Abstract
Lifestyle factors and chronic pathologic states are important contributors to interindividual variability in susceptibility to xenobiotic-induced toxicity. Nonalcoholic fatty liver disease (NAFLD) is an increasingly prevalent condition that can dramatically affect chemical metabolism. We examined the effect of NAFLD on toxicokinetics of tetrachloroethylene (PERC), a ubiquitous environmental contaminant that requires metabolic activation to induce adverse health effects. Mice (C57Bl/6J, male) were fed a low-fat diet (LFD), high-fat diet (HFD), or methionine/folate/choline-deficient diet (MCD) to model a healthy liver, steatosis, or nonalcoholic steatohepatitis (NASH), respectively. After 8 weeks, mice were orally administered a single dose of PERC (300 mg/kg) or vehicle (aqueous Alkamuls-EL620) and euthanized at various time points (1–36 hours). Levels of PERC and its metabolites were measured in blood/serum, liver, and fat. Effects of diets on liver gene expression and tissue:air partition coefficients were evaluated. We found that hepatic levels of PERC were 6- and 7.6-fold higher in HFD- and MCD-fed mice compared with LFD-fed mice; this was associated with an increased PERC liver:blood partition coefficient. Liver and serum Cmax for trichloroacetate (TCA) was lower in MCD-fed mice; however, hepatic clearance of TCA was profoundly reduced by HFD or MCD feeding, leading to TCA accumulation. Hepatic mRNA/protein expression and ex vivo activity assays revealed decreased xenobiotic metabolism in HFD- and MCD-, compared with LFD-fed, groups. In conclusion, experimental NAFLD was associated with modulation of xenobiotic disposition and metabolism and increased hepatic exposure to PERC and TCA. Underlying NAFLD may be an important susceptibility factor for PERC-associated hepatotoxicity.
Introduction
Interindividual variability is a major challenge for the assessment of human health burden associated with exposure to environmental contaminants (Zeise et al., 2013). Variability in toxicokinetics and ensuing metabolic activation of the parent chemical to relatively more toxic metabolites is often the underlying cause of the variability in adverse health responses (Bois et al., 2010). The role of genetic factors in the population variability in drug and chemical metabolism is well established (Relling and Evans, 2015); however, a variety of other factors such as sex, age, diet, and underlying disease states may also contribute to interindividual variability. Specifically, non-alcoholic fatty liver disease (NAFLD) is a growing public health burden, with a global prevalence of ∼25% (Younossi et al., 2016b) and an associated economic impact that may exceed US$900 billion over the next 10 years in the U.S. alone (Younossi et al., 2016a). NAFLD encompasses two clinically-distinct disease states: simple steatosis and the more severe non-alcoholic steatohepatitis (NASH). Experimental and human NAFLD have been shown to be associated with dysregulation in pharmaco-/toxicokinetics of a variety of chemicals, including pharmaceutical and environmental chemicals (Buechler and Weiss, 2011; Merrell and Cherrington, 2011; Naik et al., 2013; Clarke and Cherrington, 2015).
An excellent case study substance to examine the role of NAFLD as a susceptibility factor in chemical-induced adverse health effects is tetrachloroethylene (perchloroethylene; PERC). It was recently shown that genetics is a major, but not the only, determinant of interindividual variability in toxicokinetics of PERC (Cichocki et al., 2016a). The U.S. Environmental Protection Agency (EPA) in its Toxicological Review of PERC stated that preexisting disease status may contribute to variation in response to PERC through alteration of toxicokinetics of PERC and its metabolites; however, there are few studies on the contribution of disease states to PERC toxicokinetics (U.S. EPA, 2011).
PERC is a high-production volume chlorinated olefin solvent, with industrial uses in dry cleaning, metal degreasing, and as a chemical feedstock (Cichocki et al., 2016b). Because of its widespread use, high production volume, and persistence in the environment, PERC has become a ubiquitous environmental contaminant of ambient air, soil, and drinking and ground water and is one of the most commonly found contaminants at hazardous waste sites (National Research Council, 2010). Exposure to PERC in humans and animals is associated with multiple target organ toxicity, including both cancer and noncancer toxicity (U.S. EPA, 2011; Guha et al., 2012; IARC, 2013; Guyton et al., 2014; Cichocki et al., 2016b). In mice, hepatotoxicity associated with PERC exposure is thought to be mediated by trichloroacetate (TCA), an oxidative metabolite of PERC (Bull et al., 1990; Bull, 2000; Corton, 2008). Indeed, hepatic oxidative metabolism of PERC was the primary dose metric chosen by the U.S. EPA to derive an oral reference dose/concentration for the carcinogenic risk of PERC (U.S. EPA, 2011); thus, the toxicokinetics of PERC and TCA are important considerations when establishing concentration-response relationships.
Although PERC has been studied for decades, very little is known about the specific enzyme(s) involved in PERC metabolism, especially under chronic disease conditions that may have a profound impact on metabolic capacity. It is known that PERC is metabolized by human and rodent hepatic cytochrome P450s (CYPs) to TCA (Cichocki et al., 2016b). Additionally, PERC can also be conjugated with glutathione through glutathione S-transferases (GSTs) to form S-(1,2,2-trichlorovinyl)-glutathione (TCVG). TCVG can be converted in the kidney through a two-step metabolic reaction to yield S-(1,2,2-trichlorovinyl)-l-cysteine (TCVC). TCVC is a metabolite of high concern as it can be converted to cytotoxic and genotoxic electrophiles (Vamvakas et al., 1987, 1988, 1989; Irving and Elfarra, 2013) and can be detoxified to the mercapturate N-acetyl-S-(1,2,2-trichlorvinyl)-l-cysteine (NAcTCVC), which is a urinary metabolite of PERC in humans. However, little is known about the effects of NAFLD on these PERC metabolism pathways.
The objective of this study was to examine the toxicokinetics of PERC and its major metabolites in tissues from mice with varying stages of NAFLD. Concentration-time profiles were generated for PERC, TCA, TCVG, TCVC, and NAcTCVC in liver and serum. To identify mechanisms of NAFLD-associated effects on PERC toxicokinetics, mRNA sequencing and other biochemical techniques were employed.
Materials and Methods
Chemicals.
PERC (CAS 127-18-4) was purchased from Sigma Aldrich (cat. no. 270393, Batch No. SHBD9374V, purity 99.93%; St. Louis, MO). Analytical standards for S-(1,2,2-trichlorovinyl) glutathione (TCVG), 13C215N-TCVG, S-(1,2,2-trichlorovinyl)-l-cysteine (TCVC), 13C515N-TCVC, N-acetyl-S-(1,2,2-trichlorovinyl)-l-cysteine (NAcTCVC), and 13C315N-NAcTCVC were graciously provided by Dr. Avram Gold at the University of North Carolina, Chapel Hill. Purity of all standards was determined to be >95% by NMR and mass spectroscopy. All other chemicals were of the highest purity available and were acquired from commercial vendors.
Animals.
Five-week-old male C57Bl/6J mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and housed in polycarbonate cages with Sanichip hardwood chip bedding (P.J. Murphy Forest Products Corp., Montville, NJ). Mice were acclimatized for at least 1 week on a standard rodent chow containing 4% calories from fat (low-fat diet; LFD; Teklad Rodent Diet #8604; Harlan, Madison, WI). To model a healthy liver, steatosis, or NASH, mice were randomly assigned (five mice per cage) into groups fed a control LFD, a diet containing 31% fat (high-fat diet, HFD; Diet #519567; Dyets Inc., Bethlehem, PA), or a diet containing 31% kcal from fat depleted in methionine and devoid of choline and folate (methyl and choline-deficient diet, MCD; Diet #519541; Dyets Inc., Bethlehem, PA). The formulation for MCD diet used herein has been previously shown to induce NASH in C57Bl/6J mice without reduction in body weight (Tryndyak et al., 2012), which is a common deficiency of other MCD diets (Maher, 2011).
Study Design.
After 8 weeks of dietary treatments as detailed above, mice were administered a single intragastric dose of PERC (300 mg/kg in 5% Alkamuls-EL620 in saline). This dose was selected as the toxicokinetics of PERC and TCA in male mice after exposure to 150–1000 mg/kg PERC has been characterized in liver and blood (Philip et al., 2007) and was shown to be well-tolerated in acute (Philip et al., 2007) and subchronic studies (National Toxicology Program, 1977). Mice (n = 5/diet/time point) were euthanized at 1, 2, 4, 12, or 24 hours after gavage (total n = 75). In addition, 4 mice per diet group were individually placed in stainless steel metabolic cages with wire mesh bottoms (Techniplast, Chester, PA) for 36 hours for urine collection (total n = 12). Five mice per diet were treated with vehicle and euthanized at 24 hours postgavage to perform additional biochemical analyses (total n = 15). At the respected time points, animals were deeply anesthetized with pentobarbital (50 mg/kg i.p.) and euthanized via exsanguination through the vena cava, which was the site of blood collection. An additional 100 µl of whole blood was drawn from vehicle-treated animals to analyze blood:air partition coefficients. Both whole blood and serum collection via Z-gel tubes (Sarstedt, Nümbrecht, Germany) was performed. Other tissues were excised, rinsed in saline, blotted dry, weighed, and snap-frozen in liquid nitrogen. The liver left lobe was separated from the rest of the liver before freezing. A small section of liver left lobe, liver median lobe, and one kidney were fixed in formalin for histologic examination. An additional small section of liver median lobe was placed on optimal cutting temperature compound on dry ice for frozen sectioning.
Analysis of PERC in Tissues.
Details on PERC analysis in tissue homogenates are provided in the Supplemental Materials and Methods. Briefly, methanolic tissue homogenates were analyzed for PERC content via dynamic headspace gas chromatography/mass spectrometry (GC/MS).
GC/MS Analysis of TCA in Tissues.
The analysis of TCA was modified from USEPA Method (EPA 815-B-03-002). Details are provided in the Supplemental Materials and Methods. Briefly, liver and serum samples were derivatized with a methanolic esterifying reagent to generate TCA methyl ester. After liquid-liquid extraction, the derivatives were analyzed via GC/MS.
