevalence suggests that some combinations of Ad3 features contribute more potency against ovarian malignancies than do other Ad serotypes. Future research will identify which Ad3 functions increase the efficacy of the OvAd viruses against ovarian targets.20 Furthermore, comparing the potency of Ad3 on SKOV3 cells and HUVECs (Table 3; Figure S3) indicated the Ad3 parent has 4-fold more activity against SKOV3 cells than it does against HUVECs. Recombination of Ad3 with ColoAd1 generated viruses with 50-fold higher potency against high-grade serous-type ovarian cancer cells, while Ad3’s attenuation on normal cells was retained (Table 3), thus yielding tumor-specific viruses. Other groups have reported the use of Ad3 fiber knob-pseudotyping of Ad5 viruses to create ovarian virotherapeutics.28 In our experience, Ad5 is more potent on HUVECs than on SKOV3 cells (Table 3), suggesting that Ad5 does not have inherent characteristics of an ovarian oncolytic. It would be interesting to compare the Ad3/Ad5 pseudotyped virus to OvAd1 to better understand the mechanisms of potency of each virotherapy.
Both OvAd1 and OvAd2 acquired the left arm of ColoAd1. This arm of the ColoAd1 genome is identical to Ad11p except in the E2B polymerase (pol) and terminal protein (pTP) coding regions where it is Ad11p/Ad3 chimeric. Our observation that the OvAd viruses are 2–3 logs more potent on a variety of ovarian cell lines than either of the parental serotypes (Ad11p and Ad3) that recombined in the complex pattern that yielded ColoAd1 suggests that it is this chimeric E2B region inherited from ColoAd1 that is responsible for the enhanced potency of the OvAd viruses and that the mechanism of this enhanced potency is enhanced viral replication. The pol and pTP proteins form a heterodimer that binds to the terminal 18 bp of the viral genome, the minimal origin of replication. The OvAd1 and OvAd2 origins of replication are identical to those of Ad3, Ad11p, and ColoAd1. The OvAd/ColoAd1 chimeric pTP-pol heterodimer may interact more efficiently with this origin of replication than either the Ad3 or Ad11p wild-type heterodimers, as we have postulated in discussing the enhanced potency of ColoAd1.10 Furthermore, by analogy to Ad5, it is expected that the pTP-pol heterodimer interacts with NFI and NFIII/oct-1 transcription factors which also bind at the viral replication origin, interactions that directly affect viral replication efficiency,29, 30 and hence viral burst titers and potency. Since these transcription factors are cellular, they may be part of the mechanism by which the ovarian cancer cells participated in the selection of viruses with enhanced ovarian potency and tumor specificity during the directed evolution process. Thus, both the interactions of the chimeric heterodimer with the viral origin of replication and with cellular factors may contribute to larger and/or more rapid viral bursts, leading to enhanced cell lysis and viral spread.
Although equivalently potent on ovarian cancer cell monolayers in vitro, OvAd1 intriguingly proved more potent than OvAd2 (zero of seven i.p. tumors versus four of seven i.p. tumors; Figure 5) in an in vivo intraperitoneal ovarian carcinomatosis model. Given their otherwise identical genomes, the genetic basis for this functional difference maps to the 3-kb region between 10,000 and 13,060, making all functions mapping to this region of interest in discovering the mechanism of OvAd1’s apparently stronger in vivo potency against SKOV3 xenograft tumors. In this region, OvAd1 is identical to Ad11p and to ColoAd1, whereas OvAd2 is identical to Ad3. By comparison to the Ad11p (GenBank: AY163756) and Ad3 (GenBank: AY599834) genomes, the functions spanning or mapping to this region include E2B region splicing, and genes encoding Viral Associated RNA and the L1 proteins 52/55 Kda and pIIIa. Attributing OvAd1’s superior in vivo potency to “cross-serotype hybrid vigor” resulting from Ad3-serotype capsid protein interactions with gene products inherited from ColoAd1/Ad11p is one hypothesis. An alternative hypothesis is that the mechanism of OvAd1’s superior in vivo potency is due to the interactions of the 52/55 Kda protein with the viral DNA replication and packaging machinery, all of which are ColoAd1/Ad11p derived, while in the case of OvAd2 these are inter-serotype interactions (the 52/55 Kda protein derives from Ad3 while IVa2, pol, and pTP derive from ColoAd1/Ad11p). If OvAd1 continues to prove more effective than OvAd2 in larger in vivo studies, further studies to elucidate which factors are responsible for these differences in efficacy would be warranted. In addition to demonstrating improved anti-tumor activity, OvAd1 and the other group B capsid viruses (OvAd2 and ColoAd1) were better tolerated in mice showing fewer side effects than the group C capsid virus ONYX-015. This observation is consistent with previous reports comparing the tolerability of adenoviruses from Ad11 and Ad5 viruses administered into the peritoneal cavity.27 The presence of human group B receptors in transgenic mice does not appear sensitize animals to group B viruses in these studies and the difference in tolerability may be due to the response by macrophages that are responsible for clearing most of the administered dose. The improved safety profile of group B viruses in humans is currently under evaluation in a number of phase I clinical trials.
There has been concern that a patient’s innate or acquired immunity to a given virotherapy may reduce efficacy of viral agents. This possibility applies to the OvAds because they are Ad serotype 3, and seroprevalence of antibodies to Ad3 is common.31 However, it remains unclear whether anti-viral immunity enhances, or interferes with, efficacy of oncolytic virotherapy. In one study, when ovarian cancer patients were treated intraperitoneally with an Ad3 fiberknob-oncolytic (Ad5/3), the clinical data suggest this virus was able in some patients to infect and produce viral bursts despite anti-viral immunity.32 Other evidence suggests that pre-existing anti-viral immunity may enhance the therapeutic efficacy of oncolytic viruses (OV).33 In preclinical models, animals whose initial malignant xenograft had responded to oncolytic therapy were able, much later, to fully resist repeat injections of malignant cells, without additional OV administration, thus modeling OV-dependent resistance to recurrent cancer.34 Clinical experience similarly indicates that viral infection leading to tumor cell lysis may, by presenting novel combinations of viral and tumor immunogens, be able to overcome suppression of anti-tumor responses, thus aiding tumor clearance and blocking metastatic growth.7, 35, 36 Indications of tumor microenvironmental factors affecting the efficacy of oncolytic virotherapies now include many factors beyond immune functions.37 There is clear need for predictive preclinical models in which each such factor can accurately be evaluated so that we do not set aside potentially effective oncolytic therapeutics. Current efforts to develop complex co-cultures reconstituting the ovarian tumor microenvironment are promising in this context,38, 39, 40 as is the potential of hybridizing available mouse models of ovarian cancer (reviewed in Morin and Weeraratna41) with genetically diverse collaborative cross-strains to generate immunocompetent, genetically diverse ovarian cancer models.42
To investigate the effects of anti-viral immunity on oncolytic virotherapy for ovarian cancer, given that OvAd1 is serotype Ad3, it may be of interest to complete the testing and pre-clinical characterization of those Ad11p serotype viruses that were isolated in this work from the SM10 pool. Seroprevalence of anti-Ad11p antibodies is low compared with seroprevalence of anti-Ad3. Determining the in vivo efficacy of OvAd1 compared to “OvAd1-like virus bearing an Ad11p capsid” and to ColoAd1 may therefore be informative as to the role of anti-viral immunity in tumor clearing. We note that it may be possible to genomically engineer a version of OvAd1 expressing Ad11p capsid proteins for this purpose; however, such engineering may interfere with the integrity of the OvAd1-like viral particles and thus is not preferred.
The ovarian tumor-targeting oncolytic viruses described here are both potent and tumor selective in vitro and in vivo, and unlike Ad5-based oncolytic viruses, OvAd1 and OvAd2 do not generate peritoneal adhesions. Comparisons with ONYX-015 suggest that OvAd1 and OvAd2 would be safe for clinical use. The use of the basement membrane extract Matrigel appears to have been effective for the directed evolution of clinically relevant oncolytic viruses. The viral pool selected using Matrigel has a much wider therapeutic window (i.e., tumor selectivity) than the viral pool selected using standard monolayer cell culture methods. Furthermore, OvAd1, the isolate from the Matrigel pool, had somewhat greater in vivo potency than OvAd2. The data presented here warrant the further development and testing of these oncolytics for clinical use.