Liquid Chromatography/Mass Spectroscopy Analysis of TCVG, TCVC, and NAcTCVC.
The method of Luo et al. (submitted manuscript) was used for detection of TCVG, TCVC, and NAcTCVC in serum, urine, and liver tissue. Briefly, isotopically labeled internal standards (13C215N-TCVG, 13C515N-TCVC, and 13C315N-NAc-TCVC; 50 pmol each) were spiked before extraction to account for recovery and matrix effects. Liver homogenate (50 mg), urine (50 µl), or serum (50 µl) were subjected to solid-phase extraction before separation and detection via liquid chromatography/mass spectroscopy (LC-MS/MS). Calibration curves were prepared in blank tissue from C57Bl/6J mice that were subjected to the same extraction process. The concentration of analyte was determined by taking the response ratio of the analyte to the isotopically labeled internal standard.
Partition Coefficients.
Blood:air and liver:blood partition coefficients were performed using a vial equilibration method (Morris and Cavanagh, 1986). Details are provided in the Supplemental Materials and Methods. Briefly, tissues were placed in sealed vials containing PERC. After an equilibration period, headspace samples (static) were drawn into a GC/MS for detection of airborne PERC. Liver:blood partition coefficient is the ratio of liver:air and blood:air coefficients.
Body Composition.
Whole body lean and fat weights were measured in all animals before chemical exposure via an EchoMRI-100 machine (EchoMRI, Houston, TX).
Serum Clinical Chemistry.
Serum alanine aminotransferase (ALT) was measured with a commercially available kit (Sigma Aldrich) according to manufacturer’s instructions.
Histopathology.
Tissues were embedded in paraffin, sectioned at 5 µm, and stained with hematoxylin and eosin or Sirius Red according to standard protocols. Frozen sections (5 µm) were used for Oil Red O staining. Hematoxylin and eosin-stained slides were evaluated using the NAFLD activity scoring system (Kleiner et al., 2005) by a trained veterinary pathologist who was blinded to the study design and sample assignment to treatment groups.
mRNA Sequencing.
Total RNA was extracted from pulverized liver left lobe tissue from animals treated with vehicle with miRNeasy Mini kits (Qiagen, Valencia, CA). RNA was quantified with Nanodrop (Nanodrop, Wilmington, DE) and purity was assessed via Bioanalyzer (Agilent, Santa Clara, CA). All samples had an RNA integrity number >7.5 and most had an RNA integrity number >8.0. cDNA libraries from 2 µg of RNA were generated using the TruSeq Stranded mRNA Sample kit (high sample protocol; Illumina, San Diego, CA) according to the manufacturer’s instructions. Samples were pooled (7 pools of 10 samples per pool, 1 pool of 8 samples) and 50 base-pair reads were sequenced using an Illumina HiSEq.2500 instrument on 8 sequencing lanes; all samples were run on the same flow cell. A total of 1.83 billion reads were checked to trim any adapter sequences and low quality bases using Trimmomatic (Bolger et al., 2014), resulting in 1.79 billion filtered reads (98%) out of which a total of 1.74 billion filtered reads (approximately 97%) mapped to the GRcm38/mm10 assembly. Read mapping was performed using TopHat version 2.0.13 (Trapnell et al., 2009). HTSeq (Anders et al., 2015) was used to generate raw read counts per gene using the intersection-nonempty parameter to account for ambiguous read mappings.
Differential gene expression tests were then performed via the R (v 3.3.1) package DESeq2 (v 1.12.3) (Love et al., 2014) on the complete list of 18,239 genes after removing low count genes, following recommended guidelines by the authors. Normalized mRNA counts used for data analysis were an output from the DESeq2, which uses an internal normalization algorithm. Briefly, for each gene, a size factor is estimated by calculating the quotient of the raw counts in each sample divided by the raw counts of the geometric mean (all samples), which is effectively a sequencing depth ratio for each gene. The median of all of these quotients is then the relative sequencing depth of the library. This effect size is used to normalize all of the counts of each gene across all of the samples that were sequenced. To be deemed differentially expressed, log2-fold changes (compared with LFD group) and false discovery rate-adjusted P values were cut off at 0.58 and 0.1, respectively. A cut-off for log2 of 0.58 represents an approximate 1.5-fold change in expression level compared with the reference (LFD) group. Default settings in DESeq2 were used for false discovery rate adjustment (Benjamini and Hochberg, 1995). One sample from the HFD group was identified as a technical outlier and was therefore removed from further analysis.
The resulting gene expression values were used for biologic pathway analysis using the “piano” (Varemo et al., 2013) package in R (package v 1.12.0) in conjunction with the “Mouse Reactome” gene set (retrieved from www.baderlab.org on November 11, 2016). For analysis of xenobiotic-metabolizing enzymes, genes with the starting combination of letters “Abc”, “Slc”, “Gst”, “Cyp”, “Ugt”, “Sult”, “Nat”, “Ces”, “Aldh”, and “Adh” were selected from the complete list of mapped genes. A total of 618 genes were retrieved and validated as belonging to these families. Visuals of differentially expressed genes and enriched pathways were generated in R.
Western Blotting.
Liver whole tissue lysates were prepared using a commercially available reagent (T-PER, Thermo Fisher Scientific, Waltham, MA) with added HALT protease inhibitor (Thermo Fisher Scientific). Protein concentrations were determined via a bicinchonic acid assay kit (Thermo Fisher Scientific), using bovine serum albumin as standard. Reduced and denatured liver whole cell lysates (30 µg) were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis. Proteins were transferred to polyvinylidene fluoride membranes, blocked for 1 hour with Odyssey Blocking Buffer (LI-COR, Lincoln, NE) with 0.1% Tween-20 added, and probed with antibodies to goat anti-CYP3A (sc-30632, lot #F2416; Santa Cruz Biotechnology), rabbit anti-CYP2B10 (AB9916, lot #2772669; EMD Millipore, Billerica, MA), rabbit anti-CYP2C9 (ab4236, lot #GR21040-20; Abcam, Cambridge, MA), and rabbit anti-β-actin (ab8227, lot #GR288171-1; Abcam). All primary antibodies were diluted 1:1000 in blocking buffer, except β-actin, which was diluted 1:2500. Incubations were carried out overnight at 4°C. After washing, horseradish peroxidase-conjugated secondary antibodies were added (1:5000 in blocking buffer). After 90 minutes of incubation at room temperature and washing, proteins were detected via enhanced chemiluminescence. Bands were quantified via densitometry using Image Studio (LI-COR).
S9 Fraction Activity Assays.
This method was adapted from a previously established method (Lash et al., 2007). Liver tissue (100 mg) was homogenized in 4 volumes of ice-cold buffer (0.1 M potassium phosphate buffer containing 0.5 mM ethylenediaminetetraacetic acid, pH 7.4) using a bead ruptor (see above). Homogenates were centrifuged (9000 g, 20 minutes, 4°C) and supernatants were aliquoted into equal volumes. Samples were kept on ice until used (typically within 1 hour). One hundred microliters of homogenate was added to a 2-ml glass amber vial containing 400 µl of buffer supplemented with 1.5 mM (final concentration) reduced nicotinamide adenine dinucleotide phosphate (NADPH). Samples were warmed for 5 minutes at 37°C before addition of 2.5 µL of PERC (100 mM in methanol). Samples were quickly capped and incubated for 1 hour at 37°C in a water bath with frequent shaking. Reactions were terminated by adding 1.5 ml of 10% sulfuric acid in methanol (v:v) and 20 µl of 2-bromobutyric acid (550 µM, internal standard). The resulting methyl esters were extracted and analyzed via GC/MS as described above. Data were normalized to the amount of starting liver tissue.
Statistical Analysis.
GraphPad Prism (v 5.0), was used for statistical analysis for biochemical analyses. For comparisons between different disease states, analysis of variance (ANOVA) followed by Newman-Keuls post hoc test was used as a statistical test. For analysis of histopathological scoring, the Kruskal-Wallis test was employed, followed by Dunn's post hoc analysis. For all tests, a P value <0.05 was deemed statistically significant. For analysis of correlations between metabolizing enzyme transcript abundance and toxicokinetic phenotype, Pearson correlation coefficients were determined and associated P values were corrected for multiple comparisons (Benjamini and Hochberg, 1995) to derive q-values using standard R computing. For transcript abundance, normalized mRNA counts (via estimating gene-level size factors) from DESeq2 were used. Details on the algorithm used by this R package can be found in Love et al. (2014).
Data Availability.
RNA sequencing data are publically available through Gene Expression Omnibus (GEO; accession ID: GSE93132), and individual animal phenotype data are supplied as Supplemental Materials. In addition to GEO, complete lists of differentially expressed genes and enriched pathways are provided as Supplemental Materials.
Results
Diet-Associated Liver Effects.
As anticipated, animals maintained on a HFD or MCD for 8 weeks and administered a single dose of vehicle (5% Alkamuls EL-620 in saline) presented with obvious liver steatosis or steatohepatitis, respectively, as evident in histopathological analyses (Table 1, Supplemental Fig. 1). No abnormalities were observed in LFD-fed mice exposed to a single dose of vehicle. Livers from mice fed a HFD displayed mostly diffuse macrovesicular steatosis, whereas livers from mice fed an MCD displayed diffuse micro- and macrovesicular steatosis accompanied by inflammatory cell infiltration. There was pericellular collagen deposition apparent in liver tissue of MCD-fed mice, as indicated by Sirius Red-positive staining in liver sections (Supplemental Fig. 1). In addition to liver histopathology, body weight, body composition, liver- and fat-to-body weight ratios, serum ALT, and serum and liver triglycerides were measured to further characterize the underlying liver disease states in this study (Table 1). Overall, these data confirmed that animals in this study exhibited desirable NAFLD disease phenotypes, which included steatosis or NASH. MCD-fed animals in this study did not exhibit loss of body weight or body fat content.