Materials and Methods
Viruses and Cell Lines
The human Ad serotypes Ad3 (GB strain), Ad4 (RI-67 strain), Ad5 (Adenoid 75 strain), Ad9 (Hicks strain), and Ad16 (Ch. 79 strain) and the SKOV3, OVCAR3, PC-3, and HT-29 cell lines used were all purchased from the ATCC and were cultured according to ATCC directions. The chimeric virus ColoAd110 was developed in this laboratory. ONYX-015 was a gift from its developer, Onyx Pharmaceuticals. Ad11p (Slobitski strain), Ad35, and Ad40 were gifts from Dr. William S.M. Wold at St. Louis University. Other cells used were MDA-231mt1 (a cell line derivative isolated by Dr. D. Zajchowski, Berlex Laboratories, from a rapidly growing subcutaneously implanted xenograft of MDA-231 cells) and Panc1-sct (derived by Dr. S. Biroc, Berlex Laboratories, from a rapidly growing subcutaneously implanted xenograft of Panc1 cells) both previously published.10 The SKOV3-Luc cells expressing luciferase were developed by Dr. K. Fisher. Normal primary human cells isolated from tissue were used as follows. Primary normal human umbilical vein endothelial cells (HUVECs) isolated from human tissue sources were purchased from Vec Technologies and used according to their instructions.
Viral Purification and Quantification
Viral stocks were propagated on 293 cells, purified on CsCl gradients, and titered (viral particles per milliliter is the unit used throughout this report) by spectroscopy43 and by anion-exchange chromatography.19 Most Ad serotypes have distinct retention profiles when analyzed by the AIEX method used, and both purified stocks and crude lysates can be accurately titered by AIEX chromatography. Therefore, AIEX chromatography was used to determine the titer of, and to partially characterize the serotype-relatedness of, all viral stocks.19
Directed Evolution
Viral serotypes representing subgroup Ads B-F, namely Ad3, Ad4, Ad5, Ad9, Ad11p, Ad16, Ad35, Ad40, and the chimeric virus ColoAd110 were assembled into the starting viral pool. A portion of the starting pool, containing 1012 viral particles of each viral serotype, was subjected to published method for random mutagenesis by nitrous acid;44, 45 the reaction was stopped by neutralization of the nitrous acid after 2.5–3 logs of kill (loss of viral titer). A stock of Ad5 was mutagenized in parallel in order to determine the extent of mutagenesis induced by the nitrous acid. A 356-bp region of the 19 K adenovirus protein was amplified from each of 10 viral isolates from this parallel Ad5 mutagenized stock and sequenced. Sequencing showed that one in ten isolates carried a point mutation compared to non-mutagenized stocks. Extrapolation from this result (0.1 mutations in 1% of the viral genome) indicates that approximately 10 mutations were introduced on average per viral genome. The pool of mutagenized virus serotypes was termed the mutagenized pool. Another portion of the starting pool, containing 109 viral particles of each viral type, was then added to an equal amount of the mutagenized pool to create the combined pool so that the combined starting pool contained equivalent numbers of mutagenized and non-mutagenized viral particles. This combined pool was used to infect, at a multiplicity of infection (MOI) = 10, subconfluent monolayers of SKOV3. These infection conditions were chosen to invite recombination between all viral types present in the combined (mutagenized and non-mutagenized) viral pool. Viral lysates were harvested from these infected cultures at 24 and 48 hr post infection (hpi), then mixed together to produce the “recombined” pool. A fresh aliquot of the combined pool was then added to this “recombined” pool to generate the fully biodiverse viral pool used for bioselection. The biodiverse viral pool was passaged once on a sub-confluent, monolayer culture of SKOV3 cells at MOI = 10, a high particle-per-cell ratio again used to invite recombination between all virus present in the biodiverse pool. The titer of the viral lysate supernatant from this round of high viral particle-per-cell infection of subconfluent (70% confluent) SKOV3 monolayer cells was determined by AIEX chromatography, then used in a 10-fold dilution series, starting at an MOI = 0.1, to infect a series of over-confluent SKOV3 cultures grown in 6-well plates. To achieve over-confluency, SKOV3 cells were seeded at a density that allowed that cell line to reach confluency between 24 and 40 hr post seeding, and the cells were allowed to grow a total of 72 hr post seeding prior to infection. This cell density was 150,000 cells/cm2. This high cell density and prolonged growth was used to maximize confluency at time of infection, with the goal of mimicking growth conditions in human ovarian tumors. Cell culture supernatant was harvested from the well infected with the most concentrated innocula in the 10-fold dilution series that did not show any sign of cytopathic effects (CPE) at day 3 or 4 post-infection. Each harvest served as the starting material for the next passage of the virus. This process was repeated until the viral pool achieved 10 passages.