TABLE 1.
Diet-associated disease phenotypes after 8 weeks of treatment
Data were derived from vehicle-treated mice (24-hour post gavage) and are represented as mean ± S.E.M. (n = 5/group), except for liver histology score, which is represented as median (interquartile range).
| LFD | HFD | MCD | |
|---|---|---|---|
| Body weight (g) | 27.0 ± 0.2 | 33.9 ± 0.4* | 30.3 ± 0.3*,# |
| Liver to body weight ratio (%) | 4.9 ± 0.1 | 4.7 ± 0.1 | 6.2 ± 0.2*,# |
| Body weight fat (%) | 7.5 ± 0.5 | 28.4 ± 1.3* | 17.3 ± 2.4*,# |
| Body weight lean mass (%) | 88.5 ± 2.2 | 66.7 ± 1.3* | 77.0 ± 2.6*,# |
| Gonadal fat pad to body weight ratio (%) | 1.5 ± 0.2 | 4.1 ± 0.7* | 4.2 ± 0.7* |
| Serum triglycerides (mg/dl) | 29.2 ± 3.9 | 18.4 ± 5.1 | 33.6 ± 5.0 |
| Liver triglycerides (mg/g) | 11.0 ± 1.1 | 39.5 ± 2.1* | 39.4 ± 1.6* |
| Serum alanine aminotransferase activity (U/l) | 7.8 ± 0.6 | 33.6 ± 9.8 | 83.5 ± 13.3*,# |
| Total liver histology score | 0 ± 0 | 2.0 (1.0-5.0) | 8.0 (6.0-8.0)* |
P < 0.05 compared with LFD; #P < 0.05 compared with HFD groups using ANOVA with Newman-Keuls post hoc test (all parameters except for histopathological scoring, for which the Kruskal-Wallis test was employed with Dunn’s post hoc test).
Effect of NAFLD on PERC Toxicokinetics.
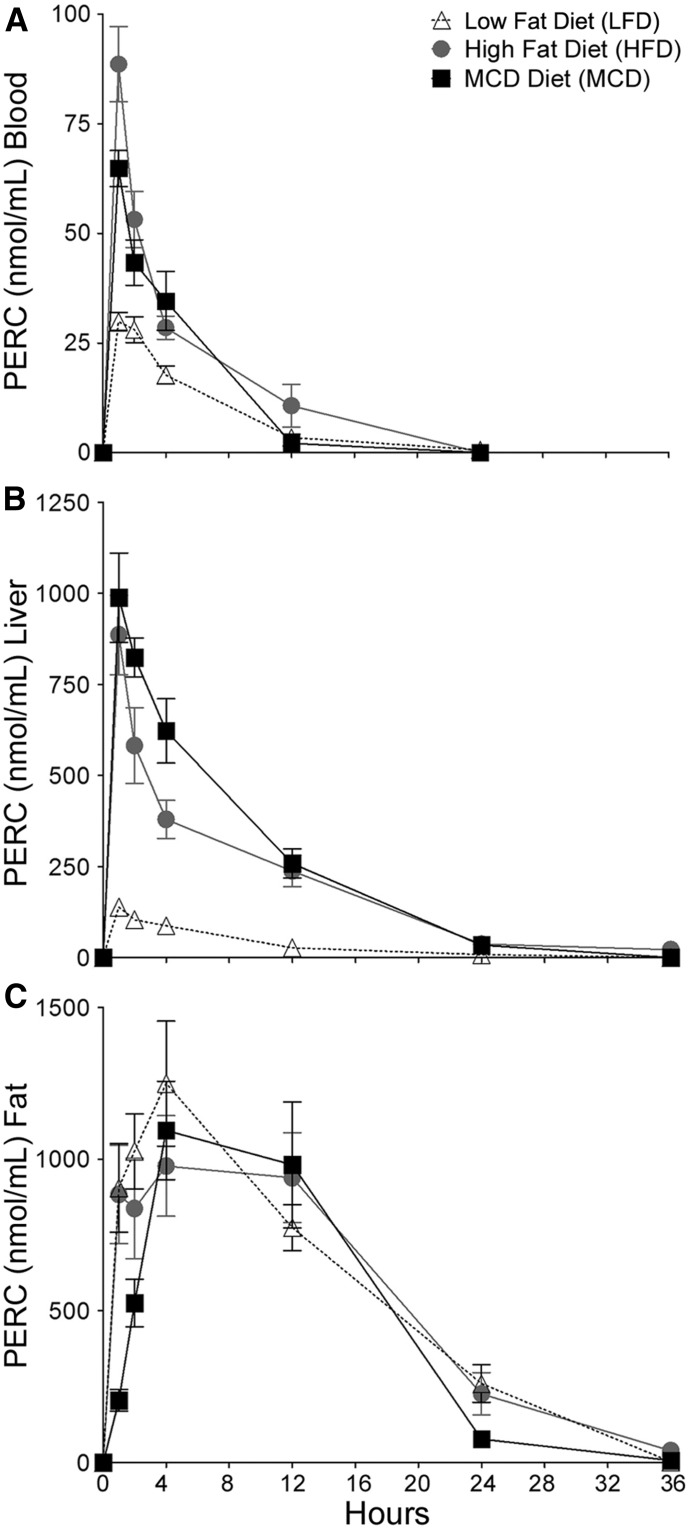
The toxicokinetics of PERC in blood, liver, and gonadal adipose tissue varied considerably after a single intragastric dose (300 mg/kg) depending on the liver disease state (Fig. 1). Table 2 lists areas under the concentration-time curve (AUCs) values for these tissues and disease states. In blood, Cmax for PERC at 1 hour postdosing was highest in HFD-fed mice (88.7 ± 8.6 nmol/ml; mean ± S.E., n = 5), followed by MCD-fed mice (64.8 ± 9.2) and LFD-fed mice (29.9 ± 2.1); PERC was undetectable in the blood by the 24-hour time point. In liver, Cmax for PERC at 1 hour postdosing was highest in MCD-fed mice (988.6 ± 122.7 nmol/g; mean ± S.E., n = 5), followed by HFD-fed mice steatosis (885.8 ± 108.7) and LFD-fed mice (139 ± 22). PERC was detectable in liver at 24 hours only in HFD- and MCD-fed groups. In the adipose tissue, PERC kinetics was not different among groups and was undetectable at 36 hours postdosing.
Fig. 1.
Concentration-time profile of PERC in (A) blood, (B) liver, and (C) fat after a single intragastric dose of PERC (300 mg/kg) after 8 weeks of treatment with different diets. Open triangles, LFD-fed mice; gray circles, HFD-fed mice; black squares, MCD-fed mice. Points represent the mean and error bars represent S.E. (n = 4 or 5/group).
TABLE 2.
AUCs for PERC and its metabolites in LFD-, HFD-, or MCD-fed mice
Partial AUCs were calculated from 0 to 36 hours and are expressed as pmol/g⋅hr for liver and fat or pmol/ml⋅hr for blood and serum. Numbers in parentheses indicate ratios of HFD or MCD to LFD for each corresponding parameter. AUCs for TCA could not be accurately calculated due to TCA not being cleared from liver or serum by the end of the experiment (36 hours postdose).
| LFD | HFD | MCD | ||
|---|---|---|---|---|
| PERC | Blood | 198 × 103 | 418 × 103 (211%) | 324 × 103 (163%) |
| Liver | 1,094 × 103 | 6,604 × 103 (604%) | 8,329 × 103 (761%) | |
| Fat | 16,075 × 103 | 19,364 × 103 (120%) | 17,252 × 103 (107%) | |
| TCVG | Serum | 26 | 8 (31%) | 15 (51%) |
| Liver | 1,058 | 355 (34%) | 296 (28%) | |
| TCVC | Serum | 101 | 111 (110%) | 75 (74%) |
| Liver | 211 | 110 (52%) | 34 (16%) | |
| NAcTCVC | Serum | 436 | 376 (86%) | 271 (62%) |
| Liver | 556 | 348 (63%) | 136 (24%) |
NAFLD Effects on PERC Absorption and Distribution.
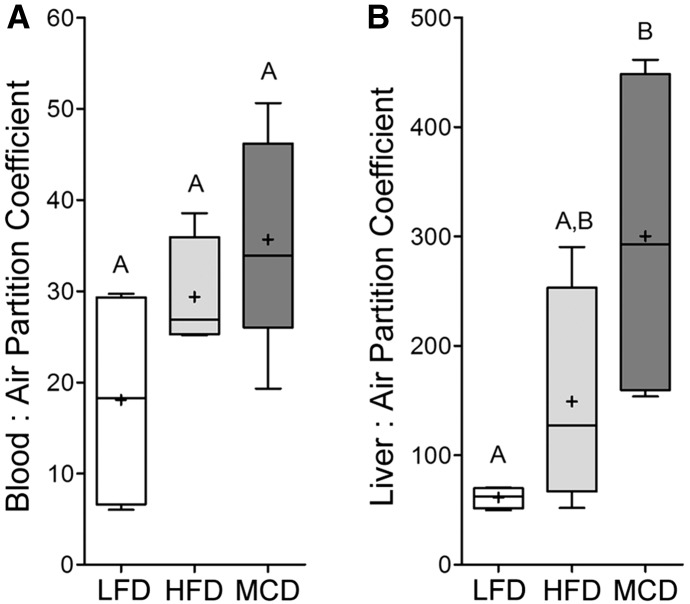
Blood:air and liver:air partitioning coefficients were measured. Blood:air partitioning coefficients were not different (P = 0.0921, ANOVA with Newman Keuls post hoc test) among groups (Fig. 2A). Values (mean ±S.E., n = 4–5) for blood:air partition coefficient were 18.1 ± 6.1, 29.4 ± 5.1, and 35.7 ± 8.4 for LFD-, HFD-, and MCD-fed mice, respectively. The values for LFD-fed mice were similar to those previously reported (Gargas et al., 1989; Gearhart et al., 1993). To determine whether lipid content of the blood, which may affect the partition coefficient, was different between groups, serum triglyceride levels were measured (Table 1). Serum triglyceride levels were similar between groups. Conversely, the liver:air partition coefficients were varying based on the underlying liver disease state (Fig. 2B). Specifically, the liver:air partition coefficients were 61.3 ± 4.9, 149 ± 50.6, and 300 ± 78.9 for LFD-, HFD-, and MCD-fed mice, respectively, with difference of the means in LFD- and MCD-fed mice being significantly different (P < 0.05, ANOVA). The values for liver:air partition coefficient in LFD-fed mice were similar to previously reported values (Gearhart et al., 1993). Liver:blood partition coefficients (a ratio of mean liver:air and blood:air partition coefficients) were 3.4, 5.1, and 8.4 for LFD-, HFD-, and MCD-fed mice, respectively, also demonstrating that PERC has greater affinity for liver tissue in mice with steatosis or NASH.