3D directed evolution using SKOV3 cells grown on growth-factor-reduced Matrigel (GFR-Matrigel)-coated tissue culture plates was done as described for directed evolution on monolayer cultures, with the exceptions that the culture dishes or wells were coated, following manufacturer’s directions, with Matrigel (Becton Dickinson Labware) at 150 μL/cm2 prior to seeding the cells. SKOV3 cells were seeded onto Matrigel-coated plates at 150,000 cells/cm2, a density that generated three-dimensional growth morphology in these cells by 24 hr post seeding (hps). The cells were infected 24–36 hps (i.e., soon after the three-dimensional growth patterns induced by Matrigel had become apparent in the cultures). One passage at MOI = 10 was done to invite recombination between all the viral genomes in the pool before starting selective passaging. Selective passaging on Matrigel was done, as described for monolayer bioselection, starting at an MOI of 0.1, followed by three 10-fold serial dilutions. At each passage, culture supernatant was harvested from the culture infected with the most concentrated innocula in the 10-fold dilution series that did not show any sign of CPE at day 3 or 4 post-infection. A total of 10 passages were performed before individual viruses were isolated and characterized from each bioselected pool.
Isolation and Characterization of Individual Directed-Evolution-Derived Viruses
Individual viruses were isolated from the 10th passage of each pool (termed SM10 [i.e., the pool that was passaged on Matrigel-based SKOV3 cultures] and SP10 [i.e., the pool that was passaged on monolayers of SKOV3], respectively) by two rounds of plaque purification on SKOV3 cells using standard methods.43 Plaques from the second round of plaque purification were deemed pure, infected cultures of A549 cells were prepared using these purified plaques, and the oncolytic potency of these culture lysates was determined by MTS assay as described.
MTS Assay
Viral potency against each cancer cell line or normal human primary cell sample was measured by MTS assay as described.10 To perform the MTS assay, cells were seeded into 96-well plates at a density determined for each cell type to generate a confluent monolayer within 24 hr. These densely seeded cells were allowed to grow for 2 additional days prior to exposure to the test virus(es). Viral lysates or stocks to be assayed for potency by MTS assay were titered by anion exchange chromatography.19 Infections of cancer cells and of human normal primary (HUVEC) cells were carried out in quadruplicate, using serial 3-fold dilutions of the viruses starting at a particle per cell ratio of 100 and ending at a particle per cell ratio of 0.005. Infected cells were incubated at 37°C, and the MTS assay was performed at the time points indicated for the individual non-malignant cells or cancer cell lines. Mock-infected cells served as negative controls and established the 100% survival point for the given assay. Each data point in any given MTS assay was assessed in quadruplicate, and IC50 values were derived from dose response curves with R2v value of ≥0.9. Each MTS assay was repeated at least twice, with consistent results.
DNA Sequencing of OvAd1 and OvAd2
DNA sequencing was performed by Commonwealth Biotechnologies of OvAd1, OvAd2, Ad3, and ColoAd1 DNA purified at Berlex Biosciences. The DNA was partially digested with Sau3 A1, shotgun cloned into pBluescriptII vector. Positive clones propagated from which plasmids were isolate and sequenced using the sequencing primers M13R and KS. Individual sequencing reactions were trimmed, edited, and assembled using Sequencher (Gene Codes). Gaps in coverage were amplified with custom oligonucleotide primers and sequenced. The 5′ and 3′ ends were sequenced directly off the ColoAd1 and Ad3 DNA.
Xenograft Model
A xenograft model of ovarian peritoneal carcinomatosis was established in MF1 nude mice by administration of 5 × 106 SKOV-3.Luc cells on day 0 (t0) into the peritoneal compartment and randomly assigned to seven groups of seven mice each. At days 3, 5, and 7, mice were administered PBS, ONYX-015, Ad11p, OvAd1, or OvAd2 via a 0.5-mL i.p. injection. All virus-treated mice received a dose of 5 × 1010 viral particles (vp) per injection for a total of 1.5 × 1011 vp/mouse. Cell growth was monitored by luciferase (in vivo imaging system [IVIS]), and survival data was taken at humane endpoint of 10% weight loss or moderate clinical symptoms.
Author Contributions
Conceived and designed the in culture experiments, I.K., T.H.; Performed the experiments to choose culture subtratum, generated and characterized by AIEX profile and MTS assay of the recombinant and selected pools as well as isolates OvAd1 and OvAd2, I.K.; Analyzed the data, I.K.; Animal studies design, K.F., T.H., L.S., N.G.; Animal studies, N.G.; Wrote the paper, I.K., T.H., K.F., M.B.
Conflicts of Interest
Irene Kuhn and Terry Hermiston are inventors on U.S. patent 8,765,463 B2 covering use of directed evolution to generate oncolytic viruses. Len Seymour, Terry Hermiston, and Kerry Fisher own stock or other financial interests in PsiOxus Therapeutics Ltd., which is developing ColoAd1 (now called enadenotucirev) for clinical use.
Acknowledgments
We thank Jean MacRobbie and other members of the Berlex Cell Core for extensive assistance with cell culture for the MTS assays. We thank Paul Harden for designing the analytical software for the MTS assays used both here and prior.10 I.K., M.B., and T.H. were supported during this research by Bayer Healthcare A.G. N.G., K.F., and L.S. acknowledge financial support from Cancer Research UK programme grant C552/A17720.
Footnotes
Supplemental Information includes three figures and can be found with this article online at http://dx.doi.org/10.1016/j.omto.2016.12.001.
Contributor Information
Irene Kuhn, Email: ikuhn@lbl.gov.
Terry Hermiston, Email: thermiston@coagulanttherapeutics.com.
Supplemental Information
References
- 1.Lowe K.A., Chia V.M., Taylor A., O’Malley C., Kelsh M., Mohamed M., Mowat F.S., Goff B. An international assessment of ovarian cancer incidence and mortality. Gynecol. Oncol. 2013;130:107–114. doi: 10.1016/j.ygyno.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 2.English D.P., Menderes G., Black J., Schwab C.L., Santin A.D. Molecular diagnosis and molecular profiling to detect treatment-resistant ovarian cancer. Expert Rev. Mol. Diagn. 2016;16:769–782. doi: 10.1080/14737159.2016.1188692. [DOI] [PubMed] [Google Scholar]
- 3.Buller R.E., Runnebaum I.B., Karlan B.Y., Horowitz J.A., Shahin M., Buekers T., Petrauskas S., Kreienberg R., Slamon D., Pegram M. A phase I/II trial of rAd/p53 (SCH 58500) gene replacement in recurrent ovarian cancer. Cancer Gene Ther. 2002;9:553–566. doi: 10.1038/sj.cgt.7700472. [DOI] [PubMed] [Google Scholar]