Fig. 2.
Blood:air (A) and liver:air (B) partition coefficients of PERC in mice fed a LFD (open box), HFD (light gray box), or MCD (dark gray box) diets for 8 weeks. Box and whisker plots are shown (+, mean; line, median; box, interquartile range; whiskers, min to max). Different letters represent different statistical groups, as determined by ANOVA with Newman-Keuls post hoc test (P < 0.05, n = 4 or 5/group).
Effects of NAFLD on Toxicokinetics of PERC Metabolites.
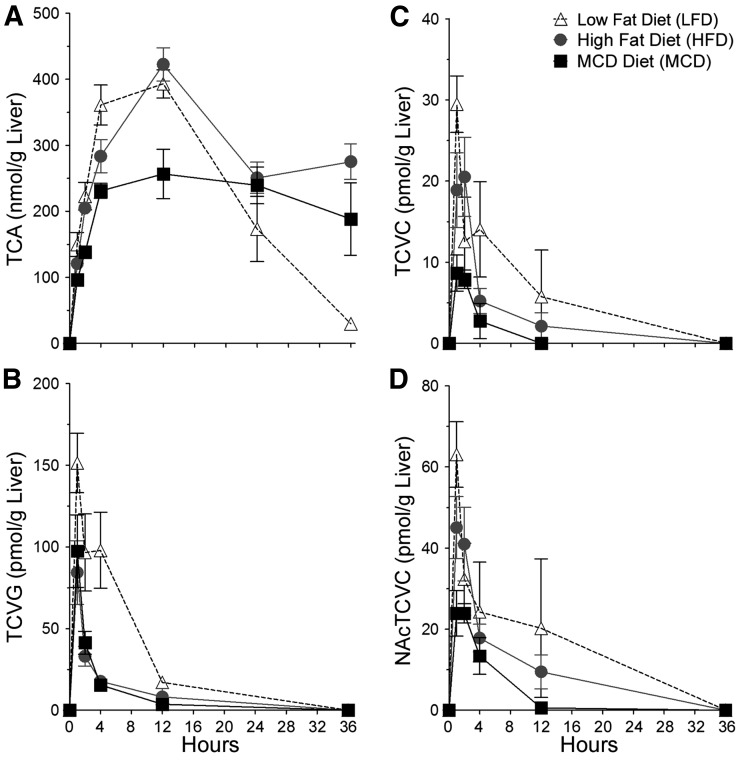
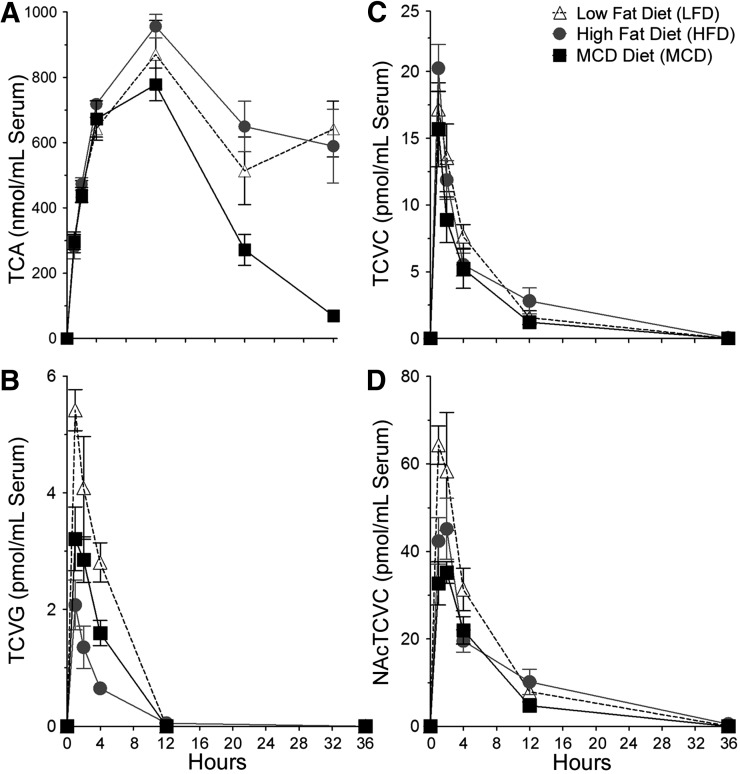
The effects of varying liver disease states on toxicokinetics of PERC metabolites was evaluated in liver (Fig. 3) and serum (Fig. 4) after a single intragastric dose of PERC (300 mg/kg). The levels of PERC metabolites in urine are shown in Table 3. Maximum liver concentrations of TCA, the major oxidative PERC metabolite, were higher in mice fed LFD or HFD compared with mice fed MCD. The values for maximum TCA concentrations in liver (mean ± S.E., n = 5/group) were 393 ± 21, 423 ± 25, and 257 ± 37 in LFD-, HFD-, and MCD-fed animals, respectively. In serum, maximum TCA concentrations were higher in MCD- and HFD-fed mice compared with LFD-fed mice. The values for maximum TCA concentrations in serum were 778 ± 51, 870 ± 105, and 956 ± 36 in LFD-, HFD-, and MCD-fed animals, respectively. Interestingly, the elimination of TCA from both liver and serum was significantly delayed in mice with NAFLD compared with healthy mice; this was concomitant with decreased amounts of TCA excreted into the urine in HFD- and MCD-fed mice compared with LFD-fed mice. Because of TCA not being cleared by the end of the experiment, our study design precluded calculations of AUCs for TCA in HFD- and MCD-fed groups.
Fig. 3.
Concentration-time profiles of TCA (A), TCVG (B), TCVC (C), and NAcTCVC (D) in liver tissue after a single intragastric dose of PERC (300 mg/kg) after 8 weeks of diet. Open triangles, LFD-fed mice; gray circles, HFD-fed mice; black squares, MCD-fed mice. Points represent the mean and error bars represent S.E. (n = 4 or 5/group).
Fig. 4.
Concentration-time profiles of TCA (A), TCVG (B), TCVC (C), and NAcTCVC (D) in serum after a single intragastric dose of PERC (300 mg/kg) after 8 weeks of diet. Open triangles, LFD-fed mice; gray circles, HFD-fed mice; black squares, MCD-fed mice. Points represent the mean and error bars represent S.E. (n = 4 or 5/group).
TABLE 3.
Urinary excretion of PERC metabolites (expressed as % of administered PERC dose)
Values represent mean ± S.E., n = 4 (LFD) or 3 (HFD, MCD) per group.
| Diet | TCA | TCVG | TCVC | NAcTCVC |
|---|---|---|---|---|
| LFD | 5.9 ± 0.9 | 6.3 ± 0.5 (x 10−5) | 8.8 ± 3.2 (x 10−4) | 7.1 ± 2.1 (x 10−3) |
| HFD | 2.9 ± 0.8 | 4.0 ± 2.8 (x 10−5) | 1.2 ± 0.4 (x 10−4) | 2.0 ± 0.8 (x 10−3) |
| MCD | 2.4 ± 0.9 | 4.4 ± 1.8 (x 10−5) | 1.0 ± 0.6 (x 10−4) | < LLOQa |
No statistically-significant changes were detected (P > 0.05; ANOVA with Newman-Keuls post hoc test).
Levels were below the lower limit of quantitation (LLOQ; 30 fmol on column).
Hepatic levels of TCVG, TCVC, and NAcTCVC were lower in HFD- or MCD-fed mice compared with LFD-fed mice; this was particularly noticeable for TCVG, where hepatic AUCs in HFD- or MCD-fed mice were approximately threefold lower compared with LFD-fed mice (Table 2). A similar trend for TCVG and NAcTCVC was observed in serum (except that serum AUCs were similar between HFD- and LFD-fed mice for NAcTCVC), but serum concentrations of TCVC were similar between all groups at all time points. Levels of TCVG, TCVC, and NAcTCVC were lower in HFD- and MCD-fed mice compared with LFD-fed mice.
NAFLD-Associated Effects on Liver Transcriptome and PERC Metabolism.