- 4.Aghi M., Martuza R.L. Oncolytic viral therapies—the clinical experience. Oncogene. 2005;24:7802–7816. doi: 10.1038/sj.onc.1209037. [DOI] [PubMed] [Google Scholar]
- 5.Vasey P.A., Shulman L.N., Campos S., Davis J., Gore M., Johnston S., Kirn D.H., O’Neill V., Siddiqui N., Seiden M.V., Kaye S.B. Phase I trial of intraperitoneal injection of the E1B-55-kd-gene-deleted adenovirus ONYX-015 (dl1520) given on days 1 through 5 every 3 weeks in patients with recurrent/refractory epithelial ovarian cancer. J. Clin. Oncol. 2002;20:1562–1569. doi: 10.1200/JCO.2002.20.6.1562. [DOI] [PubMed] [Google Scholar]
- 6.Bourke M.G., Salwa S., Harrington K.J., Kucharczyk M.J., Forde P.F., de Kruijf M., Soden D., Tangney M., Collins J.K., O’Sullivan G.C. The emerging role of viruses in the treatment of solid tumours. Cancer Treat. Rev. 2011;37:618–632. doi: 10.1016/j.ctrv.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Hartkopf A.D., Fehm T., Wallwiener D., Lauer U. Oncolytic virotherapy of gynecologic malignancies. Gynecol. Oncol. 2011;120:302–310. doi: 10.1016/j.ygyno.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 8.Liu T.C., Galanis E., Kirn D. Clinical trial results with oncolytic virotherapy: a century of promise, a decade of progress. Nat. Clin. Pract. Oncol. 2007;4:101–117. doi: 10.1038/ncponc0736. [DOI] [PubMed] [Google Scholar]
- 9.Burke J.M., Lamm D.L., Meng M.V., Nemunaitis J.J., Stephenson J.J., Arseneau J.C., Aimi J., Lerner S., Yeung A.W., Kazarian T. A first in human phase 1 study of CG0070, a GM-CSF expressing oncolytic adenovirus, for the treatment of nonmuscle invasive bladder cancer. J. Urol. 2012;188:2391–2397. doi: 10.1016/j.juro.2012.07.097. [DOI] [PubMed] [Google Scholar]
- 10.Kuhn I., Harden P., Bauzon M., Chartier C., Nye J., Thorne S., Reid T., Ni S., Lieber A., Fisher K. Directed evolution generates a novel oncolytic virus for the treatment of colon cancer. PLoS ONE. 2008;3:e2409. doi: 10.1371/journal.pone.0002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barcellos-Hoff M.H., Aggeler J., Ram T.G., Bissell M.J. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105:223–235. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aggeler J., Ward J., Blackie L.M., Barcellos-Hoff M.H., Streuli C.H., Bissell M.J. Cytodifferentiation of mouse mammary epithelial cells cultured on a reconstituted basement membrane reveals striking similarities to development in vivo. J. Cell Sci. 1991;99:407–417. doi: 10.1242/jcs.99.2.407. [DOI] [PubMed] [Google Scholar]
- 13.Petersen O.W., Rønnov-Jessen L., Howlett A.R., Bissell M.J. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weigelt B., Lo A.T., Park C.C., Gray J.W., Bissell M.J. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res. Treat. 2010;122:35–43. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenny H.A., Lal-Nag M., White E.A., Shen M., Chiang C.Y., Mitra A.K., Zhang Y., Curtis M., Schryver E.M., Bettis S. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat. Commun. 2015;6:6220. doi: 10.1038/ncomms7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng L., Hu X., Huang Y., Xu G., Yang J., Li L. In vivo bioengineered ovarian tumors based on collagen, matrigel, alginate and agarose hydrogels: a comparative study. Biomed. Mater. 2015;10:015016. doi: 10.1088/1748-6041/10/1/015016. [DOI] [PubMed] [Google Scholar]