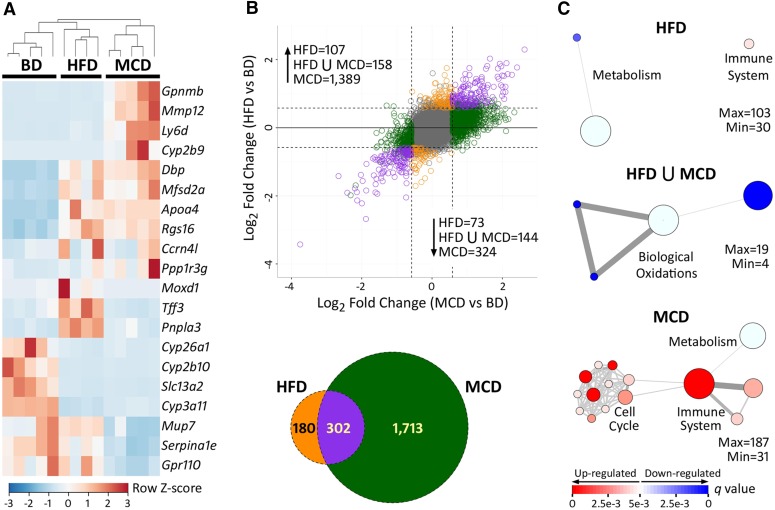
To further characterize the molecular mechanisms that may be underlying NAFLD-associated modulation of PERC metabolism, RNA sequencing was performed on liver tissue of LFD-fed mice or mice fed HFD or MCD. For this analysis, only mice treated with a single dose of vehicle were used to best approximate “basal” conditions. Principal component (Supplemental Fig. 2) and unsupervised hierarchical clustering analysis (Fig. 5A) of the transcriptomic data showed clear separation of the diet-specific groups. A total of 482 and 2015 genes (Fig. 5B; see Supplemental Files 1 and 2 for complete lists) were differentially expressed between mice fed a HFD or MCD, respectively, compared with LFD-fed mice. Furthermore, there were 1131 genes that were differentially expressed in MCD-fed mice as compared with HFD-fed mice (Supplemental File 3). Gene set enrichment analysis revealed that pathways involved in the immune response were significantly induced in both HFD- and MCD-fed animals, whereas cellular metabolism, specifically “biological oxidations,” were repressed by both HFD and MCD diets (Fig. 5C). Complete lists of significantly enriched pathways are provided as Supplemental Files 4–6.
Fig. 5.
Effect of diet on hepatic gene transcription. All analyses were conducted on vehicle-treated animals (24-hour time point) after 8 weeks of LFD, HFD, or MCD diets. (A) Heatmap of unsupervised hierarchical clustering of the top 20 most variable genes (as determined by Z-scores). Dendrogram represents statistical clustering among columns (sample IDs). Column keys are colored by diet (gray, LFD; orange, HFD; green, MCD). (B) Plot of fold-changes (log2, compared with LFD) of individual transcripts in HFD- and MCD-fed animals. A Venn diagram is also shown for comparative purposes. To be deemed differentially expressed, transcripts needed to have an absolute value of log2-fold change > 0.58 (represented by dotted lines) and an FDR-adjusted q-value < 0.1. Transcripts that did not meet these requirements are colored gray. Differentially expressed genes are color coded by whether they were changed by HFD (orange), MCD (green), or both (purple) diets. (C) Network plots of gene sets enriched by HFD (orange, top), both (purple, middle), and MCD (green, bottom) diets. Red circles represent pathways that were significantly induced, whereas blue circles represent repressed pathways (FDR-adjusted q-value < 0.005). The size of the circle is proportional to the number of genes that are enriched in that pathway. The thickness of connecting lines between circles is proportional to the number of common genes between those two pathways. At least 10 genes needed to be common between pathways in order for a line to be drawn. A color key is provided at the bottom.
Effects of NAFLD on Expression of Xenobiotic-metabolizing Enzymes in Liver.
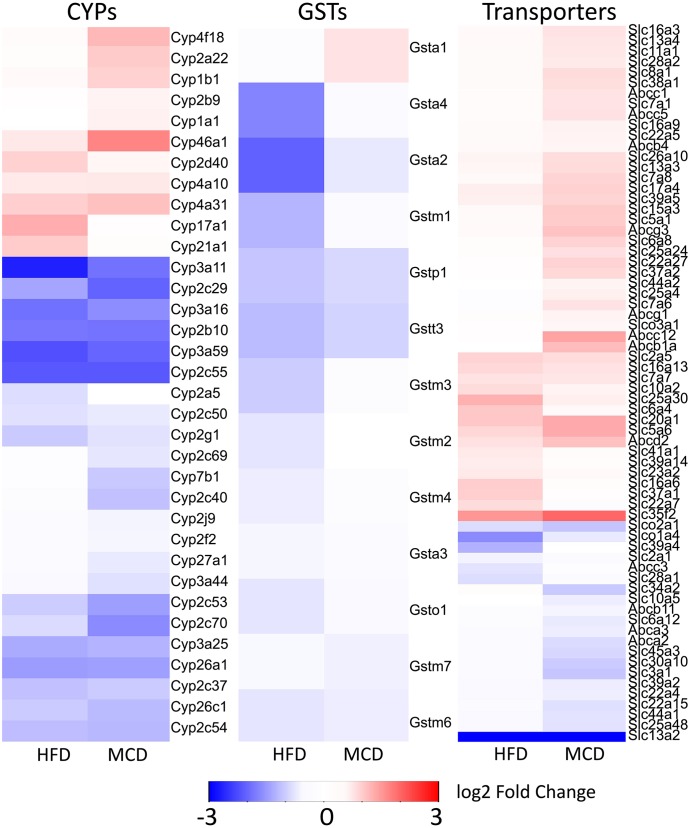
Little is known about specific enzymes that are responsible for PERC metabolism and most of the metabolic pathways are inferred from a related chemical trichloroethylene (Cichocki et al., 2016b). Underlying disease states of NAFLD had a major impact on PERC and TCA toxicokinetics and transcriptomic data showed significant repression of biologic oxidative metabolism in HFD- and MCD-fed mice. Therefore, we interrogated a potential role of xenobiotic-metabolizing enzymes in NAFLD-associated disruption of PERC toxicokinetics. We have examined differential expression of the genes in Cyp, Gst, Abc transporter, and Slc/Slco transporter families due to their potential contribution to PERC metabolism. As reported previously (Canet et al., 2014), we found that most CYPs, except for the Cyp4a family involved in lipid metabolism, were downregulated by experimental NAFLD (Fig. 6). Multiple GSTs were also repressed, with Gsta1 being the only exception in MCD-fed mice.
Fig. 6.
Heatmaps of differentially expressed genes from the CYP, GST, and transporter (Slc, Slco, and ABC) families. The fold changes (log2) compared with LFD-fed animals are represented by colors (see color key). Blue indicates significantly repressed, red indicates significantly induced. All red- or blue-colored genes had an FDR-adjusted q-value < 0.1 in at least one of the diet groups. If q > 0.1 or if log2-fold change < 0.58, the gene is colored white.
Interestingly, levels of multiple GSTs were increased in MCD-fed mice compared with HFD-fed mice. The pattern of xenobiotic transporter expression was diet dependent. We observed that the number of differentially expressed xenobiotic transporters were higher in MCD-fed mice compared with HFD-fed mice. Furthermore, in both HFD- and MCD-fed mice, there were more transporters that were upregulated compared with downregulated. Expression of Slc35f2 and Slc13a2 were notably higher and lower, respectively, in HFD- and MCD-fed mice compared with LFD-fed controls. We did not observe any xenobiotic efflux transporter genes that decreased in expression in both HFD- and MCD-fed mice. Furthermore, we did not observe any xenobiotic uptake transporter genes that increased in expression in both HFD- and MCD-fed mice.
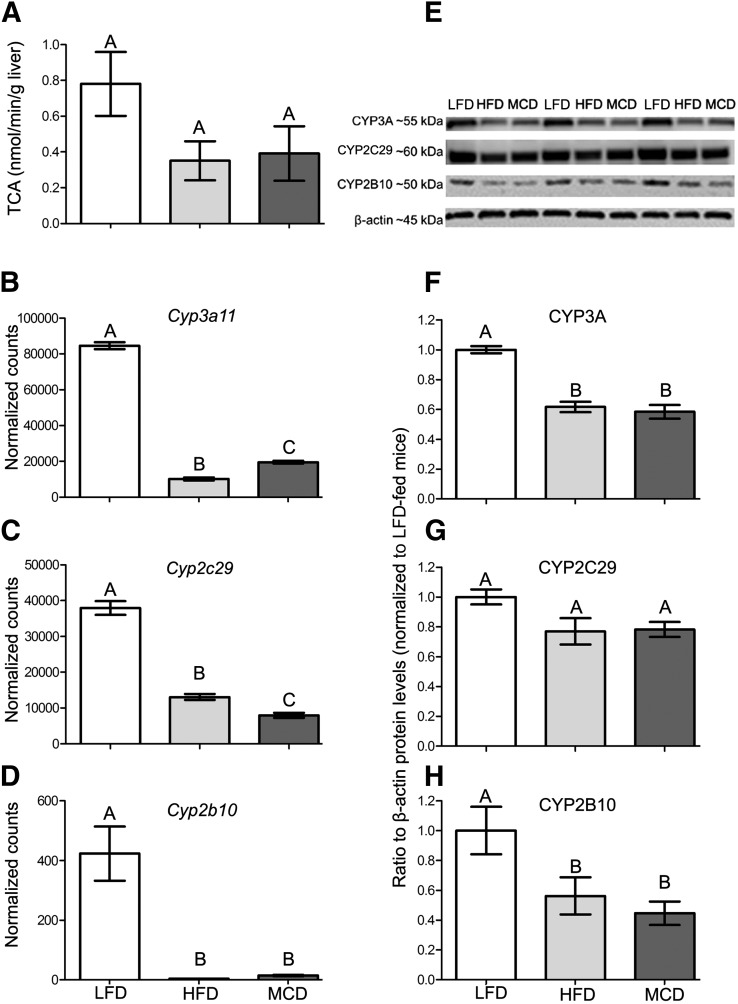
Next, we performed a correlation analysis among differentially expressed metabolism genes and PERC and TCA pharmacokinetic parameters. We found that 17 CYP450 genes were significantly negatively correlated with hepatic PERC AUC and positively correlated with TCA levels at 4 hours (Pearson’s r > 0.70, q < 0.05; Supplemental Table 1). The 4-hour time point was chosen because TCA levels differed among the three groups at this time point. Of particular interest (Supplemental Fig. 3, Fig. 7) were the highly expressed Cyp2c29 and Cyp3a11 genes, along with Cyp2b10, the mouse ortholog of CYP2B6, which was shown to metabolize PERC (White et al., 2001). No CYP450 genes were identified that correlated with hepatic TCA levels at 12 or 24 hours (data not shown). Mean hepatic PERC AUC and TCA levels at 4 hours were strongly negatively correlated to each other (Pearson’s r < −0.99, q < 0.01).
Fig. 7.
Effects of diets on PERC metabolism to TCA and cytochrome P450 enzymes. (A) Metabolic activity, as assessed by oxidation of PERC to TCA in ex vivo metabolism assays using S9 fractions from vehicle-treated LFD-, HFD-, or MCD-fed mouse liver tissue. Hepatic mRNA (B—D) and protein (E—H) levels of Cyp3A11, Cyp2C29, and Cyp2B10 in LFD-, HFD-, and MCD-fed vehicle-treated mice. The corresponding Western blots for data in (F–H) are shown in (E). Different letters represent different statistical groups, as determined by ANOVA with Newman-Keuls post hoc test (P < 0.05, n = 4 or 5/group for mRNA and activity assays, n = 3/group for protein quantification).
To determine whether the observed changes in Cyp2c29, Cyp3a11, and Cyp2b10 mRNA expression were associated with changes in protein expression and activity, we performed Western blotting and ex vivo activity assays using hepatic S9 fractions. These results, along with the mRNA results, are displayed in Fig. 7. Although not statistically significant among the different diet groups (P = 0.137, ANOVA), the formation of TCA from PERC in hepatic S9 incubations was almost twofold greater in LFD-fed mice compared with HFD- or MCD-fed mice. TCA formation (nmol/min/g liver) in LFD-, HFD-, and MCD-fed mice was 0.781 ± 0.179, 0.352 ± 0.109, and 0.392 ± 0.153 (mean ± S.E.M.), respectively. The hepatic protein expression of CYP3A and CYP2B10 was significantly decreased by approximately 40–50% in HFD- or MCD-fed mice compared with LFD-fed mice, which agreed with our mRNA data. CYP2C29 expression was decreased in HFD- and MCD-fed mice by about 20% compared with LFD-fed controls; however, this was not a statistically significant finding (P = 0.081, ANOVA).
Discussion
We previously showed that interindividual variability in toxicokinetics of PERC varies significantly across a population of inbred mice (Cichocki et al., 2016a). In this study, PERC was used as a model toxicant to investigate the contribution of underlying NAFLD to interindividual variability in toxicokinetics of environmental chemicals. PERC is an ideal case study chemical because its metabolism is proposed to play a critical role in its mode of toxicity, and thus alteration of metabolism will likely alter PERC-associated adverse health effects.
NAFLD carries significant public health and economic burden (Younossi et al., 2016a,b). Experimental NAFLD has been shown to alter the metabolism of a variety of different xenobiotics (Buechler and Weiss, 2011; Merrell and Cherrington, 2011; Naik et al., 2013; Clarke and Cherrington, 2015), including environmental chemicals (Canet et al., 2012), which can have a consequence on pharmaco- and toxicodynamics (Hardwick et al., 2014; Dzierlenga et al., 2015). However, a detailed examination of the effects of NAFLD on toxicokinetics of an environmental chemical and its metabolites in multiple tissues has not yet been reported. Because delivered dose of both PERC and its major metabolites may contribute to target organ-specific toxicity (Lash and Parker, 2001; Guyton et al., 2014; Cichocki et al., 2016b), such an examination is warranted for this ubiquitous environmental contaminant.
PERC is a volatile chlorinated solvent with almost no solubility in water. As such, it is not surprising that it has a high affinity for adipose tissue, as indicated by its high fat:blood partition coefficient of >1000; (Gargas et al., 1989; Gearhart et al., 1993). Because there are no data on blood:air and liver:air partitioning of PERC in liver disease models, our data contribute significantly to the understanding of pharmacokinetics of this important environmental chemical in a human population. Here, we demonstrate that fat-laden livers from mice with steatosis or NASH have an increased liver:air and liver:blood partition coefficient relative to healthy mice, indicating that PERC will preferentially adsorb to liver tissue in mice with NAFLD. It is likely that the same pattern will hold true in humans with NAFLD as well.
The observed effects of NAFLD on liver lipid homeostasis and liver:blood partitioning of PERC were associated with significant effects on toxicokinetics of PERC and its metabolites. The nearly sevenfold increase in PERC exposure in animals with NASH (compared with healthy animals) was coincident with an approximate 35% decrease in maximum hepatic TCA concentration. The decreased level of TCA in the livers of mice with NASH may have been a consequence of multiple biologic factors. The time-to-maximum concentration of PERC in blood and multiple tissues was similar among all three diet groups, suggesting that the gut absorption of PERC was unaffected by diet. The increased retention of PERC in the lipids in the liver would likely slow its metabolism and make it temporarily unavailable for oxidation. Indeed, there was lesser TCA in the liver at early time points in mice with NASH compared with healthy mice. Another consequence of increased liver fat content in NAFLD could be a decreased affinity of TCA, a water-soluble chemical that is ionized at physiologic pH. Disease state-specific physiologically based pharmacokinetic modeling that builds upon the current PBPK model for PERC (Chiu and Ginsberg, 2011) may potentially be a useful tool in the future to better understand the contribution of these physiologic parameters to PERC toxicokinetics in individuals with or without NAFLD. The data from this study will be critical for such modeling.
The decreased expression of multiple CYPs in liver tissue of mice with NASH may also contribute to decreased hepatic oxidative capacity and TCA production. Although PERC is a ubiquitous environmental contaminant and high-production volume chemical, the specific CYPs involved in its metabolism in the liver are largely uncharacterized. Here, we show that in mice with steatosis or NASH, there is a significant decrease in CYP2B10 expression at both the mRNA and protein level. CYP2B10 is the mouse ortholog of human CYP2B6, which has been shown to oxidize PERC in human lymphoblastoid MCL-5 cells (White et al., 2001). The potential for CYP2B10 to contribute to PERC metabolism is further supported by strong correlation of Cyp2b10 levels with PERC liver AUC and TCA levels at 4 hours postgavage. Similar to the findings with CYP2B10, we also observed decreases in Cyp2c29 and Cyp3a11, two highly expressed hepatic CYPs, at the mRNA and protein level. Expression of both Cyp2c29 and Cyp3a11 also exhibited strong correlations with PERC liver AUC and TCA levels at 4 hours postgavage. Furthermore, Cyp2b10 and Cyp3a11 were two of the top 20 most variably expressed genes among the three diet groups.
Compared with LFD-fed mice, we observed no significant effect on the capacity for liver tissue derived from HFD- or MCD-fed mice to metabolize PERC to TCA ex vivo. We found that liver tissue from LFD-fed male C57Bl/6J formed 780 pmol TCA/min/g of tissue. Our findings in LFD-fed mice are comparable with the previously reported values of 421 pmol of PERC/min/g liver being metabolized to water-soluble metabolites (Reitz et al., 1996), although strain differences (C57Bl/6J versus B6C3F1) and substrate concentration (2.5 versus 0.068 mM) and overall extraction and analytical procedures may account for the approximate twofold difference. Collectively, our mRNA, protein, and activity data suggest that NAFLD-associated modulation of xenobiotic-metabolizing enzymes contribute to interindividual variability in PERC metabolism in mice. Furthermore, because PERC is a small lipophilic chemical, it is likely a promiscuous substrate for multiple oxidative enzymes, and our data confirm the complexity of the oxidative metabolic pathways for PERC.
The expression of hepatic transporters in experimental NASH was previously characterized in great detail (Canet et al., 2014). In the current study, a total of 68 genes from the SLC or ABC transporter families were found to be differentially expressed between mice with NAFLD and healthy mice. Because PERC is a lipophilic chemical, it does not require xenobiotic transporters to enter the cell. However, TCA will not easily diffuse across the cell membranes and will therefore likely require active transport; however, little is known about specific transporters that may facilitate movement of TCA across membranes in various tissues. We did not find that expression of efflux transporters was negatively correlated with TCA levels or that expression of uptake transporters was positively correlated with TCA levels. Thus, the potential role of xenobiotic transporters in TCA toxicokinetics remains unknown.
In addition to oxidation, PERC undergoes conjugation via GSTs to form conjugative metabolites that are mutagenic and nephrotoxic (Cichocki et al., 2016b). However, quantitation of these metabolites has not been previously possible because of their low abundance and quick clearance from tissue and their unsuitability for gas chromatography. We recently developed a LC-MS/MS method with capability to detect 1 pmol metabolite/g of tissue, with 50 mg or less of tissue input (Luo et al., submitted manuscript). Here, for the first time, we apply that method to investigate toxicokinetics of TCVG, TCVC, and NAcTCVC in liver and serum of mice exposed to PERC.
From a mass-balance perspective TCVG, TCVC, and NAcTCVC are not significant contributors to PERC metabolism in the mouse, because their peak levels in liver were less than 0.05% of TCA levels and they were quickly cleared from both liver and serum. These findings are similar to those with trichloroethylene (Yoo et al., 2015). Interestingly, we found that liver and serum levels of all three glutathione conjugation pathway metabolites of PERC were generally reduced in mice with NAFLD compared with healthy mice. This was associated with a decreased expression of a number of genes in the Gst family, which is in agreement with previously published results showing decreased GST activity from tissue samples from patients with varying stages of NAFLD (Hardwick et al., 2010). In humans, NAFLD may be associated with an increased risk for chronic kidney disease (Musso et al., 2014; Targher et al., 2014). Because NAFLD may be a risk factor for the development of chronic kidney disease and because it alters the toxicokinetics of nephrotoxic PERC metabolites, follow up studies on the effects of experimental NAFLD on PERC-associated nephrotoxicity are warranted.
Our study is not without limitations. First, we only exposed animals to a single dose of PERC, which is fairly high and is not likely to occur in environmental exposure scenarios. Extrapolating these data to environmentally relevant exposure levels may be a source of uncertainty (Bois et al., 1996, 2000; Yang et al., 2006). Furthermore, because PERC oxidative metabolism is saturable (Buben and O'Flaherty, 1985; Green et al., 1990), the types of kinetics observed in our study may not be fully representative of what occurs in environmentally exposed humans. However, in male human volunteers exposed to occupationally relevant concentrations of PERC (∼70-140 ppm, nominal air concentration) via inhalation for 4 hours, ∼1% of the absorbed PERC dose was recovered as TCA in the urine (Monster et al., 1979). This is on the same order of magnitude as observed in our study in mice (∼6%, see Table 3). In male human volunteers exposed to 1 ppm PERC for 6 hours, recovery of urinary TCA was only ∼0.4% of the absorbed PERC dose (Chiu et al., 2007). Characterization of the effect of NAFLD on PERC toxicokinetics at both low and high concentrations is worthy of further study.