- 17.Bacchetti S., Graham F. Inhibition of cell-proliferation by an adenovirus vector expressing the human wild type-p53 protein. Int. J. Oncol. 1993;3:781–788. doi: 10.3892/ijo.3.5.781. [DOI] [PubMed] [Google Scholar]
- 18.Li B., Chen D., Li W., Xiao D. 20(S)-Protopanaxadiol saponins inhibit SKOV3 cell migration. Oncol. Lett. 2016;11:1693–1698. doi: 10.3892/ol.2016.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn I., Larsen B., Gross C., Hermiston T. High-performance liquid chromatography method for rapid assessment of viral particle number in crude adenoviral lysates of mixed serotype. Gene Ther. 2007;14:180–184. doi: 10.1038/sj.gt.3302851. [DOI] [PubMed] [Google Scholar]
- 20.Strauss R., Sova P., Liu Y., Li Z.Y., Tuve S., Pritchard D., Brinkkoetter P., Möller T., Wildner O., Pesonen S. Epithelial phenotype confers resistance of ovarian cancer cells to oncolytic adenoviruses. Cancer Res. 2009;69:5115–5125. doi: 10.1158/0008-5472.CAN-09-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garber K. China approves world’s first oncolytic virus therapy for cancer treatment. J. Natl. Cancer Inst. 2006;98:298–300. doi: 10.1093/jnci/djj111. [DOI] [PubMed] [Google Scholar]
- 23.Kirn D. Clinical research results with dl1520 (Onyx-015), a replication-selective adenovirus for the treatment of cancer: what have we learned? Gene Ther. 2001;8:89–98. doi: 10.1038/sj.gt.3301377. [DOI] [PubMed] [Google Scholar]
- 24.Zheng K., Kitazato K., Wang Y. Viruses exploit the function of epidermal growth factor receptor. Rev. Med. Virol. 2014;24:274–286. doi: 10.1002/rmv.1796. [DOI] [PubMed] [Google Scholar]
- 25.Lin T., Gu J., Zhang L., Davis J.J., Huang X., Cabbini G., Ji L., Fang B. Enhancing adenovirus-mediated gene transfer in vitro and in vivo by addition of protamine and hydrocortisone. J. Gene Med. 2003;5:868–875. doi: 10.1002/jgm.427. [DOI] [PubMed] [Google Scholar]
- 26.Zeimet A.G., Marth C. Why did p53 gene therapy fail in ovarian cancer? Lancet Oncol. 2003;4:415–422. doi: 10.1016/s1470-2045(03)01139-2. [DOI] [PubMed] [Google Scholar]
- 27.Thoma C., Bachy V., Seaton P., Green N.K., Greaves D.R., Klavinskis L., Seymour L.W., Morrison J. Adenovirus serotype 11 causes less long-term intraperitoneal inflammation than serotype 5: implications for ovarian cancer therapy. Virology. 2013;447:74–83. doi: 10.1016/j.virol.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 28.Kanerva A., Zinn K.R., Chaudhuri T.R., Lam J.T., Suzuki K., Uil T.G., Hakkarainen T., Bauerschmitz G.J., Wang M., Liu B. Enhanced therapeutic efficacy for ovarian cancer with a serotype 3 receptor-targeted oncolytic adenovirus. Mol. Ther. 2003;8:449–458. doi: 10.1016/s1525-0016(03)00200-4. [DOI] [PubMed] [Google Scholar]
- 29.Mul Y.M., Verrijzer C.P., van der Vliet P.C. Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J. Virol. 1990;64:5510–5518. doi: 10.1128/jvi.64.11.5510-5518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mysiak M.E., Bleijenberg M.H., Wyman C., Holthuizen P.E., van der Vliet P.C. Bending of adenovirus origin DNA by nuclear factor I as shown by scanning force microscopy is required for optimal DNA replication. J. Virol. 2004;78:1928–1935. doi: 10.1128/JVI.78.4.1928-1935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogels R., Zuijdgeest D., van Rijnsoever R., Hartkoorn E., Damen I., de Béthune M.P., Kostense S., Penders G., Helmus N., Koudstaal W. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 2003;77:8263–8271. doi: 10.1128/JVI.77.15.8263-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim K.H., Dmitriev I.P., Saddekni S., Kashentseva E.A., Harris R.D., Aurigemma R., Bae S., Singh K.P., Siegal G.P., Curiel D.T., Alvarez R.D. A phase I clinical trial of Ad5/3-Δ24, a novel serotype-chimeric, infectivity-enhanced, conditionally-replicative adenovirus (CRAd), in patients with recurrent ovarian cancer. Gynecol. Oncol. 2013;130:518–524. doi: 10.1016/j.ygyno.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong A.W., Senzer N., Cerullo V., Templeton N.S., Hemminki A., Nemunaitis J. Oncolytic viruses for induction of anti-tumor immunity. Curr. Pharm. Biotechnol. 2012;13:1750–1760. doi: 10.2174/138920112800958913. [DOI] [PubMed] [Google Scholar]
- 34.Benencia F., Courrèges M.C., Fraser N.W., Coukos G. Herpes virus oncolytic therapy reverses tumor immune dysfunction and facilitates tumor antigen presentation. Cancer Biol. Ther. 2008;7:1194–1205. doi: 10.4161/cbt.7.8.6216. [DOI] [PubMed] [Google Scholar]
- 35.Kim Y., Clements D.R., Sterea A.M., Jang H.W., Gujar S.A., Lee P.W. Dendritic cells in oncolytic virus-based anti-cancer therapy. Viruses. 2015;7:6506–6525. doi: 10.3390/v7122953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naik J.D., Twelves C.J., Selby P.J., Vile R.G., Chester J.D. Immune recruitment and therapeutic synergy: keys to optimizing oncolytic viral therapy? Clin. Cancer Res. 2011;17:4214–4224. doi: 10.1158/1078-0432.CCR-10-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ilkow C.S., Marguerie M., Batenchuk C., Mayer J., Ben Neriah D., Cousineau S., Falls T., Jennings V.A., Boileau M., Bellamy D. Reciprocal cellular cross-talk within the tumor microenvironment promotes oncolytic virus activity. Nat. Med. 2015;21:530–536. doi: 10.1038/nm.3848. [DOI] [PubMed] [Google Scholar]
- 38.Chen J., Wang J., Chen D., Yang J., Yang C., Zhang Y., Zhang H., Dou J. Evaluation of characteristics of CD44+CD117+ ovarian cancer stem cells in three dimensional basement membrane extract scaffold versus two dimensional monocultures. BMC Cell Biol. 2013;14:7. doi: 10.1186/1471-2121-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutmacher D.W., Cukierman E. Engineering of tumor microenvironments. Adv. Drug Deliv. Rev. 2014;79-80:1–2. doi: 10.1016/j.addr.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 40.White E.A., Kenny H.A., Lengyel E. Three-dimensional modeling of ovarian cancer. Adv. Drug Deliv. Rev. 2014;79-80:184–192. doi: 10.1016/j.addr.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morin P.J., Weeraratna A.T. Genetically-defined ovarian cancer mouse models. J. Pathol. 2016;238:180–184. doi: 10.1002/path.4663. [DOI] [PubMed] [Google Scholar]
- 42.Durrant C., Tayem H., Yalcin B., Cleak J., Goodstadt L., de Villena F.P., Mott R., Iraqi F.A. Collaborative Cross mice and their power to map host susceptibility to Aspergillus fumigatus infection. Genome Res. 2011;21:1239–1248. doi: 10.1101/gr.118786.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tollefson A., Hermiston T., Wold W.S. Preparation and titration of cesium chloride banded adenovirus stock. In: Wold W., editor. Adenovirus Methods and Protocols. Humana Press; 1999. pp. 1–9. [Google Scholar]
- 44.Williams J.F., Gharpure M., Ustacelebi S., McDonald S. Isolation of temperature-sensitive mutants of adenovirus type 5. J. Gen. Virol. 1971;11:95–101. doi: 10.1099/0022-1317-11-2-95. [DOI] [PubMed] [Google Scholar]
- 45.Klessig D.F. Isolation of a variant of human adenovirus serotype 2 that multiplies efficiently on monkey cells. J. Virol. 1977;21:1243–1246. doi: 10.1128/jvi.21.3.1243-1246.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.