Although the dose of PERC used in this study is much greater than expected human exposures, previous reports on humans occupationally exposed to 50 ppm PERC (nominal air concentration) via inhalation show that urinary levels of NAcTCVC were approximately 0.05% of those of TCA (Birner et al., 1996), which were similar to what we observed in our study (0.1%). Our results are dissimilar to those observed in rats and mice exposed to 800 mg/kg PERC via gavage, whereby the urinary level of NAcTCVC was 1–2% of TCA (Dekant et al., 1986), which may have been due to differences in administered dose (800 versus 300 mg/kg), strain (NMRI versus C57Bl/6J), sex (female versus male), vehicle (corn oil versus aqueous Alkamuls-El 620), or analytical methodology (GC/MS after derivatization versus LC-MS/MS).
Although we made a number of important discoveries about the effect of NAFLD on toxicokinetics of PERC, information that will be of great relevance to human health assessments, we also note that very limited experimental data are available for the toxicological effects of environmental chemicals in subjects with NAFLD. Most of the studies of the effect of NAFLD on xenobiotic metabolism involve drugs, a topic that has been extensively reviewed (Buechler and Weiss, 2011; Merrell and Cherrington, 2011; Naik et al., 2013; Clarke and Cherrington, 2015). One study examined the effects of NAFLD induced by an MCD diet on hepatotoxicity due to environmental chemical exposure, using a single dose of carbon tetrachloride in rats (Donthamsetty et al., 2007). Similar to PERC, carbon tetrachloride is a lipophilic chlorinated solvent with a history of being used as a dry-cleaning solvent until its replacement by PERC in the late 1940s (Doherty, 2000). Donthamsetty et al. (2007) reported that the sensitivity to carbon tetrachloride-induced hepatotoxicity in rats was not due to differences in metabolism of the parent chemical to a reactive intermediate (based on data from in vivo covalent binding of radiolabeled chemical to hepatic proteins and expression of CYP2E1), but rather that inhibited tissue repair mechanisms in steatotic livers contributed to sensitivity to toxicity. However, the authors did not evaluate levels of parent chemical in liver tissues. Because carbon tetrachloride is highly lipophilic, it is conceivable that, similar to our observations with PERC, it would have increased affinity for liver tissue in subjects with NAFLD. On the basis of the findings of Donthamsetty et al. (2007), there is potential that NAFLD would contribute to susceptibility to PERC-associated toxicity; thus, follow up studies of longer duration are needed.
In conclusion, we report for the first time that experimental NAFLD in mice is associated with an increased hepatic deposition of PERC, a lipophilic environmental chemical, and increased hepatic retention of TCA, a hepatotoxic metabolite of PERC. We also report for the first time the toxicokinetics of TCVG, TCVC, and NAc-TCVC in the mouse, including animals that are healthy and with varying degrees of fatty liver disease, and provide evidence that experimental NAFLD alters conjugative metabolism of PERC. Furthermore, different toxicokinetic phenotypes are observed in mild compared with severe stages of disease, which may have important implications for susceptibility to PERC-associated toxicity. Taken together, the potential for accumulation of ubiquitous lipophilic xenobiotics and their metabolites in a major toxicological target tissue in individuals with common underlying diseases such as NAFLD may be an important consideration in future public health assessments of environmental toxicants.
Acknowledgments
The authors acknowledge Drs. Anthony H. Knap, Terry Wade, and Stephen Sweet from the Geochemical and Environmental Research Group at Texas A&M University for technical assistance with analytical chemistry analyses. The authors also thank Drs. John House and Fred A. Wright from the Department of Statistics at North Carolina State University for consultations regarding gene expression analysis.
Abbreviations
- ANOVA
analysis of variance
- AUC
area under the concentration-time curve
- CYP
cytochrome P450
- EPA
United States Environmental Protection Agency
- GC/MS
gas chromatography/mass spectrometry
- GST
glutathione-S-transferase
- HFD
high-fat diet
- LC-MS/MS
liquid chromatography/mass spectroscopy
- LFD
low-fat diet
- MCD
methionine/choline/folate-deficient diet
- NAcTCVC
N-acetyl-S-(1,2,2-trichlorvinyl)-l-cysteine
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- PERC
perchloroethylene, tetrachloroethylene
- TCA
trichloroacetate
- TCVC
S-(1,2,2-trichlorovinyl)-l-cysteine
- TCVG
S-(1,2,2-trichlorovinyl)-glutathione
Authorship Contributions
Participated in research design: Cichocki, Pogribny, Chiu, Threadgill, and Rusyn.
Conducted experiments: Cichocki, Furuya, Luo, and Iwata.
Contributed analytical tools: Konganti and McDonald.
Performed data analysis: Cichocki.
Wrote or contributed to the writing of the manuscript: Cichocki and Rusyn.
Footnotes
J.A.C. was a recipient of a National Research Service Award through the National Institutes of Health National Institute of Environmental Health Sciences [Grant F32 ES026005]. This work was supported, in part, by a cooperative agreement STAR [RD83561202] from U.S. EPA to Texas A&M University. The views expressed in this paper are those of the authors and do not necessarily reflect the views or policies of the NIH, the U.S. EPA, or the U.S. Food and Drug Administration.
Part of this work was presented at the following meeting: Cichocki JA, Furuya S, Konganti K, Luo YS, McDonald TJ, Iwata Y, Chiu WA, Threadgill DW, Pogribny IP, Rusyn I (2016) Impact of nonalcoholic fatty liver disease on toxicokinetics of tetrachloroethylene in mice. Lone Star Chapter of the Society of Toxicology Annual Meeting; 2016 Oct 21-22; Waco, TX.
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Anders S, Pyl PT, Huber W. (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B 57:289–300. [Google Scholar]
- Birner G, Rutkowska A, Dekant W. (1996) N-acetyl-S-(1,2,2-trichlorovinyl)-L-cysteine and 2,2,2-trichloroethanol: two novel metabolites of tetrachloroethene in humans after occupational exposure. Drug Metab Dispos 24:41–48. [PubMed] [Google Scholar]
- Bois FY. (2000) Statistical analysis of Clewell et al. PBPK model of trichloroethylene kinetics. Environ Health Perspect 108 (Suppl 2):307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bois FY, Gelman A, Jiang J, Maszle DR, Zeise L, Alexeef G. (1996) Population toxicokinetics of tetrachloroethylene. Arch Toxicol 70:347–355. [DOI] [PubMed] [Google Scholar]
- Bois FY, Jamei M, Clewell HJ. (2010) PBPK modelling of inter-individual variability in the pharmacokinetics of environmental chemicals. Toxicology 278:256–267. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buben JA, O’Flaherty EJ. (1985) Delineation of the role of metabolism in the hepatotoxicity of trichloroethylene and perchloroethylene: a dose-effect study. Toxicol Appl Pharmacol 78:105–122. [DOI] [PubMed] [Google Scholar]
- Buechler C, Weiss TS. (2011) Does hepatic steatosis affect drug metabolizing enzymes in the liver? Curr Drug Metab 12:24–34. [DOI] [PubMed] [Google Scholar]
- Bull RJ. (2000) Mode of action of liver tumor induction by trichloroethylene and its metabolites, trichloroacetate and dichloroacetate. Environ Health Perspect 108 (Suppl 2):241–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RJ, Sanchez IM, Nelson MA, Larson JL, Lansing AJ. (1990) Liver tumor induction in B6C3F1 mice by dichloroacetate and trichloroacetate. Toxicology 63:341–359. [DOI] [PubMed] [Google Scholar]
- Canet MJ, Hardwick RN, Lake AD, Dzierlenga AL, Clarke JD, Cherrington NJ. (2014) Modeling human nonalcoholic steatohepatitis-associated changes in drug transporter expression using experimental rodent models. Drug Metab Dispos 42:586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet MJ, Hardwick RN, Lake AD, Kopplin MJ, Scheffer GL, Klimecki WT, Gandolfi AJ, Cherrington NJ. (2012) Altered arsenic disposition in experimental nonalcoholic fatty liver disease. Drug Metab Dispos 40:1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WA, Ginsberg GL. (2011) Development and evaluation of a harmonized physiologically based pharmacokinetic (PBPK) model for perchloroethylene toxicokinetics in mice, rats, and humans. Toxicol Appl Pharmacol 253:203–234. [DOI] [PubMed] [Google Scholar]
- Chiu WA, Micallef S, Monster AC, Bois FY. (2007) Toxicokinetics of inhaled trichloroethylene and tetrachloroethylene in humans at 1 ppm: empirical results and comparisons with previous studies. Toxicol Sci 95:23–36. [DOI] [PubMed] [Google Scholar]
- Cichocki JA, Furuya S, Venkatratnam A, McDonald TJ, Knap AH, Wade T, Sweet S, Chiu WA, Threadgill DW, Rusyn I. (2016a) Characterization of variability in toxicokinetics and toxicodynamics of tetrachloroethylene using the Collaborative Cross mouse population. Environ Health Perspect. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki JA, Guyton KZ, Guha N, Chiu WA, Rusyn I, Lash LH. (2016b) Target organ metabolism, toxicity, and mechanisms of trichloroethylene and perchloroethylene: key similarities, differences, and data gaps. J Pharmacol Exp Ther 359:110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Cherrington NJ. (2015) Nonalcoholic steatohepatitis in precision medicine: Unraveling the factors that contribute to individual variability. Pharmacol Ther 151:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corton JC. (2008) Evaluation of the role of peroxisome proliferator-activated receptor alpha (PPARalpha) in mouse liver tumor induction by trichloroethylene and metabolites. Crit Rev Toxicol 38:857–875. [DOI] [PubMed] [Google Scholar]
- Dekant W, Metzler M, Henschler D. (1986) Identification of S-1,2,2-trichlorovinyl-N-acetylcysteine as a urinary metabolite of tetrachloroethylene: bioactivation through glutathione conjugation as a possible explanation of its nephrocarcinogenicity. J Biochem Toxicol 1:57–72. [DOI] [PubMed] [Google Scholar]
- Doherty RE. (2000) A history of the production and use of carbon tetrachloride, tetrachloroethylene, trichloroethylene and 1,1,1-trichloroethane in the United States: Part 1 - Historical background; Carbon tetrachloride and tetrachloroethylene. Environmental Forensics 1:69–81. [Google Scholar]
- Donthamsetty S, Bhave VS, Mitra MS, Latendresse JR, Mehendale HM. (2007) Nonalcoholic fatty liver sensitizes rats to carbon tetrachloride hepatotoxicity. Hepatology 45:391–403. [DOI] [PubMed] [Google Scholar]
- Dzierlenga AL, Clarke JD, Hargraves TL, Ainslie GR, Vanderah TW, Paine MF, Cherrington NJ. (2015) Mechanistic basis of altered morphine disposition in nonalcoholic steatohepatitis. J Pharmacol Exp Ther 352:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargas ML, Burgess RJ, Voisard DE, Cason GH, Andersen ME. (1989) Partition coefficients of low-molecular-weight volatile chemicals in various liquids and tissues. Toxicol Appl Pharmacol 98:87–99. [DOI] [PubMed] [Google Scholar]
- Gearhart JM, Mahle DA, Greene RJ, Seckel CS, Flemming CD, Fisher JW, Clewell HJ., 3rd (1993) Variability of physiologically based pharmacokinetic (PBPK) model parameters and their effects on PBPK model predictions in a risk assessment for perchloroethylene (PCE). Toxicol Lett 68:131–144. [DOI] [PubMed] [Google Scholar]
- Green T, Odum J, Nash JA, Foster JR. (1990) Perchloroethylene-induced rat kidney tumors: an investigation of the mechanisms involved and their relevance to humans. Toxicol Appl Pharmacol 103:77–89. [DOI] [PubMed] [Google Scholar]
- Guha N, Loomis D, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Baan R, Mattock H, Straif K, International Agency for Research on Cancer Monograph Working Group (2012) Carcinogenicity of trichloroethylene, tetrachloroethylene, some other chlorinated solvents, and their metabolites. Lancet Oncol 13:1192–1193. [DOI] [PubMed] [Google Scholar]
- Guyton KZ, Hogan KA, Scott CS, Cooper GS, Bale AS, Kopylev L, Barone S, Makris SL, Glenn B, Subramaniam RP, et al. (2014) Human health effects of tetrachloroethylene: key findings and scientific issues. Environ Health Perspect 122:325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick RN, Clarke JD, Lake AD, Canet MJ, Anumol T, Street SM, Merrell MD, Goedken MJ, Snyder SA, Cherrington NJ. (2014) Increased susceptibility to methotrexate-induced toxicity in nonalcoholic steatohepatitis. Toxicol Sci 142:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick RN, Fisher CD, Canet MJ, Lake AD, Cherrington NJ. (2010) Diversity in antioxidant response enzymes in progressive stages of human nonalcoholic fatty liver disease. Drug Metab Dispos 38:2293–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC (2014) IARC Monographs on the evaluation of carcinogenic risks to humans (Vol. 106): Trichloroethylene, tetrachloroethylene and some other chlorinated agents. International Agency for Research on Cancer, Lyon. [PMC free article] [PubMed]
- Irving RM, Elfarra AA. (2013) Mutagenicity of the cysteine S-conjugate sulfoxides of trichloroethylene and tetrachloroethylene in the Ames test. Toxicology 306:157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, et al. Nonalcoholic Steatohepatitis Clinical Research Network (2005) Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41:1313–1321. [DOI] [PubMed] [Google Scholar]
- Lash LH, Parker JC. (2001) Hepatic and renal toxicities associated with perchloroethylene. Pharmacol Rev 53:177–208. [PubMed] [Google Scholar]
- Lash LH, Putt DA, Huang P, Hueni SE, Parker JC. (2007) Modulation of hepatic and renal metabolism and toxicity of trichloroethylene and perchloroethylene by alterations in status of cytochrome P450 and glutathione. Toxicology 235:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher JJ. (2011) New insights from rodent models of fatty liver disease. Antioxid Redox Signal 15:535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell MD, Cherrington NJ. (2011) Drug metabolism alterations in nonalcoholic fatty liver disease. Drug Metab Rev 43:317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monster AC, Boersma G, Steenweg H. (1979) Kinetics of tetrachloroethylene in volunteers; influence of exposure concentration and work load. Int Arch Occup Environ Health 42:303–309. [DOI] [PubMed] [Google Scholar]
- Morris JB, Cavanagh DG. (1986) Deposition of ethanol and acetone vapors in the upper respiratory tract of the rat. Fundam Appl Toxicol 6:78–88. [DOI] [PubMed] [Google Scholar]
- Musso G, Gambino R, Tabibian JH, Ekstedt M, Kechagias S, Hamaguchi M, Hultcrantz R, Hagström H, Yoon SK, Charatcharoenwitthaya P, et al. (2014) Association of non-alcoholic fatty liver disease with chronic kidney disease: a systematic review and meta-analysis. PLoS Med 11:e1001680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik A, Belič A, Zanger UM, Rozman D. (2013) Molecular Interactions between NAFLD and Xenobiotic Metabolism. Front Genet 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (2010) Review of the Environmental Protection Agency’s Draft IRIS Assessment of Tetrachloroethylene, The National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- National Toxicology Program (1977) Bioassay of tetrachloroethylene for possible carcinogenicity. Natl Cancer Inst Carcinog Tech Rep Ser 13:1–83. [PubMed] [Google Scholar]
- Philip BK, Mumtaz MM, Latendresse JR, Mehendale HM. (2007) Impact of repeated exposure on toxicity of perchloroethylene in Swiss Webster mice. Toxicology 232:1–14. [DOI] [PubMed] [Google Scholar]
- Reitz RH, Gargas ML, Mendrala AL, Schumann AM. (1996) In vivo and in vitro studies of perchloroethylene metabolism for physiologically based pharmacokinetic modeling in rats, mice, and humans. Toxicol Appl Pharmacol 136:289–306. [DOI] [PubMed] [Google Scholar]
- Relling MV, Evans WE. (2015) Pharmacogenomics in the clinic. Nature 526:343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targher G, Chonchol MB, Byrne CD. (2014) CKD and nonalcoholic fatty liver disease. Am J Kidney Dis 64:638–652. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryndyak VP, Latendresse JR, Montgomery B, Ross SA, Beland FA, Rusyn I, Pogribny IP. (2012) Plasma microRNAs are sensitive indicators of inter-strain differences in the severity of liver injury induced in mice by a choline- and folate-deficient diet. Toxicol Appl Pharmacol 262:52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA (2011) Toxicological Review of Tetrachloroethylene (CAS No. 127-18-4), in Support of Summary Information on the Integrated Risk Information System (IRIS), U.S. Environmental Protection Agency, Washington, DC. [Google Scholar]
- Vamvakas S, Dekant W, Berthold K, Schmidt S, Wild D, Henschler D. (1987) Enzymatic transformation of mercapturic acids derived from halogenated alkenes to reactive and mutagenic intermediates. Biochem Pharmacol 36:2741–2748. [DOI] [PubMed] [Google Scholar]
- Vamvakas S, Dekant W, Henschler D. (1989) Genotoxicity of haloalkene and haloalkane glutathione S-conjugates in porcine kidney cells. Toxicol In Vitro 3:151–156. [DOI] [PubMed] [Google Scholar]
- Vamvakas S, Elfarra AA, Dekant W, Henschler D, Anders MW. (1988) Mutagenicity of amino acid and glutathione S-conjugates in the Ames test. Mutat Res 206:83–90. [DOI] [PubMed] [Google Scholar]
- Väremo L, Nielsen J, Nookaew I. (2013) Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res 41:4378–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White IN, Razvi N, Gibbs AH, Davies AM, Manno M, Zaccaro C, De Matteis F, Pähler A, Dekant W. (2001) Neoantigen formation and clastogenic action of HCFC-123 and perchloroethylene in human MCL-5 cells. Toxicol Lett 124:129–138. [DOI] [PubMed] [Google Scholar]
- Yang HT, Chen YH, Chiu WC, Huang SY. (2006) Effects of consecutive high-dose alcohol administration on the utilization of sulfur-containing amino acids by rats. J Nutr Biochem 17:45–50. [DOI] [PubMed] [Google Scholar]
- Yoo HS, Bradford BU, Kosyk O, Shymonyak S, Uehara T, Collins LB, Bodnar WM, Ball LM, Gold A, Rusyn I. (2015) Comparative analysis of the relationship between trichloroethylene metabolism and tissue-specific toxicity among inbred mouse strains: liver effects. J Toxicol Environ Health A 78:15–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi ZM, Blissett D, Blissett R, Henry L, Stepanova M, Younossi Y, Racila A, Hunt S, Beckerman R. (2016a) The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 64:1577–1586. [DOI] [PubMed] [Google Scholar]
- Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. (2016b) Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64:73–84. [DOI] [PubMed] [Google Scholar]
- Zeise L, Bois FY, Chiu WA, Hattis D, Rusyn I, Guyton KZ. (2013) Addressing human variability in next-generation human health risk assessments of environmental chemicals. Environ Health Perspect 121:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA sequencing data are publically available through Gene Expression Omnibus (GEO; accession ID: GSE93132), and individual animal phenotype data are supplied as Supplemental Materials. In addition to GEO, complete lists of differentially expressed genes and enriched pathways are provided as Supplemental Materials.